Abstract
Understanding the regulatory networks which control specific macrophage phenotypes is essential in identifying novel targets to correct macrophage mediated clinical disorders, often accompanied by inflammatory events. Since mesenchymal stromal cells (MSCs) have been shown to play key roles in regulating immune functions predominantly via a large number of secreted products, we used a fractional factorial approach to streamline experimental evaluation of MSC mediated inflammatory macrophage regulation. Our macrophage reprogramming metrics, human bone marrow MSC attenuation of macrophage pro-inflammatory M1 TNFα secretion and simultaneous enhanced expression of the M2 macrophage marker, CD206, were used as analysis endpoints. Objective evaluation of a panel of MSC secreted mediators indicated that PGE2 alone was sufficient in facilitating macrophage reprogramming, while IL4 only provided partial reprogramming. Inhibiting stromal cell PGE2 secretion with Indomethacin, reversed the macrophage reprogramming effect. PGE2 reprogramming was mediated through the EP4 receptor and indirectly through the CREB signaling pathway as GSK3 specific inhibitors induced M1 macrophages to express CD206. This reprogramming pathway functioned independently from the M1 suppression pathway, as neither CREB nor GSK3 inhibition reversed PGE2 TNF-α secretion attenuation. In conclusion, fractional factorial experimental design identified stromal derived PGE2 as the factor most important in facilitating macrophage reprogramming, albeit via two unique pathways.
Keywords: fractional factorial design, macrophage, mesenchymal stromal cells, PGE2
Introduction
Macrophages exhibit tremendous phenotypic plasticity which is regulated by their physiological or pathological environments (Kigerl et al., 2009). These phenotypic states can broadly be categorized as pro-inflammatory M1 and anti-inflammatory M2. When the transitions between these phenotypic states are not regulated appropriately, chronic inflammation ensues which can lead to permanent tissue damage (Sindrilaru et al., 2011). Deleterious macrophage function observed in several diseases supports the notion that macrophage phenotypes play an integral role in neoplastic and pathological progression (Dayan et al., 2011; Kigerl et al., 2009; Nemeth et al., 2009; Ruffell et al., 2012; Sindrilaru et al., 2011). Therefore, understanding the regulation of macrophage plasticity is essential in order to identify novel therapeutic targets.
A growing body of evidence supports the fact that stromal cells, and MSCs in particular, selectively control the development of immune cell phenotypes in both primary and secondary lymphoid organs (Parsonage et al., 2005). For example, MSCs prevented dendritic cell differentiation as well as pro-inflammatory cytokine secretion (Jiang et al., 2005; Nauta et al., 2006). Furthermore, MSCs have been found to regulate macrophage inflammatory functions in vivo (Fujiu et al., 2011; Nemeth et al., 2009). Thus, understanding MSC/macrophage interactions may provide novel insights into the cellular and molecular interactions which govern macrophage reprogramming events.
Several in vitro studies demonstrated monocyte differentiation to M2 phenotypes when co-cultured with MSCs (Kim and Hematti, 2009; Zhang et al., 2010). While the M1 macrophage stage is an essential component of normal wound healing (Lucas et al., 2010; Nemeth et al., 2009), most observed macrophage pathologies manifest in an inappropriate transition of M1 to M2 phenotypes. Therefore, we focused our studies on MSC regulation of the M1 to M2 transition.
MSC intercellular control is regulated largely by a very large number of MSC secreted products, which can act either individually or synergistically. Therefore, given the enormous number of potential experimental variables, the analysis of MSC mediated macrophage M1 to M2 transition was streamlined by incorporating fractional factorial experimental design to identify single or combinatorial factor effects. The macrophage cultures were exposed to 13 of the most relevant MSC immunomodulatory factors at their biologically secreted levels. Whereas, exploring the effects of each of these factors and their combinations requires 8,192 wells for a single biological replicate, 1/64 fractioning of the factorial experimental design can reduce the experimental design to only 128 wells without losing any information from single factors and 2 factor combination effects. Using this approach our studies demonstrated that the M1–M2 transition was driven by MSC secretion of the single factor PGE2, which potentiated CREB transcriptional regulation indirectly through GSK-3α inhibition and that following PGE2 activation, the PGE2/EP4 axis controls downstream MSC macrophage reprogramming.
Methods
Reagents
IL4, MCSF, IL-6, HGF, IL-10, IL-13, TNF-αR were purchased from Peprotech (Rocky Hill, NJ), GMCSF, TGFb-1 were purchased from R&D (Minneapolis, MN) and IGF-1 from Lonza (Basel, Switzerland). GSK3 inhibitors, Lithium chloride and SB415286, CREB inhibitors, pamoic acid and RO-31-8220, COX2 inhibitor Indomethacin and LPS were purchased from Sigma-Aldrich (St. Louis, MO). EP receptor antagonists, L161982 and Ah6809, and PGE2 were purchased from Cayman Chemical (Ann Arbor, MI). Rabbit polyclonal Abs to CD206 and α-Tubulin were purchased from Abcam (Cambridge, MA). Rabbit monoclonal Abs to phosphorylated CREB (ser-133), GSK-α (ser-21), GSK3-β (ser-9), and IL-6 (tyr-641) as well as to CREB, GSK3-α, GSK3-β, and IL-6 were purchased from Cell Signaling (Boston, MA).
Mesenchymal Stem/Stromal Cells
All cell cultures were incubated in a humidified 37°C and 5% CO2 environment. Human MSC (MSC) were purchased from Texas A&M at passage 1 and cultured as previously described (Parekkadan et al., 2007). Briefly, hMSC were cultured in MEM-α (Gibco) medium, containing no deoxy and ribo nucleosides, supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Lawrenceville, GA), 1 ng/mL basic fibroblast growth factor (bFGF) (Gibco), 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco). hMSC were plated at 5,000 cells/cm2 and allowed to proliferate to 70% confluence (approximately 4–5 days) before passaging. Only hMSC at passages 2 through 5 were used in experiments.
M1 Macrophage Cultures
Peripheral blood mononuclear cells were collected from blood of healthy donors (Blood Center of New Jersey) by density gradient centrifugation using Ficoll at a density of 1.077 (GE Healthcare, Rahway, NJ). Monocytes were isolated to high purity (>90%) by magnetic cell sorting using anti-CD14 coated beads (per manufacturer recommendation; Miltenyi Biotec, Auburn, CA). 107 CD14+ monocytes were cultured on 175 cm2 flasks (BD) in RPMI (Gibco). RPMI was supplemented with 10% fetal bovine serum (FBS) (Gibco), 100 U/mL Penicillin (Gibco), 100 μg/mL streptomycin (Gibco) and 400 mM L-glutamine (Gibco). Monocytes were cultured for 2 h and then washed three times with PBS to remove non-adherent cells. Monocytes were then cultured for 7 days in culture medium supplemented with 5 ng/mL GMCSF. On the 7th day of culture, macrophages were washed once with PBS and detached with trypsin-EDTA (Gibco) for 30 min at room temperature. Cells were re-suspended in RPMI, counted and re-plated at 1 × 105 cells/mL in a 24-well plate (Corning, NY) and allowed to attach overnight. The following day cells were used for experimentation. This macrophage subset has been shown to express a repertoire of M1 macrophage phenotypes, which has been validated through epigenetic (Satoh et al., 2010), genetic (Fleetwood et al., 2007; Szanto et al., 2010), and protein (Verreck et al., 2004, 2006) characterization.
MSC M1 Macrophage Co-Culture
Macrophage/MSC co-cultures were established with 8 μm transwell inserts (Corning). The transwell membrane excluded MSC migration into the macrophage compartment. Macrophages were treated with 1 μg/mL LPS from Escherichia coli (serotype 055:B5, Sigma-Aldrich) and MSC at various cell concentrations (2 × 103, 2 × 104, and 105 cells/mL) within transwell inserts. COX-2 blocking was performed under the identical culture conditions, but in the presence of Indomethacin (10 μM). The cultures were incubated for 2 or 5 days, after which culture supernatants were collected and macrophages fixed with 4% paraformaldehyde (PF).
Fractional Factorial Design
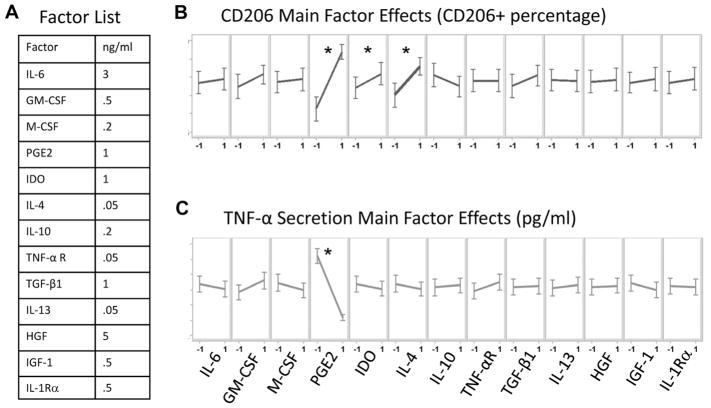
Fractional factorial experimental designs have been successfully utilized to explore the main and interaction effects of several factors on desired biological assayed parameters while minimizing the required experimental conditions in biological experimentation (Zupke et al., 1998). Herein, 13 MSC secreted immunomodulatory candidates (IL-6, GM-CSF, M-CSF, PGE2, IDO, IL-4, IL-10, IL-13, IL-1Rα, TNF-α receptor 1 (TNF-αR1), IGF-1, HGF, and TGF-β1) and their concentrations were selected based on prior experimental work and previous publications as described in Figure 2A. A 1/64 fractional factorial experiment was chosen to bring the runs of a complete, 1/2 or 1/4 factorial to a biological feasible 128 run experiment. The 1/64 fractional factorial experiment was designed and performed based on Montgomery’s suggestions (Montgomery, 1994) using 128 experimental conditions, where two levels for each of the 13 factors were evaluated as described in supplementary Table SI, where “−1” corresponds to the absence of the factor and “1” corresponds to the presence of the factor at the MSCs secreted concentration (Fig. 2A). The fractional factorial design utilized the following confounding rules and aliasing to evaluate the main effects of all 13 factors mentioned above as well as the two factor interactions between them.
Figure 2.
Fractional factorial evaluation of MSC factor effects on M1 function. A: List of MSC secreted factors and their concentrations evaluated in the study. A 128-condition experiment was designed to identify factors which may mediate MSC regulation of M1 macrophages. Conditions were made, spiked with LPS and transferred to M1 macrophage 96 well cultures. After 48 h cells were immunostained for CD206 and TNF-α levels were measured via ELISA. B and C: Illustrate main factor effects on the metrics CD206 and TNF-α, respectively. −1 and 1 designate the absence or presence of the factor, respectively. The slope of the line conveys the magnitude of change in the presence of a particular factor. Differences were considered statistically significant based on a confidence interval of 95% (P < 0.05). PGE2 and IL4 demonstrated the ability to significantly promote CD206. However, only PGE2 was able to significantly attenuate TNF-α secretion. *P < 0.05 represent statistical significant based on one-way ANOVA
Confounding rules:
Alias structure (up to 2 factor interactions):
Each of the 128 different experimental conditions was developed within 96 well plates and then spiked with LPS. The cocktail array was then transferred to M1 macrophages previously seeded in 96 well plates at a density of 105 cells/mL, for 48 h incubation period. Following the 48 h incubation, supernatants were collected for secreted TNF-α ELISA detection and the cells paraformaldehyde fixed for CD206 immunostaining (N = 4). The data from the factorial experiment were analyzed using statistical analysis software 9.2 (SAS Institute, Inc., Cary, NC) where one-way analysis of variance (ANOVA) was performed to evaluate the statistical difference of the means of main effects and of two factor interactions on CD206+ cells (Supplementary Table SII) and secreted TNF-α levels (Supplementary Table SIII). The residuals for the %CD206+ and secreted TNF-α models were calculated as the difference between the experimentally observed mean value and the model predicted value for each of the 128 DOE combinations to generate the plot of residuals as indicated in supplementary Figures 1 and 2, respectively. The residuals exhibited lack of correlation by visual inspection as well as by a correlation values nearing zero and regression coefficient of determination (R2) values approaching zero when the data are fitted to a linear or 2nd order polynomial models as indicated in the supplementary Figures S1 and S2. The lack of correlation in the residuals validates the DOE based model analysis described in Equations (1) and (2).
Macrophage PGE2 and Blocking Studies
Cytotoxic thresholds for Lithium chloride, SB415286, pamoic acid and RO-31-8220 were determined by titration experiments for each inhibitor. Macrophages were pre-treated with 20 μM SB415286 (1 h), 40 mM Lithium Chloride (1 h), 100 μM Leflunomide (1 h), and 1 μM RO-31-8220 (10 min). Inhibitors as opposed to siRNA were used to study the targets of interest due to the technical and immunogenic difficulties associated with monocyte transduction (Muhlebach et al., 2005). Macrophages were treated with cocktails containing various combinations of LPS, PGE2, and IL4 in the presence of the different inhibitors for 48 h before supernatant collection for ELISA and cells PF fixed for CD206 immunostaining. Conditions for macrophage protein immunoblotting studies were established identically except protein extraction was performed after 0.25, 1, 4, and 24 h of culture.
Immunostaining and Cytokine Measurement
Macrophages were immunostained as previously described (Kim and Hematti, 2009) with Rabbit pAb to CD206 (1 μg/mL). Images were acquired using an Olympus IX81 spinning disc confocal microscope and stereology was performed using Slidebook software (Olympus, Center Valley, PA). To analyze CD206 positive cells within the M1 population, images were thresholded based on levels determined from isotype controls wells. There was a slight basal expression of CD206 in the M1 culture, however after induction with either PGE2 or IL4 a significant percentage of the population expressed high levels of CD206. The threshold for CD206 expressing cells was set by taking the average basal expression of CD206 in the M1 population and adding one standard deviation. All CD206 percentages represent the percentage of the population above basal levels. Macrophage TNF-α or IL12 secretion was used to characterize M1 function. Immunocytology markers are not able to clearly capture the macrophage transition from M1 to M2 and the markers that suffice to assess M1 to M2 transition in mice differ from human macrophages (Geissmann et al., 2010). Supernatants were analyzed via ELISA for IL-10, TNF-α, IL-12 (Biolegend, San Diego, CA), and PGE2 (Ann Arbor, MI).
Immunoblotting
Whole macrophage extracts were obtained and protein levels quantitated by BCA protein Assay kit (Thermo Scientific, Waltham, MA). Forty micrograms of cell lysates were fractioned on 10% polyacrylamide gels (Bio-Rad, Hercules, CA) by SDS–PAGE and transferred to nitrocellulose membranes (Bio-Rad). Nitrocellulose membranes were blocked for 90 min with 5% BSA (phosphorylated protein) or 5% milk (total protein). Membranes were then incubated with specific antibodies overnight and the following day enhanced chemiluminescence was used for detection. Membranes were stripped for 15 min with a solution of 1.5% (w/v) glycine, 0.1% (w/v) SDS, 1% tween and brought to a pH of 2.2. Membranes were washed twice with PBS, twice with TBST and then incubated with primary antibody to α-Tubulin overnight. The next day enhanced chemiluminescence was used for detection.
Statistical Analysis
Each data point represents the mean of three or more experiments (each with biological triplicates), and the error bars represent the standard deviation from the mean, unless otherwise specified. Statistical significance was determined using one or two way ANOVA analysis with 95% confidence intervals.
Results
MSC Facilitate M1 Macrophage Reprogramming to M2 Phenotypes
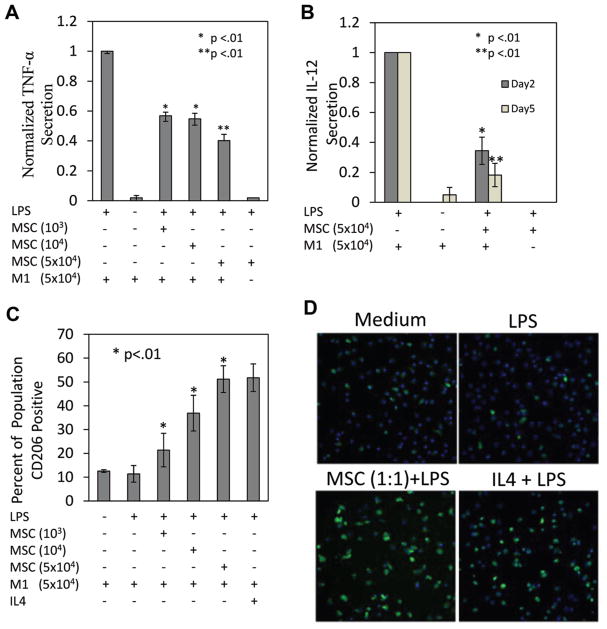
To investigate the regulatory effects of MSCs on macrophage function, monocyte derived M1 macrophages were obtained from human blood donors by CD14+ monocyte isolation and GMCSF (5 ng/mL) differentiation. M1 macrophages were plated within transwell inserts, which prevented cell–cell contact and enabled us to study only the effect of MSC secreted products on the macrophages. MSC were cultured with the macrophages at ratios of 1:1, 1:5, and 1:50 (MSC:M1) in the presence of LPS. In the absence of this activating factor, MSC effects were not induced (not shown). After 48 h of culture, TNF-α was assessed via ELISA and found to be significantly reduced at all M1: MSC ratios tested (Fig. 1A). We next assessed M1 macrophage IL-12 secretion at 2 and 5 days after LPS stimulation, focusing on the 1:1 ratio, which resulted in the highest TNF-α attenuation. Following LPS stimulation macrophage IL-12 secretion was maintained at steady levels for at least 5 days. MSC attenuated IL-12 secretion ~60% on day 2 and ~80% by day 5 (Fig. 1B).
Figure 1.
MSC attenuate macrophage pro-inflammatory cytokine secretion. A: At day 2 M1 macrophage TNF-α secretion was diminished with MSC co-culture. There was a concentration dependent reduction in TNF-α, maximum attenuation (55% reduction) was achieved at a: MSC/M1 ratio of 1:1. *Represents statistical significance relative to LPS and ** to MSC/M1 ratios of 1:50 and 1:5. B: MSCs attenuate M1 macrophage IL12 (p40) secretion at days 2 and 5 after LPS stimulation. *Represents statistical significance relative to LPS and ** to MSC/M1 day 2 cultures. C: MSC promote M1 macrophages stimulated with LPS to express CD206. At day 2 MSC co-culture resulted in a significant increase in the percentage of macrophages expressing CD206 relative to LPS and medium conditions. There was a dose dependent increase in the percentage of CD206 expressing cells which plateaued at ~50%. IL4 (10 ng/mL) promoted CD206 expression to approximately the same level as PGE2. *,**Represent statistical significance relative to LPS. D: Images depicting the elevated levels of CD206 observed when M1 macrophages are treated with MSC and IL4.
Having demonstrated that MSC can attenuate M1 macrophage secretion, we evaluated the expression of CD206, an M2 marker, to determine if MSC facilitated macrophage M2 reprogramming. Initially, ~10% of the M1 population, in basal or LPS treated conditions, was CD206+. When increased numbers of MSC were introduced into the cultures, a steady increase in CD206+ cells in the macrophage population was observed (Fig. 1C). IL-4, a cytokine which promotes M2 macrophage phenotypes (Stein et al., 1992), was used as a positive control. IL-4 (10 ng/mL) treatment induced ~50% of the population to express CD206 (Fig. 1C and D). Similarly, MSC administered to the culture at a 1:1 ratio induced ~50% of the M1 population to express CD206 (Fig. 1C and D). Increases in the ratio of MSC to macrophages did not lead to a further increase in the percentage of CD206+ cells (data not shown). Consistently, with either MSC or IL4 treatment, ~ 40% of the population was not induced to express CD206.
Fractional Factorial Design Determination of MSC Mediator(s) of M1 Reprogramming
To objectively assess the effects of MSC secreted factors and their combinations on the attenuation of M1 macrophage inflammatory TNF-α cytokine secretion and increase in M2 CD206 expression, a 2 level 1/64 fractional factorial experiment was performed using a panel of 13 MSC secreted immune mediators at the approximate level MSC secrete the factor in vitro (Fig. 2A and Supplementary Table SI). Following 48 h incubation of M1 macrophages with each of the 128 MSC-secreted factor experimental conditions, the CD206+ cells and the secreted TNF-α levels were analyzed. Using the statistical analytical system (SAS) software, a one-way ANOVA was performed to identify single factors and 2 factor interactions that are able to statistically significantly alter the CD206+ cells and the secreted TNF-α levels as indicated in supplementary Figures 2 and 3. The relative contribution of these factors to the change in the M1 and M2 markers is demonstrated in Figure 2 and is indicated by the following predictive model equations (the models fit statistics is represented in supplementary Tables SIIb and SIIIB):
| (1) |
Predictive model for the statistically significant contribution to macrophage CD206+ percentage by a single factor or by the interaction of any two factors where the concentration of each factor is normalized to be between −1 and 1. The statistical significance of the factors main and interaction effects was determined by one-way ANOVA (α = 0.05, Supplementary Table SIIA). Statistically not significant factors main effects were included in the model if they are part of a statistically significant interaction effect to preserve a proper model hierarchy. The predictive model statistical fit information is presented in Supplementary Table SIIB.
Out of the 13 factors examined, only three candidate factors led to a statistically significant increase in the percentage of CD206+ macrophages in the culture: PGE2, IDO, and IL-4 (Fig. 2B and supplementary Table SIIA). Based on the predictive model for CD206+ percentage described in Equation (1), PGE2 is the factor that contributed the most to the CD206+ percentage increase by a single factor (~50% of the total contribution to CD206 increase by single factors). While IL-4 contributed to ~25% increase in CD206+ when present as a single factor to the culture, this contribution is dramatically diminished when IL-4 is present in the culture together with PGE2, as indicated by the negative interaction contribution of the two in Equation (1). In addition, the other statistically significant two factors interaction effects resulted in a decrease or only in a minor increase in CD206+ percentage emphasizing the significant role of PGE2 as a single factor contributor to the increase in the M2 macrophages marker, CD206+.
| (2) |
Predictive model for the statistically significant contribution to macrophage TNF-α reduction by a single factor or by the interaction of any two factors where the concentration of each factor is normalized to be between −1 and 1. The statistical significance of the factors main and interaction effects was determined by one-way ANOVA (α = 0.05, Supplementary Table SIIIA). Statistically not significant factors main effects were included in the model if they are part of a statistically significant interaction effect to preserve a proper model hierarchy. The predictive model statistical fit information is presented in Supplementary Table SIIB.
Out of the 13 factors examined, PGE2 was the only single factor that led to a statistically significant reduction in secreted TNF-α in the culture (Fig. 2C and supplementary Table SIIIA). Interestingly, factors that were able to significantly increase CD206+ percentage, namely IL-4 and IDO, did not lead to a statistically significant decrease in TNF-α secretion (Fig. 2C, Supplementary Table SIIIA). Overall, the results suggest that within the panel evaluated, PGE2 alone was able to attenuate M1 macrophage TNF-α secretion and promote M2 macrophage CD206 expression.
MSC PGE2 Secretion Mediates Macrophage Inflammatory Action and Reprogramming
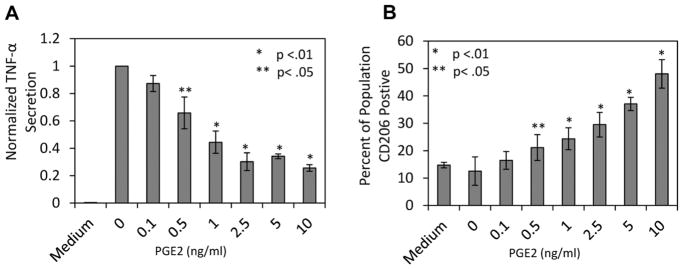
Having established PGE2 as the most likely MSC secreted factor responsible for M1 reprogramming, experiments were designed to examine the PGE2 concentration dependence in attenuation of M1 secretion and promotion of CD206 expression. M1 macrophages were stimulated with LPS in the presence of several different concentrations of PGE2. TNF-α ELISA quantitation indicated that there was a dose dependent attenuation of TNF-α secretion with increasing PGE2 concentrations. However, at and above 2.5 ng/mL, PGE2 attenuation was ~75% (Fig. 3A). The percentage of macrophages that was CD206+ correlated with PGE2 dose as well. However, CD206 expression plateaued at 10 ng/mL (Fig. 3B).
Figure 3.
PGE2 attenuates M1 TNF-α secretion and promotes M1 CD206 expression. M1 macrophages were stimulated with LPS and immediately treated with increasing concentrations of PGE2, except for the none treated control labeled Medium. At day 2 macrophage (A) TNF-α secretion and (B) CD206 expression were quantified. Increasing concentration of PGE2 attenuated M1 macrophage TNF-α secretion, which plateaued at 2.5 ng/mL at a reduction of ~75%. The percentage of CD206 expressing macrophages displayed the reverse relationship to increasing PGE2 concentration, however displayed a more linear response, and reached a maximum value of ~50%. *,**Represent statistical significance of P < 0.01 and P < 0.05 relative to LPS conditions respectively.
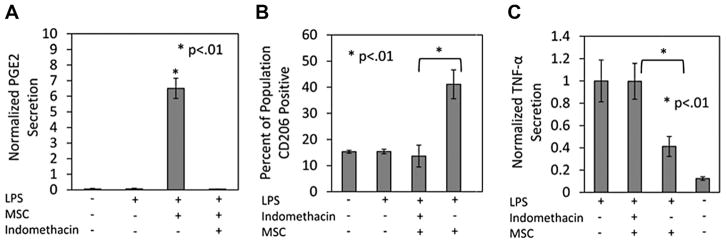
Having determined that PGE2 can modulate macrophages, the effect of MSC derived PGE2 secretion on macrophage plasticity was evaluated. Supernatants were harvested from MSC/macrophage co-cultures and PGE2 was measured. Secreted PGE2 was not detected in macrophage supernatants in either basal or stimulated conditions (Fig. 4A). Conversely, in supernatants collected from MSC/macrophage co-cultures, PGE2 was detected at a concentration of ~7.5 ng/mL (Fig. 4A). Adding Indomethacin, a factor known to inhibit PGE2 synthesis (Kuroda and Yamashita, 2003), prevented MSC secretion of PGE2 (Fig. 4A). We next assessed the ability of MSC to attenuate macrophage TNF-α and promote CD206 elevation in the presence of Indomethacin. Our results indicated that both CD206 promotion (Fig. 4B) and TNF-α attenuation (Fig. 4C) were completely reversed in the presence of Indomethacin.
Figure 4.
MSC PGE2 secretion promotes macrophage reprogramming. A: Supernatant from M1: MSC co-cultures treated with LPS (1 μg/mL) were assessed for PGE2 concentration. M1 macrophages in basal and stimulated conditions did not produce PGE2. However, in the presence of MSC ~6.5 ng/mL was detected. When a COX2 inhibitor, indomethacin (10 μM), is administered to the co-culture MSC PGE2 secretion is eliminated. *Represents statistical significance (P < 0.01) relative to LPS, medium and indomethacin. B: CD206 expression and (C) TNF-α secretion were assessed with indomethacin present in the MSC:M1 co-culture. The inhibition of MSC PGE2 secretion prevented the percent increase of CD206 expressing cells as well as the attenuation of TNF-α. *Represents statistical significance (P < 0.01) between MSC:M1 conditions in the presence and absence of indomethacin.
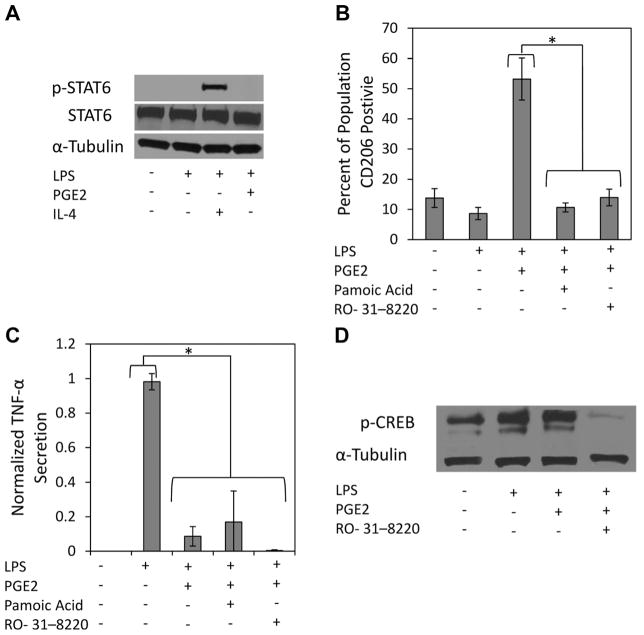
PGE2 Effects on Macrophage Function Are Mediated Through CREB Signaling
The transcription of M2 macrophage associated genes has been shown to be mediated by STAT6 transcription factor binding (Gordon, 2003). Therefore, we assessed p-STAT6 expression and found that M1 macrophage p-STAT6 was only detected with IL-4 treatment (Fig. 5A). Therefore, a different signaling mechanism was explored to explain PGE2 regulation of macrophage plasticity. Recently, cAMP response element-binding (CREB) has been implicated in macrophage reprogramming (Ruffell et al., 2009) and PGE2 mediated cAMP elevation has been shown to activate CREB through phosphorylation of ser-133 (Bullock and Habener, 1998). Therefore, experiments were designed to assess whether CREB is involved in PGE2 macrophage regulation. Two CREB inhibitors were identified, Pamoic Acid and RO-31-8220. Pamoic acid (PA) is known to prevent p-CREB/CREB binding protein (CBP) binding, which is imperative for target gene induction (Best et al., 2004). RO-31-8220 is a protein kinase inhibitor, which prevents phosphorylation of CREB at ser-133 (Ruffell et al., 2009). PA and RO-31-8220 both prevented PGE2 promotion of CD206 (Fig. 5B), but did not alter PGE2 attenuation of TNF-α (Fig. 5C).
Figure 5.
PGE2 promotes increased M1 macrophage CD206 expression through CREB signaling. A: M1 macrophages were stimulated with LPS (1 μg/mL) and treated with IL-4 (10 ng/mL) or PGE2 (10 ng/mL) for 15 min before protein extraction. Immunoblotting for phosphorylated STAT6 at tyr-641 showed that PGE2, unlike IL4, does not promote STAT6 activation. B: M1 macrophages were pre-incubated with RO-31-8220 for 10 min, before stimulation and treatment with PGE2 (10 ng/mL). Pamoic acid was administered at the time of LPS stimulation and PGE2 treatment. Inhibition of CREB signaling with Pamoic acid or RO-31-8220 prevented MSC promotion of M1 CD206 expression, but had no effect on (C) TNF-α secretion. D: Immunoblotting for phosphorylated CREB, at ser-133 15 min after LPS stimulation and treatment with PGE2 did not show increased expression of pCREB. LPS stimulation alone was able to significantly increase p-CREB expression. Immunoblotting confirmed RO-31-8220 inhibition of CREB activation.
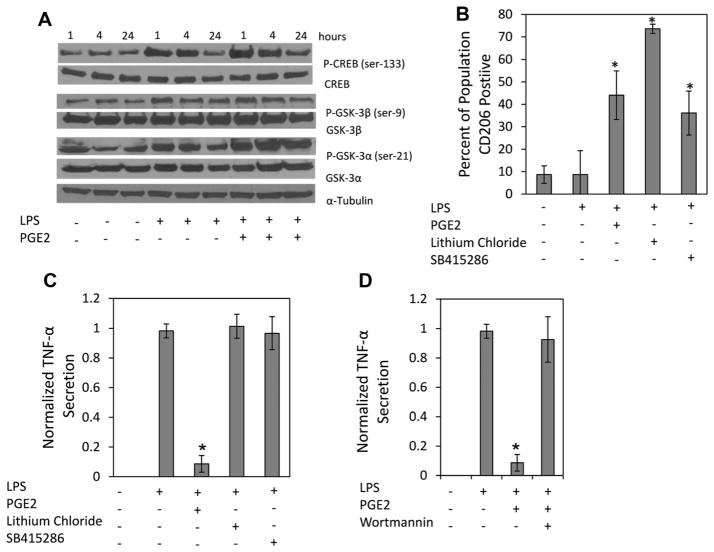
Next, immunoblotting for pCREB was performed to determine if PGE2 treatment led to increased CREB phosphorylation at ser-133. Macrophages stimulated with LPS exhibited enhanced pCREB expression above basal levels, which was not enhanced by PGE2 after 15 min of treatment (Fig. 5D). This suggests that reprogramming occurs through a mechanism independent of direct CREB phosphorylation. Phosphorylation of Ser-129, in addition to the ser-133, by glycogen synthase kinase 3 (GSK3) inhibits CREB binding affinity to CRE promoter regions (Grimes and Jope, 2001). GSK3 activity is negatively regulated by several factors, two of which PGE2 has been shown to induce, PI3K and cAMP (Fang et al., 2000; Moore and Willoughby, 1995). It was therefore posited that over time, PGE2 treatment would phosphorylate GSK3, preventing its inhibition of CREB. M1 macrophages stimulated with LPS and treated with PGE2 were cultured for 24 h. Protein was extracted at 1, 4, and 24 h for immunoblotting. PGE2 treatment resulted in prolonged and enhanced phosphorylation of GSK3-α at serine 21 compared to all other conditions (Fig. 6A). In contrast, GSK3-β did not display differences in phosphorylation at ser-9 with PGE2 treatment (Fig. 6A). CREB de-phosphorylation was slightly diminished by 24 h with PGE2 treatment (Fig. 6A). At no time point was phosphorylation of STAT6 observed in any condition except IL-4, which was present at 1 h, but undetectable by 4 h (data not shown).
Figure 6.
PGE2 inhibits GSK-3α and leads to increased M1 CD206 expression. A: Protein samples from M1 macrophages stimulated with LPS and treated with PGE2 (10 ng/mL) for 1, 4, and 24 h were immunoblotted for GSK, CREB, and STAT6 phosphorylation. PGE2 treatment increased the expression of phosphorylated GSK-3α at ser-21 over 24 h. No significant differences were observed in the expression of phosphorylated GSK-3β at ser-9 in any condition. PGE2 treatment modestly prevented CREB de-phosphorylation overtime. B and C: M1 macrophages were pre-treated with GSK inhibitors, lithium chloride (40 mM) and SB415286 (40 μM), for 1 h before LPS stimulation. Two days of LPS stimulation occurred in the presence of the inhibitors and led to an increase in the percentage of (B) CD206 positive cells in the M1 macrophage population, but had no effect on (C) TNF-α Secretion. D: M1 macrophage were stimulated with LPS and treated with PGE2 in the presence of a PI3k inhibitor, which reversed PGE2 attenuation of M1 macrophage TNF-α secretion. *Represents statistical significance (P < 0.01) relative to LPS conditions.
To test the role of GSK3 in regulating M1 macrophage function GSK3 inhibitors, lithium chloride and SB415286, were cultured with M1 macrophages stimulated with LPS. Inhibition of GSK3 led to elevation of CD206 after 48 h (Fig. 6B). However, GSK3 inhibition did not result in TNF-α attenuation (Fig. 6C). Since the PI3K pathway has been found to regulate pro-inflammatory cytokine secretion (Fukao et al., 2002), M1 macrophages were stimulated with LPS, and treated with PGE2 in the presence and absence of wortmannin, a specific PI3K inhibitor. Wortmannin reversed PGE2 TNF-α attenuation (Fig. 6D), but had no effect on CD206 promotion (data not shown).
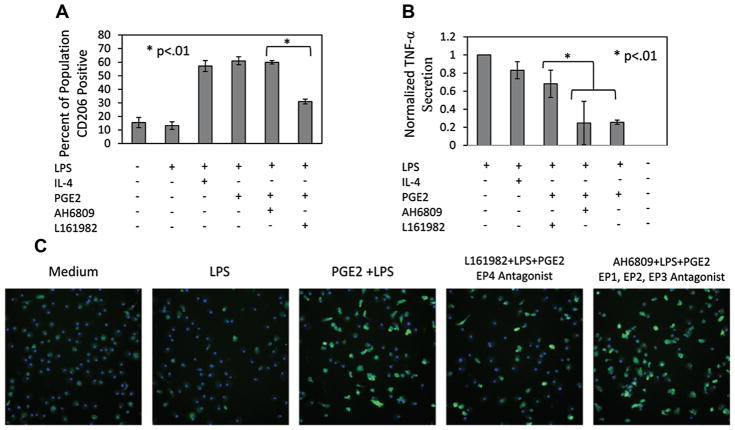
PGE2 EP4 Axis Regulates Macrophage Inflammatory Phenotype
In order to identify the specific PGE2 receptor responsible for M1 macrophage regulation, antagonists to EP receptors were employed. Recently, PGE2 binding to EP4 has been found to result in the downstream activation of the PI3k pathway (Moore and Willoughby, 1995). Therefore, we anticipated that PGE2–EP4 interactions would specifically be responsible for M1 macrophage regulation. Specific antagonists (L161982) to EP4 were tested. Macrophages were incubated for 48 h in the presence of LPS with IL4 or PGE2 as well as with and without EP receptor antagonists. The data indicate that blocking EP4 results in significant reduction in the number of CD206+ cells (~30%) (Fig. 7A and C). However, blockade of the other receptors did not inhibit PGE2 promotion of CD206 (Fig. 7A). Likewise, EP4 antagonists inhibited PGE2 attenuation of TNF-α (Fig. 7B). Consistently, IL4 promoted CD206, but did not significantly attenuate TNF-α (Fig. 7A and B).
Figure 7.
PGE2 mediates M1 macrophage reprogramming through the EP4 receptor. LPS stimulated M1 macrophages were treated with IL4 (10 ng/mL), PGE2 (10 ng/mL), PGE2 with an EP4 antagonist (L161982) or a general antagonist to the other EP receptors (AH6809). A: The increase in the percentage of CD206 positive cells previously observed with PGE2 treatment was diminished with an EP4 antagonist. Antagonists to the other EP receptors did not interfere with PGE2 promotion of CD206. B: PGE2 attenuation of TNF-α was also reversed with an EP4 antagonist. Blocking the other EP receptors did not reverse PGE2 TNF-α attenuation. C: Representative images of CD206 depicting the reduction of percent CD206 positive cells when the EP4 receptors is blocked during PGE2 treatment.
Discussion
We have incorporated fractional factorial experimental design to identify the MSC secreted factor(s) which regulate inflammatory macrophage reprogramming. Fractional factorial experimental designs have been previously implemented by our lab (Zupke et al., 1998) as well as by others (Chen et al., 2011; Jaynes et al., 2013) to examine the in vitro main and interaction effects of up to 6 examined factors on selected biological assayed parameters.
In the present studies, we have utilized the approach to improve experimental efficiency to study MSC secreted factors which regulate cellular functions. Our results indicate that fractional factorial design enabled us to successfully assess single and combinatorial effects of 13 MSC secreted factors, thereby reducing experimental conditions from 8,192 to only 128. We demonstrated that MSC can promote reprogramming of human monocyte derived M1 macrophages predominantly via PGE2 secretion alone (Verreck et al., 2004).
MSC co-culture both attenuated M1 macrophage pro-inflammatory cytokine levels and promoted M2 marker expression. This approach enabled us to investigate downstream regulation, which was dictated by PGE2 binding to the macrophage EP4 receptor and activation of CREB regulatory programs indirectly, via the inhibition of GSK-3α. Furthermore, this mode of PGE2 action differed from IL-4 which was not able to attenuate M1 macrophage secretion but mediated expression of M2 associated transcripts via STAT6 regulatory networks.
PGE2 represents one of the initial biochemical cues in an inflammatory cascade and has been considered by many as a pro-inflammatory mediator. However, there is accumulating support for anti-inflammatory PGE2 function from experimental models of colitis and peritonitis (Gilroy et al., 1999, 2003; Rajakariar et al., 2007; Reuter et al., 1996). Our data suggest that through PGE2, MSC facilitate M1 macrophage reprogramming to M2 like cells. MSC transplantation in sepsis models have also been shown to facilitate macrophage reprogramming through PGE2 (Nemeth et al., 2009). Furthermore, others have also measured MSC PGE2 secretion and implicated PGE2 to be responsible for several of the immunomodulatory effects MSC impart (Bouffi et al., 2010; Hegyi et al., 2012; Hsu et al., 2013). Therefore, stromal cell PGE2 regulation of macrophage function may represent a regulatory checkpoint in an innate immune response. The use of non-specific inhibition of Cox activity with Indomethacin does not exclude the possibility that other lipid mediators may be partly responsible for MSC regulation of macrophages. We have begun to address this question by evaluating PDG2 in our system, and thus far we have observed neither attenuation of TNF-α secretion nor promotion of CD206 expression using this mediator (data not shown). Using additional fractional factorial experiments in the future we will continue to evaluate other inhibitors to the PGE2 pathway as well as other lipid mediators to determine if they are responsible for the programming effect observed in MSC-macrophage cultures.
Our observations are consistent with previous reports (Kim and Hematti, 2009; Maggini et al., 2010; Zhang et al., 2010) which have suggested that MSC promote M2 macrophage phenotypes. The identification of PGE2 as an important mediator of macrophage attenuation consistent with Maggini et al. who used thioglycolate induced peritoneal macrophages. On the other hand, the studies of Zhan et al. indicated that IL-6 and GMCSF were responsible for MSC promotion of M2 phenotypes. IL-6 and GMCSF did not facilitate macrophage reprogramming in our system, which may be attributed to differences in macrophage maturity or species specificity (Haniffa et al., 2007). Nevertheless, our studies suggest that PGE2 can reprogram differentiated human peripheral blood derived macrophages.
We observed that downstream CREB GSK3 interactions mediated PGE2 induced M1 reprogramming, as opposed to the traditional IL4 dependent STAT-6 pathway (Szanto et al., 2010). The STAT-6 pathway was not found to be phosphorylated upon PGE2 treatment in our system, which further supports the lack of IL4 involvement in MSC regulation of M1 reprogramming. Accumulating evidence supports CREB’s involvement in immune cell functions, specifically, as a transcription factor responsible for mediating anti-inflammatory function (Hu et al., 2006; Ruffell et al., 2009; Wen et al., 2010). PGE2 has been shown to inhibit GSK3 activity (Jing et al., 2004), consistent with the GSK3 inhibition observed here. Specific inhibitors to GSK3 activity, Lithium chloride and SB415286, were able to promote CD206 expression in LPS stimulated M1 macrophages. In general, there is a considerable evidence that GSK3 plays a significant role in the regulation of innate and adaptive immune responses (Beurel et al., 2010). GSK3 activity negatively regulates the production of anti-inflammatory mediators such as IL-10 and IFN-β (Hu et al., 2006; Rehani et al., 2009; Wang et al., 2008), and therefore it may be an important regulatory checkpoint in macrophage reprogramming which could be exploited for therapy.
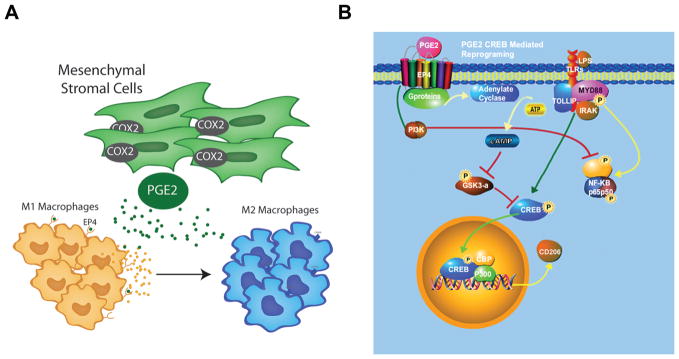
In summary, using fractional factorial experimental design, our studies have identified a PGE2 as an MSC secreted factor which can reprogram M1 macrophages to M2 like cells. This reprogramming effect is largely mediated by MSC PGE2 binding to the EP4 receptor on M1 macrophages (Fig. 8A). Lastly, PGE2 induction of macrophage reprogramming involves two pathways (1) CREB signaling, via inhibition of GSK3 activity (2) and PI3K signaling, which leads to inhibition of pro-inflammatory cytokine secretions (Fig. 8B).
Figure 8.
MSC PGE2 secretion reprograms M1 via two independent pathways to M2 macrophages. A: MSCs secrete PGE2, which binds to the EP4 receptors on M1 macrophages and facilitates their transition to M2 macrophages. B: PGE2 binding to EP4 receptors activates two independent pathways (1) the PI3k pathway which inhibits NF-kβ simulation of pro-inflammatory cytokines such as TNF-α and IL12 and (2) the cAMP pathway which leads to the inhibition of GSK and subsequent activation of CREB signaling, which leads to the transcription of genes associated with the M2 macrophage phenotype.
Supplementary Material
Acknowledgments
Contract grant sponsor: New Jersey Commission on Brain Injury Research
Contract grant number: BI1775
Contract grant sponsor: New Jersey Commission on Spinal Cord Injury
Research Grant numbers: SCC2865; NIH R21; NS070175
Contract grant sponsor: Rutgers-UMDNJ NIH Biotechnology Training Grant
Contract grant number: T32 GM00833923
These studies were partially supported by funding from the following grants: New Jersey Commission on Brain Injury Research (BI1775), New Jersey Commission on Spinal Cord Injury Research (SCC2865, NIH R21, NS070175). J.B. and N.N. were partially supported by Rutgers-UMDNJ NIH Biotechnology Training Grant (T32 GM00833923).
Footnotes
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Best JL, Amezcua CA, Mayr B, Flechner L, Murawsky CM, Emerson B, Zor T, Gardner KH, Montminy M. Identification of small-molecule antagonists that inhibit an activator: Coactivator interaction. Proc Natl Acad Sci USA. 2004;101:17622–17627. doi: 10.1073/pnas.0406374101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurel E, Michalek SM, Jope RS. Innate and adaptive immune responses regulated by glycogen synthase kinase-3 (GSK3) Trends Immunol. 2010;31:24–31. doi: 10.1016/j.it.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouffi C, Bony C, Courties G, Jorgensen C, Noel D. IL-6-dependent PGE2 secretion by mesenchymal stem cells inhibits local inflammation in experimental arthritis. PLoS ONE. 2010;5:e14247. doi: 10.1371/journal.pone.0014247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock BP, Habener JF. Phosphorylation of the cAMP response element binding protein CREB by cAMP-dependent protein kinase A and glycogen synthase kinase-3 alters DNA-binding affinity, conformation, and increases net charge. Biochemistry. 1998;37:3795–3809. doi: 10.1021/bi970982t. [DOI] [PubMed] [Google Scholar]
- Chen Y, Bloemen V, Impens S, Moesen M, Luyten FP, Schrooten J. Characterization and optimization of cell seeding in scaffolds by factorial design: Quality by design approach for skeletal tissue engineering. Tissue Eng Part C Methods. 2011;17:1211–1221. doi: 10.1089/ten.tec.2011.0092. [DOI] [PubMed] [Google Scholar]
- Dayan V, Yannarelli G, Billia F, Filomeno P, Wang XH, Davies JE, Keating A. Mesenchymal stromal cells mediate a switch to alternatively activated monocytes/macrophages after acute myocardial infarction. Basic Res Cardiol. 2011;106:1299–1310. doi: 10.1007/s00395-011-0221-9. [DOI] [PubMed] [Google Scholar]
- Fang X, Yu SX, Lu Y, Bast RC, Jr, Woodgett JR, Mills GB. Phosphorylation and inactivation of glycogen synthase kinase 3 by protein kinase A. Proc Natl Acad Sci USA. 2000;97:11960–11965. doi: 10.1073/pnas.220413597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleetwood AJ, Lawrence T, Hamilton JA, Cook AD. Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: Implications for CSF blockade in inflammation. J Immunol. 2007;178:5245–5252. doi: 10.4049/jimmunol.178.8.5245. [DOI] [PubMed] [Google Scholar]
- Fujiu K, Manabe I, Nagai R. Renal collecting duct epithelial cells regulate inflammation in tubulointerstitial damage in mice. J Clin Invest. 2011;121:3425–3441. doi: 10.1172/JCI57582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Tanabe M, Terauchi Y, Ota T, Matsuda S, Asano T, Kadowaki T, Takeuchi T, Koyasu S. PI3K-mediated negative feedback regulation of IL-12 production in DCs. Nat Immunol. 2002;3:875–881. doi: 10.1038/ni825. [DOI] [PubMed] [Google Scholar]
- Geissmann F, Gordon S, Hume DA, Mowat AM, Randolph GJ. Unravelling mononuclear phagocyte heterogeneity. Nat Rev Immunol. 2010;10:453–460. doi: 10.1038/nri2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy DW, Colville-Nash PR, McMaster S, Sawatzky DA, Willoughby DA, Lawrence T. Inducible cyclooxygenase-derived 15-deoxy(Delta) 12–14PGJ2 brings about acute inflammatory resolution in rat pleurisy by inducing neutrophil and macrophage apoptosis. FASEB J. 2003;17:2269–2271. doi: 10.1096/fj.02-1162fje. [DOI] [PubMed] [Google Scholar]
- Gilroy DW, Colville-Nash PR, Willis D, Chivers J, Paul-Clark MJ, Willoughby DA. Inducible cyclooxygenase may have anti-inflammatory properties. Nat Med. 1999;5:698–701. doi: 10.1038/9550. [DOI] [PubMed] [Google Scholar]
- Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- Grimes CA, Jope RS. CREB DNA binding activity is inhibited by glycogen synthase kinase-3 beta and facilitated by lithium. J Neurochem. 2001;78:1219–1232. doi: 10.1046/j.1471-4159.2001.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniffa MA, Wang XN, Holtick U, Rae M, Isaacs JD, Dickinson AM, Hilkens CM, Collin MP. Adult human fibroblasts are potent immunoregulatory cells and functionally equivalent to mesenchymal stem cells. J Immunol. 2007;179:1595–1604. doi: 10.4049/jimmunol.179.3.1595. [DOI] [PubMed] [Google Scholar]
- Hegyi B, Kudlik G, Monostori E, Uher F. Activated T-cells and pro-inflammatory cytokines differentially regulate prostaglandin E2 secretion by mesenchymal stem cells. Biochem Biophys Res Commun. 2012;419:215–220. doi: 10.1016/j.bbrc.2012.01.150. [DOI] [PubMed] [Google Scholar]
- Hsu WT, Lin CH, Chiang BL, Jui HY, Wu KK, Lee CM. Prostaglandin E2 potentiates mesenchymal stem cell-induced IL-10 + IFN-gamma+ CD4+ regulatory T cells to control transplant arteriosclerosis. J Immunol. 2013;190:2372–2380. doi: 10.4049/jimmunol.1202996. [DOI] [PubMed] [Google Scholar]
- Hu X, Paik PK, Chen J, Yarilina A, Kockeritz L, Lu TT, Woodgett JR, Ivashkiv LB. IFN-gamma suppresses IL-10 production and synergizes with TLR2 by regulating GSK3 and CREB/AP-1 proteins. Immunity. 2006;24:563–574. doi: 10.1016/j.immuni.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Jaynes J, Ding X, Xu H, Wong WK, Ho CM. Application of fractional factorial designs to study drug combinations. Stat Med. 2013;32:307–318. doi: 10.1002/sim.5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang XX, Zhang Y, Liu B, Zhang SX, Wu Y, Yu XD, Mao N. Human mesenchymal stem cells inhibit differentiation and function of monocyte-derived dendritic cells. Blood. 2005;105:4120–4126. doi: 10.1182/blood-2004-02-0586. [DOI] [PubMed] [Google Scholar]
- Jing H, Yen JH, Ganea D. A novel signaling pathway mediates the inhibition of CCL3/4 expression by prostaglandin E2. J Biol Chem. 2004;279:55176–55186. doi: 10.1074/jbc.M409816200. [DOI] [PubMed] [Google Scholar]
- Kigerl KA, Gensel JC, Ankeny DP, Alexander JK, Donnelly DJ, Popovich PG. Identification of two distinct macrophage subsets with divergent effects causing either neurotoxicity or regeneration in the injured mouse spinal cord. J Neurosci. 2009;29:13435–13444. doi: 10.1523/JNEUROSCI.3257-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hematti P. Mesenchymal stem cell-educated macrophages: A novel type of alternatively activated macrophages. Exp Hematol. 2009;37:1445–1453. doi: 10.1016/j.exphem.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda E, Yamashita U. Mechanisms of enhanced macrophage-mediated prostaglandin E2 production and its suppressive role in Th1 activation in Th2-dominant BALB/c mice. J Immunol. 2003;170:757–764. doi: 10.4049/jimmunol.170.2.757. [DOI] [PubMed] [Google Scholar]
- Lucas T, Waisman A, Ranjan R, Roes J, Krieg T, Muller W, Roers A, Eming SA. Differential roles of macrophages in diverse phases of skin repair. J Immunol. 2010;184:3964–3977. doi: 10.4049/jimmunol.0903356. [DOI] [PubMed] [Google Scholar]
- Maggini J, Mirkin G, Bognanni I, Holmberg J, Piazzon IM, Nepomnaschy I, Costa H, Canones C, Raiden S, Vermeulen M, Geffner JR. Mouse bone marrow-derived mesenchymal stromal cells turn activated macrophages into a regulatory-like profile. PLoS ONE. 2010;5:e9252. doi: 10.1371/journal.pone.0009252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery DC. Design and analysis of experiments. 6. New York: John Wiley & Sons; 1994. [Google Scholar]
- Moore AR, Willoughby DA. The role of cAMP regulation in controlling inflammation. Clin Exp Immunol. 1995;101:387–389. doi: 10.1111/j.1365-2249.1995.tb03123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlebach MD, Wolfrum N, Schule S, Tschulena U, Sanzenbacher R, Flory E, Cichutek K, Schweizer M. Stable transduction of primary human monocytes by simian lentiviral vector PBj. Mol Ther. 2005;12:1206–1216. doi: 10.1016/j.ymthe.2005.06.483. [DOI] [PubMed] [Google Scholar]
- Nauta AJ, Kruisselbrink AB, Lurvink E, Willemze R, Fibbe WE. Mesenchymal stem cells inhibit generation and function of both CD34+-derived and monocyte-derived dendritic cells. J Immunol. 2006;177:2080–2087. doi: 10.4049/jimmunol.177.4.2080. [DOI] [PubMed] [Google Scholar]
- Nemeth K, Leelahavanichkul A, Yuen PS, Mayer B, Parmelee A, Doi K, Robey PG, Leelahavanichkul K, Koller BH, Brown JM, Hu X, Jelinek I, Star RA, Mezey E. Bone marrow stromal cells attenuate sepsis via prostaglandin E(2)-dependent reprogramming of host macrophages to increase their interleukin-10 production. Nat Med. 2009;15:42–49. doi: 10.1038/nm.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekkadan B, van Poll D, Suganuma K, Carter EA, Berthiaume F, Tilles AW, Yarmush ML. Mesenchymal stem cell-derived molecules reverse fulminant hepatic failure. PLoS ONE. 2007;2:e941. doi: 10.1371/journal.pone.0000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonage G, Filer AD, Haworth O, Nash GB, Rainger GE, Salmon M, Buckley CD. A stromal address code defined by fibroblasts. Trends Immunol. 2005;26:150–156. doi: 10.1016/j.it.2004.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakariar R, Hilliard M, Lawrence T, Trivedi S, Colville-Nash P, Bellingan G, Fitzgerald D, Yaqoob MM, Gilroy DW. Hematopoietic prostaglan-din D2 synthase controls the onset and resolution of acute inflammation through PGD2 and 15-deoxyDelta12 14 PGJ2. Proc Natl Acad Sci USA. 2007;104:20979–20984. doi: 10.1073/pnas.0707394104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehani K, Wang H, Garcia CA, Kinane DF, Martin M. Toll-like receptor-mediated production of IL-1Ra is negatively regulated by GSK3 via the MAPK ERK1/2. J Immunol. 2009;182:547–553. doi: 10.4049/jimmunol.182.1.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter BK, Asfaha S, Buret A, Sharkey KA, Wallace JL. Exacerbation of inflammation-associated colonic injury in rat through inhibition of cyclooxygenase-2. J Clin Invest. 1996;98:2076–2085. doi: 10.1172/JCI119013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffell B, Affara NI, Coussens LM. Differential macrophage programming in the tumor microenvironment. Trends Immunol. 2012;33:119–126. doi: 10.1016/j.it.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffell D, Mourkioti F, Gambardella A, Kirstetter P, Lopez RG, Rosenthal N, Nerlov C. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc Natl Acad Sci USA. 2009;106:17475–17480. doi: 10.1073/pnas.0908641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T, Honma K, Matsuyama T, Yui K, Tsujimura T, Standley DM, Nakanishi K, Nakai K, Akira S. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11:936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- Sindrilaru A, Peters T, Wieschalka S, Baican C, Baican A, Peter H, Hainzl A, Schatz S, Qi Y, Schlecht A, Weiss JM, Wlaschek M, Sunderkotter C, Scharffetter-Kochanek K. An unrestrained proinflammatory M1 macrophage population induced by iron impairs wound healing in humans and mice. J Clin Invest. 2011;121:985–997. doi: 10.1172/JCI44490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: A marker of alternative immunologic macrophage activation. J Exp Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szanto A, Balint BL, Nagy ZS, Barta E, Dezso B, Pap A, Szeles L, Poliska S, Oros M, Evans RM, Barak Y, Schwabe J, Nagy L. STAT6 transcription factor is a facilitator of the nuclear receptor PPARgamma-regulated gene expression in macrophages and dendritic cells. Immunity. 2010;33:699–712. doi: 10.1016/j.immuni.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreck FA, de Boer T, Langenberg DM, Hoeve MA, Kramer M, Vaisberg E, Kastelein R, Kolk A, de Waal-Malefyt R, Ottenhoff TH. Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco)bacteria. Proc Natl Acad Sci USA. 2004;101:4560–4565. doi: 10.1073/pnas.0400983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreck FA, de Boer T, Langenberg DM, van der Zanden L, Ottenhoff TH. Phenotypic and functional profiling of human proinflammatory type-1 and anti-inflammatory type-2 macrophages in response to microbial antigens and IFN-gamma- and CD40L-mediated costimulation. J Leukoc Biol. 2006;79:285–293. doi: 10.1189/jlb.0105015. [DOI] [PubMed] [Google Scholar]
- Wang H, Garcia CA, Rehani K, Cekic C, Alard P, Kinane DF, Mitchell T, Martin M. IFN-beta production by TLR4-stimulated innate immune cells is negatively regulated by GSK3-beta. J Immunol. 2008;181:6797–6802. doi: 10.4049/jimmunol.181.10.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen AY, Sakamoto KM, Miller LS. The role of the transcription factor CREB in immune function. J Immunol. 2010;185:6413–6419. doi: 10.4049/jimmunol.1001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QZ, Su WR, Shi SH, Wilder-Smith P, Xiang AP, Wong A, Nguyen AL, Kwon CW, Le AD. Human gingiva-derived mesenchymal stem cells elicit polarization of m2 macrophages and enhance cutaneous wound healing. Stem Cells. 2010;28:1856–1868. doi: 10.1002/stem.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupke CA, Stefanovich P, Berthiaume F, Yarmush ML. Metabolic effects of stress mediators on cultured hepatocytes. Biotechnol Bioeng. 1998;58:222–230. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.