Abstract
Alfin1 cDNA encodes a putative transcription factor associated with NaCl tolerance in alfalfa (Medicago sativa L.). The recombinant protein binds DNA in a sequence-specific manner, including promoter fragments of the NaCl-inducible gene MsPRP2. Alfin1 function was tested in transgenic alfalfa under the control of the 35S promoter in the sense and antisense orientations with the endogenous MsPRP2 as a reporter gene. Calli overexpressing Alfin1 were more resistant to growth inhibition by 171 mm NaCl than vector-transformed controls, whereas calli expressing Alfin1 in the antisense orientation were more sensitive to NaCl inhibition. Transgenic plants overexpressing Alfin1 in the sense orientation grew well. In contrast, the antisense transgenic plants grew poorly in soil, demonstrating that Alfin1 expression is essential for normal plant development. Transgenic calli and plant roots overexpressing Alfin1 showed enhanced levels of endogenous MsPRP2 mRNA accumulation. However, MsPRP2 mRNA accumulation was also regulated in a tissue-specific manner, as shown in leaves of transgenic plants overexpressing Alfin1. These results suggest that Alfin1 acts as a transcriptional regulator in plants and regulates MsPRP2 expression in alfalfa. Alfin1 overexpressing transgenic plants showed salinity tolerance comparable to one of our NaCl-tolerant plants, indicating that Alfin1 also functions in gene regulation in NaCl tolerance.
Plants and cells adapt to changes in the ionic environment as a result of salinity and drought through temporal or sustained regulation of a large number of genes (for review, see Bohnert et al., 1995; Ingram and Bartels, 1996; Bray, 1997), but the molecular mechanisms responsible for this regulation have remained elusive. We have documented coordinated gene regulation in long-term acquired NaCl tolerance in alfalfa (Medicago sativa L.) and rice (Winicov et al., 1989; Winicov, 1991, 1996) and have been interested in defining a functional role for a putative transcription factor, Alfin1, in the altered gene expression in NaCl-tolerant alfalfa (Winicov, 1993; Bastola et al., 1998).
A relatively small number of transcription factors have been identified to date that bind to promoter elements in genes regulated by NaCl/drought stress (for review, see Ingram and Bartels, 1996; Shinozaki and Yamaguchi-Shinozaki, 1997; Winicov and Bastola, 1997), and much of the information has been gene specific. A more complex view of transcriptional regulation is implied by the requirement of a coupling element for stress regulation of the barley HVA22 gene containing the ABA response element (Shen et al., 1996) and the combined role of myc and myb transcriptional activators in ABA- and dehydration-inducible expression of a promoter region of the rd22 gene (Abe et al., 1997). The potential interactions of various factors is compounded further in that transcription factors such as myc and myb belong to extensive multigene families with tissue-specific expression patterns. Nevertheless, recent reports have shown that ectopic expression of transcriptional activators can result in changes in plant responses to cold (Jaglo-Ottosen et al., 1998) and disease resistance (Cao et al., 1998) and changes in metabolic products in plants (Tamagnone et al., 1998) and cultured cells (Grotewold et al., 1998) by affecting the levels of expression of endogenous genes, indicating the possibility of testing the function of individual transcription factors.
Alfin1 cDNA encodes a novel member of the zinc-finger family of proteins, and its modulation in NaCl tolerance makes it an interesting target for manipulation in plants. It contains sequence information for adjacent Cys-4 and His/Cys-3 zinc-finger domains that appear to bind adjacent G-rich triplet motifs in DNA (Bastola et al., 1998). It also contains an acidic region characteristic of DNA-binding proteins that interact with other proteins (Kakidani and Ptashne, 1988) and therefore is likely to function as a transcription factor in plants. Alfin1 is expressed predominantly in roots, appears to be unique or a low-copy gene in the alfalfa genome, and shows conservation among such diverse plants as alfalfa, rice, and Arabidopsis (Winicov and Bastola, 1997). These characteristics, in addition to in vitro binding to promoter fragments of the root-specific MsPRP2 gene that is also NaCl inducible (Winicov and Deutch, 1994; Deutch and Winicov, 1995), suggested that it may have a significant function in plant-root gene expression and contribute to gene regulation in NaCl tolerance.
To test the functions of Alfin1, we made constructs of the Alfin1 cDNA in the sense and antisense orientations, driven by the strong CaMV 35S promoter, transformed alfalfa, and looked for MsPRP2 expression as a potential reporter for Alfin1 activity in vivo. The antisense transformants demonstrated that normal Alfin1 transcript levels were essential for plant development in soil. However, antisense transformation only minimally affected callus growth on control medium. Nonetheless, increased or decreased Alfin1 expression in the transformed callus correlated positively with relative growth in NaCl-containing medium in culture. In addition, we were able to monitor the mRNA levels of the endogenous alfalfa MsPRP2 gene. In this paper we report that Alfin1 overexpression in transgenic plants led to MsPRP2 accumulation in callus and roots, suggesting that Alfin1 acts as a transcriptional regulator in plants and plays an important role in MsPRP2 expression in alfalfa. Because transgenic plants overexpressing Alfin1 also showed improved NaCl tolerance, comparable to our NaCl-tolerant plant previously regenerated from cell culture, Alfin1 expression must play an important regulatory role that can provide enhanced NaCl tolerance in alfalfa.
MATERIALS AND METHODS
Plant Material
Alfalfa (Medicago sativa L. cv Regen S) cell lines were maintained on SH growth medium (Schenk and Hildebrandt, 1972) in continuous light with and without 171 mm NaCl, as described previously (Winicov et al., 1989; Winicov and Button, 1991). Because of the autotetraploid genotype of alfalfa, all experiments were performed with the parent control plant labeled no. 1, which represents the NaCl-sensitive wild type. All transformations were done with material from this plant or the NaCl-tolerant mutant no. 9, originally selected and regenerated from no. 1 (Winicov, 1991). The NaCl-sensitive parent and NaCl-tolerant plants regenerated from the NaCl-tolerant cell cultures (Winicov, 1991) were maintained in the greenhouse and propagated by cuttings. The influence of NaCl on plant growth was determined on replicate rooted cuttings of plants established in Conetainers in perlite and watered daily with one-quarter-strength Hoagland solution (Hoagland and Arnon, 1938), with or without the indicated concentrations of NaCl, as described previously (Winicov, 1991). All plant material was harvested at the same time of day.
Recombinant Plasmid Construction
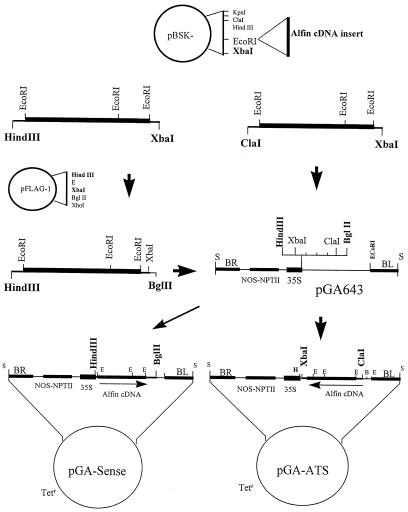
The full-length coding Alfin1 clone (pA50) consists of a 904-bp fragment of Alfin1 cDNA (accession no. L07291) in pBluescript SK− (Stratagene). It contains a 30-bp 5′-untranslated leader, a complete 771-bp coding sequence, and 103 bp of the 3′-untranslated region, including the translation termination codon (Winicov, 1993). This cDNA fragment was cloned in the sense and antisense orientations in the MCS of the binary expression vector pGA643 (An et al., 1988), as shown in Figure 1.
Figure 1.
Schematic representation of Alfin1 sense and antisense constructs used in transformation of alfalfa. Restriction sites are as follows: E, EcoRI; H, HindIII; B, BglII; and S, SalI. BR and BL are T-DNA right and left borders, respectively (An et al., 1988).
To generate the sense construct, the 939-bp HindIII-XbaI fragment from pBluescript SK− was first subcloned in pFLAG (International Biotechnologies Inc., New Haven, CT), designated as PF-pA50, to gain a restriction site suitable for cloning the cDNA fragment in pGA643. The 957-bp HindIII-BglII fragment from PF-pA50 containing Alfin1 cDNA was then ligated to pGA643 in the MCS 3′ to the CaMV 35S promoter to give pGA-sense. This clone was predicted to give the complete Alfin1-coding transcript, but unlike the endogenous Alfin1 mRNA it carried additional sequences from the vector in its 3′-untranslated region.
To generate the antisense construct (pGA-ATS), the 944-bp ClaI-XbaI fragment from pA50 (pBluescript SK−) was ligated directly into the pGA643 MCS. Although another ClaI site has been reported upstream of the MCS in pGA643, we found that only the ClaI site in the MCS, indicated in Figure 1, was cut by the enzyme.
The plasmids pGA-sense, pGA-ATS (antisense), and pGA643 (vector) were propagated in Escherichia coli strain MC1000 (a gift from Dr. G. An, Washington State University, Pullman) in the presence of tetracycline. The freeze-thaw method, as described by An et al. (1988), was used to transform Agrobacterium tumefaciens LBA 4404 (Hoekema et al., 1983) with the recombinant binary plasmid. Transformed colonies were selected on 12 mg/L rifampicin and 6 mg/L tetracycline. Recombinant transformed colonies were identified by colony hybridization using the Alfin1 670-bp EcoRI fragment from pA50 (Sambrook et al., 1989).
Plant Transformation
Alfalfa NaCl-sensitive wild-type parent plant no. 1 (Winicov, 1991) leaves were transformed by A. tumefaciens cocultivation on SH growth medium, including 2 mg/L 2,4-D and 2 mg/L kinetin (Schenk and Hildebrandt, 1972), and supplemented with 50 μm acetosyringone (Aldrich) for 30 to 60 min at room temperature. One of the successful transformations was carried by cocultivating A. tumefaciens carrying the pGA-ATS with immature ovaries from the NaCl-tolerant alfalfa IW9 line (Winicov, 1991). After 2 to 6 d on callus medium the explants were transferred to selection medium (SH medium supplemented with 300 mg/L carbenicillin and 100 mg/L kanamycin) and incubated for 3 to 4 weeks. The resistant calli were subcultured on the selection medium on a monthly basis. Plants were regenerated from the transformed calli on SH medium (without hormones) supplemented with 100 mg/L kanamycin. Plants with well-defined shoots and roots were transferred to peat moss and subsequently to soil.
DNA Extraction and PCR Analysis
Genomic DNA was extracted from 0.5 g of frozen callus or leaves using DNAzol genomic DNA isolation reagent (Molecular Research Center, Inc., Cincinnati, OH), as described by the manufacturer. PCR was carried out in a 25-μL total reaction containing 250 ng of genomic DNA, 1× PCR buffer (50 mm KCl, 10 mm Tris-HCl, pH 9.0, and 0.1% Triton X-100), 100 μm deoxynucleoside triphosphates, 0.2 μm each of the forward (primer common to all PCR analyses in this section = 5′ CCA CTA ATT CGT CCT GCT GG 3′) and the reverse sequence primers (Midland Certified Reagent Co., Midland, TX) (PS for sense [5′ CCA GTC CCT CTC CTG CAT TC 3′], PA for antisense [5′ GGA CAA GGT GCA ACC TGT GG 3′], and PG for vector [5′ AAG TGT GCT TGA GCT CGG TC 3′]), and 0.25 unit of Taq polymerase (Promega). The forward sequence primer was from position 2432 bp and the reverse primer was from 3404 bp for the pGA-vector, 3356 bp for the pGA-sense, and 3359 bp for the pGA-antisense DNA sequence of the T-DNA right border. This combination of PCR primers gave 973-, 926-, and 928-bp products, respectively.
The Gene Amp PCR System (model 2400, Perkin-Elmer) was programmed for an initial denaturing temperature of 94°C for 4 min, a second denaturing temperature of 94°C for 1 min, an annealing temperature of 62°C for 90 s, and an extension temperature of 72°C for 1 min. The reaction was carried out for 35 cycles. An additional extension at 72°C followed for 7 min after completion of the final cycle.
RNA Extraction and Blot Analysis
Total RNA was extracted from roots and shoots containing both leaves and stems from plants grown for 17 d with and without 128 mm NaCl, or callus grown for 1 month with and without 171 mm NaCl, and analyzed under high-stringency hybridization and wash conditions, as described previously (Winicov and Deutch, 1994; Winicov and Krishnan, 1996). Northern analysis for Alfin1 was done with the 670-bp EcoRI large fragment from pA50; for MsPRP2, the probe was the EcoRI fragment from pA9 (Winicov and Deutch, 1994); the constitutively expressed Msc27 was probed with the PstI fragment (Gyorgyey et al., 1991); and the 763-bp EcoRI-BglII fragment from pGA643 (the region between the 3′ end of the MCS and the T-DNA left border) was used to detect transgenic Alfin1 expression. Gel-purified fragment probes were labeled with [32P]dCTP using the random primer-extension system (DuPont-NEN).
RESULTS
Alfalfa Calli Transformed with Sense and Antisense Alfin1
NaCl-sensitive alfalfa cells were transformed with pGA-sense, pGA-ATS (antisense), and the vector pGA643. Many kanamycin-resistant lines were isolated from independent transformations in three different experiments. A total of 22 independent transformed lines were obtained with pGA-sense, 14 independent lines were obtained with pGA-ATS, and comparable numbers were obtained using the empty vector pGA643. No consistent differences in cell growth were observed between transformants of the different constructs, although significant growth differences could be seen between independently transformed cell lines. Only transformed calli showing good growth on kanamycin were further maintained and analyzed. Kanamycin-resistant transformants were confirmed by PCR to carry the appropriate inserts (data not shown).
The influence of transformation with Alfin1 was measured by alfalfa callus growth on SH medium with and without 171 mm NaCl, as shown in Table I. Two NaCl-sensitive cell lines (1,1 and 1,5) were independently initiated in culture. They showed 92% and 84% growth inhibition by NaCl, respectively, as measured by an NaCl-dependent increase in callus wet weight after 4 weeks of growth. The 1,1 cells transformed with pGA-sense showed less growth inhibition by NaCl than those transformed by the pGA643 vector alone. In contrast, 1,5 cells transformed with the pGA-ATS appeared to grow somewhat more slowly on the control medium and were more sensitive to growth inhibition by NaCl than the pGA643 vector-transformed cells. These results were consistent with our hypothesis that Alfin1 helps to maintain cellular functions in our NaCl-tolerant alfalfa. However, none of the sense transformants was able to grow as well on 171 mm NaCl as on the control SH medium.
Table I.
Cell growth of transformed and untransformed alfalfa cell lines
| Cell Line | Kanamycin | Growtha
|
|
|---|---|---|---|
| 0 NaCl | 171 mm NaCl | ||
| g wet wt/plate | |||
| 1,1-Untransformed | − | 5.49 ± 0.81 | 0.90 ± 0.47 |
| (n = 2) | (n = 3) | ||
| 1,1-t-Vector (3)b | + | 4.34 ± 1.35 | 1.08 ± 0.20 |
| (n = 4) | (n = 6) | ||
| 1,1-t-Alfin1-sense (6)b | + | 5.06 ± 1.13 | 1.63 ± 0.38 |
| (n = 7) | (n = 9) | ||
| 1,5-Untransformed | − | 5.36 ± 0.84 | 1.30 ± 0.48 |
| (n = 3) | (n = 3) | ||
| 1,5-t-Vector (2)b | + | 3.83 ± 0.27 | 1.25 ± 0.27 |
| (n = 6) | (n = 6) | ||
| 1,5-t-Alfin1-antisense (4)b | + | 3.39 ± 0.91 | 0.93 ± 0.23c |
| (n = 7) | (n = 6) | ||
Growth (means ± sd) after 4 weeks on SH medium ± 171 mm NaCl, using an initial inoculum of about 0.1 g/callus and five calli/plate. n = number of plates.
Number in parentheses, Number of different individual transformants included in test.
Brown, dead callus.
Overexpression of Alfin1 in Transgenic Callus Increases MsPRP2 mRNA Levels
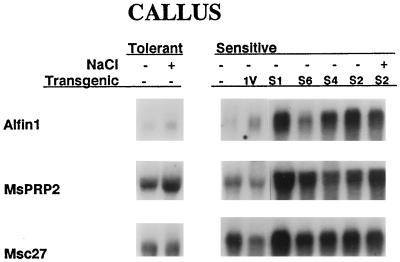
Alfin1 expression was determined in the pGA-sense-transformed callus by northern analysis of total RNA using the constitutively expressed Msc27 gene probe to monitor RNA concentrations in each lane. In Figure 2, the results show clearly that Alfin1 expression was greatly enhanced in the S1, S2, S4, and S6 pGA-sense-transformed cell lines compared with untransformed and vector-transformed cells. Some variability in the levels of expression was observed between different transformants, consistent with the prevalent variability resulting from independent transformation events. Concurrent with the enhanced Alfin1 expression in the transgenic cells we also found significantly increased levels of MsPRP2 transcripts. The levels of MsPRP2 transcripts found in pGA-sense-transformed cells were higher than those found in NaCl-tolerant cells grown in the presence of NaCl, and we could not detect further NaCl-induced enhancement of the high levels of MsPRP2 mRNA accumulation in the pGA-sense-transformed callus. Because recombinant Alfin1 was shown to bind to promoter fragments of MsPRP2 in vitro (Bastola et al., 1998), the enhanced levels of endogenous MsPRP2 transcripts in callus overexpressing Alfin1 suggest that Alfin1 regulates alfalfa MsPRP2 expression in vivo.
Figure 2.
Northern analysis of Alfin1 and MsPRP2 expression in control and transgenic calli from Alfin1 sense transformants. Lanes 1 and 2, RNA isolated from untransformed NaCl-tolerant callus grown with or without 171 mm NaCl for 4 weeks; lane 3, RNA isolated from untransformed NaCl-sensitive callus; lane 4, RNA isolated from NaCl-sensitive callus transformed with pGA vector (1V); lanes 5 to 9, RNA isolated from NaCl-sensitive callus transformed with Alfin1 sense construct (S1, S2, S4, and S6 are independently transformed lines); and lane 9, RNA isolated from S2-transformed callus grown in 171 mm NaCl. Each lane contained 10 μg of total RNA. Each blot was hybridized sequentially with the following probes: Alfin1, the large EcoRI fragment (Fig. 1); MsPRP2, the carboxy-terminal and 3′-untranslated region fragment (Winicov and Deutch, 1994); and Msc27, the fragment of a constitutively expressed alfalfa gene.
Phenotype of Alfin1 Sense and Antisense Transgenic Plants
To investigate the molecular and growth characteristics influenced by Alfin1 numerous plants were regenerated from pGA-sense-transformed calli and calli transformed with the vector alone. Three pGA-sense-transformed plants, regenerated from independent transformations events, were maintained for molecular and growth studies. All three plants grew well, flowered, and set seed. The sense transformants appeared normal, although young leaves were somewhat broader than those from the parent plant and appeared to senesce somewhat earlier.
Calli transformed with the pGA-ATS construct regenerated shoots readily, but root development was poor. Treatment of the regenerating shoots with 5 μm naphthalene acetic acid gave some root development, but none of the dozen plantlets transferred to soil survived for more than 2 weeks. Only one pGA-ATS-transformed plant survived in soil for about 6 months, but it remained severely dwarfed in both root and shoot growth. These results strongly indicated that Alfin1 antisense expression was deleterious to growth and root formation and that Alfin1 transcripts were necessary for plant development in soil, although antisense did not have a similar impact on callus growth in normal SH medium.
Overexpression of Alfin1 in Transgenic Plants Increases MsPRP2 mRNA Levels in Roots
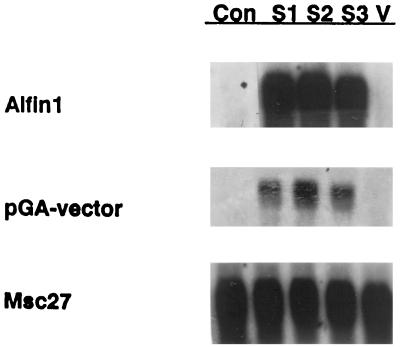
Three of the primary transformed plants with pGA-sense constructs were analyzed for tissue-specific expression of the Alfin1 transgene and its putative target gene MsPRP2. Gel-blot analysis of leaf total RNA from soil-grown plants shown in Figure 3 confirmed that the pGA-sense-transformed plants showed high levels of Alfin1 mRNA expressed from Alfin1 under the control of the CaMV 35S promoter, in contrast to the untransformed parent plant. The presence of the transgene transcripts was demonstrated by probing of the same blot with the BglII/EcoRI fragment of the pGA643 vector, which is adjacent to the 3′ end of Alfin1 cDNA and is apparently transcribed in Alfin1 sense mRNA in the transformants.
Figure 3.
Northern analysis of Alfin1 expression in control and transgenic plants from Alfin1 sense transformants. RNA was isolated from leaves of control and transgenic plants. Lane Con, No. 1 control NaCl-sensitive parent plant for all transformations; lanes S1, S2, and S3, plants transformed with the Alfin1 sense construct and regenerated from transformed callus; and lane V, vector-transformed plant. Each blot was hybridized sequentially with the following probes: Alfin1, large EcoRI fragment (Fig. 1); pGA-vector, EcoRI/BglII fragment from pGA643 to show readthrough of the Alfin1 transgene; and Msc27, fragment of a constitutively expressed alfalfa gene. Each lane contained 10 μg of total RNA.
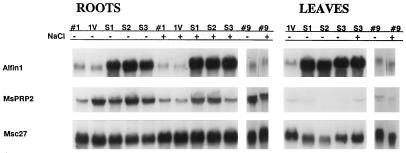
Figure 4 shows similar results from the Alfin1-overexpressing transgenic plants grown in one-quarter-strength Hoagland solution. The MsPRP2 transcript levels increased in the roots of the Alfin1-overexpressing plants (Fig. 4). The vector-transformed plant no. 1 showed somewhat increased levels of MsPRP2 mRNA in roots, but this level was not maintained in the presence of NaCl. In fact, the MsPRP2 mRNA levels were comparable from NaCl-grown control no. 1 and the vector-transformed no. 1 plants. In contrast, the three transgenic plants overexpressing Alfin1 maintained proportionately higher levels of MsPRP2 mRNA in roots after growing for 17 d on 128 mm NaCl-supplemented one-quarter-strength Hoagland solution. The mRNA profiles from NaCl-tolerant no. 9 plants are shown for a comparison. Whereas high levels of Alfin1 mRNA were found in both roots and leaves because of the 35S promoter control of the transgene, Alfin1 overexpression had a negligible effect on MsPRP2 transcript levels in leaves of transgenic plants grown on one-quarter-strength Hoagland solution. NaCl treatment did not further enhance the MsPRP2 mRNA levels in the transgenic plants, as shown in Figure 4. These results support the Alfin1 functional role in MsPRP2 expression primarily in roots and indicate that additional tissue-specific factors contribute to the differences observed in MsPRP2 mRNA levels between roots and leaves.
Figure 4.
Northern analysis of Alfin1 and MsPRP2 expression in control and transgenic plants from Alfin1 sense transformants grown in one-quarter-strength Hoagland solution with or without 128 mm NaCl. RNA was isolated from roots and leaves of control plants and plants tested for NaCl tolerance described in Table II legend. Lanes #1, Parent wild-type control; lanes 1V, control transformed with empty vector; lanes #9, NaCl-tolerant plant regenerated from NaCl-tolerant callus; and lanes S1, S2, and S3, parent no. 1 transformed with pGA-sense. The blot was hybridized sequentially with probes as described for Figure 2. Each lane contained 10 μg of total RNA.
Effect of Alfin1 Overexpression on the NaCl-Tolerance Characteristics of the Transgenic Plants
To determine if Alfin1 overexpression had an effect on the NaCl-tolerance phenotype of the transgenic plants, we compared the growth characteristics of the three pGA-sense-expressing transgenic plants with those of the wild-type NaCl-sensitive parent plant (no. 1), vector-transformed plants, and our previously regenerated NaCl-tolerant plant IW9 (Winicov, 1991). Tolerance was measured as relative new growth obtained from established transgenic and control plants that had been cut back to the crown and then treated for 17 d with 128 mm NaCl. As shown in Table II, one parental control and one vector-transformed plant died from the NaCl treatment. All pGA-sense-expressing transgenic plants and our NaCl-tolerant IW9 plants survived and grew two to three times as well as the parent and vector-transformed controls. It is important to note that IW9 had maintained its significant NaCl-tolerant characteristics for more than 9 years after propagation by cuttings in the greenhouse. All three independently regenerated transgenic plants overexpressing Alfin1 showed growth characteristics similar to or better than those of our NaCl-tolerant IW9. Vector-transformed controls were as NaCl sensitive as the parent plant. These results are consistent with data from another experiment, which tested tolerance to 171 mm NaCl in plants established for only 1 week. That experiment showed 14%, 43%, 57%, and 86% survival of no. 1 (parent), sense-1, sense-2, and IW9 plants, respectively, after 9 d of NaCl treatment. The results from both experiments indicate that Alfin1 overexpression can provide increased NaCl tolerance in alfalfa.
Table II.
Growth properties of Alfin1-sense-transformed plants on 128 mm NaCl
| Plant | Survival | New Leaf Growth | Percentage |
|---|---|---|---|
| g/plant | |||
| No. 1 (parent) | 4 /5 | 0.56 ± 0.32 | 100 |
| No. 1 + vector | 3 /4 | 0.42 ± 0.32 | 75 |
| No. 1 + sense-1a | 7 /7 | 1.40 ± 0.17 | 250 |
| No. 1 + sense-2a | 7 /7 | 1.85 ± 0.23 | 330 |
| No. 1 + sense-3a | 3 /3 | 1.45 ± 0.32 | 259 |
| IW9b | 7 /7 | 1.10 ± 0.18 | 196 |
Multiple rooted cuttings from each plant were established in individual Conetainers in perlite for 6 weeks and grown on one-quarter-strength Hoagland solution. All shoots were then cut back to the crown. Growth was continued from that point on one-quarter-strength Hoagland solution supplemented with 128 mm (0.75%) NaCl. The newly regrown shoots were harvested and weighed after 17 d. Data are means ± sd.
Plants sense-1, sense-2, and sense-3 correspond to S1, S2, and S3 shown in Figure 4.
NaCl-tolerant plant regenerated after selection in tissue culture from parent plant no. 1 (Winicov, 1991).
Alfin1 and MsPRP2 steady-state mRNA levels were determined for the NaCl-treated and control plants at the time of the harvest described in Table II and are shown in Figure 4. The S1, S2, and S3 pGA-sense transgenic plants had high levels of Alfin1 and MsPRP2 mRNA in roots, but not in shoots, regardless of growth in 128 mm NaCl, although some NaCl-dependent decrease in MsPRP2 mRNA levels is apparent in the S3 transgenic plant. The MsPRP2 transcript levels appear to be higher in the pGA-sense transgenic plants than in our NaCl-tolerant plant no. 9. Although Table II shows significant differences in the NaCl tolerance of the plants at 128 mm NaCl after 17 d, we did not detect comparable levels of NaCl inducibility of MsPRP2 mRNA accumulation (Fig. 4), as had been seen in plants treated with 171 mm NaCl for 7 d (Winicov and Deutch, 1994). Whether this difference was due to the lower NaCl concentration or to plant adjustment after a longer time of growth in NaCl will have to be determined and correlated with levels of MsPRP2 protein accumulation when plants are grown for prolonged periods in NaCl.
DISCUSSION
Overexpression of Alfin1 was engineered in transgenic callus and alfalfa plants under the control of the strong CaMV 35S promoter. Our previous experiments suggested that Alfin1 was likely to function as a transcription factor, since we had shown sequence-specific DNA binding of the recombinant protein in vitro and specific binding to promoter fragments of the MsPRP2 gene from alfalfa (Bastola et al., 1998). In this paper we are able to show in callus and plants overexpressing Alfin1 a concomitant increase in the endogenous MsPRP2 mRNA levels, indicating that the Alfin1 gene product regulates MsPRP2 expression in vivo from its normal promoter. These results are consistent with our prediction that Alfin1 is a transcription factor, regulating plant gene expression, and acts in a dominant fashion in overexpressing transgenic plants.
Although Alfin1 was expressed from the 35S promoter in both roots and leaves, significant MsPRP2 transcript induction from its natural promoter in the transgenic plants was detected in callus and roots, the tissues in which Alfin1 is primarily expressed (Bastola et al., 1998). Small differences in MsPRP2 mRNA induction by Alfin1 overexpression were observed in leaves of soil-grown plants (data not shown) but not in plants grown on one-quarter-strength Hoagland solution, suggesting subtle variation due to the nutritional state of the plants. The differential response in leaves and roots to high levels of Alfin1 mRNA could result from the presence of a transcriptional or posttranscriptional inhibitor of MsPRP2 transcript accumulation in leaves or may indicate the requirement for additional root-specific transcription factors for high levels of expression from the MsPRP2 promoter. Additional experiments should differentiate between these two possibilities. The callus complement of participating factors in MsPRP2 expression appears similar to that of the root, because Alfin1 overexpression led to a significant increase of MsPRP2 transcripts in callus culture.
The plant phenotype of pGA-ATS transformants was striking in its inability to sustain growth in soil, especially since we observed no substantially altered phenotype in antisense-expressed callus grown on SH medium. These results suggested a low level of redundancy for Alfin1 function and demonstrated that maintenance of Alfin1 expression was essential for root development and plant growth in soil. Another function affected by Alfin1 antisense expression could be root-shoot communication via the vascular system, which suggests that the Alfin1 protein may regulate other genes in addition to MsPRP2. On the other hand, overexpression of Alfin1 showed no major visible phenotype, even though it was inappropriately expressed in the shoot.
Because Alfin1 was first cloned from NaCl-tolerant alfalfa callus (Winicov, 1993), our demonstration of improved NaCl tolerance in the transgenic plants overexpressing Alfin1 significantly associates the product of this gene with improved NaCl tolerance. However, its relationship to the mutation(s) that allowed the regeneration of our NaCl-tolerant plants, such as IW9 (Winicov, 1991), remains unclear. Transgenic plants have been engineered in a number of laboratories to overexpress single genes, which are known to be up-regulated by NaCl/drought stress in prokaryotes or plants with incremental improvements in NaCl tolerance (Tarcyznski et al., 1993; Kishor et al., 1995; Pilon-Smits et al., 1995; Xu et al., 1996). However, NaCl tolerance has also been considered to be a quantitative trait (Foolad and Jones, 1993), and the molecular mechanisms by which plants could acquire improved long-term NaCl tolerance, involving the regulation of many genes, are still not understood (for review, see Winicov and Bastola, 1997; Winicov, 1998). Therefore, the possible function of transcription factors associated with stress responses has been of significant interest.
It has been shown that both myc and myb proteins function as transcriptional activators in the rd22 gene, which is induced by ABA and dehydration (Abe et al., 1997). Many of the NaCl- and drought-induced genes are also induced by ABA, and ABA response element-binding proteins have been cloned (Guiltinan et al., 1990). Other genes responding to NaCl/drought stress and cold are induced in an ABA-independent manner involving the cis-acting DRE (DNA regulatory element) (Yamaguchi-Shinozaki and Shinozaki, 1994). Recently, the CBF1 protein (Stockinger et al., 1997), which has been shown to recognize the DRE, was shown to function in enhancing freezing tolerance (Jaglo-Ottosen et al., 1998) in Arabidopsis. These findings suggest that the phenotypic changes involving altered gene expression and resistance to stress might be manipulated through the relevant transcription factors.
Transgenic manipulation of Alfin1 expression, therefore, is of interest because we have demonstrated Alfin1 to be a regulatory gene that can influence the expression of MsPRP2 in a specific manner. An interesting result of the enhanced Alfin1 expression in our transgenic plants was the finding that these plants demonstrated enhanced NaCl tolerance. It is likely that, as a transcriptional regulator, Alfin1 also influenced the regulation of other genes in our transformed plants, which could have contributed to the enhanced NaCl tolerance observed in our transgenic plants. Table III shows that many of the genes that have been shown to be up-regulated by NaCl/drought stress also contain Alfin1-binding motifs in their promoters. At present, we do not know if any of these other genes are differentially regulated in our Alfin1-overexpressing plants, but we might expect to see changes in their expression if Alfin1 had a general regulatory role in NaCl tolerance.
Table III.
Alfin1-binding sites found in NaCl/drought stress-induced promoter sequences
| Gene and Plant | Sequence | Ref. |
|---|---|---|
| MsPRP2, alfalfa | −299 5′ GTGGGG 3′ −289 | Bastola et al. (1998) (AFO28841) |
| HVA1, barley ABA response element 2 | −93 5′ GTGGCG 3′ −87 | Straub et al. (1994) (X78205) |
| Atmyb2, Arabidopsis | −559 5′ GAAGTG 3′ −555 | Urao et al. (1993) (D14712) |
| −461 5′ GTGTGG 3′ −435 | ||
| −222 5′ GCCGTG 3′ −217 | ||
| rab28, maize | −378 5′ GTCGTGCAG 3′ −360 | Pla et al. (1991) (X59138) |
| salT, rice | −1451 5′ GTGCAG 3′ −1446 | Claes et al. (1990) (Z25811) |
| −843 5′ GTGACG 3′ −828 | ||
| Osmotin, tobacco | −1447 5′ GTGGTG 3′ −1442 | Ragothama et al. (1993) (S68111) |
| −596 5′ GTGGTG 3′ −591 | ||
| −471 5′ GTGGAG 3′ −466 | ||
| CDeT27-45, resurrection plant | −703 5′ GTGTGGGCG 3′ −695 | Michel et al. (1993) (X69883) |
Selection of potential Alfin1-binding sites was made for the coding strand on the basis of at least two adjacent triplets, one of which is GTG and the other of which is bordered by a G as defined by in vitro Alfin1 binding (Bastola et al., 1998). Additional sites (not shown) were found on the noncoding strand in many of these gene promoters. Numbers in parentheses indicate accession numbers.
All sequences identified are relative to the first ATG codon.
Future experiments will determine the extent and specificity of plant gene regulation by Alfin1 and the extent to which enhanced Alfin1 expression could be useful in manipulating plant growth tolerance of environmental conditions.
ACKNOWLEDGMENT
We thank B. Mitchell for excellent greenhouse and laboratory help.
Abbreviations:
- CaMV
cauliflower mosaic virus
- MCS
multiple cloning site
- SH
Schenk and Hildebrandt
Footnotes
This work was supported in part by a Hatch grant from the Nevada Agricultural Experiment Station, by the National Science Foundation Experimental Program to Stimulate Competitive Research, Women in Science and Engineering, and by the National Research Initiative Competitive Grants Program (grant no. 9401235 to I.W).
LITERATURE CITED
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G, Ebert P, Mitra A, Ita S. Binary vectors. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual, Section A, Chapter 3. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1988. pp. 1–19. [Google Scholar]
- Bastola DR, Pethe VV, Winicov I. Alfin1, a novel zinc-finger protein in alfalfa roots that binds to promoter elements in the salt-inducible MsPRP2 gene. Plant Mol Biol. 1998;38:1123–1135. doi: 10.1023/a:1006081926699. [DOI] [PubMed] [Google Scholar]
- Bohnert HJ, Nelson DE, Jensen RG. Adaptations to environmental stresses. Plant Cell. 1995;7:1099–1111. doi: 10.1105/tpc.7.7.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray EA. Plant responses to water deficit. Trends Plant Sci. 1997;2:48–54. [Google Scholar]
- Cao H, Li X, Dong X. Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc Natl Acad Sci USA. 1998;95:6531–6536. doi: 10.1073/pnas.95.11.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claes B, Dekeyser R, Villarroel R, Van den Bulcke M, Bauw G, Van Montagu M, Caplan A. Characterization of a rice gene showing organ-specific expression in response to salt stress and drought. Plant Cell. 1990;2:19–27. doi: 10.1105/tpc.2.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutch CE, Winicov I. Post-transcriptional regulation of a salt-inducible alfalfa gene encoding a putative chimeric proline-rich cell wall protein. Plant Mol Biol. 1995;27:411–418. doi: 10.1007/BF00020194. [DOI] [PubMed] [Google Scholar]
- Foolad MR, Jones RA. Mapping salt-tolerance genes in tomato (Lycopersicon esculentum) using trait-based marker analysis. Theor Appl Genet. 1993;87:184–192. doi: 10.1007/BF00223763. [DOI] [PubMed] [Google Scholar]
- Grotewold E, Chamberlin M, Snook M, Siame B, Butler L, Swenson J, Maddock S, St. Clair G, Bowen B. Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell. 1998;10:721–740. [PMC free article] [PubMed] [Google Scholar]
- Guiltinan MJ, Marcotte WR, Quatrano RS. A plant leucine zipper protein recognizes an abscisic acid response element. Science. 1990;250:267–270. doi: 10.1126/science.2145628. [DOI] [PubMed] [Google Scholar]
- Gyorgyey J, Gartner A, Nemeth K, Magyar Z, Hirt H, Heberle-Bors E, Dudits D. Alfalfa heat shock genes are differentially expressed during somatic embryogenesis. Plant Mol Biol. 1991;16:999–1007. doi: 10.1007/BF00016072. [DOI] [PubMed] [Google Scholar]
- Hoagland DT, Arnon DI. The water culture method for growing plants without soil. Calif Agric Exp Stn Circ. 1938;347:1–39. [Google Scholar]
- Hoekema A, Hirsch PR, Hooykaas PJJ, Schilperoort RA. A binary plant vector strategy based on separation of the vir- and T-region of A. tumefaciens Ti plasmid. Nature. 1983;303:179–180. [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Kakidani H, Ptashne M. Gal4 activates gene expression in mammalian cells. Cell. 1988;52:161–167. doi: 10.1016/0092-8674(88)90504-1. [DOI] [PubMed] [Google Scholar]
- Kishor PBK, Hong Z, Miao G-H, Hu C-A, Verma SPS. Over-expression of Δ1-pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol. 1995;108:1387–1394. doi: 10.1104/pp.108.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel D, Salamini F, Bartels D, Dale P, Baga M, Szalay A. Analysis of a desiccation and ABA-responsive promoter isolated from the resurrection plant Craterostigma plantagineum. Plant J. 1993;4:29–40. doi: 10.1046/j.1365-313x.1993.04010029.x. [DOI] [PubMed] [Google Scholar]
- Pilon-Smits EAH, Ebskamp MJM, Paul MJ, Jeuken MJW, Weisbeek PJ, Smeekens SCM. Improved performance of transgenic fructan-accumulating tobacco under drought stress. Plant Physiol. 1995;107:125–130. doi: 10.1104/pp.107.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pla M, Gomez J, Goday A, Pages M. Regulation of the abscisic acid-responsive gene rab28 in maize viviparous mutants. Mol Gen Genet. 1991;230:394–400. doi: 10.1007/BF00280296. [DOI] [PubMed] [Google Scholar]
- Raghothama G, Liu D, Nelson DE, Hasegawa PM, Bressan RA. Analysis of an osmotically regulated pathogenesis-related osmotin gene promoter. Plant Mol Biol. 1993;23:1117–1128. doi: 10.1007/BF00042346. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schenk RU, Hildebrandt AC. Medium and techniques for induction of growth of monocotyledonous and dicotyledonous plant cell cultures. Can J Bot. 1972;50:199–204. [Google Scholar]
- Shen Q, Zhang P, Ho T-HD. Modular nature of abscisic acid (ABA) response complexes: composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell. 1996;8:1107–1119. doi: 10.1105/tpc.8.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene expression and signal transduction in water-stress response. Plant Physiol. 1997;115:327–334. doi: 10.1104/pp.115.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub PF, Shen Q, Ho T-hD. Structure and promoter analysis of an ABA- and stress-regulated barley gene HVA1. Plant Mol Biol. 1994;26:617–630. doi: 10.1007/BF00013748. [DOI] [PubMed] [Google Scholar]
- Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C. The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell. 1998;10:135–154. doi: 10.1105/tpc.10.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarczynski MC, Jensen RG, Bohnert HJ. Stress protection of transgenic tobacco by production of the osmolyte mannitol. Science. 1993;259:508–510. doi: 10.1126/science.259.5094.508. [DOI] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell. 1993;5:1529–1539. doi: 10.1105/tpc.5.11.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winicov I. Characterization of salt tolerant alfalfa (Medicago sativa L.) plants regenerated from salt tolerant cell lines. Plant Cell Rep. 1991;10:561–564. doi: 10.1007/BF00232511. [DOI] [PubMed] [Google Scholar]
- Winicov I. cDNA encoding putative zinc finger motifs from salt-tolerant alfalfa (Medicago sativa L.) cells. Plant Physiol. 1993;102:681–682. doi: 10.1104/pp.102.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winicov I. Characterization of rice (Oryza sativa L.) plants regenerated from salt-tolerant cell lines. Plant Sci. 1996;113:105–111. doi: 10.1007/BF00232511. [DOI] [PubMed] [Google Scholar]
- Winicov I. New molecular approaches to improving salt tolerance in crop plants. Ann Bot. 1998;82:703–710. [Google Scholar]
- Winicov I, Bastola DR. Salt tolerance in crop plants: new approaches through tissue culture and gene regulation. Acta Physiol Plant. 1997;19:435–449. [Google Scholar]
- Winicov I, Button JD. Induction of photosynthesis gene transcripts by sodium chloride in a salt tolerant alfalfa cell line. Planta. 1991;183:478–483. doi: 10.1007/BF00194267. [DOI] [PubMed] [Google Scholar]
- Winicov I, Deutch CE. Characterization of a cDNA clone from salt-tolerant alfalfa cells that identifies salt inducible root specific transcripts. J Plant Physiol. 1994;144:222–228. [Google Scholar]
- Winicov I, Krishnan M. Transcriptional and post-transcriptional activation of genes in salt-tolerant alfalfa cells. Planta. 1996;200:397–404. [Google Scholar]
- Winicov I, Waterborg JH, Harrington RE, McCoy TJ. Messenger RNA induction in cellular salt tolerance of alfalfa (Medicago sativa) Plant Cell Rep. 1989;8:6–11. doi: 10.1007/BF00735767. [DOI] [PubMed] [Google Scholar]
- Xu D, Duan X, Wang B, Hong B, Ho T-HD, Wu R. Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 1996;110:249–257. doi: 10.1104/pp.110.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]