SUMMARY
Linking putatively pathogenic variants to the tissues they affect is necessary for determining the correct diagnostic workup and therapeutic regime in undiagnosed patients. Here, we explored how gene expression across healthy tissues can be used to infer this link. We integrated 6,665 tissue-wide transcriptomes with genetic disorder knowledge bases covering 3,397 diseases. Receiver-operating characteristics (ROC) analysis using expression levels in each tissue and across tissues indicated significant but modest associations between elevated expression and phenotype for most tissues (maximum area under ROC curve = 0.69). At extreme elevation, associations were marked. Upregulation of disease genes in affected tissues was pronounced for genes associated with autosomal dominant over recessive disorders. Pathways enriched for genes expressed and associated with phenotypes highlighted tissue functionality, including lipid metabolism in spleen and DNA repair in adipose tissue. These results suggest features useful for evaluating the likelihood of particular tissue manifestations in genetic disorders. The web address of an interactive platform integrating these data is provided.
INTRODUCTION
Human tissues and organs comprise a mixture of different cell types, each executing a distinctive transcriptional program. Individually, each cell contributes to the global transcriptional landscape that drives tissue functionality. Genetic variants that alter gene products or influence gene expression can affect tissue function. However, linking genomic variants to phenotype, e.g., affected tissues, remains a challenge (MacArthur et al., 2014). In this regard, multi-tissue transcriptomic data from healthy individuals, offer valuable information for understanding the functional roles of genes and perhaps their involvement in disease. One hypothesis is that elevated gene expression in healthy tissues indicates functional significance in the tissue. Therefore, deleterious germline variants are more likely to affect tissues where the genes with variants are highly expressed in normative conditions. Oellrich et al. linked wild type expression to the phenotypes observed across tissues in knockout mice; however, they also identified circumstances where gene expression and phenotype were spatially separated (Oellrich et al., 2014). Barshir et al. demonstrated that a majority of disease-associated genes were expressed at elevated levels in the affected tissues for a set of 233 genes with deleterious germline mutations (Barshir et al., 2014). Antanaviciute et al. showed that on average, expression levels of disease genes were significantly higher in affected tissues compared to unaffected ones (Antanaviciute et al., 2015). As such, prioritizing de novo mutation candidates in patients with congenital heart diseases based on expression levels in the heart, successfully distinguished cases from controls (Zaidi et al., 2013). To this end, gene expression levels have been incorporated into workflows that aim to prioritize disease-associated variants from genome and exome sequencing such as CANDID (Hutz et al., 2008), geneTIER (Antanaviciute et al., 2015) and Endeavour (Tranchevent et al., 2016).
Associations between highly expressed genes and phenotypes, i.e., affected organs and tissues, is obvious in some cases. For instance, alpha and beta myosin heavy chain genes (MYH6 and MYH7) and cardiac troponin (TNNI3) are almost exclusively expressed in heart and implicated in various cardiac diseases (Fatkin et al., 2014). Synuclein alpha and beta (SNCA and SNCB), associated with Parkinson’s disease and dementia (Online Mendelian Inheritance in Man (OMIM) IDs: 163890 and 602569), are highly expressed in multiple brain regions and nervous tissue. Among the genes associated with genetic disorders affecting multiple tissues, cystic fibrosis transmembrane conductance regulator (CFTR) is mainly expressed in pancreas, gastro-intestinal track, salivary glands, and lung. Disruptive mutations in CFTR are causally associated with life-threatening pathologic conditions such as fibrotic cysts in the pancreas, inflammation in the lungs and sinus, and other exocrine organs (OMIM ID: 602421). In this case, disease phenotypes correlate well with the organs and tissues expressing CFTR. Many genes however, including a large portion of known disease genes, are expressed in most tissues. For such genes, cross-tissue transcriptional activity may provide limited explanation with regard to the affected tissues (Barshir et al., 2014; Greene et al., 2015; Melé et al., 2015). Furthermore, in some cases, expression profiles “contradict” our knowledge of disease manifestation. For instance, Salt-Inducible Kinase 1 (SIK1, OMIM ID: 605705) is expressed at its highest level in skin and the lowest in brain. Yet mutations in SIK1 lead to severe developmental epilepsy without a known skin phenotype (Hansen et al., 2015). HNRNPDL associated with Limb-Girdle Muscular Dystrophy (OMIM ID: 609115) has lower expression in skeletal muscle than most other tissues (see the Genotype-Tissue Expression Project (GTEx) portal for gene expression levels across tissues).
Discrepancies between expression levels and phenotypes may be, in part, addressed by looking beyond the expression of single genes. Lage and colleagues demonstrated that the mean expression of protein complexes including disease genes, was elevated in affected tissues (Lage et al., 2008). An extension of this study also revealed high co-expression between the disease genes and the other genes forming the protein complexes (Börnigen et al., 2013). Greene et al. recently demonstrated an important role for tissue-specific co-expression networks in disease and utilized their approach to refine disease gene associations from genome-wide association studies (Greene et al., 2015). Barshir and colleagues demonstrated that tissue-specific protein interactions can shed light on the affected tissues due to germline mutations (Barshir et al., 2014).
To systematically assess the relationship between gene expression levels and affected tissues in rare genetic disorders, we integrated tissue-wide transcriptome profiles with genetic disorder knowledge databases. We analyzed genes in a two-dimensional space, where the expression level of a gene in one tissue is compared with its expression in other tissues and with the expression levels of other genes within that tissue. Our results show that expression in each dimension separately and more so in their combination, can inform the likelihood of disease phenotypes in rare genetic disorders. As a source of gene expression, we used RNA-seq data from the GTEx project (GTEx Consortium, 2015; Melé et al., 2015; Rivas et al., 2015). Genetic disorder information was extracted from OMIM, a catalogue summarizing phenotypes for thousands of predominantly Mendelian disorders and their associated genes and variants. However, OMIM is principally a corpus of unstructured text, making it difficult to systematically extract the phenotypes of each disease. We bridged this gap using information from the Human Phenotype Ontology (HPO) which constitutes a hierarchical structure of terms describing human phenotypes (Köhler et al., 2014). HPO terms have conveniently been mapped to their relevant OMIM records in a combination of computational and manual efforts, enabling the systematic association between diseases and their phenotypes. In this study we used the GTEx dataset in conjunction with the OMIM and HPO to evaluate the relationship between gene expression and phenotype, and created an interactive online platform to further explore this data.
RESULTS
Tissues affected in genetic disorders from the OMIM database
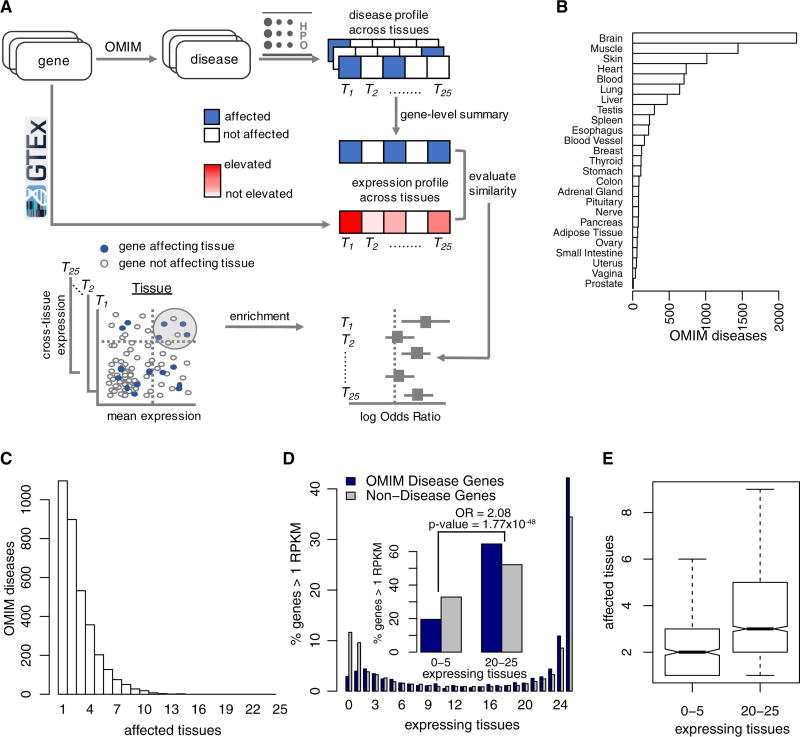
Using information extracted from the OMIM compendium, we inked 4,508 diseases with 3,483 genes. Based on specific HPO terms chosen to represent each tissue, we linked 3,397 diseases and 2,747 genes (denoted as “disease genes” hereafter) with the tissues they affect (see Methods and Supplementary Tables S1 and S2). In the current study, we included the following 25 tissues: adipose tissue, adrenal gland, blood, blood vessel, brain, breast, colon, esophagus, heart, liver, lung, muscle, nerve (tibial), ovary, pancreas, pituitary, prostate, skin, small intestine, spleen, stomach, testis, thyroid, uterus and vagina, for which RNA-seq data are available from GTEx.
Disease-tissue connections were skewed across tissues, as 67% of connections attributed to five organs: brain, muscle, skin, heart and blood (Figure 1A). The majority of diseases in OMIM (74%, 2,527 of 3,397) affected 3 or fewer tissues out of the 25 tissues included in this study (Figure 1B), illustrating tissue-specific manifestation for genetic disorders (Barshir et al., 2014; Goh et al., 2007). On the other end of the spectrum, several diseases mapped to multiple tissues. For instance, CHARGE Syndrome (OMIM ID: 214800) and Smith-Lemli-Opitz Syndrome (OMIM ID: 270400) each affected 14 tissues included in our analysis. When a single gene was associated with multiple diseases affecting different tissues, we created a gene-centric mapping by connecting each gene to the totality of tissues affected by all associated diseases (Supplementary Table S3).
Figure 1. Disease manifestation and gene expression across tissues.
(A–B) Based on information gleaned from the Human Phenotype Ontology, distributions of 3,397 OMIM diseases across the tissues and organs they affect (A) and across the number of tissues and organs affected in each disease (B). (C) Distribution of 19,644 protein coding genes (coloured grey) and 2,747 disease genes (coloured dark blue) across the number of tissues in which they are expressed (mean RPKM ≥ 1). The insert combines bars from ≤ 5 tissues and ≥ 20 tissues to demonstrate enrichment of disease genes in the latter. (D) Boxplots representing the number of affected tissues by genes expressed in ≥ 20 tissues and ≤ 5 tissues. All analyses here are limited to the 25 tissues included in this study.
Expression patterns of disease genes
We used 6,665 GTEx samples to create a matrix of expression levels for 19,644 protein-coding genes across 25 tissues (Supplementary Table S4 lists the samples included in this study). Based on reads per kilo-base per million mapped reads (RPKM), we calculated the mean expression level of each gene in each tissue. The number of genes expressed in each of the 25 tissues at a level of mean RPKM ≥ 1 was bimodal, i.e., the majority of genes were either expressed in a small number of tissues or in most tissues. This supports findings in previous reports (Barshir et al., 2014; Jongeneel et al., 2005; Melé et al., 2015) (Figure 1C). At this threshold, 53.9% of the genes were expressed in ≥ 20 tissues (denoted as “ubiquitously expressed”) and 30.9% were expressed in ≤ 5 tissues (denoted as “specifically expressed”). Of note, approximately 10% of genes were not expressed in any of the 25 tissues at the same threshold. These genes may either express in the tissues not included in our analysis, exclusively express during fetal and early developmental periods or express under specific conditions such as disease.
Of 2,747 disease genes in our analysis (Supplementary Table S5), 1,771 genes were expressed in ≥ 20 tissues (16.7% of all ubiquitously expressed genes) and 536 were expressed in ≤ 5 tissues (8.8% of all specifically expressed genes) suggesting a disproportionately high number of ubiquitously expressed genes among disease genes (Odds Ratio (OR) 2.08, 95% confidence interval (CI) 1.87–2.3 and Fisher’s exact p-value 1.77 × 10−48, Figure 1C). Changing the threshold of expression level from ≥1 RPKM to higher thresholds increased the number of specifically expressed (≤ 5 tissues) genes and reduced the number of ubiquitously expressed (≥ 20 tissues) genes. However, the relative over-representation of disease genes among ubiquitously expressed ones was consistently observed at different thresholds, as enumerated in Table S1. These results were not sensitive to changes in thresholds of ≥ 20 and ≤ 5 for specific and ubiquitous grouping, respectively (data not shown). Finally, we computed the number of affected tissues for ubiquitously and specifically expressed genes. Not surprisingly, the number of affected tissues was higher for ubiquitously expressed genes compared to that of specifically expressed genes (Kolmogorov-Smirnov two-sided p-value = 1.01 × 10−14, Figure 1D). In summary, disease genes were enriched among ubiquitously expressed genes and the number of tissues expressing disease genes was positively associated with the number of affected tissues.
Expression levels of disease genes in affected tissues
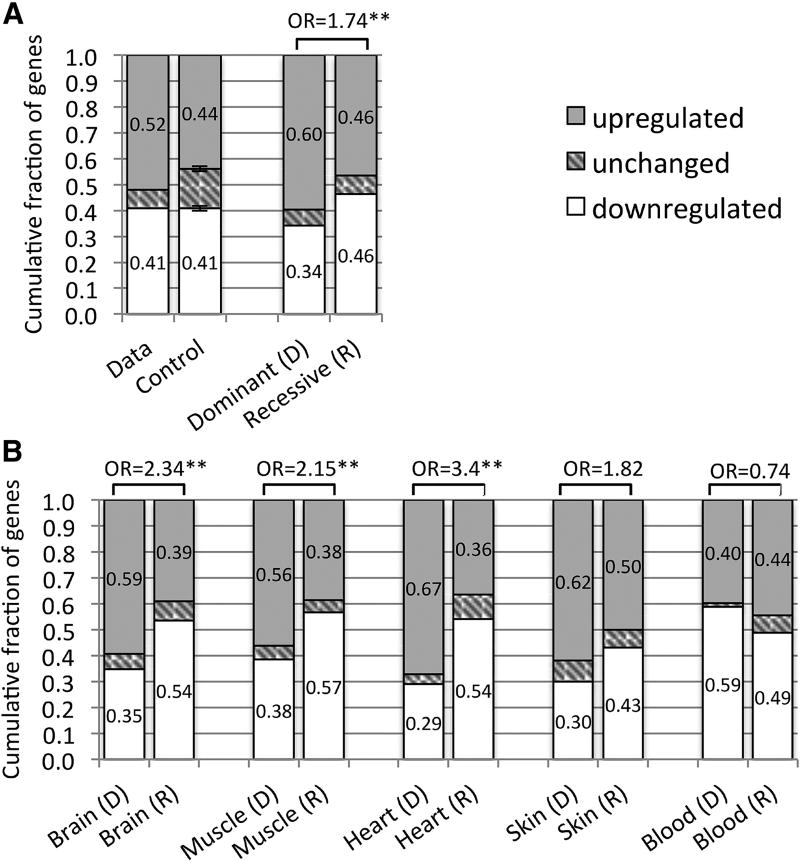
Although disease genes are often expressed ubiquitously across tissues, their relative expression levels could be informative. To investigate this assumption, we compared the expression levels of disease genes in their affected and unaffected tissues. For each gene, we divided 6,665 GTEx samples into those from affected and unaffected tissues and compared their expression levels using a Wilcoxon rank sum test (see Methods and Supplementary Table S6). Out of 1,823 genes that mapped to three or less affected tissues (through the HPO), more genes were up regulated (52%) than down regulated (41%). To test the significance of these proportions, we constructed a random control model where the affected tissue(s) were randomly chosen 1,000 times for each gene and recomputed the fraction of up and down regulated genes. The fraction of up regulated genes was significantly larger in our data compared to the control (one sample T-test p-value < 0.001, Figure 2A,). This finding also supports previous observations (Antanaviciute et al., 2015; Barshir et al., 2014).
Figure 2. Up and down regulated genes in affected versus unaffected tissues.
(A) Fractions of up and down regulated disease genes in affected tissues compared to unaffected tissues are shown for our data and for a random control model, including disease genes affecting up to 3 tissues (n=1,823, left bars). These fractions are also shown for subsets of genes associated with autosomal dominant (n=461) and autosomal recessive (n=743) disorders (right bars). (B) Comparing up and down regulation of genes associated with autosomal dominant (D) and autosomal recessive (R) disorders across different tissues. In these analyses up and down regulation were determined by dividing 6,665 GTEx samples into those from affected and unaffected tissues and comparing their expression using a Wilcoxon rank sum test. Fractions of genes in each group are inscribed in the bars (rounded to 2 decimal points). Odds ratios (OR) comparing up and down regulated genes in dominant and recessive disorders are indicated (“*” and “**” correspond to p-values < 0.01 and < 0.001 respectively).
We further divided these genes into those linked with autosomal dominant (AD) and autosomal recessive (AR) disorders according to the HPO. Out of 461 genes exclusively associated with AD disorders, 60% were up regulated whereas 34% were down regulated in the affected tissue(s). For 743 AR genes, the fractions of up and down regulated genes were not different. As such, enrichment of up regulated genes in affected tissues was significantly different between AD and AR diseases (OR 1.7 95% CI 1.35–2.24, Fisher’s exact p-value 5.76×10−6, Figure 2A). Further analysis by tissue suggested that the difference between AD and AR was mostly driven by diseases affecting specific tissues such as brain and muscle, but not blood (Figure 2B, using tissues with ≥ 50 genes in AD and AR categories).
In some tissues relatively few genes may dominate expression, pancreas and blood being extreme examples (Melé et al., 2015). Since GTEx experiments were conducted at a set read depth for all tissue samples, cross-tissue comparisons with these tissues could be biased, especially for low expressed genes. To address this potential bias, we substituted the RPKM expression values of each gene with their rank in the sample, and recalculated fractions of up and down regulated genes (Supplementary Figure S1). Another possible bias may stem from the different numbers of samples available for each tissue. All brain tissues combined constituted the largest group (n=889), outnumbering the smallest group of uterus samples (n=70) by more than 12-fold. Therefore, we recomputed this analysis using the 70 expression values from the samples closest to the median expression of each gene (see Methods). Although the number of up and down regulated genes varied between these methods, the enrichment of up regulated genes in affected versus unaffected tissues was consistent. Moreover, AD genes maintained a stronger signal than AR genes both globally and for each tissue separately across these methods (Supplementary Figures S1–2, see Methods).
Linking elevated cross-gene and cross-tissue expression levels to phenotype
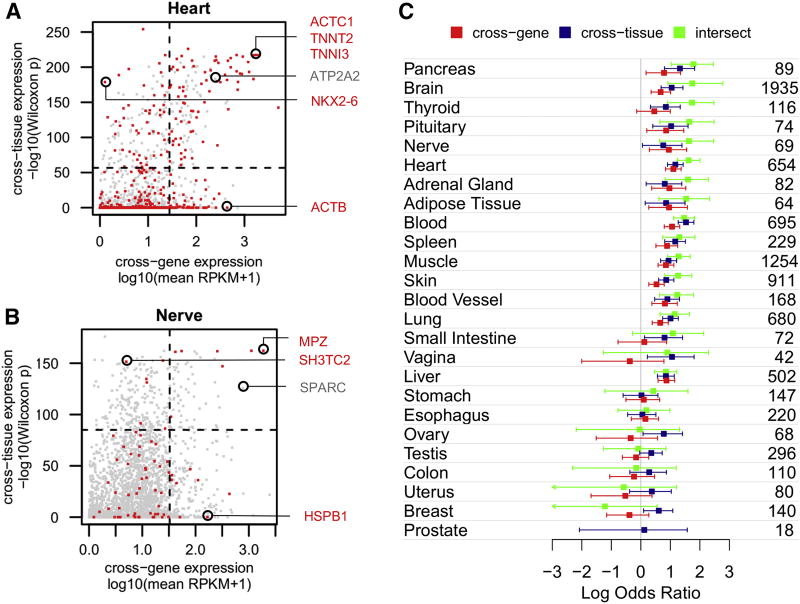
The expression level of a gene in a tissue can be compared either with the expression levels of other genes in the same tissue (“cross-gene”) or with the expression levels of the same gene across other tissues (“cross-tissue”). Using 6,665 GTEx samples from 25 tissues, we compared cross-gene and cross-tissue expression levels of 2,747 disease genes in each tissue. Cross-gene expression was represented by the mean expression of all GTEx samples within a tissue. Cross-tissue expression was estimated by comparing the expression level of a gene in one tissue with its expression in all other tissues. Specifically, we compared the expression levels in GTEx samples from one tissue with expression levels in samples from all other tissues using a Wilcoxon rank sum test. For each gene we repeated this procedure 25 times (one for each tissue) and recorded the −log10 p-values derived from the test. The p-values were computed to recognize elevated expression (see Methods and Supplementary Table S7). Cross-gene and cross-tissue expression levels for 2,747 disease genes are illustrated for heart and peripheral nervous tissue (Figures 3A and 3B). It is noteworthy that disease genes affecting these tissues could have low mean expression (cross-gene) but high cross-tissue expression, e.g., NKX2-6 in heart and SH3TC2 in nervous tissue, or vice versa, e.g., ACTB in heart and HSPB1 in nervous tissue. In both tissues, some genes with extreme values on both cross-gene and cross-tissue axes, e.g., ACTC1, TNNT2 and TNNI3 in heart and MPZ in nervous tissue, have been implicated in cardiac and peripheral nervous system diseases, respectively (Supplementary Table S3). However, there are many examples in which both elevated cross-tissue and cross-gene expression do not correspond to disease manifestation. For instance, mutations in ATP2A2, a gene elevated in heart, have been implicated in a skin disorder (OMIM ID: 124200) but not in cardiac disease (Sakuntabhai et al., 1999). Similarly, the SPARC gene elevated in peripheral nervous tissue, affects the central nervous system but not the peripheral one (OMIM ID: 616507).
Figure 3. Cross-gene and cross-tissue expression.
Cross-gene and cross-tissue expression measures are plotted for 2,747 disease genes in heart (A) and nervous tissue (B). Genes associated with phenotypes affecting each tissue are colored red; other disease genes are colored grey. Cross-gene expression corresponds to mean expression of all GTEx samples from one tissue. Cross-tissue expression corresponds to the –log10(p-value) derived from a Wilcoxon rank sum test comparing expression of a gene in one tissue with its expression in all other tissues (computed to identify elevated expression – see Methods). Circled genes are discussed in the text. Horizontal and vertical dashed lines mark the top 10% value on each axis, dividing the plot into quadrants. (C) Odds ratios representing enrichment of genes associated with phenotypes in each tissue versus those that are not, are computed using the top 10% of genes on the cross-gene axis (colored red), cross-tissue axis (colored dark blue) and their intersect (colored green). The numbers of genes associated with a phenotype in each tissue are shown on the right.
We began to quantify expression-phenotype relationships by systematically examining the genes at the extreme points of expression. For each tissue, we computed the enrichment of phenotype-causing genes among the top 10% of genes on the cross-gene and/or cross-tissue axes (shown as dashed lines in Figures 3A and 3B). For 14 out of the 25 tissues, ORs demonstrated overrepresentation of phenotype-causing genes in the top 10% of genes using cross-gene or cross-tissue measures (i.e., lower CI of log2(OR) > 0 and Fisher’s exact p-values < 0.05). Genes with both high cross-gene and high cross-tissue values (referred to as “intersect”) showed stronger enrichment for 12 of these tissues. These enrichments were greater than 5-fold (ORs > 5, Fisher’s exact p-values < 0.001) in pancreas, brain, thyroid, pituitary, nervous tissue and heart. Supplementary Figure S3 illustrates cross-gene and cross-tissue expression levels of disease genes in all tissues.
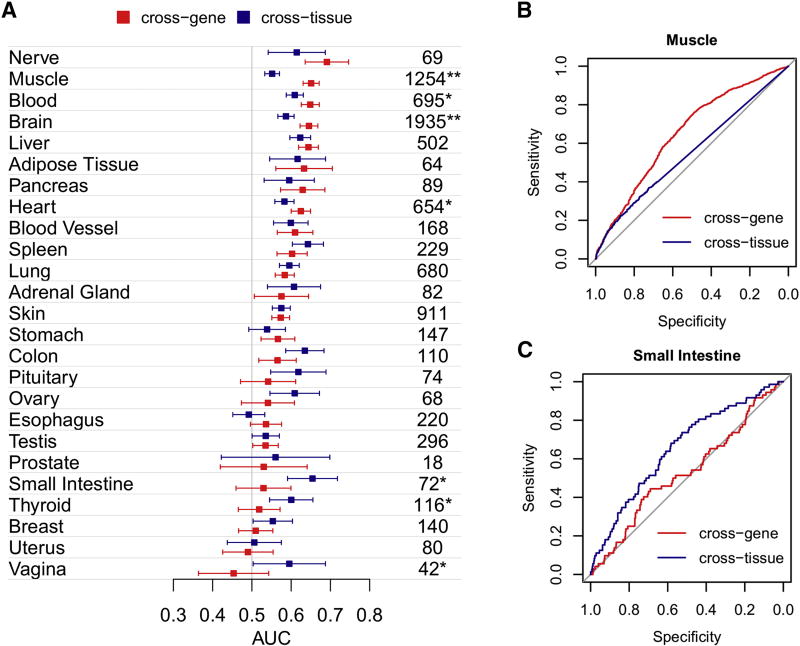
To expand this analysis we performed a receiver operating characteristic (ROC) analysis so that all genes and not only the top 10% were included. Here, an area under the ROC curve (AUC) of > 0.5 indicates that elevated expression in a tissue increases the chance to observe a phenotype in that tissue. Figure 4 summarizes the AUCs computed for all tissues using cross-gene and cross-tissue expression separately. Overall, 15 of the 25 tissues presented a non-random relationship (i.e., a lower bound AUC CI > 0.5) between elevated expression levels of disease genes and the affected tissues using cross-gene and cross-tissue measures (Figure 4A). The tissues identified here agreed with those identified using only the top 10% of expressed genes, with the exception of pituitary and with the addition of colon and testis. The maximum AUC of 0.69 (95% CI 0.64–0.75) was observed for nervous tissue using cross-gene expression. Significant differences between AUCs using cross-gene and cross–tissue expression measures were observed for muscle, brain, blood, heart, small intestine, thyroid and vagina (DeLong’s p-values < 0.01, indicated in Figure 4A). For example, muscle demonstrated a 10% increase using cross-gene expression (DeLong’s p-value 2.6×10−17, Figure 4B) whereas small intestine demonstrated a 13% increase using the cross-tissue measure (DeLong’s p-value 0.002, Figure 4C). Overall, AUCs demonstrated a weak (relatively small AUCs) but significant (p-values < 0.01) link between elevated expression and phenotype for a majority of tissues.
Figure 4. Linking expression and phenotype.
(A) AUCs quantifying the strength of expression-phenotype relationships are shown for each tissue with 95% confidence intervals based on cross-gene (colored red) and cross-tissue (colored dark blue) expression measures. Numbers of genes associated with a phenotype in each tissue are shown on the right. Significant differences between cross-gene and cross-tissue measures are indicated with an asterisk (“*” and “**” correspond to p-values < 0.01 and < 0.001 respectively). ROC curves for Muscle (B) and Small Intestine (C) demonstrate differences between cross-gene and cross-tissue measures.
To assess the robustness of our results, we recalculated cross-tissue p-values using additional methods described above. Specifically, we substituted RPKM expression values with gene ranks and used an equal sample size across all tissues (Supplementary Tables S8–9). We chose a non-parametric ranking test over conventional differential expression tools since the latter are designed to compare groups, each relatively homogenous. In our analyses however, heterogeneous expression profiles from multiple tissues were grouped together. Nonetheless, we also calculated p-values using limma-voom (Law et al., 2014) instead of the Wilcoxon rank sum test (see Methods and Supplementary Table S10). A comparison between the p-values that were calculated using the different methods is shown in Supplementary Figures S4–6. Ranking expression data mainly impacted the tissues where few genes dominated the global expression repertoire (e.g., blood, muscle and pancreas, Supplementary Figure S4). Using an equal but relatively small number of samples for each tissue resulted in lower statistical power limiting the range of p-values (Supplementary Figure S5). The relationship between p-values from our original approach and from limma-voom was non-linear for most tissues, which suggested a general property of comparing parametric (limma-voom) and non-parametric (Wilcoxon rank-sum) tests. Discordant p-values observed in brain tissue could be the result of combining heterogeneous brain regions. Cerebellum, for example, is known to have a unique expression pattern compared to other brain regions (GTEx Consortium, 2015). Indeed, a separate analysis of cerebellum expression showed a similar relationship to that observed in other tissues (Supplementary Figure S6). Most importantly, these differences did not significantly alter the level of agreement between expression and phenotype for most tissues, supporting the robustness of our results (Supplementary Figure S7).
Web Interface
To facilitate the integration of gene expression and phenotype, we created an online platform where a set of genes can be projected onto cross-gene and cross-tissue expression space in each tissue. We linked this platform to the full set of HPO terms, such that genes associated with any phenotype can be analyzed: http://e2p.dbmi.hms.harvard.edu.
Mapping diseases to sub-organ tissues
Although GTEx includes expression profiles of sub-organ regions (e.g., multiple brain regions), systematic mapping of diseases to these specific regions through OMIM and HPO is challenging. Nonetheless, we attempted to make this distinction in some cases. For instance, we classified brain phenotypes into those mapped or not mapped to cerebellum (using HPO terms containing the string “cerebellar” or “cerebellum” – see Methods). We found that cerebellum expression levels of 527 genes associated with cerebellum phenotypes matched slightly better than the collection of all other brain diseases with mixed expression (Supplementary Figure S8). Similarly, we singled out genes associated with cardiac atrial (159 genes) and ventricular (285 genes) phenotypes (using keywords associated with each – see Methods), and matched them with GTEx expression from atrial appendage and left ventricle respectively. Here the specific tissues did not match better than the combined set of heart diseases and mixed expression (Supplementary Figure S9, and Supplementary Tables S11–12). Our online platform enables a view of expression in all sub-organ tissues.
Enriched biological processes of disease genes in each tissue
To further explore expression-phenotype relationships, we categorized 2,747 disease genes into four groups based on expression (mean RPKM ≥ 1) and the presence of a phenotype in each tissue (see Methods and Supplementary Table S13). Groups are denoted by “E” for expression or “P” for phenotype and with “+” or “-” signs to indicate presence or absence respectively (Supplementary Figure S10A). For instance, E+P- group represents genes that are expressed in a tissue but, where no information linking the genes to a phenotype in the same tissue have been reported. Combining all tissues, we generated a global summary of the genes in each of the four groups and their intersections (Supplementary Figure S10B). Notably, no single gene was unique to any one of the four groups, and 487 genes were found in all four groups. In each tissue, we then performed a Gene Ontology (GO) enrichment analysis for each of the four groups separately. We searched for enriched terms in the GO Biological Process category with only disease genes as a background (see Methods). The full list of enriched categories is presented in Supplementary Table S14.
Genes expressed in their affected tissues (E+P+) were likely to participate in tissue-specific processes such as steroid biosynthesis for adrenal gland, immune pathways for blood, neural and synaptic pathways for brain, and metabolic processes in liver. Of note, we observed several pathways in this group that have not been widely described. For instance, excision repair cross-complementing genes and POLD1, involved in DNA replication and repair, were expressed and associated with the diseases in adipose tissue. This observation could explain recent findings linking DNA damage and metabolic disease (Shimizu et al., 2014). In spleen, the E+P+ group was enriched with the genes involved in lipid transport and metabolism suggesting a role for spleen in lipid metabolism (Asai et al., 1988; Fatouros et al., 1995). An interesting observation for E+P+ genes was that compared to cardiac muscle, skeletal muscle was associated with diverse metabolic pathways. The genes in the E+P- group could, in part, suggest undiscovered phenotypes. Alternatively, such genes may be functionally redundant or only required under certain conditions. The majority of tissues in this group were enriched with diverse metabolic pathways. This supports the hypothesis suggesting redundant metabolic pathways (Wang and Zhang, 2009).
Genes causing phenotype but not expressed (E-P+) constituted the smallest group (919 genes, Supplementary Figure S10). These genes warrant an explanation as to the observed phenotype. One possibility would be that the affected tissues are not a direct outcome of the mutated gene but rather a secondary effect caused by disorders in other tissues. Among 919 genes, 592 (64.4%, Supplementary Figure S10) are associated with phenotypes in the other tissues, i.e., they also belong to E+P+ group. This could account for comorbidities affecting multiple tissues. Additionally, the genes in this group might only be expressed under certain conditions. For instance, genes from the blood coagulation and wound healing processes, activated during bleeding and injury, were enriched for blood in this group. Furthermore, genes that are not expressed in adult tissues could play important roles, such as those highly expressed at specific time windows during development. We did not include gene expression profiles from developing organs and tissues in this study as all GTEx samples included in our analysis were collected from adults (ages 20–70). For instance, mutations in DCX are implicated in X-linked lissencephaly-1 (OMIM ID: 300067), causing mental retardation and seizures. However, this gene is expressed at low levels (mean RPKM ≤ 1 in GTEx) in multiple regions of adult brains. Conversely, expression values from the Human Brain Transcriptome (HBT) project (Kang et al., 2011) show that DCX is highly expressed in the developing brain, and that its expression level decreases with age (Supplementary Figure S11). The final group of genes that are not expressed and do not present a phenotype in the tissue (E-P-) were enriched for pathways unrelated to their steady state tissue function including general immune response pathways in brain, muscle and testis, and neurologic system processes in almost all tissues excluding brain and pituitary.
DISCUSSION
Genomics remains a long way from being able to predict disease phenotype ab initio from novel mutations. Nonetheless, the accumulation of genome-scale data has permitted incremental advance. Specifically, whole exome sequencing (WES) has considerably broadened our perspective on genetic causes of diseases and on an increasing fraction of variants that do not appear to be associated with any clinical phenotype (Chen et al., 2016; Lek et al., 2016). Furthermore, the clinical imperative to look for disease manifestations in the correct tissue for any new potentially deleterious variant discovered in individuals, continues to grow with the increased clinical use of WES.
At a whole organismal level, predicting the existence of any disease phenotype based on sequence variation has been approached through breaches of constraints on conservation and biochemistry (Adzhubei et al., 2013; Moreau and Tranchevent, 2012; Sim et al., 2012; Thusberg et al., 2011), protein interaction network properties (Barabási et al., 2011), guilt-by-association in multi-omic studies (Lee et al., 2011), and patterns of gene expression (Butler et al., 2015) to name but a few. Here, we examined the extent to which cross-tissue transcriptional activity of genes implicated in disease can inform which tissues are affected in the disease. We focused on rare genetic disorders where genetics has large effects on phenotype. The plausibility of extending our current approach to common diseases is unclear (Blair et al., 2013; Manolio et al., 2009). To the best of our knowledge, a comprehensive analysis integrating gene expression and phenotypes of rare genetic disorders has not previously been performed at this scale.
Potential mechanisms for dominant inheritance include gain of function mutations, reduced gene dosage (haploinsufficiency) and dominant negative effect of mutated proteins (Wilkie, 1994). For instance Zhong et al. showed that dominant disorders were enriched for in-frame mutations likely to produce a defective protein that could interfere with the normal allele, as compared to truncating mutations that eliminate the protein (Zhong et al., 2009). In line with these findings we observe a stronger tendency of genes associated with dominant diseases to have elevated expression in their affected tissues. This may indicate the sensitivity of highly expressed genes to dosage change (Figure 2).
There are several limitations in our study. In the context of gene expression, the dynamic nature of transcriptional activity must be considered. Gene expression levels change rapidly under pathophysiological changes and along with development, maturation and aging. We used tissue-wide gene expression profiles from generally healthy adults between 20–70 years old, while the majority of Mendelian disorders onset early in life and are possibly linked to gene expression programs specific to fetal development (Wilber et al., 2011). Indeed, under disease conditions such as heart failure, reactivation of fetal gene expression program is observed (Chien and Olson, 2002). Expanding our approach to organ specific developmental gene expression profiles such as the HBT dataset (Kang et al., 2011) could inform of fetal gene programs; however, such datasets are not available for most human tissues. Inevitably, a gene expression snapshot at a single time point and under specific conditions conveys a narrow scope of information. Other factors limiting the power of this study are partial knowledge of disease-causing genes and of the affected tissues. Efforts to systematically map diseases to HPO terms have contributed greatly. However, in addition to incomplete annotation, disease manifestations vary largely across age, gender and ethnicity and cannot be summarized trivially. System level processes may also have a crucial role in determining the affected tissues. A dysfunctional pathway in one tissue could initiate the perturbation of reactive pathways that propagate and affect other organs and tissues. Primary signs and symptoms in patients may reflect the secondary effect of causal pathways in different tissues. For instance, genes that regulate body weight do not necessarily regulate energy metabolism or pathways in adipose tissue but rather exert an effect through altered activity in brain (Locke et al., 2015). Lastly, severe disease-causing genetic variants and associated genes are absent in genetic disorder databases due to embryonic lethality and low fitness (Filges and Friedman, 2015). These limitations suggest future directions for sample collection and measurement to enhance the links between genetic variants and their clinical manifestations. Specifically, extending GTEx to include developing tissues, or at least cognate ex vivo organoids from multiple individuals (Camp et al., 2015) should shed more light on the incomplete links between gene variants and tissue-specific disease manifestation.
Notwithstanding these limitations, our findings suggest several features of gene expression that could be incorporated into evaluating likelihood of disease manifestation and gene function in specific tissues. These include those tissues particularly susceptible such as pancreas and central nervous system, patterns of expression across and within tissues, disease transmission model and extremes in expression distribution. Existing disease-prediction models can be augmented with these insights, and our online application can facilitate discoveries in this domain.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Isaac Kohane (isaac_kohane@harvard.edu).
METHOD DETAILS
Mapping genes to diseases and diseases to tissues
OMIM text files were downloaded from the OMIM catalog: www.omim.org, (on 21/01/2016) and parsed with perl scripts to extract the associations between 4,508 phenotypes and 3,483 genes. All evidence codes linking genes to diseases were included. Using the Human Phenotype Ontology (HPO) browser (http://human-phenotype-ontology.github.io) we searched for terms representing 25 GTEx tissues. For each term we recorded the OMIM entries linked with it. For example, the term “Abnormality of adipose tissue” (HP:0009124) was linked to 139 OMIM entries. Supplementary Table S1 lists the HPO terms used for each tissue. Our choice of HPO terms used to represent each tissue was determined by the medical expertise of the authors, however the online platform accompanying this paper enables users to make their own selection. 5,295 OMIM entries mapped to at least one tissue. Of these, 3,449 entries were associated with at least one disease gene spanning a total of 2,808 genes. Intersecting these genes with the genes included in our GTEx expression dataset (see below) resulted in 2,747 genes associated with 3,397 diseases. Four disease genes (DCAF8, IDS, SHOX, CSF2RA) were represented multiple times in GTEx. In these cases we chose the expression profile with the highest expression levels across a majority of tissues. To generate mappings of affected tissue sub-regions we used the HPO text files (downloaded on 23/01/2017) to construct a graph (tree) representing the hierarchical structure of HPO terms (using the hp.obo file). We identified the HPO terms matching keywords and extracted those terms and their sub-terms (i.e., sub-component in the graph) using the R igraph package. Specifically, for cerebellum we retrieved 76 such terms using the string “cerebell” (ignoring case). Similarly for heart we queried the words “atrium” or “atrial” (42 terms) and “ventricle” or “ventricular” (195 terms). For heart we limited our search to the branch of the term: “Abnormality of the cardiovascular system” (HP:0001626) in the HPO graph. A list of the HPO terms for the tissue sub-regions is presented in Supplementary Table S11. Finally, we used the phenotype_annotation.tab file to connect the terms with their diseases and crossed this information with our data presented in Supplementary Table S12.
Tissue-wide transcriptome profiles
Gene level RPKM values from 8,555 GTEx samples were downloaded from the GTEx portal (www.gtexportal.org) using version 6 files (GTEx_Analysis_v6_RNA-seq_RNA-SeQCv1.1.8_gene_rpkm.gct). In our analysis we included 7,051 samples that were included by the GTEx consortium in the expression Quantitative Trait Loci (eQTL) analysis (GTEx_Analysis_V6_eQTLInputFiles_geneLevelNormalizedExpressionMatrices.tar.gz). Since we were interested in tissues and organs affected in disease, we further filtered out cell line samples, arriving at a final set of 6,665 samples spanning 44 specific tissue types. In our analysis, we used the GTEx broad tissue categories (defined in GTEx_Data_V6_Annotations_SampleAttributesDS.txt), which combines multiple specific tissue types into broader groups. For example multiple specific brain regions are combined to the single tissue “Brain”. Thus 44 specific tissues in our data map to 25 broader tissue types as presented in Supplementary Table S4. We focused on 19,644 protein coding genes excluding all other gene types as defined in the gene model used by the GTEx consortium, a patched version of GENCODE v19 (file: gencode.v19.genes.patched_contigs.gtf.gz). For each gene, we computed the mean expression level of samples from each of the 25 broad tissue types.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data normalization
For statistical tests RPKM values were transformed to log10 scale and one was added so that 0 RPKM values could be log transformed (log10(RPKM+1)). Quantile normalization was performed separately for each tissue using the R package preprocessCore.
Cross-tissue gene expression
Using 6,665 GTEx samples, for each gene we compared the expression of samples from a single tissue with all other tissues using the Wilcoxon rank sum test from the standard stats package in R. We repeated this procedure 25 times, once for each tissue comparing each gene’s expression in one tissue with its expression in all other tissues. The direction of the difference was determined by setting the alternative hypothesis to “greater” for up regulation (and “less” for down regulation). We used the negative log10 p-values from this test as our measure of cross-tissue expression. When p-values were zero and therefore transformed to infinity (Inf in R) we set the negative log10 p-value to be just above the next largest value. The results for the “greater” alternative are presented in Supplementary Table S7. To evaluate p-values for specific tissue sub-regions, we singled out the samples of the specific sub-region and compared their expression with all other samples.
To demonstrate the robustness of our results, we recomputed the cross-tissue p-values where the GTEx gene expression values were substituted for their rank within each sample (Supplementary Table S8). Additionally, p-values were computed using the same number of samples for each tissue. This was achieved by selecting the 70 (based on the lowest number of samples in uterus) representative samples for each gene with the closest expression to the median value of all samples (Supplementary Table S9). Finally we computed limma-voom p-values by executing the “voom” function from the R “limma” package and extracting the p-values. Since we are evaluating the level of up regulation, the p-values of genes with negative fold changes were set to 1 (Supplementary Table S10). Our choice for limma-voom was because it could easily handle thousands of samples as opposed to most other tools. As input to limma-voom we used the GTEx raw counts raw counts matrix (GTEx_Analysis_v6p_RNA-seq_RNA-SeQCv1.1.8_gene_reads.gct).
For comparing expression in affected versus unaffected tissues we performed a similar procedure as above, only merging samples from all the affected tissues into one group and comparing them with the expression of all samples of the unaffected tissues. Here we corrected the p-values for multiple hypothesis (using the R function p.adjust with the “fdr” option).
Receiver-Operating Characteristic (ROC) analysis
Receiver Operating Characteristics (ROC) analysis was performed using the pROC packge in R. This package implements the method described in (Carpenter and Bithell, 2000) to compute confidence intervals for the area under the ROC curve. To compare two ROC curves, the roc.test function was used which implements the DeLong method to assign a p-value (DeLong et al., 1988).
GO enrichment analysis
For Gene Ontology enrichment analysis, we used GO terms from the Molecular Signature Database (MSigDB) C5 - Biological Process (file: c5.bp.v5.1.symbols.gmt, downloaded on 05/18/2016). Crossing 2,747 disease genes with 6,178 genes in the MSigDB C5 Biological Process file, resulted in 1,494 shared genes. These genes served as the background for the GO enrichment analysis. For each tissue we divided the disease genes into the four groups as described in the Results section and performed a hypergeometric test for each of the four groups in all 25 tissues. GO terms were enriched if their corrected hypergeometric p-value was < 0.01 and the number of intersecting genes was ≥ 3 (Supplementary Tables S13 and S14).
ADDITIONAL RESOURCES
Description: http://e2p.dbmi.hms.harvard.edu
To facilitate the integration of gene expression and phenotype, we created an online platform where a set of genes can be projected onto cross-gene and cross-tissue expression space in each tissue from the GTEx data. We linked this platform to the full set of HPO terms such that genes associated with any phenotype can be analyzed.
Supplementary Material
Acknowledgments
We thank Susanne Churchill, Inbar Fried and Daria Prilutsky for their comments on the manuscript. We thank all the people involved in creating and maintaining high quality resources such as OMIM, HPO and GTEx.
FUNDING
This work was funded by 5P50MH106933-03 and 5U01HG007530-03. S.W.K. was supported by NIH R01MH107205 and P50MH094267.
Footnotes
AUTOUR CONTRIBUTIONS
A.F., S.W.K. and I.S.K. conceived the study. A.F. performed the analyses and drafted the manuscript. S.W.K. and I.S.K. edited the paper. B.A. developed the web interface. All authors read and approved the manuscript.
References
- Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet Chapter 7, Unit7.20. 2013 doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antanaviciute A, Daly C, Crinnion LA, Markham AF, Watson CM, Bonthron DT, Carr IM. GeneTIER: prioritization of candidate disease genes using tissue-specific gene expression profiles. Bioinformatics. 2015;31:2728–2735. doi: 10.1093/bioinformatics/btv196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai K, Kuzuya M, Naito M, Funaki C, Kuzuya F. Effects of splenectomy on serum lipids and experimental atherosclerosis. Angiology. 1988;39:497–504. doi: 10.1177/000331978803900602. [DOI] [PubMed] [Google Scholar]
- Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barshir R, Shwartz O, Smoly IY, Yeger-Lotem E. Comparative analysis of human tissue interactomes reveals factors leading to tissue-specific manifestation of hereditary diseases. PLoS Comput Biol. 2014;10:e1003632. doi: 10.1371/journal.pcbi.1003632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair DR, Lyttle CS, Mortensen JM, Bearden CF, Jensen AB, Khiabanian H, Melamed R, Rabadan R, Bernstam EV, Brunak S, et al. A nondegenerate code of deleterious variants in Mendelian loci contributes to complex disease risk. Cell. 2013;155:70–80. doi: 10.1016/j.cell.2013.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börnigen D, Pers TH, Thorrez L, Huttenhower C, Moreau Y, Brunak S. Concordance of gene expression in human protein complexes reveals tissue-specificity and pathology. Nucleic Acids Res. 2013;41:e171. doi: 10.1093/nar/gkt661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Wang K, Marshall JD, Naggert JK, Rethmeyer JA, Gunewardena SS, Manzardo AM. Coding and noncoding expression patterns associated with rare obesity-related disorders: Prader-Willi and Alström syndromes. Advances in Genomics and Genetics. 2015;2015:53–75. doi: 10.2147/AGG.S74598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A. 2015;112:15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter J, Bithell J. Bootstrap confidence intervals: when, which, what? A practical guide for medical statisticians. Stat Med. 2000;19:1141–1164. doi: 10.1002/(sici)1097-0258(20000515)19:9<1141::aid-sim479>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Chen R, Shi L, Hakenberg J, Naughton B, Sklar P, Zhang J, Zhou H, Tian L, Prakash O, Lemire M, et al. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat Biotechnol. 2016;34:531–538. doi: 10.1038/nbt.3514. [DOI] [PubMed] [Google Scholar]
- Chien KR, Olson EN. Converging pathways and principles in heart development and disease: CV@CSH. Cell. 2002;110:153–162. doi: 10.1016/s0092-8674(02)00834-6. [DOI] [PubMed] [Google Scholar]
- DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- Fatkin D, Seidman CE, Seidman JG. Genetics and disease of ventricular muscle. Cold Spring Harb Perspect Med. 2014;4:a021063. doi: 10.1101/cshperspect.a021063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatouros M, Bourantas K, Bairaktari E, Elisaf M, Tsolas O, Cassioumis D. Role of the spleen in lipid metabolism. Br J Surg. 1995;82:1675–1677. doi: 10.1002/bjs.1800821230. [DOI] [PubMed] [Google Scholar]
- Filges I, Friedman JM. Exome sequencing for gene discovery in lethal fetal disorders--harnessing the value of extreme phenotypes. Prenat Diagn. 2015;35:1005–1009. doi: 10.1002/pd.4464. [DOI] [PubMed] [Google Scholar]
- Goh K-I, Cusick ME, Valle D, Childs B, Vidal M, Barabási A-L. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene CS, Krishnan A, Wong AK, Ricciotti E, Zelaya RA, Himmelstein DS, Zhang R, Hartmann BM, Zaslavsky E, Sealfon SC, et al. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet. 2015;47:569–576. doi: 10.1038/ng.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J, Snow C, Tuttle E, Ghoneim DH, Yang C-S, Spencer A, Gunter SA, Smyser CD, Gurnett CA, Shinawi M, et al. De novo mutations in SIK1 cause a spectrum of developmental epilepsies. Am J Hum Genet. 2015;96:682–690. doi: 10.1016/j.ajhg.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutz JE, Kraja AT, McLeod HL, Province MA. CANDID: a flexible method for prioritizing candidate genes for complex human traits. Genet Epidemiol. 2008;32:779–790. doi: 10.1002/gepi.20346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongeneel CV, Delorenzi M, Iseli C, Zhou D, Haudenschild CD, Khrebtukova I, Kuznetsov D, Stevenson BJ, Strausberg RL, Simpson AJG, et al. An atlas of human gene expression from massively parallel signature sequencing (MPSS) Genome Res. 2005;15:1007–1014. doi: 10.1101/gr.4041005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AMM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S, Doelken SC, Mungall CJ, Bauer S, Firth HV, Bailleul-Forestier I, Black GCM, Brown DL, Brudno M, Campbell J, et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42:D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage K, Hansen NT, Karlberg EO, Eklund AC, Roque FS, Donahoe PK, Szallasi Z, Jensen TS, Brunak S. A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes. Proc Natl Acad Sci U S A. 2008;105:20870–20875. doi: 10.1073/pnas.0810772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Blom UM, Wang PI, Shim JE, Marcotte EM. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011;21:1109–1121. doi: 10.1101/gr.118992.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, et al. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau Y, Tranchevent L-C. Computational tools for prioritizing candidate genes: boosting disease gene discovery. Nat Rev Genet. 2012;13:523–536. doi: 10.1038/nrg3253. [DOI] [PubMed] [Google Scholar]
- Oellrich A, Sanger Mouse Genetics Project. Smedley D. Linking tissues to phenotypes using gene expression profiles. Database (Oxford) 2014;2014:bau017. doi: 10.1093/database/bau017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas MA, Pirinen M, Conrad DF, Lek M, Tsang EK, Karczewski KJ, Maller JB, Kukurba KR, DeLuca DS, Fromer M, et al. Human genomics. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science. 2015;348:666–669. doi: 10.1126/science.1261877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuntabhai A, Burge S, Monk S, Hovnanian A. Spectrum of novel ATP2A2 mutations in patients with Darier’s disease. Hum Mol Genet. 1999;8:1611–1619. doi: 10.1093/hmg/8.9.1611. [DOI] [PubMed] [Google Scholar]
- Shimizu I, Yoshida Y, Suda M, Minamino T. DNA damage response and metabolic disease. Cell Metab. 2014;20:967–977. doi: 10.1016/j.cmet.2014.10.008. [DOI] [PubMed] [Google Scholar]
- Sim N-L, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- Tranchevent L-C, Ardeshirdavani A, ElShal S, Alcaide D, Aerts J, Auboeuf D, Moreau Y. Candidate gene prioritization with Endeavour. Nucleic Acids Res. 2016;44:W117–W121. doi: 10.1093/nar/gkw365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhang J. Abundant indispensable redundancies in cellular metabolic networks. Genome Biol Evol. 2009;1:23–33. doi: 10.1093/gbe/evp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilber A, Nienhuis AW, Persons DA. Transcriptional regulation of fetal to adult hemoglobin switching: new therapeutic opportunities. Blood. 2011;117:3945–3953. doi: 10.1182/blood-2010-11-316893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie AO. The molecular basis of genetic dominance. J Med Genet. 1994;31:89–98. doi: 10.1136/jmg.31.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Simonis N, Li Q-R, Charloteaux B, Heuze F, Klitgord N, Tam S, Yu H, Venkatesan K, Mou D, et al. Edgetic perturbation models of human inherited disorders. Mol Syst Biol. 2009;5:321. doi: 10.1038/msb.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.