Abstract
The ability to directly image and quantify drug-target engagement and drug distribution with subcellular resolution in live cells and whole organisms is a prerequisite to establishing accurate models of the kinetics and dynamics of drug action. Such methods would thus have far-reaching applications in drug development and molecular pharmacology. We recently presented one such technique based on fluorescence anisotropy, a spectroscopic method based on polarization light analysis and capable of measuring the binding interaction between molecules. Our technique allows the direct characterization of target engagement of fluorescently labeled drugs, using fluorophores with a fluorescence lifetime larger than the rotational correlation of the bound complex. Here we describe an optimized protocol for simultaneous dual-channel two-photon fluorescence anisotropy microscopy acquisition to perform drug-target measurements. We also provide the necessary software to implement stream processing to visualize images and to calculate quantitative parameters. The assembly and characterization part of the protocol can be implemented in 1 d. Sample preparation, characterization and imaging of drug binding can be completed in 2 d. Although currently adapted to an Olympus FV1000MPE microscope, the protocol can be extended to other commercial or custom-built microscopes.
INTRODUCTION
Previously, the lack of methods that would allow for direct and quantitative measurement of drug-target engagement in live cells with temporal resolution considerably limited our ability to better understand the molecular pharmacology of small-molecule drugs and consequently hampered drug development efforts.
New microscopy approaches allow for single-cell resolution imaging to resolve the pharmacokinetics (PK) and pharmacodynamics (PD) of drugs in vivo, thus overcoming these methodological limitations. Imaging the transport, inter- and intra-localization, and subsequent cellular effects of drugs has helped demonstrate several key biological barriers to drug effectiveness1, including inadequate drug delivery2, heterogeneity in drug-target interaction and the presence of multidrug resistance3, that are not easily observable using other imaging methods. Thus, single-cell PK/PD microscopy is well suited to high-content elucidation of drug behavior in different diseases.
Here, we describe our technology for measuring drug distribution and drug-target engagement in vitro4. The imaging technique we describe offers high spatial and temporal resolution while allowing the quantitative measurement and imaging of the fraction of the amount of drug bound to its target within cells. The potential of the technique is demonstrated using a cancer drug currently in phase III development as an example of how it can be used.
Advantages of the protocol as compared with alternative methods
The methods that are traditionally used to assess drug-target engagement include the cellular thermal shift assay (CETSA)5, which has been used to measure engagement of unlabeled drugs with target proteins in cells and soluble protein fractions extracted from cell lysates. Thermal proteome profiling6, an extension of CETSA combined with quantitative mass spectrometry, has also been used to monitor drug–target interaction effects on the thermal profile of a cellular proteome in intact cells. Other methods aimed at measuring drug behavior have also been used, including positron emission tomography (PET)7,8 and mass spectrometry imaging (MSI)9. Despite their ability to provide information regarding target engagement, these techniques either lack the necessary cellular resolution (PET) or the potential to make temporal measurements in vivo (MSI) that are ideally required.
Directly imaging drug delivery and PD effects has now become possible. Recent reviews1,10 elaborate how fluorescent companion imaging drugs and/or genetic reporters can be used10. Here we describe an imaging platform based on nonlinear two-photon excitation of fluorescence. This imaging platform offers several advantages over other microscopy imaging techniques11. Specifically, it offers extended imaging penetration depth, which enables imaging deep within tissue and tumors in physiologically relevant contexts. In addition, the low-scattering component of the near-IR excitation source substantially reduces the tissue scattering properties, which can be detrimental to the determination of fluorescence anisotropy. These characteristics contribute in making the technique directly translatable to in vivo studies in animals and humans, and contrast with other imaging modalities such as, for example, light-sheet microscopy. In addition, low phototoxicity enables long imaging times, facilitating the measurement of drug-target engagement in real time, even over extended periods of time (hours). The nonlinear nature of the effect also confines the excitation within a sub-femtoliter region, which is in stark contrast to other imaging modalities such as confocal and charge-coupled device (CCD)-based wide-field microscopy12.
Although CCD-based wide-field microscopy can easily be implemented, it presents two issues. First, it has poor axial resolution as compared with two-photon imaging. This makes it very difficult to accurately quantify target engagement when imaging in solution in the presence of free unbound or unspecifically bound fluorescent molecules. Second, CCD-based wide-field microscopy cannot be used in in vivo studies.
Basic theory of fluorescence anisotropy with one- and two-photon excitation
Fluorescence anisotropy has been widely adopted in several screening assays for drug development and pharmacological studies, and, if used in combination with fluorescently labeled drugs, it can allow for the determination of the degree of interaction of a drug with its target or large compounds. When used for two-photon imaging microscopy, it offers the capability to quantify drug–target interaction intracellularly with high temporal and spatial resolution.
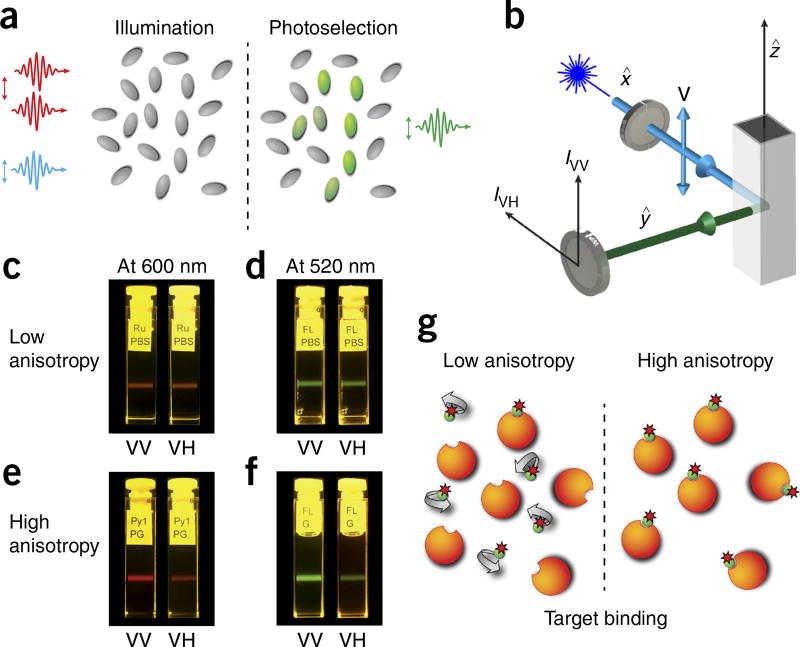
The basic theory of fluorescence anisotropy is illustrated in Figure 1. The excitation, via polarized light, of randomly distributed molecules results in a nonrandom distribution of the excited molecules’ transition moments (photoselection process, Fig. 1a). If the excitation light is polarized along the z axis (vertical polarization), molecules with transition moments oriented in the same direction will be preferentially excited. The probability of excitation is then proportional to cos2 θ, where θ is the angle between the transition moment of the molecule and the z axis. As a result, the fluorescence intensity observed through a polarizer oriented along the vertical direction is three times stronger than the intensity observed through a polarizer oriented along the (horizontal) x or y axis (Fig. 1a,b). This is true for molecules with parallel orientation of absorption and emission transition moments, and with absent molecular rotations or other depolarization processes. The fluorescence anisotropy r for the cylindrical symmetry is then defined as
| (1) |
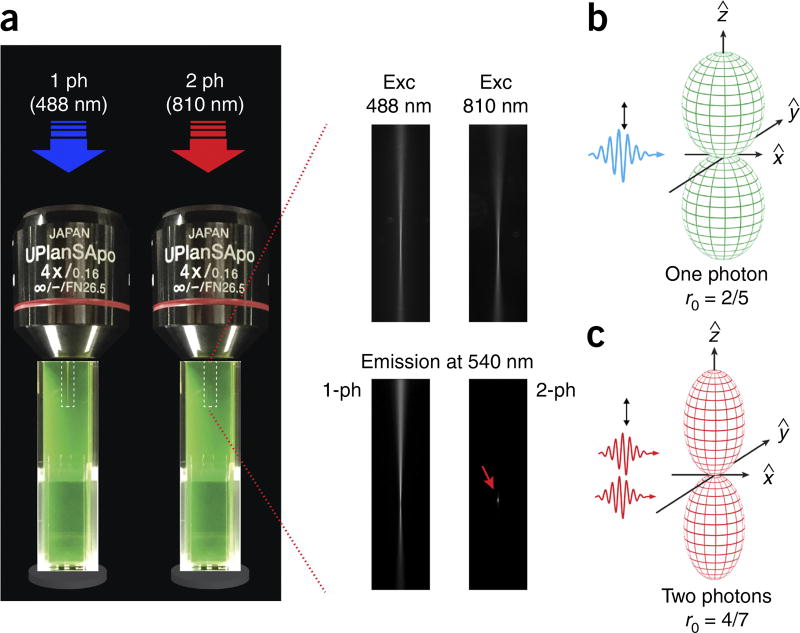
where I is the intensity of the light, V is vertical and H is horizontal. The first subscript symbol refers to the polarization of the excitation light and the second to the orientation of the analyzer (Fig. 1b). During two-photon excitation (Fig. 2a), instead, two low-energy photons interact simultaneously with a molecule, exciting the fluorophore to its singlet state. In this case, the probability of the excitation is no longer proportional to cos2 θ (Fig. 2b) but instead is proportional to cos4 θ (Fig. 2c). The higher photoselection value will result in higher fluorescence anisotropy values13,14.
Figure 1.
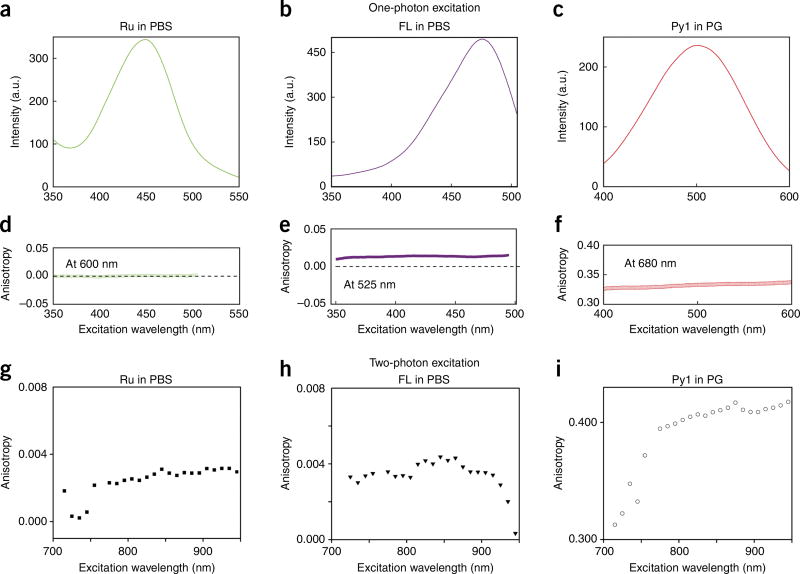
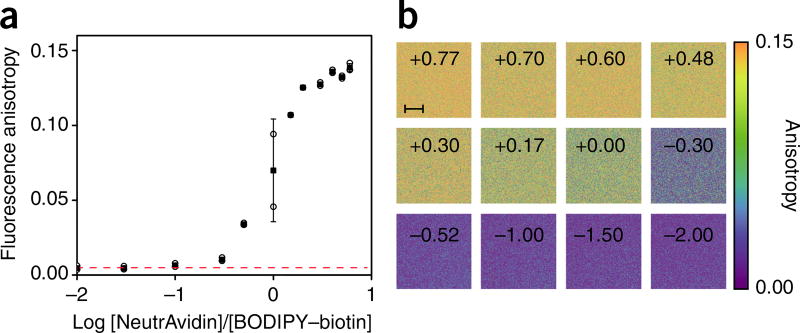
Fluorescence anisotropy principles and measurements. (a) Polarized light selectively excites fluorophores aligned along a preferential axis. If the rotation of the molecule occurs on a time scale that is shorter than its fluorescence lifetime, the fluorescence emission will present a certain degree of anisotropy. (b) Standard configuration for fluorescence anisotropy measurements. A sample is excited with vertically polarized light (IV). The intensity of the emitted fluorescence is analyzed through a polarizer oriented parallel (IVV) and perpendicular (IVH) to the incident excitation vector field. x̂, ŷ, ẑ indicate the unit vectors. (c–f) Images of fluorescence anisotropy for different dyes presenting low and high anisotropy. (c) RU PBS (ruthenium tris(bipyridyl) [Ru(bipy)3] in PBS). (d) FL PBS (fluorescein in PBS). (e) Py1 PG (pyridine 1 in propylene glycol). (f) FL G (fluorescein in glycerol). (g) Upon binding to their target, fluorescently labeled small-molecule drugs increase their fluorescence anisotropy value.
Figure 2.
One-photon versus two-photon excitation. (a) Comparison between one- and two-photon excitation and emission beam profiles in a fluorescein solution. For two-photon excitation, the emission is confined within a smaller focal point (red arrow in lower right-hand image), allowing for accurate optical sectioning imaging and accurate probing of volumes on the order of or below a femtoliter. A dry 4× objective was used for the demonstration of one- and two-photon fluorescence in order to distance the two-photon spot from the edge of the cuvette. (b,c) Photoselection function for one-photon (b) and two-photon (c) excitation. The fundamental anisotropy (r) for one-photon excitation is equal to 2/5. Two-photon excitation yields a strongly aligned population with a photoselection function proportional to cos4 θ. The fundamental anisotropy in this case is higher and is equal to 4/7. Exc, excitation; ph, photon.
In solutions or within intracellular environments, the rotational diffusion of the excited molecules tends to scramble the orientation of the emission dipoles, removing the preferential direction present in the emission fluorescence13,14. If the molecule rotation occurs on a time scale shorter than its fluorescence lifetime, the fluorescence emission will be isotropic (r = 0, Fig. 1c,d). If, instead, the molecule rotation occurs on a time scale much slower with respect to the fluorescence lifetime, a strong degree of anisotropy will be present (Fig. 1e,f).
The observed steady-state fluorescence anisotropy (r) will depend on depolarization processes occurring during the lifetime of the excited molecules. A dominant depolarization process is the rotational diffusion of the fluorescent molecules, which changes the direction of their transition moments. In the anisotropy notation, it has an elegant form:
| (2) |
where r0 is the fundamental anisotropy, τ is the lifetime of the fluorophore and Θ is the correlation time of the rotational diffusion. The rotational correlation time is given by
| (3) |
where η is the viscosity, T is the temperature in Kelvin, kB is the Boltzmann constant and V is the molecular volume of the dye or dye conjugate.
In contrast to polarization degrees, the fluorescence anisotropy can be directly written in additive form. The total anisotropy rTot of N species can then be expressed as
| (4) |
where ri is the anisotropy of the ith individual specie and fi is its fraction. This simple relation allows an immediate resolution of bound and unbound fluorophores.
Although the anisotropy of a fluorophore can be affected by different cellular environmental factors, it is largely defined by its intrinsic properties, such as its fundamental anisotropy, size and fluorescence lifetime. If the small fluorophore becomes heavier—e.g., following binding to a much slower-rotating molecule such as a protein—there will be an increase in fluorescence anisotropy (with respect to its value in free solution). Therefore, by titration and measurements of fluorescence anisotropy, binding isotherms can be reconstructed, and owing to the additive property (equation 4), the measurements can provide the fraction-weighted sum of the two possible states (bound and unbound)1.
To test receptor/binding quantification through fluorescence anisotropy imaging (FAIM), we first illustrate a fluorescence anisotropy binding assay of a BODIPY–biotin solution titrated against NeutrAvidin, before moving to a small-molecule drug and in vitro cell imaging.
The strategy used for drug labeling is based on direct attachment of the fluorescent dye to a functional group that is not relevant to the biological activity of the parent compound or corresponding analogs. This can include, but is not limited to, reaction of primary or secondary amines with fluorescent dyes functionalized with isothiocyanate, or active esters such as succinimidyl (N-hydroxysuccinimidyl (NHS)) esters or tetrafluorophenyl esters. Alternatively, carboxylates can be coupled to amino-functionalized dyes or, where applicable, fluorophores can be attached through click chemistry. In all cases, it is critical to confirm in appropriate assays that the fluorescently labeled drug retains functional activity in the settings. A detailed protocol for the synthesis of a fluorescently labeled drug, poly(ADP-ribose)polymerase (PARP) inhibitor (PARPi-FL), is given in the Supplementary Methods.
Several considerations must be taken into account when choosing the labeling fluorophore. Fluorescent dyes with short (e.g., Cy3) or long fluorescence lifetimes are not appropriate for fluorescence anisotropy measurements of small-molecule binding events. A suitable fluorophore, and the one here used, is BODIPY, which presents a fluorescence lifetime (ca. 4.0 ns) larger than the rotational correlation of the bound complex, making it an excellent choice for polarization-based assays. Importantly, BODIPY dyes, unlike most other fluorescent dyes, are not charged and are generally highly cell permeable, which is a prerequisite for intracellular imaging in live cells. BODIPY dyes presents a high extinction coefficient (EC; >80,000 cm−1 M−1), a high fluorescence quantum yield and a large two-photon cross-section. If alternative dye options are pursued, all the aforementioned characteristics must be satisfied. Although in this protocol we describe the use of a fluorescently labeled drug PARPi-FL, the method is generalizable to other fluorescently tagged small-molecule drugs (see Table 1 for a list of published examples).
TABLE 1.
Tagged small-molecule drugs.
The imaging technique illustrated in this article, when used in combination with fluorescently labeled drugs, offers the ability to obtain quantitative pharmacological binding information about diverse molecular interactions in real time at the microscopic level, in live cells (Fig. 3), discriminating between the different bound and unbound states of the drug4.
Figure 3.
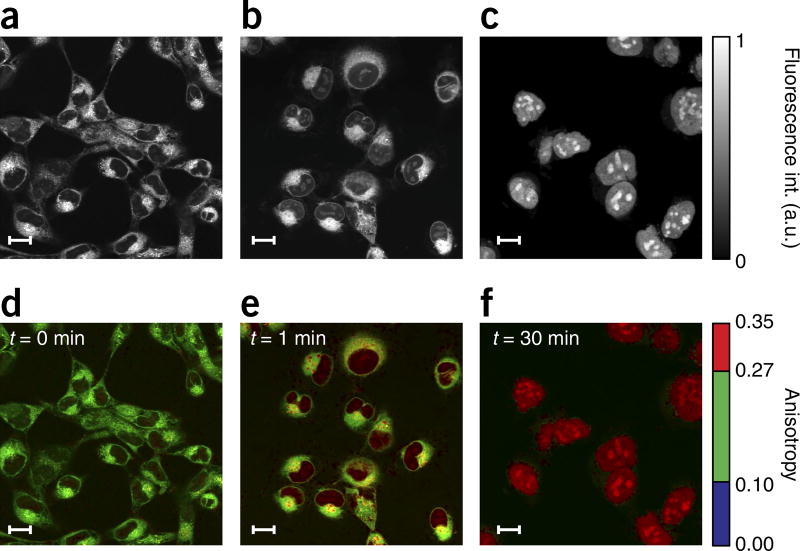
Results from HT1080 cells incubated in PARPi-FL. Fluorescence intensity (a–c) and weighted fluorescence anisotropy images (d–f) for HT1080 cells incubated in PARPi-FL (1 µM) for 1 min. (a,d) HT1080 cells incubated for 1 min in PARPi-FL with no washing. (b,e) 1 min after washing. (c,f) 30 min after washing. Following washing, the cytoplasmic PARPi-FL is cleared, whereas the nuclear, bound drug remains present. Because PARP1 is a relatively large molecule (~120 kDa), a considerable change in fluorescence anisotropy is present between the free and the target-bound PARPi-FL (Fig. 1g). Two-photon fluorescence anisotropy microscopy allows the quantification of the exact fraction of drug bound to its target in the cell. In the images, an anisotropy threshold is assigned to distinguish among the different states (bound/unbound) of the drug. When the cells are loaded, we observe nonspecifically bound drug in the cytoplasm, whereas the bound drug is present in the nucleoli, where PARP accumulates. The weighted fluorescence anisotropy images are color-mapped using the weighted RGB color-mapping scheme illustrated in the Experimental design section. Scale bars, 17 µm.
In addition to drug-target engagement studies, the protocol that we describe here can also be applied in other fields and for other uses, such as in obtaining spatially resolved images of fluorescence anisotropy of fluid-phase fluorophores in the cytoplasm15, studying the viscous properties of intracellular aqueous compartments16, studying the order of photosensitizers in nuclear envelopes17, directly monitoring DNA digestion in any region of live cells in real time18, imaging enzyme activity19 and probing rotational mobility of the fluorescent molecules and their interaction with their surroundings20,21. Although in the protocol we describe a hardware setup based on two-photon microscopy, other groups have demonstrated that different implementations are possible for FAIM22, such as CCD-based wide-field microscopy12, confocal microscopy19,21 and light-sheet microscopy23, as well as steady-state and time-resolved anisotropy22. We recommend several papers that provide a comprehensive review of this subject22,24–27.
Current limitations of the technique
This procedure has allowed measurements to be made that would have been difficult to make otherwise4. However, in its current implementation, there is also room for improvement. Perhaps most importantly, the acquisition time necessary for obtaining a drug–response curve in live cells is on the order of several hours, meaning it is not well suited for large-scale compound screening protocols. Rather, the method is better suited to understanding the biological behavior of lead compounds. New implementations based on high-throughput configurations28, combined with well plates or microfluidic-based microsystems, could potentially accelerate imaging and quantification capabilities. Second, the method relies on the availability of a cell-permeable fluorescently labeled companion imaging probe with ideal imaging and biological properties. We and others have developed a series of such agents, but each one requires considerable optimization and validation before use29.
It is important to note that the modification of a small-molecule drug by attachment and/or modification of functional groups can alter its physicochemical properties and target selectivity. As such, the fluorescently labeled drug might exhibit differential PK, cellular uptake, and target selectivity as compared with those of the parent compound. Therefore, the parent drug should always be used in a ligand displacement experiment to validate that specific binding of the fluorescent tracer can be displaced. Furthermore, using a set of fluorescent analogs that are labeled with chemically distinct fluorophore classes or labeled at orthogonal positions will be beneficial in further validating the specificity of the probe compounds.
Experimental design
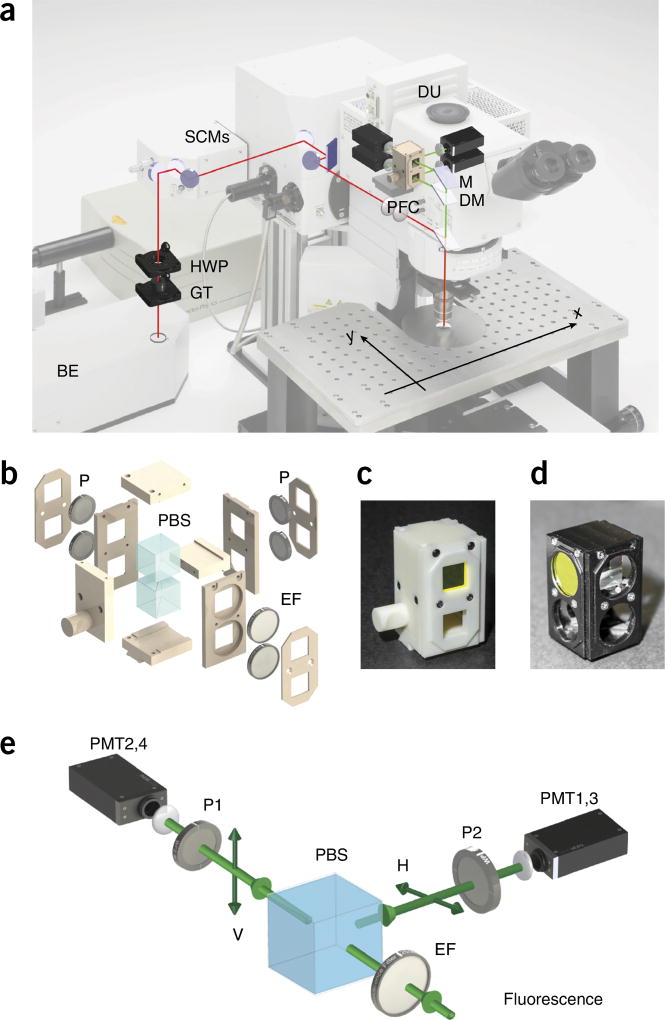
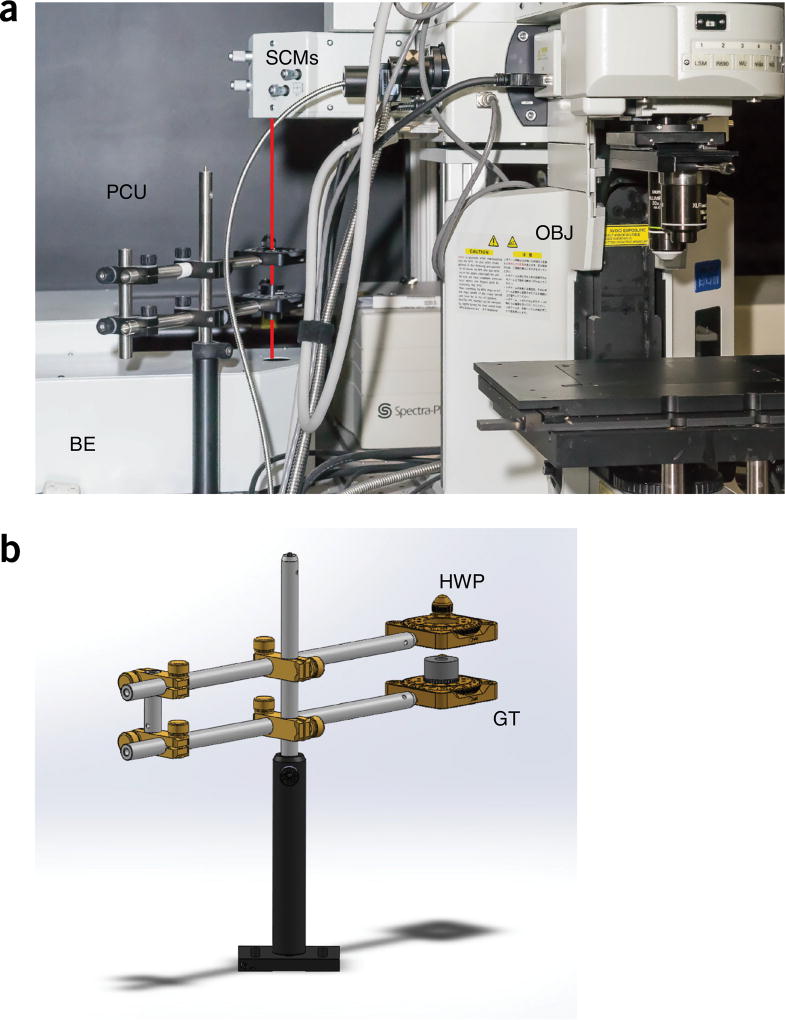
In this article, we provide step-by-step instructions on how to modify and adapt a commercially available microscope (Fig. 4a) so it can perform FAIM. Two-photon laser scanning microscopy is used because nonlinear imaging offers extended imaging penetration depth, reduced scattering, increased spatial and axial resolution (Fig. 2a), limited phototoxicity and reduced out-of-plane photobleaching as compared with other microscopy imaging modalities11. In the setup, light from a Ti:Sapphire laser is linearly polarized along a fixed predetermined axis, using a polarization control unit (Figs. 4 and 5), and focused through a water-immersion objective onto the imaging sample. Fluorescence is epi-collected (i.e., light is collected through the same objective used for excitation) in a non-descanned mode (Fig. 4a) and, using a polarization filter cube (PFC) (Fig. 4b–d), light is spectrally filtered and separated into two orthogonal states of polarization parallel and perpendicular with respect to the excitation light (Fig. 4e). Both light components are then simultaneously detected by two separate photomultiplier tubes (PMTs) within the detection unit (DU) (Fig. 4a), and the fluorescence anisotropy image is calculated according to equation 1. Here we provide guidelines on how to assemble the polarization control unit (PCU; 3D view included in STL format in Supplementary Data 1) and provide specifications for building the PFC (3D-printing STL files are included in Supplementary Data 2). We also illustrate procedures for alignment and calibration, and provide standard references for calibration and details with regard to imaging phantoms. Software to implement stream processing for calculation and visualization of fluorescence anisotropy images during time-lapse acquisition is also included (Supplementary Software). Data sets for testing the software are provided (Supplementary Data 3). Finally, we demonstrate the application of the method for determining accurate isotherm binding curves, as well as provide the necessary steps to follow and to quantify detailed live-cell imaging of drug-target engagement, using the fluorescently labeled drug PARPi-FL. A detailed protocol for the synthesis of the drug is provided in the Supplementary Methods.
Figure 4.
Setup overview. (a) Optical path overview of the two-photon fluorescence anisotropy imaging system and the imaging reference plane orientation xy. Adapted with permission from the Olympus FV1000 microscope brochure. (b) 3D exploded view of the PFC. (c,d) Views of the final assembled 3D-printed (c) and machined (d) polarization filter cube. See Supplementary Data 2. (e) Schematic principle view of the optical path within the DU. BE, beam expander; DM, dichroic mirror; DU, detection unit; EF, emission filter; GT, Glan-Thompson polarizer; HWP, half-wave plate; M, mirror; P, polarizer; PBS, polarization beam splitter; PFC, polarization filter cube; PMT, photomultiplier; SCM, steering combining mirror.
Figure 5.
Polarization control unit overview. Overview of the polarization control unit and optical components. (a) Overview of the microscope. The red line indicates the optical path between the BE and the microscope body, through the PCU. (b) 3D drawing of the PCU. BE, beam expander; GT, Glan-Thompson polarizer; HWP, half-wave plate; OBJ, objective; PCU, polarization control unit; SCM, steering combining mirror. See Supplementary Data 1.
Fluorescently labeled drug
We describe how to use PARPi-FL as an example fluorescently labeled drug. PARPi-FL (ref. 30; Supplementary Fig. 1) is a green fluorescent imaging tracer that provides PARP-specific contrast when injected intravenously31. Inhibitors of poly(ADP-ribose) polymerase (PARPis) are of considerable interest as anticancer agents, given their role in synthetic lethality32–39. Over the past decades, a series of PARPis have been developed and translated to the clinical arena, and one of them, the small molecule olaparib (AZD2281), was recently approved for clinical use by the US Food and Drug Administration (FDA) for prostate cancer40. Fluorescent derivatives of small-molecule PARPis are also of diagnostic relevance, because PARP is upregulated in a number of different types of cancer41–47.
Intrigued by the central role of PARP1 in DNA repair and encouraged by the uniformly strong expression in malignancies, we hypothesized in 2010 that intracellular PARP could be visualized via a two-step bio-orthogonal imaging process48. In 2012, we first used PARPi-FL. Although other imaging agents possessing similar affinities can be synthesized, efficient wash-in and wash-out kinetics, driven by superior tissue permeability, seem to be fundamental to and characteristic of the in vivo imaging success seen with PARPi-FL49,50. We have also demonstrated that PARPi-FL can be used for oral cancer screening, and its high tissue permeability allows topical application and efficient contrast generation51. The detailed protocol for the synthesis of PARPi-FL is given in the Supplementary Methods. In addition to fluorescent PARP inhibitors, our group has also synthesized a large number of other tagged small-molecule drugs. (See Table 1 for a list of published examples and details of how to obtain further information about these drugs.)
Microscope setup
Our imaging system (Figs. 4a and 5a) is based on a commercially available upright imaging microscope (Olympus FV1000MPE) equipped with several laser lines provided by two solid-state diode lasers, a multiline argon laser for confocal imaging, and a mode-locked Ti:Sapphire femtosecond tunable laser (Mai Tai DeepSee, 690–1,050-nm tuning range) with adjustable dispersion compensation for two-photon and second harmonic generation microscopy. Filter cubes equipped with different dichroic mirrors (DMs) and emission filter (EF) sets allow for the simultaneous acquisition of four fluorescence channels at four distinct PMTs (Fig. 4a). The microscope electronics and acquisition are controlled by the proprietary Olympus FluoView software installed on the main (data acquisition) PC.
To process the fluorescence data during acquisition, a secondary PC (data processing PC) is needed. Software is provided (Supplementary Software) to perform stream processing during time-lapse imaging, for raw fluorescence image filtering and for fluorescence anisotropy image calculation and visualization.
A three-axis xyz translation stage is needed for accurate positioning during longitudinal imaging within the perfusion chamber. A high-numerical-aperture (NA) water-immersion objective with a 2-mm working distance is recommended for high-resolution imaging and two-photon imaging.
Although objectives with low NA do not introduce depolarization effects, it is recommended to use high-NA objectives to increase two-photon excitation efficiency. Depending on their quality, design, magnification and NA, a fluorescence depolarization study could be necessary to test for and eventually compensate for these effects52–54. For the objective used in this protocol, acquisitions over a field of view (FOV) of ~100 µm guarantee a proper polarization distribution within the FOV itself4.
Although we describe how to use the Olympus FV1000MPE microscope here, this protocol can be easily adapted to most commercially available or custom-designed imaging systems. This is because the hardware arrangements in the different microscopes are fairly similar and the necessary components can be easily exchanged in these systems. Obviously, the 3D-printed or machined polarization beam splitter (PBS) filter cube must be configured to match the proprietary slider specifications of each commercial system. However, the detection principle is the same for each one of them. Detection is generally performed on at least two fluorescence channels and typically through the use of a DM, which can be replaced by a beam splitter (as done here) or a wire grid polarizer (25.0-mm square wire grid linear polarizer, Edmund Optics) if the hardware geometry is more stringent.
Systems based on non-descanned parallel detection for hyper-spectral imaging could present some challenges, depending on their design, particularly if spectral separation is achieved using gratings instead of prisms. In addition, microscopy systems equipped with detection schemes based on single-photon counting (SPC)55 (not yet particularly common for laser scanning microscopy, even though SPC has been successfully demonstrated for fluorescence anisotropy measurements56) could be problematic. The difficulty arises from the presence of afterpulses55, which constitute a problem for imaging detection, particularly affecting low-level signals following large-amplitude pulses. Only recently has SPC found applications in a few specialized studies57–61, but its use for FAIM would require a separate study.
The protocol described here is valid for an upright platform, which is recommended for an in vivo setting. However, the protocol could also be applied to inverted setups. In inverted setups, the angle of the waveplate (or how the single orthogonally polarized components of the fluorescence signal are collected on the detectors) must be altered on the basis of how the optical path within the microscope is designed.
The microscope modifications that are necessary to perform fluorescence anisotropy measurements can be easily removed in a few seconds by sliding the Glan-Thompson (GT) polarizer out of the optical beam path and replacing the PBS cube with the standard filter cube present by default in any system. The software we describe here can be used for processing data acquired with other commercial or custom-made microscopes if data are saved in .tif format and a naming convention followed by the FluoView software is used.
Polarization beam splitter filter cube
We have explored two different methods for fabricating the PFC: 3D printing (Fig. 4b,c) and machining (Fig. 4d). Thus, if either a machine shop or a 3D printer is available, it should be easy to implement the protocol. Machining is a traditional manufacturing method that includes milling, drilling and computer numerical control (CNC) machining processes. Traditionally, machining provides very high accuracy and is best suited to the production of high-precision parts for microscope components. Because each single machining process (e.g., drilling a hole) leads to additional costs and because complicated designs cannot be easily manufactured or involve many-step processes, it is generally recommended to design parts that are as simple as possible.
3D printing is a rapidly growing manufacturing process known in the past as rapid prototyping. It is generally fast and cost-effective, and can make complex parts without additional cost and difficulty. Currently, the maximum precision that typical 3D printing can achieve is hundreds of micrometers, and material selection is also limited to specific materials such as plastic and resin. However, it is expected that the resolution of 3D printing will continue to further improve with next-generation printers. For comparison purposes, we have therefore manufactured the PFC using both techniques, and we provide STL files for the two manufacturing routes (Supplementary Data 2).
The PFC (Fig. 4b) consists of a sliding holder containing two polarizers, an EF and a polarizing cube beam splitter. It replaces the common EFs’ sliding cube holder, which is positioned after the DM and directly before the detecting photomultipliers (Fig. 4a,e). Considering the cost, 3D printing is therefore a good choice for this manufacturing application.
3D printing often results in cumulative errors because of varying conditions such as temperature, and there is also a little variation in the size of the commercial filters and cube beam splitters. Therefore, when sketching the PFC for 3D printing (Fig. 4b), it is desirable to design the filter cube with a little larger tolerance in the components containing the filters and the cube than the specification given for the tolerance of the 3D printer. Here, for example, the circular hole space for the filters was designed with a 0.5-millimeter tolerance, which is approximately twice the precision of the 3D printer that we used. This tolerance is not a problem for the alignment of the optics, which at the detection stage is not sensitive to small variations.
System characterization
After two-photon excitation, fluorescence emission is epi-collected by the objective, separated into two orthogonal polarization components and focused on two PMTs (Fig. 4a,e).
The DMs and imaging objective affect both transmissivity and reflectivity, in addition to affecting collection and focusing of the different polarization components of the light in different ways. It is therefore important to make sure that the alignment of the optical components is performed properly by placing them accurately in the light path. First, the polarization controller unit (PCU) (Fig. 5) must be aligned accurately to ensure a very high extinction ratio between the two orthogonal polarization components. This will ensure that the incoming excitation state of polarization is highly polarized along one preferential axis. Then the extinction ratio of the PBS positioned within the polarizer cube filter must be measured and the polarizers must be aligned to avoid any mismatch that could alter the readings. A differential detector sensitivity (G-factor) can be determined using two methods. In the first method, the optical properties of all components with respect to polarization transmissivity and reflectivity are measured, as described by Lakowicz13,14 and Dix62. An alternative method uses a series of fluorophores as polarization standards63,64. Fluorescein in aqueous solution, because of its moderate lifetime of ~4 ns and short correlation time, exhibits a very low value of fluorescence polarization. However, a well-established number has not yet been determined, which makes it difficult to use it as a valid standard. An alternative is ruthenium tris(bipyridyl) [Ru(bipy)3], which has a very long-lived excited state, resulting in effectively a zero fluorescence polarization value across its entire excitation band63. By fine-tuning the PMT voltage of the two acquisition channels, the condition for calibration can be then determined.
The calibration procedure for the appropriate EF should be tested. In our protocol, we have used two different optical filters covering a combined range from 495 to 630 nm. Calibration in the region of 495–540 nm can be done with fluorescein. Calibration from 575 to 630 nm can be done using ruthenium tris(bipyridyl) [Ru(bipy)3]. With fluorescein, attention needs to be paid to controlling the pH of the final solution; unfortunately, alternatives in that emission range are not yet well established.
Measurements of the standards should be carried out before and at the end of each imaging session. This ensures that fluorescence anisotropy values are consistent and can be compared. In addition, it must be ensured that the fluorophores are used at concentrations that avoid quenching and inner filter effects63. This is particularly true for fluorescein.
The use of fluorescence anisotropy standards characterized by high fluorescence anisotropy values is also highly recommended, because the presence of depolarization effects will tend to reduce their values. As a standard, we have chosen a solution of pyridine 1 in propylene glycol (PG). In addition, measurements of a solution of fluorescein dissolved at different ratios of glycerol/water can be used to monitor the accuracy of the alignment and to establish the sensitivity for detecting changes in anisotropy across a large range of values.
Image acquisition and processing
Two separate fluorescence anisotropy images are obtained that are combined numerically according to equation 1. Each image, recorded by two different PMTs, presents a certain amount of signal-to-noise ratio (SNR) depending on several imaging and fundamental parameters, particularly when the PMTs are operating at high voltages. The noise superimposed on the collected images propagates in a nonlinear way to give rise to fluorescence anisotropy images that exhibit considerable noise levels. Even if measurement errors in the raw fluorescence images may not appear severe, fluorescence anisotropy calculation leads to a more complex form of noise, which may exceed the fluorescence anisotropy signal. In such a situation, features within fluorescence anisotropy images are fairly undistinguishable and the underlying information unusable.
To design viable methods that are capable of avoiding or at least reducing information loss, it is important to understand the nature of the errors related to the collected fluorescence images. The quality of the fluorescence anisotropy images is directly related to the signal intensity, as in the first approximation the major contribution to noise in raw images is due to the discrete nature of light, which is properly described by a Poisson distribution. Therefore, to ameliorate the quality of the images, it is sufficient to increase the number of photons collected. To achieve this goal, several strategies can be adopted: adjusting the laser power, increasing the dwell time, and image averaging are common ways used by microscopists to increase the quality of the images in terms of SNR. However, all these methods may lead to photobleaching or cellular damage. Furthermore, both dwell-time increase and image averaging hamper the speed of acquisition, limiting the possibility of imaging fast dynamic events. An alternative solution relies on the use of image-processing techniques. Instead of operating on the calculated anisotropic images, image processing is applied to the raw fluorescence images before computing the fluorescence anisotropy. This strategy avoids bias in fluorescence anisotropy estimates64. Applying filtering to the raw fluorescence images is also preferable, as modeling of noise statistics in anisotropy images is more complicated, requiring the implementation of much more complex algorithms. Although numerical filters may lead to loss of spatial/temporal resolution, the benefits of such procedures usually overcome this limitation.
Gaussian and median filters are examples of linear and nonlinear filtering techniques, respectively, and are widely used in image processing. Gaussian filtering has been exploited to reduce noise in FAIM65. The median filter can also be effectively implemented, as it reduces pixel noise by applying a nonlinear transformation on groups of neighboring pixels. The filter is often used in image processing when outliers are present and has the advantage of preserving edges better than the Gaussian filter. However, its kernel size should be generally limited, as artifacts can potentially be introduced by the nonlinear computations involved.
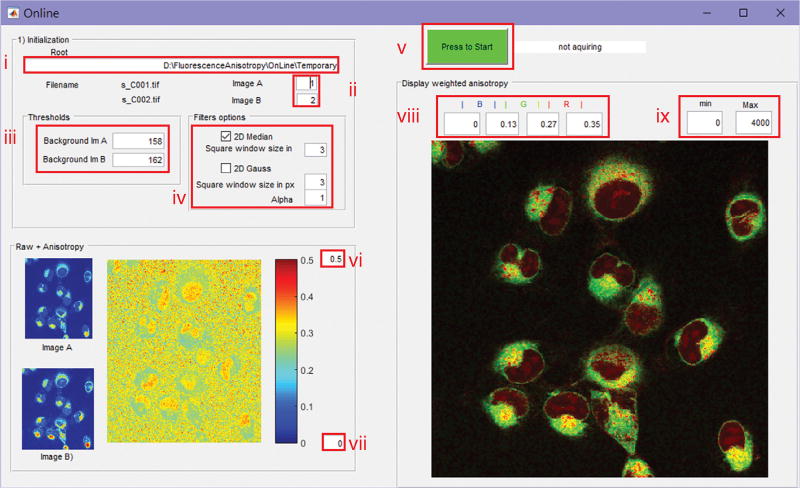
Herein we suggest the use of a combination of these two filters rather than choosing one over the other, and their optional implementation is available in both the OnLine and OffLine software (Supplementary Software).
We therefore recommend keeping the kernel sizes as small as possible (3 × 3) to benefit from the nonlinear features of the median filter (e.g., against outliers), leaving the task of smoothing the noise linearly to a subsequent Gaussian filter. However, although the Gaussian filter is capable of smoothing the signal, leading to images lacking filter-induced artifacts, it is far less effective against outliers.
The Gaussian filter in our software is implemented as a weighted average of pixels falling within a prefixed window. It is important to emphasize that the filter may induce oversmoothing if its design parameters are not chosen properly. In fact, an optimal choice of parameters provides the best tradeoff between smoothing and image-features preservation. The Gaussian filter also exhibits an important property (separability) that is particularly useful when fast processing is required. In computers, Gaussian filters are implemented as numerical convolution. By exploiting the associative property of convolution, the filtering can be reduced to two sequentially applied 1D convolutions. In practice, 1D convolution is applied along image rows, and then along columns. In addition, the outcome does not change if the order is switched—i.e., if the convolution is applied along image columns and then along rows. This method has important advantages, as the number of computations is in general considerably reduced if two 1D filters are applied sequentially rather than using a 2D Gaussian kernel directly. In the OffLine/OnLine software provided (Supplementary Software), we exploited the sequential applied filter implementation to achieve faster stream processing for anisotropy visualization during fluorescence image acquisitions.
Visualization mapping
Two-photon fluorescence anisotropy microscopy can provide accurate spatial and temporal maps of fluorescence anisotropy distributions in living cells. However, the direct computation and visualization of the fluorescence anisotropy arising from raw fluorescence images often leads to uninformative results. The cause is primarily attributed to the presence of noise in the original fluorescence images (Fig. 6). Because fluorescence anisotropy is a ratiometric measurement, the calculated image is prone to appearing noisier than the collected fluorescence images from which it is computed (Fig. 6).
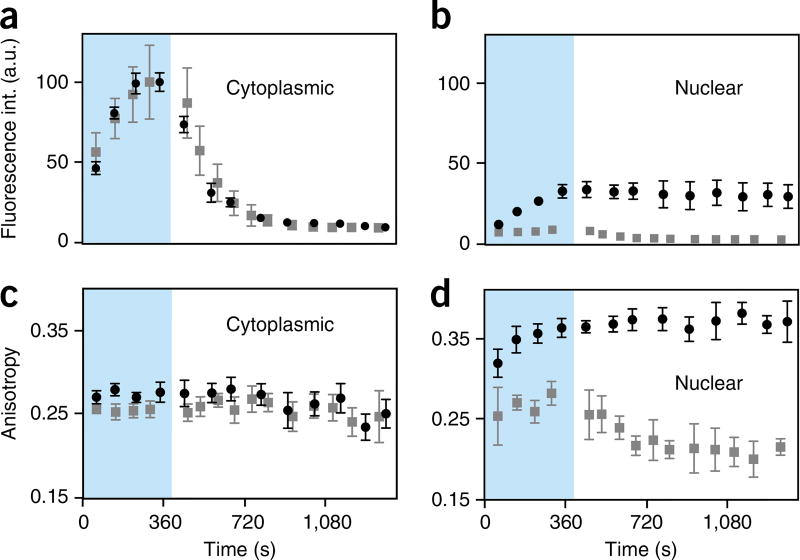
Figure 6.
Noise contribution to fluorescence anisotropy images. Noise contribution for raw and weighted fluorescence anisotropy images of HT1080 cells incubated in PARPi-FL (1 µM) for 1 min. (a,b) After 30 min of washing. (c,d) Before washing. The noise contribution in the raw fluorescence images propagates in a nonlinear way to give rise to fluorescence anisotropy images that exhibit considerably higher noise levels. Averaging or image processing improves the SNR of the fluorescence intensity images and consequently the calculated raw and weighted fluorescence anisotropy images. In a and c, fluorescence intensity images acquired at the two PMTs (left and right columns, respectively) are averaged over 1 or 16 acquisitions (top and bottom rows, respectively). In b and d, raw and weighted fluorescence anisotropy images (left and right columns, respectively) are calculated for intensity images averaged over 1 or 16 acquisitions (top and bottom rows, respectively) shown in a and c. Insets indicate zoomed areas. Scale bars, 17 µm. Int., intensity.
Unsatisfactory results are also obtained under regular imaging conditions—i.e., with sufficient SNR. By properly tuning the acquisition parameters, the SNR of the images may be increased—for instance, by averaging over multiple acquisitions or by increasing the integration time. In addition, denoising of the acquired fluorescence images can be exploited. However, in general, because fluorescence anisotropy images provide information on the degree of anisotropy whereas fluorescence intensity images contain detailed structural information, the two representations may present a low degree of correlation (Fig. 6a–d). To overcome the lack of structural information present within the fluorescence anisotropy images, different information fusion methods can be implemented. The same issue is present, for example, in fluorescence lifetime imaging microscopy, in which one approach for merging lifetime and fluorescence image features into a single image representation can be realized by implementing a method that maps functional and structural features into the hue–saturation–value (HSV) color space66. For our representation, we used a similar approach, but we exploited the red–green–blue (RGB) space rather than the HSV space to merge data.
The merging algorithm used in our visualization software splits the pixels’ anisotropy values into three separate clusters. Manually selected thresholds are used for this. Blue, green and red correspond to the clusters with lowest, intermediate and highest values, respectively. However, the presence of noise in the anisotropy image and the choice of hard thresholding for coloring clusters have the disadvantage of leading to artifacts. To overcome this issue, discontinuities are replaced with smoother transitions. Color intensity is then modulated (weighting) upon the total fluorescence intensity. As a result, colors appear dark if they originate from pixels with low fluorescence intensity values (statistically less relevant), whereas they appear bright in the opposite case. The advantage of the proposed procedure is twofold: (i) weighting acts somehow as an image filter, as pixels with the highest noise levels in anisotropy are mapped to the darkest levels in intensity; and (ii) weighting allows the clustered anisotropy and fluorescence images to be merged. This leads to a single image that contains both the functional and structural information. With manually selected thresholding, the cluster with the highest values is determined by measuring the fluorescence anisotropy distribution after a few minutes of washing (here specifically after 10 min) such that all the fluorescence signal present within the cells is due to drug molecules specifically bound to their targets. The intermediate cluster can be calculated by measuring the fluorescence anisotropy distribution during the loading phase of the drug. The cluster with the lowest values is assigned to all anisotropy values below those of the intermediate-values cluster. A possible alternative way to assign the highest and the intermediate clusters would be to perform washing and competitive experiments to determine the anisotropy values corresponding to the bound and unbound states within the nucleus and the cytoplasm.
Software
We acquire two-photon fluorescence images using an Olympus laser scanning microscope system (FV1000MPE) in combination with the proprietary Olympus FluoView software.
The fluorescence anisotropy images can then be displayed in stream processing while the raw fluorescence images are acquired in time-lapse modality (using the OnLine software) or during a postacquisition (using OffLine) phase. Whereas the OnLine program is designed to be used as an add-on to the FluoView software to provide fluorescence anisotropy visualization during scanning, the OffLine program can be used on previously gathered images. The OffLine program features an export function for saving processed images.
The software is intended to work in combination with the Olympus FluoView software with data saved in the .oif format, but it can be implemented with any other imaging acquisition software, as long as the data are saved in .tif format and the same naming convention as is used for the Olympus FluoView software is followed (refer to the Olympus FluoView manual for detailed instructions).
Even though other imaging systems are controlled by different software, the parameter selection is always similar, if not exactly the same, for all of them.
The two software programs we use exploit user-friendly graphical user interfaces (GUIs) and are designed to compute and display both fluorescence anisotropy and weighted fluorescence anisotropy. The GUIs also offer the user the choice to apply basic image processing to the raw fluorescence images before computing the fluorescence anisotropy, in order to reduce noise and optimize the visualization results.
It is better to avoid running the OnLine program on the same PC as the one controlling the microscope (data acquisition PC) to avoid problems in the acquisition that could be due to limited available memory or due to the number of other programs concurrently running on it. We recommend instead sharing a temporary folder from the data acquisition PC over the network while running the OnLine software on a secondary PC (data processing PC). To achieve faster transfer speed (1 Gbps) between the two PCs, a direct crossover Ethernet cable could be considered as an alternative to a network connection.
The image processing and elaboration necessary for the computation of the fluorescence anisotropy images can be performed using different commercially available or open-source software packages (e.g., Visual C++, Python, MATLAB). We have decided to write our software in MATLAB and to build it into a stand-alone executable in binary format. For the software to run on computers without the MATLAB platform installed, a stand-alone set of shared libraries that enables the execution of the compiled MATLAB applications can be downloaded (MATLAB Runtime, R2015a v8.5, 64-bit). Both 32- and 64-bit versions of the MATLAB Runtime installer can be directly downloaded from the MathWorks website.
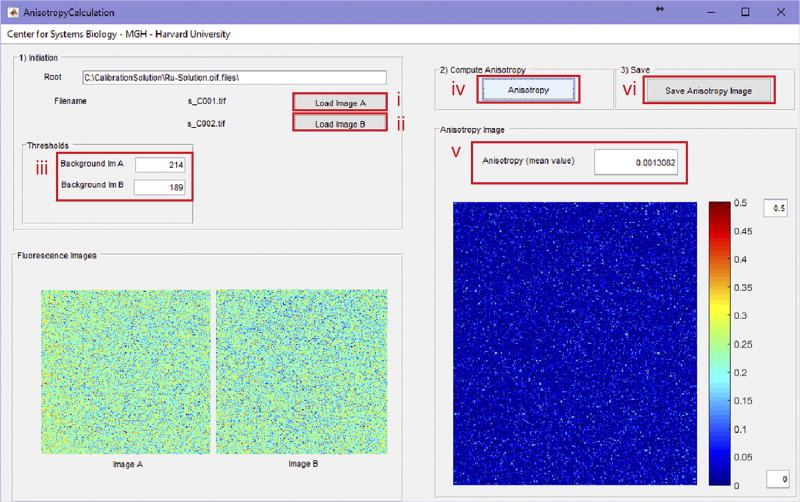
In addition to the OnLine and OffLine software (Supplementary Software), we provide a program to calculate the noise background for each photodetector (BackgroundEstimation), which needs to be subtracted to correctly calculate the fluorescence anisotropy67. It is important to note that the correction of the background contribution to a fluorescence anisotropy measurement cannot be obtained by direct subtraction of the background anisotropy from the sample anisotropy. Instead, measurements of the parallel and perpendicular components of the background signal must be subtracted from the respective measurements of the sample67.
We also provide a program (AnisotropyCalculation, Supplementary Software) to calculate the average fluorescence anisotropy over an image. This last program calculates the average anisotropy over an acquired set of images, and is particularly useful when measuring dye solutions for standard calibration purposes or for obtaining binding isotherms.
Acquisition parameters
Different acquisition parameters must be selected depending on whether fluorescent solutions or live cells are imaged. When imaging standard calibrating solutions or dyes, we recommend acquisition with image sizes of 256 × 256, and with pixel integration times between 20 and 100 µs, depending on the quantum efficiency and concentration of the fluorophore. Laser power should be kept between 0.1 and 5 mW, as measured at the exit of the objective. A zoom factor of 13× is also recommended. For cell imaging, typical image sizes are 512 × 512 or 1,024 × 1,024, with a dwell time of 20 µs per pixel. A zoom factor 3× is preferable, in order to avoid NA-induced depolarization effects at the edges of the imaging FOV. Image averaging or noise filtering is also recommended if the SNR of the images is low.
MATERIALS
REAGENTS
Ruthenium-tris(2,2-bipyridyl) dichloride, tris(2,2-bipyridyl)ruthenium(II) chloride hexahydrate (Sigma-Aldrich, cat. no. 50525-27-4)
Rhodamine B, analytical standard (Sigma-Aldrich, cat. no. 79754-25MG)
Fluorescein, reference standard (Thermo Fisher Scientific, cat. no. F1300)
Glycerol (Sigma-Aldrich, cat. no. 49781-500ML)
Pyridine 1; LDS 698 [2-[4-[4-(dimethylamino)phenyl]-1,3-butadienyl]-1-ethylpyridinium monoperchlorate; (Exciton, cat. no. 2-92).
PBS (Sigma-Aldrich, cat. no. P5493)
Ethanol (Sigma-Aldrich, cat. no. 24102)
InSpeck Green (505/515) Microscope Image Intensity Calibration Kit (6 µm; Thermo Fisher Scientific, cat. no. I14785)
Autofluorescent plastic slides (Chroma Technology, cat. no. 92001)
Poly-l-lysine solution, 0.1% (wt/vol) in H2O (Sigma-Aldrich, cat. no. P8920)
Poly(ethylene glycol) 300 (PEG 300; Sigma-Aldrich, cat. no. 90878)
BODIPY–biotin synthesis
Dichloromethane (DCM; Fisher Scientific, cat. no. BDH23373)
Methanol (MeOH; VWR, cat. no. BDH20864)
Milli-Q water (EMD Millipore Milli-Q Integral System; Thermo Fisher Scientific, cat. no. ZRXQ005T0)
Acetonitrile (MeCN; VWR, cat. no. BDH83640)
N,N-diisopropylethylamine (DIPEA; Sigma-Aldrich, cat. no. D125806)
BODIPY 493/503 succinimidyl ester (BODIPY-SE, Thermo Fisher Scientific, cat. no. D2191)
Biotin-X cadaverine trifluoroacetic acid salt (biotin-amine; Thermo Fisher Scientific, cat. no. B1596)
NeutrAvidin protein (10 mg; Thermo Fisher Scientific, cat. no. 31000)
Sodium sulfate, anhydrous (Na2SO4, Fisher Scientific, cat. no. S-421-1)
Purity Flash Cartridge, 12 g, granular silica gel, 20- to 45-µm particle size, 60 Å (Sorbent Technologies, cat. no. CFC-52300-012-18)
Thin-layer chromatography (TLC) plates, Silica XG w/UV254 (Sorbent Technologies, cat. no. 4434126)
Tween 20 (50 ml; Sigma-Aldrich, cat. no. P9416-50ML)
Cell line and cell culture
Any suitable cell line of interest can be used. The procedure illustrated in the protocol is optimized for human HT1080 fibrosarcoma cells (ATCC, cat. no. CCL-121) ! CAUTION It is important to observe all the necessary precautions when working with human cell lines. In addition, experiments with human cell lines must be in compliance with and approved by the relevant institutional and governmental review committees. ▲ CRITICAL STEP Cells must be regularly checked to ensure that they are authentic and that they are not contaminated with mycoplasma.
Dulbecco’s Modified Eagle Medium (DMEM; Thermo Fisher Scientific, cat. no. 21063-029)
Trypsin–EDTA solution (10×; Sigma-Aldrich, cat. no. 59418C)
FBS (Thermo Fisher Scientific, cat. no. 26140079)
Penicillin–streptomycin (Thermo Fisher Scientific, cat. no. 15140122)
EQUIPMENT
Equipment for closed-bath imaging chamber
Large closed-bath imaging chambers (Warner Instruments, cat. no. RC-21BR)
PE10 tubing (0.011-inch i.d., 0.025-inch o.d.; Scientific Commodities, cat. no. BB31695-PE/1)
Round cover glass, no. 1 thickness (Warner Instruments, cat. no. 64-0705 (CS-25R))
Polyethylene tubing, PE-160, 10 feet (Warner Instruments, cat. no. 64-0755)
MPP Series perfusion minimanifold, 2-to-1 ports (Warner Instruments, cat. no. 64-0212)
Syringes, 10 ml, BD Luer-Lok tips (Becton-Dickinson, cat. no. 309301)
Tubing to/from vacuum trap (Fisher Scientific)
Vacuum outlet
1-liter Filter flask, with side arm (Gilson, cat. no. GW-74)
Neoprene no. 8 stopper for 1-liter filter Flask (Gilson, cat. no. GWA-1)
3/8-inch Clear vinyl tubing (Gilson, cat. no. WT-4R)
1/4-inch Clear vinyl tubing (Gilson, cat. no. WT-8R)
Silicon grease (Warner Instruments, cat. no. 64-0378)
Animal temperature controller (World Precision Instruments, cat. no. ATC2000)
Large heating plate with built-in RTD sensor (World Precision Instruments, cat. no. 61840)
Equipment for dye sample preparation and mounting
Fisherbrand cover glasses: rectangles (Fisher Scientific, cat. no. 12-545M)
Fisherbrand economy plain glass microscope slides (Fisher Scientific, cat. no. 12-550-A3)
CoverWell imaging chamber gaskets (Thermo Fisher Scientific, cat. no. C18160)
Heated ultrasonic cleaning bath (Fisher Scientific, Branson, cat. no. 15-336-110)
Eppendorf microcentrifuge tubes (Fisher Scientific, cat. no. 05-402-25)
3D printing and assembly
3D printer (uPrint; Stratasys)
Production-grade thermoplastic (ABSplus; Stratasys)
3D printing and CNC machining service (Proto Labs)
SolidWorks software (Dassault Systèmes SolidWorks)
General-purpose tap for closed-end hole threading, M3 thread, 1 needed (McMaster, cat. no. 8305A52)
Fixed T-handle tap wrench, 2–3/4-inch length, 1 needed (McMaster, cat. no. 2546A23)
Flat-head Phillips machine screws, M3, 8-mm length, 6 needed (McMaster, cat. no. 91698A304)
Pan-head Phillips machine screws, M3, 8-mm length, 6 needed (McMaster, cat. no. 92005A118)
STL file for 3D printing (Supplementary Data 2).
Optical components
ET Series Multiphoton Emission Filter, 495–540 nm (Chroma Technologies, cat. no. BA495-540HQ)
ET Series Multiphoton Emission Filter, 575–630 nm (Chroma Technologies, cat. no. BA575-630HQ)
25-mm VIS, polarizing cube beam splitter (Edmund Optics, cat. no. 49-002). Alternatively, a 25.0-mm square wire grid linear polarizer (Edmund Optics, cat. no. 47-102) can be used.
Two unmounted linear polarizers, 500–720 nm (Thorlabs, cat. no. LPVISB100)
Glan-Thompson (GT) calcite polarizer, 650–1,000 nm (Newport, cat. no. 10GT04AR.16)
Three 360° Continuous Rotation Stages, 1 in Aperture, Coarse & Fine Adj. (Newport, cat. no. RSP-1T)
Two mounting bases (Thorlabs, cat. no. BA2)
One 1/2-inch-diameter optical post, 2-inch length (Thorlabs, cat. no. TR2)
Four 1/2-inch-diameter optical posts, 12-inch length (Thorlabs, cat. no. TR12)
Two 12-inch post holders (Thorlabs, cat. no. PH6)
Five right-angle clamps for 1/2-inch-diameter posts (Newport, cat. no. CA-1)
Unmounted linear polarizer, 650–2,000 nm (Thorlabs, cat. no. LPNIR100)
1/2-inch-diameter mounted achromatic half-wave plate (Thorlabs, cat. no. AHWP05M-980)
Standard iris, 20 mm (Thorlabs, cat. no. D20S)
Laser safety goggles, 35% visible-light transmittance (Newport, cat. no. LV-R14.T1Q01)
IR sensor card, 700–1,600 nm (Newport, cat. no. F-IRC4)
Optical power meter (Newport, cat. no. 1918-R)
918D silicon wand detector, 400–1,100 nm (Newport, cat. no. 918-ST-SL)
Computers
For data processing (data processing PC): Precision Tower Workstation (Dell, cat. no. T7810) with Intel Xeon Processor E5-2637, 3.5 GHz, 64 bit, with a NVIDIA GeForce GTX 1080 graphics card, 128 GB RAM, a fast-drive PCI 512 GB SSD Samsung 950 Pro, running Windows 10
For data acquisition (data acquisition PC): HP Z420 Workstation, Intel Xeon E5-1620, 3.70 GHz, 64 bit, running Windows 7
Optical setup
Spectrofluorometer equipped with polarizers (Varian, model Cary Eclipse)
Microscope setup
Olympus FV1000MPE (Olympus)
25× water-immersion objective (25×, 1.05 NA, 2-mm working distance; Olympus, model no. XLPLN25XWMP2)
Femtosecond pulsed laser (Mai Tai DeepSee, 690–1,050-nm tuning range; SpectraPhysics).
Software
MATLAB software for data analysis (MATLAB 2014, MathWorks)
MATLAB Runtime (R2015a 8.5, 32/64 bits; MathWorks, downloadable from https://www.mathworks.com/products/compiler/mcr/index.html)
FluoView (Olympus)
Accompanying protocol software (Supplementary Software)
Accompanying data sets (Supplementary Data 3)
Cell culture
Incubator for cell culture (Thermo Fisher Scientific, Forma Steri-Cycle)
Centrifuge tubes, 15/50 ml (Greiner Bio-One, cat. no. 188271/227261)
Cell culture flow cabinet (Thermo Fisher Scientific, cat. no. 1300 Series A2 Class II)
Falcon polystyrene microplates, six-well, tissue-culture-treated (Fisher Scientific, cat. no. 08-772-33)
Corning cell culture flask, 75 cm2 (Sigma-Aldrich, cat. no. CLS430641)
BODIPY–biotin synthesis
Stirring plate (IKA, cat. no. 0003810001)
1-dram vial, borosilicate glass (e.g., Fisher Scientific, cat. no. 03-339-25B)
Aluminum foil
Magnetic stir bar, Teflon coated, 5 mm × 2 mm (e.g., Bel-Art, cat. no. F371190005)
Disposable pipette (e.g., Fisher Scientific, cat. no. 13-678-20A)
Rotary evaporator (e.g., Büchi, model no. Rotavapor R-300)
Mass balance (Mettler Toledo)
Test tubes, 16 × 100 mm (e.g., Fisher Scientific, cat. no. 14-961-29)
UV lamp (e.g., UVGL-25; UVP, cat. no. 95-0021-12)
Kimax media storage bottle (Cole Parmer, cat. no. UX-34523-24)
Reversed-phase analytical column (5 µm, 300 Å, 10 × 250 mm; Phenomenex, model no. Jupiter C18)
UFLC system, consisting of a degasser (Shimadzu, cat. no. DGU-20A3), pump (Shimadzu, cat. no. LC-20AB), detector (Shimadzu, cat. no. SPD-M20A), module (Shimadzu, cat. no. CBM-20A) and a fraction collector (Shimadzu, cat. no. FRC-10A)
Disposable borosilicate glass tubes (Fisher Scientific, cat. no. 14-961-26)
Reversed-phase analytical column (10 µm, 100 Å, 4.6 × 250 mm; Waters, model no. Atlantis T3 C18)
LCMS system, consisting of a degasser (Shimadzu, cat. no. DGU-20A5), pump (Shimadzu, cat. no. LC-20AD), autosampler (Shimadzu, cat. no. SIL-20AC), detector (Shimadzu, cat. no. SPD-20AV), module (Shimadzu, cat. no. CBM-20A), nitrogen generator (Shimadzu, cat. no. NM32LA) and a liquid chromatograph mass spectrometer (Shimadzu, cat. no. LCMS-2020)
Autosampler vials (Fisher Scientific, cat. no. 03-377-45)
NMR spectrometer (Bruker, model no. Ultrashield 600 Plus)
-
NMR sample tubes (Sigma-Aldrich, cat. no. Z274526)
! CAUTION All chemicals used in this protocol are potentially harmful. Hence, this protocol should be carried out in a well-vented chemical fume hood while wearing proper personal protective equipment (gloves, lab coat and eye protection).
REAGENT SETUP
Culture medium
DMEM supplemented with 10% (vol/vol) FBS and 1% (vol/vol) penicillin/streptomycin is used as the cell culture medium for human HT1080 fibrosarcoma cells. This solution can be stored for a month at 4 °C.
▲ CRITICAL Alternative media may be needed for different cells.
Ru calibration solution
The Ru calibration solution is a 100 mM solution of ruthenium tris(bipyridyl) [Ru(bipy)3] in PBS. Use ultrasonic dissolution to reach solubility. The solution is used as a standard for low anisotropy measurements at 600 nm. This solution can be stored for a month at 4 °C.
Py1-PG calibration solution
The Py1-PG calibration solution is a solution of pyridine 1 in PG. The addition of PG increases the solution viscosity (~35 cP), slowing down rotation and increasing the overall anisotropy. The OD value (absorption at maximum in a 1-cm cuvette) of the solution equals 0.8. The extinction coefficient of Py1 is ~40,000, and its concentration is 0.02 mM. This solution can be stored for a month at 4 °C.
Fl calibration solution
The Fl calibration solution is a solution of fluorescein dissolved in a 0.1 M NaOH solution at a 10 µM concentration. Use DMSO and ultrasonic dissolution to reach solubility. The solution is used as a standard for low anisotropy measurements at 517 nm and to measure the response of the PMTs at different voltage settings. This solution can be stored for a month at 4 °C.
Fluorescein–glycerol (95%) calibration solution
The fluorescein–glycerol (95% (wt/wt)) calibration solution is a solution of fluorescein dissolved in a 0.1 M NaOH solution at a 10 µM concentration, mixed in glycerol. Use ultrasonic dissolution to reach solubility. This solution should be freshly prepared.
RhBMeth calibration solution
The RhBMeth calibration solution is a 20 mM solution of rhodamine B in methanol. The solution is used to measure the response of the PMTs at different voltage settings. This solution can be stored for a month at 4 °C.
Preparation of solutions for fluorescence anisotropy measurements as a function of viscosity
Starting from a solution of fluorescein dissolved in a 0.1 M NaOH solution at a 1 mM concentration, prepare different glycerol–water mixtures in the range from 5% to 95% (wt/wt). Use a warm bath sonicator to reach uniform solubility. This solution should be freshly prepared.
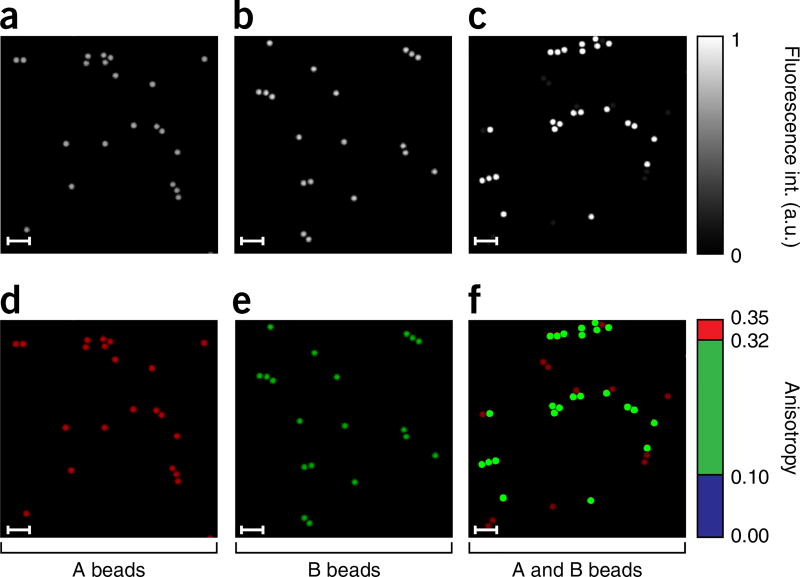
Preparation of fluorescent microspheres for fluorescence anisotropy imaging phantom
Select beads (10 µl) from the Microscope Image Intensity Calibration Kit, from the two vials corresponding to the 100% (B beads) and 10% (A beads) relative intensity concentrations. Prepare two aqueous solutions (1% (vol/vol) in dH2O) and sonicate them for 3 min. Prepare a third mixed solution by mixing the two solutions just prepared in a 1:1 ratio (A beads/B beads). Take three coverslips, clean them with ethanol and immerse them for 10 s in poly l-lysine solution. Wash in dH2O and let them dry. Apply 100 µl of the three different solutions on three distinct coverslips. Allow 1 h for the beads to adhere to the poly-l-lysine coating, and remove the remaining water with a pipette. Flip the coverslips upside down, and let them dry in a tilted position. Place an imaging chamber gasket on top of a glass microscope slide, fill it with dH2O and position the coverslips with the beads facing the dH2O. ▲ CRITICAL Wait for 30 min before imaging to let the coverslips stabilize in position. This phantom should be freshly prepared.
Preparation of seeding coverslips
Wash six coverslips in 70% (vol/vol) ethanol for 1 h. Rinse them with dH2O and let them dry in air before sterilizing them in the autoclave. Dilute the poly-l-lysine solution to a 1:10 ratio with deionized water, place the sterilized coverslips in a six-well plate and cover them with the solution, incubating for 24 h (37 °C, 5% (wt/vol) CO2). The coverslips are now ready to be used for cell seeding. Coverslips should be freshly prepared before use.
Preparation of the fluorescence anisotropy binding assay
For the synthesis of the BODIPY–biotin (N-(5-(3-(5,5-difluoro-1,3,7,9-tetramethyl-5H-4λ4,5λ4-dipyrrolo[1,2-c:2′,1′-f][1,3,2]diazaborinin-10-yl)propanamido)pentyl)-6-(5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanamido)hexanamide), weigh out 2.95 mg (7.07 µmol) of BODIPY 493/503 NHS ester and 4.42 mg (7.78 µmol) of biotin-X cadaverine trifluoroacetic acid salt in a 1-dram vial. Cover the vial with aluminum foil and add a magnetic stir bar. Add 300 µl of dichloromethane; place the vial on a magnetic stirrer and turn on the stirrer. Add 1.6 µl of N,N-diisopropylethylamine and cap the vial. After 1 h of stirring at room temperature, (25 °C), concentrate the contents under vacuum using a rotary evaporator. Purify the crude product by silica gel flash chromatography. Equilibrate the flash cartridge with water/MeCN/MeOH in a 1:1:1 (vol/vol/vol) ratio. Dissolve the crude product in 500 µl of water/MeCN/MeOH (1:1:1 (vol/vol/vol) ratio) and load it onto the flash cartridge. Elute with water/MeCN/MeOH in a 1:1:1 (vol/vol/vol) ratio.
Analyze fractions by TLC and liquid chromatography mass spectrometry (LCMS) and combine the clean product fractions. Concentrate under vacuum using a rotary evaporator. These solutions should be freshly prepared. ▲ CRITICAL BODIPY dyes can be prone to degradation on silica gel. Using water/MeCN/MeOH (1:1:1 (vol/vol/vol) ratio) as eluent proved to be well suited.
Preparation of the NeutrAvidin solution
Reconstitute NeutrAvidin (MW, 60 kDa) with ultrapure water and dilute it to 1 mg/ml in PBS and 0.1% (vol/vol) Tween 20. This solution should be freshly prepared. ▲ CRITICAL The biotin-binding protein contains four identical subunits.
0.1% trifluoroacetic acid in water (mobile phase A)
In a Kimax media storage bottle, measure 900 ml of Milli-Q water and add 900 µl of trifluoroacetic acid. Mix the contents thoroughly. Store this solution at 20 °C for no more than 6 d.
0.1% trifluoroacetic acid in acetonitrile (mobile phase B)
In a Kimax media storage bottle, measure 900 ml of acetonitrile (HPLC grade) and add 900 µl of trifluoroacetic acid. Mix the contents thoroughly. Store this solution at 20 °C for no more than 6 d.
0.05% formic acid in water (mobile phase C)
In a Kimax media storage bottle, measure 900 ml of Milli-Q water and add 450 µl of formic acid. Mix the contents thoroughly. Store this solution at 20 °C for no more than 6 d.
0.05% formic acid in acetonitrile (mobile phase D)
In a Kimax media storage bottle, measure 900 ml of acetonitrile (LCMS grade) and add 450 µl of formic acid. Mix the contents thoroughly. This solution should be freshly prepared.
Fluorescently labeled drug
We use PARPi-FL, as discussed in the introduction. Preparation of PARPi-FL is described in detail in the Supplementary Methods.
EQUIPMENT SETUP
Laser scanning microscopy system
The two-photon laser scanning microscopy system that we use for FAIM is based on a commercially available Olympus FV1000MPE confocal and multiphoton microscope controlled by the Olympus FluoView software. Assembly is described in the Procedure.
HPLC
For semipreparative HPLC, a Phenomenex Jupiter C18 reversed-phase analytical column (5 µm, 300 Å, 10 × 250 mm) on a Shimadzu UFLC system consisting of a degasser (DGU-20A3), pump (LC-20AB), detector (SPD-M20A), module (CBM-20A) and a fraction collector (FRC-10A) should be used. The fraction collector should be equipped with disposable borosilicate glass tubes. The linear gradient of water (0.1% trifluoroacetic acid, mobile phase A) and acetonitrile (0.1% trifluoroacetic acid, mobile phase B) should start with 5% and reach 95% of mobile phase B over 15 min with a flow rate of 3.0 ml/min; this should be followed with 95% of mobile phase B for 3 min. The UV detection should be set to 280 nm. For analytical HPLC, a Waters Atlantis T3 C18 reversed-phase analytical column (10 µm, 100 Å, 4.6 × 250 mm) using the same equipment setup with the same linear gradient and a flow rate of 1.0 ml/min should be used.
Liquid chromatography mass spectrometry
For LCMS, a Waters Atlantis T3 C18 reversed-phase analytical column (5 µm, 100 Å, 4.6 × 100 mm) on a Shimadzu LCMS system consisting of a degasser (DGU-20A5), pump (LC-20AD), autosampler (SIL-20AC), detector (SPD-20AV), module (CBM-20A), nitrogen generator (NM32LA) and a liquid chromatograph mass spectrometer (LCMS-2020) should be used. The autosampler should be equipped with a Shimadzu Prominence autosampler vial containing the substance solution (<1 µg/ml). The linear gradient of water (0.05% formic acid, mobile phase C) and acetonitrile (0.05% formic acid, mobile phase D) should start with 5% and reach 95% of mobile phase D over 6 min with a flow rate of 0.5 ml/min, and should be followed with 95% of mobile phase D for 2 min. The UV detection should be set to 280 nm. The injection volume of the sample should be 10 µl.
NMR spectroscopy
For NMR spectroscopy, a Bruker Ultrashield 600 Plus and NMR sample tubes from Wilmad LabGlass should be used. The sample (0.8–0.9 mg) should be dissolved in DMSO-d6 (600 µl) and a NMR sample tube should be charged with the resulting PARPi-FL solution. Then, the NMR vial should be placed into the autosampler of the NMR spectrometer and default settings for acquiring 13C NMR and 1H NMR spectra should be chosen.
PROCEDURE
Assembly of the polarization control unit ● TIMING 3–4 h
-
1|
Turn on all the microscope controllers, the Mai Tai femtosecond pulsed laser and the computer controlling the Olympus microscope (data acquisition PC). Launch the Olympus FluoView control software. Because this article discusses the modification and adaptation of the commercially available Olympus FV1000MPE laser scanning microscope, it is recommended that the user be familiar with the system’s user manuals for precise start-up procedures.
! CAUTION This protocol requires open beam alignment while working with a Mai Tai Class IV high-power laser operating in the IR spectral range, which is extremely dangerous to the eye. It is very important to take appropriate safety precautions. Refer to the laser safety section of the Mai Tai laser for the proper recommendations. Wear protective goggles at all times during the waveplate and the GT alignment procedures (Steps 1–12). It is also recommended, during Mai Tai alignment, to work with the lowest power possible and with the room lights on.
-
2|
In the ‘Image Acquisition Control’ window, set the FluoView software for two-photon acquisition modality (red box 1, Supplementary Fig. 2).
-
3|
To perform the alignment of the PCU (Figs. 4a and 5a,b), open the laser shutter in the ‘Acquisition Setting’ window (red box 1, Supplementary Fig. 3). The laser beam must pass through the acousto-optic modulator positioned right before the beam expander (BE) (Figs. 4a and 5a). For this to occur, it is necessary to run an acquisition through the FluoView software.
▲ CRITICAL STEP If you are operating the microscope with light present in the room, attention must be paid in order to avoid saturating or perhaps damaging the PMTs. In the ‘Image Acquisition Control’ window, set the PMT voltages to zero (red boxes 2,3, Supplementary Fig. 2).
-
4|
Press the ‘XY’ button in the ‘Image Acquisition Control’ window (red box 4, Supplementary Fig. 2) to start a single image acquisition. In the ‘Acquisition Setting’ window within the ‘Mode’ setting, select single-point scanning (red box 2, Supplementary Fig. 3) and position the point in the center of the acquired image (red boxes 1,2, Supplementary Fig. 4). This will ensure that during acquisition the laser is not scanned across the back-aperture of the imaging objective but will instead remain fixed in position over time.
▲ CRITICAL STEP This option should be set only during this phase of the protocol. For dye and cell imaging, acquisition should be set to square imaging.
-
5|
Press the ‘Time’ button (red box 5, Supplementary Fig. 2) in the ‘Image Acquisition Control’ window to activate the time-lapse acquisition option, and in the ‘TimeScan’ section of the ‘Acquisition Setting’ window, select ‘FreeRun’ in the ‘Interval’ field (red box 3, Supplementary Fig. 3), and set an appropriate value N for the number of frames to be acquired (typically 10,000) (red box 4, Supplementary Fig. 3). In the ‘Mode’ settings window, choose an integration time of 100 µs (red box 5, Supplementary Fig. 3), and in the ‘Size’ settings window, choose an image size of 256 × 256 (red box 6, Supplementary Fig. 3). In the ‘Microscope’ settings window, select the proper objective specifications (red box 7, Supplementary Fig. 3).
▲ CRITICAL STEP Depending on the amount of RAM available, the number of frames (N) could be limited.
-
6|
Make sure that the laser shutter is OFF, and unscrew the lens tube cover connecting the BE (Fig. 4a) to the steering combining mirrors (SCMs; Fig. 4a).
! CAUTION Make sure to wear protective goggles.
-
7|
In the ‘Image Acquisition Control’ window, press the ‘XY’ button (red box 4, Supplementary Fig. 2) to start an acquisition and open the shutter of the laser.
▲ CRITICAL STEP In the ‘Acquisition Setting’ window, within the ‘Laser’ settings area, set the parameters of the two-photon excitation laser such that it operates at the appropriate wavelength and it is set at low power (1–5%) (red boxes 1 and 8, Supplementary Fig. 3).
-
8|
Insert the GT polarizer (Figs. 4a and 5b) into a 360° continuous rotation stage mounted at the exit of the BE and block one of two orthogonal linear polarization components.
▲ CRITICAL STEP Pay attention that the alignment of the beam does not change as a result of the insertion of the GT polarizer. Use a pinhole mounted at the entry of the SCMs to facilitate the adjustments.
▲ CRITICAL STEP Because we found the light to be elliptically polarized on different imaging systems, we recommend blocking the minor axis to increase the excitation power and to rotate the axis using the half-wave plate (HWP).
-
9|
Insert the HWP (Figs. 4a and 5b) into a 360° continuous rotation stage and mount it following the GT polarizer (Fig. 5b, Supplementary Data 1). As above, pay attention to ensure that its insertion into the optical path does not change the alignment of the beam.
! CAUTION During alignment procedures, use the IR sensor card to trace the beam in space along its path.
-
10|
Press the ‘Stop’ button (red square 6, Supplementary Fig. 2) in the ‘Image Acquisition Control’ window, to stop the acquisition and turn the laser shutter OFF. Mount the near-IR linear polarizer into a 360° continuous rotation stage and position it under the imaging objective (OBJ in Fig. 5a). Remove the objective and align the polarizer such that its orientation is parallel to the microscope y axis.
-
11|
Position the power meter below the linear polarizer, and start an acquisition by pressing the ‘XY’ button (red square 4, Supplementary Fig. 2) in the ‘Image Acquisition Control’ window. The laser shutter will open. Read the power as measured at the exit of the polarizer, after properly setting the acquisition parameters of the power meter. The settings are dependent on the particular model used and the reader can refer to its user manual.
-
12|
Rotate the HWP to make sure that there is correct rotation of the optical axis and a good extinction ratio between detection at the two orthogonal directions (ŷ and x̂). Graph the measured intensity as a function of angle. In our case, we measured a better than 1:100 ratio (Supplementary Fig. 5).
▲ CRITICAL STEP Make sure that there is proper alignment at the wavelengths intended to be used for the proper fluorophore excitation.
Assembly of the polarization filter cube ● TIMING 1–2 h
-
13|
Choose the appropriate material for manufacture of the PFC, such as production-grade thermoplastic.
-
14|
3D-print all the components for the PFC using the STL files included with the article, or have the filter cube manufactured by a machining service (PFC, Fig. 4b–d, Supplementary Data 2).
▲ CRITICAL STEP Although we use our own printer (uPrint, Stratasys), several 3D-printing and CNC machining services are available online, such as Proto Labs.
-
15|
Assemble the PFC as indicated in the exploded view in Figure 4b.
▲ CRITICAL STEP In this work, we present data collection and analysis of fluorescence anisotropy performed using only two channels (i.e., a single PBS, Fig. 4b).
-
16|
Position the PBS inside the PFC on the top part of it, carefully orienting it such that the horizontal component of the light is oriented toward PMT 3. Alternatively, if the PBS is mounted on the bottom part of the cube, orientate the horizontal component of the light toward PMT 1 (Fig. 4e). In this configuration, the PFC and the PBS replace the filter cube holder and the DM indicated in the ‘LightPath & Dyes’ window (dashed square and red box 1, Supplementary Fig. 6). Set the light paths accordingly.
-
17|
Depending on the fluorescent dye of interest, insert appropriate band-pass filters. For the experiment demonstrated herein, we used two different fluorescence EFs centered at 600 and 517 nm. Position the EFs as indicated in the PFC’s exploded view and images (Fig. 4b–d).
▲ CRITICAL STEP If you are using the 3D-printed version of the PFC, it is important to take particular care when inserting and tightening the fixing screws, in particular, the ones holding the EFs, which can be replaced several times depending on the particular experiment. Excessive force over time can lead to a cracking of the threaded fittings.
-
18|
Insert the linear polarizer (P) filters, selecting their orientation to match the transmitted and reflected components of the PBS.
System calibration ● TIMING 4–6 h
▲ CRITICAL This part of the protocol is critical to proper measurement of fluorescence polarization. While it is not, in principle, important to determine the absolute true value of fluorescence polarization, it is critical to make sure that the systems can reproduce the same results over the course of an entire imaging session and, more importantly, that it is consistent over a period of months/years. It is therefore important to make sure that, in general, the microscope is maintained in good alignment and to check that the values of two-photon fluorescence polarization are reproducible using appropriate calibration solutions. It is also imperative that the system has reached stability in terms of laser mode-locking, electronics and PMT detector noise. On the basis of our experience, we recommend a period of ~2 h.
▲ CRITICAL As fluorescence polarization calibrating solutions, we consider in our protocol three dyes covering two distinct EFs and two different ranges (high and low) of fluorescence anisotropy.
-
19|
Mount 180 µl of Ru calibration solution in an imaging chamber gasket positioned over a glass microscope slide, and seal the gasket with a round microscope coverslip. Also mount 180 µl of dH2O in a similar imaging chamber gasket.
▲ CRITICAL STEP Let the imaging chamber gasket stabilize in position before imaging (~30 min).
-
20|
Insert into the top part of the PFC the EF centered at 600 nm, so that the two orthogonal components of the polarization are detected by PMT 3 (perpendicular) and PMT 4 (parallel), respectively.
▲ CRITICAL STEP The filter can be inserted alternatively into the bottom part of the PFC. In this case, PMT 1 and PMT 2 must be considered.
-
21|
Turn on the microscope controllers, laser subsystems and the data acquisition PC. Launch the FluoView control software on the data acquisition PC, and, in the ‘Acquisition Settings’ window, set the acquisition modality for two-photon imaging (red box 1, Supplementary Fig. 2). Tune the Ti:Sapphire laser to 750 nm and set the excitation laser power at 15% in the ‘Laser’ section. Set a dwell time of 20 µs per pixel, choose a zoom factor of 13 and set the scan area to 256 × 256 pixels. Select the 25× objective. In the ‘Image Acquisition Control’ window, mark the checkboxes for PMT 3 and PMT 4 and set the voltage values to 558 and 629.
▲ CRITICAL STEP The voltage numbers are unique for each system and depend on the PMTs and their alignment with respect to the imaging and collecting lenses.
▲ CRITICAL STEP Noise on the detection channels changes greatly over time, reflecting the SNR of the images. More importantly, because the dark noise must be properly subtracted from the acquired fluorescence images, the noise must be measured periodically. It is therefore important before taking any measurement to make sure that the Ti:Sapphire laser has been turned ON for 1 h in order to stabilize and that all the electronic components of the microscope have been turned ON for at least 2 h. In this way, the noise will remain approximately constant over the course of the entire experiment.
▲ CRITICAL STEP Because of the presence of the GT polarizer, the real power at the objective is approximately half of that before its insertion (i.e., 15% in the modified configuration corresponds to roughly 7% in the standard microscope configuration).
-
22|
Take a measurement of the background noise. Turn the laser shutter to OFF, and start an acquisition by pressing the ‘XY’ button in the ‘Image Acquisition Control’ window.
-
23|
Launch the BackgroundEstimation software (provided with the article) and in the GUI, press the button ‘Load Image A’ (‘Load Image B’) (red boxes 1,2 Supplementary Fig. 7) to load the images corresponding to PMT 3 (PMT 4). Press the button ‘Run Computation’ (red box 3, Supplementary Fig. 7) to measure the background noise for the two acquisition channels PMT 3 and PMT 4 (red boxes 4,5 Supplementary Fig. 7).
▲ CRITICAL STEP For our system, we found that the background noise is equal to the dark noise. This may not be true for other systems and will ultimately depend on the quality of the dichroics and filters present. Use the dH2O-filled imaging chamber gasket prepared at Step 19 to determine the background signal.
-
24|
Place the mounted Ru calibration solution under the imaging objective, and take a measurement of the fluorescence on the two channels PMT 3 and PMT 4. Adjust the power excitation in order to achieve a good SNR for both images. The solution presents a low anisotropy value across a large excitation spectrum (Fig. 7).
▲ CRITICAL STEP Avoid acquisition of fluorescence images with saturated pixels, which could produce artifacts in the calculation of the fluorescence anisotropy.
? TROUBLESHOOTING
-
25|
Launch the AnisotropyCalibration software (provided as Supplementary Software) to calculate the average anisotropy value of the solution. In the GUI, press the buttons ‘Load Image A’ (‘Load Image B’) (red boxes i,ii, Fig. 8) to load the fluorescence images acquired with PMT 3 and PMT 4. In the ‘Thresholds’ settings window (red box iii of Fig. 8), enter the threshold values for the background subtraction calculated above, and press the ‘Anisotropy’ button (red box iv, Fig. 8) to calculate the average fluorescence anisotropy (red box v, Fig. 8) and save the fluorescence anisotropy image (red box vi, Fig. 8).
-
26|
Adjust the voltage of PMT 4 to set the anisotropy equal to zero. These voltage settings should be used for all fluorescence anisotropy measurements.
▲ CRITICAL STEP Fluorescence anisotropy measurements may require different ranges of PMT voltages depending on the fluorophore or fluorescently labeled drug used and the absolute abundance of the fluorophore or the target. It is therefore recommended at this point to calibrate the system at several voltages.
▲ CRITICAL STEP Fluorescence anisotropy is calculated at each pixel and may vary in total intensity depending on the features of the image, particularly when imaging cells. It is important to make sure that the PMTs exhibit a linear behavior across the entire detection range.
-
27|
Mount 180 µl of RhBMeth calibration solution in an imaging chamber gasket positioned over a glass microscope slide, and seal the gasket with a round microscope coverslip. Use the same settings as the ones selected before within the FluoView GUI. Collect a series of measurements and, as excitation power, choose a series of values such that the fluorescence signal goes from zero (after background subtraction) to saturation.
-
28|
With a power meter, measure the total excitation power at the exit of the objective, and plot on a log–log scale the two-photon-induced fluorescence as a function of excitation power, calculated as the average of the entire fluorescence image. Calculate the fitting parameter and make sure that the PMTs are behaving linearly and in the same way for the different voltage settings (Fig. 9a–c).
▲ CRITICAL STEP For our imaging systems, we found that the PMTs behaved linearly across their entire range of detection at different voltage settings. In addition, when plotted on a log–log scale, the fitted slopes of the measured values of two-photon fluorescence intensity versus excitation intensity are in accordance with the values reported in the literature (1.99 and 1.97 for fluorescein and rhodamine B, respectively, at 755-nm excitation)68.
? TROUBLESHOOTING
-
29|
To test the settings and calibrate the system in the 517-nm emission range, mount 180 µl of Fl calibration solution in an imaging chamber gasket positioned over a glass microscope slide, and seal the gasket with a round microscope coverslip. The solution presents a low anisotropy value across a large excitation spectrum (Fig. 7b,e,h).
-
30|
Insert into the top part of the PFC the EF centered at 517 nm, so that the two orthogonal components of the polarization are detected by PMT 3 and PMT 4.
-
31|
Repeat the steps described above (Steps 21–26) to set the proper voltage settings necessary to achieve zero fluorescence anisotropy and to measure the linearity of the detecting PMTs in the 517-nm fluorescence emission range (Steps 27 and28) (Fig. 9d–f).
Figure 7.
Optical characterization of three calibration solutions, ruthenium tris(bipyridyl) [Ru(bipy)3] in PBS (Ru in PBS), fluorescein in PBS (FL in PBS), pyridine 1 in propylene glycol (Py1 in PG). (a–f) One-photon excitation spectra (a–c) and one-photon fluorescence anisotropy emission (d–f) as a function of excitation wavelength for emissions at 600 nm (a,d), 525 nm (b,e), and 680 nm (c,f). (g–i) Two-photon fluorescence anisotropy as a function of wavelength for the corresponding dyes, for detection at 600 nm (g,h) and 517 nm (i).
Figure 8.
GUI interface of the AnisotropyCalculation program. The program is used to calculate the average fluorescence anisotropy of an acquired set of images.
Figure 9.
PMT intensity responses across their dynamic ranges at different voltage settings. The responses are measured at different detection wavelengths using different calibration solutions. (a–c) Rhodamine B. (d–f) fluorescein. Voltage settings for PMT3 and PMT4 are 498 and 559 V, respectively (a,d); 558 and 629 V, respectively (b,e); 625 and 706, respectively (c,f). PMT3 is shown in red, and PMT4 is shown in black. Red and blue arrows indicate the proper vertical axes (left or right) for each fitted data set.
High anisotropy measurements ● TIMING 2 h
▲ CRITICAL It is important to verify that after the system calibration part of the protocol, measurements of samples presenting high values of anisotropy are reproducible over the course of different experiments. This guarantees that fluorescence anisotropy images collected during the same imaging session or on different days can be compared and used for further analysis across their entire range of anisotropy values. For this purpose, we need a convenient solution that can be easily accessed and routinely used. We chose to work with a solution of pyridine 1 in PG (Py1-PG calibration solution), with fluorescence emission in the 600-nm range. Alternative samples that can be used include commercially available autofluorescent plastic slides. Bear in mind that the reproducibility across slides from different stocks is not guaranteed.
-
32|
Mount 180 µl of Py1-PG calibration solution in an imaging chamber gasket positioned over a glass microscope slide, and seal the gasket with a round microscope coverslip.
-
33|
Insert into the top part of the PFC the EF centered at 600 nm, so that the two orthogonal components of the polarization are detected by PMT 3 and PMT 4.
-
34|
Repeat the steps described above (Steps 21–25) using the PMT voltages, and collect images. Calculate the average value of fluorescence anisotropy using the AnisotropyCalibration software, and keep record of its value at the wavelength of interest for future comparisons over time (Fig. 7c,f,i).
▲ CRITICAL STEP If discrepancies between measurements are present, it is recommended to perform a realignment of the optical components, perform another calibration or prepare a new solution. Temperature variability can also cause problems when measuring fluorescence anisotropy, so extreme care must be taken to keep the imaging room at a constant temperature.
? TROUBLESHOOTING
Measurements of fluorescence anisotropy at increasing values of viscosity ● TIMING 4 h
-
35|
To monitor the accuracy of the alignment and the reproducibility of the system over an extended range of fluorescence anisotropy values and over time, perform measurements of anisotropy of a solution of fluorescein dissolved at different ratios of glycerol/water, prepared as described in the Reagent Setup. This also serves as a method of checking accuracy at another emission wavelength range (520 nm) with respect to the Py1-PG calibration solution. Prepare a series of dilutions and, for each stock solution, calculate the average anisotropy (Reagent Setup).
-
36|
Mount 180 µl of each stock solution in an imaging chamber gasket, as described in Step 19.
-
37|
Insert the EF centered at 517 nm into the top part of the PFC, and tune the laser to 755 nm.
-
38|
Repeat Steps 21–25 and collect images for each stock solution. Calculate the average value of fluorescence anisotropy using the AnisotropyCalibration software, after proper background subtraction, and plot the values as a function of glycerol concentration (Fig. 10). Keep the results obtained as reference for future comparisons over time.
▲ CRITICAL STEP Take multiple measurements for each stock solution by mounting the solution in different imaging chambers and not by repeating the measurement for the same imaging chamber.
? TROUBLESHOOTING
Figure 10.
Measurements of fluorescein at varying viscosities. Standard measurements used to perform the system calibration and determine its sensitivity across a large range of fluorescence anisotropy values, for a solution of fluorescein dissolved in a glycerol–water mixture. (a) The graph shows that the fluorescence anisotropy of the solution is increasing as a function of glycerol content (i.e., viscosity). Individual measurements (open circles), averages (black squares) and standard deviations of the means (bars) are indicated. N = 3. (b) Fluorescence anisotropy images of the stock solutions at different values of glycerol concentration. Average anisotropy is indicated within the images. Images are color-mapped using a MATLAB hot colormap, for better visualization across the entire range of anisotropy. Images are filtered for noise (2D Gauss, window size = 21 pixels, α = 3). Scale bar, 10 µm.
Measurements of fluorescence anisotropy for receptor and ligand binding quantification ● TIMING 4 h
▲ CRITICAL Measure the fluorescence anisotropy of 15 µM BODIPY–biotin with various concentrations of NeutrAvidin titrated against it. Choose the titration points (reaction volume equal to 30 µl) such that they are equally distributed around the estimated dissociation constant (Kd value).
-
39|
Make up solutions of 15 µM BODIPY–biotin with various concentrations of NeutrAvidin. Place the solutions in separate Eppendorf tubes on ice to avoid protein degradation over the course of the preparation of the titration and experiment. This quantifies receptor binding.
▲ CRITICAL STEP The concentration of the labeled ligand is determined by the sensitivity of the microscope and the acquisition parameters necessary to measure high-SNR fluorescence images.
▲ CRITICAL STEP Add 0.1% (vol/vol) Tween 20 to the BODIPY–biotin solution to prevent the dye from sticking to the surface of the coverslip of the imaging chamber gasket.
-
40|
Remove each Eppendorf tube from the ice and incubate at room temperature for 10 min to reach equilibrium before measuring. Use a pipette to mix the solution repeatedly.
-
41|
Mount the titrating solution in an imaging chamber gasket and insert into the top part of the PFC the EF centered at 517 nm.
-
42|
Repeat Steps 21–25 and collect three measurements for each titrating solution. Using the AnisotropyCalibration software, calculate the average value of fluorescence anisotropy, after proper background subtraction.
-
43|
Plot the values of the fluorescence anisotropy for all titrating points as a function of the log of the receptor (NeutrAvidin) concentration normalized for the BODIPY–biotin concentration (Fig. 11). The anisotropy of the free and bound states is determined by the top and bottom part of the semi-log binding isotherm. Using equation 4, the measured anisotropy can be directly converted into the fraction of bound versus free BODIPY–biotin.
▲ CRITICAL STEP The number of binding sites per NeutrAvidin molecule is equal to four; therefore, remember to rescale the receptor concentration.
Figure 11.
Fluorescence anisotropy measurements for receptor/binding quantification. (a) Fluorescence anisotropy of a 15 µM solution of BODIPY–biotin titrated against NeutrAvidin. The anisotropy of the free and bound states is determined by the top and bottom part of the semi-log binding isotherm. Owing to the fluorescence anisotropy additive property (equation 4), the measurements can be directly converted to the fraction-weighted sum of the two possible states (bound versus free BODIPY–biotin). Individual measurements (open circles), averages (black squares), and standard deviations of the means (bars) are indicated. Red dashed line indicates the fluorescence anisotropy value for free BODIPY–biotin. N = 3.
(b) Fluorescence anisotropy images of the stock solutions at different titration points (indicated in black in the boxes). Images are color-mapped using a perceptually balanced color scheme for better visualization across the entire anisotropy range. Scale bar, 10 µm.
Imaging of phantoms ● TIMING 1 h
▲ CRITICAL Before proceeding with cell imaging, it is recommended to perform FAIM tests on phantoms presenting different degrees of anisotropy and distinct morphological features.
-
44|
Prepare a series of coverslips with fluorescent microspheres as indicated in the Reagent Setup section.
-
45|
Take several images, adjusting the zoom factor and the imaging plane. Choose the proper values for laser intensity and integration time, to avoid image saturation and guarantee good SNR.
-
46|
Postprocess the data as described in Step 75 and make sure that there is good separation in fluorescence anisotropy between the two populations of beads (Fig. 12).
▲ CRITICAL STEP If the two populations cannot be clearly resolved, the calibration and alignment of the imaging system must be repeated.
? TROUBLESHOOTING
Figure 12.
Imaging phantom for fluorescence anisotropy imaging. (a–f) Fluorescence intensity (a–c) and fluorescence anisotropy (d–f) images of two separate (a,b,d,e) and mixed (c,f) bead populations with different relative intensity concentrations. At a high fluorophore concentration (B beads), homo-FRET takes place, reducing the effective value of fluorescence anisotropy69. The two bead populations can then be resolved on the basis of their fluorescence anisotropy values. Scale bars, 17 µm.
Real-time two-photon fluorescence anisotropy measurement and visualization of drug-target engagement in live cells ● TIMING 30 min
-
47|
Prepare appropriate fluorescently labeled drug; for example, we use PARPi-FL, prepared as described in the Supplementary Methods. The following steps describe how to use PARPi-FL.
-
48|
Let the stock solution of PARPi-FL warm to room temperature for 10 min.
▲ CRITICAL STEP Handle the tracer with care in the dark.
-
49|
Set up an amber glass vial (4 ml) on the balance and tare the balance.
-
50|
Charge the amber glass vial with PARPi-FL (1.0 mg).
-
51|
Dissolve the PARPi-FL with PEG 300 (1.0 ml).
▲ CRITICAL STEP It may be necessary to warm the sample at 37 °C for 1 min.
-
52|
Transfer 50 µl of the PARPi-FL solution to a microcentrifuge tube.
▲ CRITICAL STEP Immediately close the tube and store it in the dark at 20 °C for up to 12 h.
-
53|
Repeat Step 52 until no more stock solution remains.
■ PAUSE POINT Store aliquots of PARPi-FL at −80 °C for up to 12 months.
-
54|
Before imaging, let the PARPi-FL aliquot warm to room temperature for 10 min.
▲ CRITICAL STEP The color of the solution should be green. It may be necessary to warm the sample at 37 °C for 1 min.
-
55|
Prepare 1-ml aliquots of inhibitor at a 1 µM concentration in culture medium.
Preparation of adherent cells for fluorescence anisotropy imaging ● TIMING 24 h
-
56|
Incubate the cells in the culture medium, in a T125 flask at 37 °C with 5% (wt/vol) CO2 in humidified air.
-
57|
When the cells reach 80% confluence, trypsinize them with 0.5% trypsin until all cells have detached, place the cells in a 50-ml Falcon tube and deactivate the enzymes by adding 2× cell culture medium.
-
58|
Centrifuge the cell suspension at 300g for 3 min at 25 °C and resuspend at a density of 67,000 cells per ml in medium. Count the cells using a standard hemocytometer.
-
59|
Prepare a six-well plate by placing the seeding coverslip into each well and adding 3 ml of the cell suspension directly to the surface of the coverslip.
▲ CRITICAL STEP Care must be taken to ensure that the cell suspension is added only to the surface of the coverslip. If more medium is required, then add some 4–6 h after plating the cells.
-
60|
Incubate the plated cells for 24 h before imaging (37 °C, 5% (wt/vol) CO2), ensuring qualitatively that the cells are at 80% confluency on the coverslip.
▲ CRITICAL STEP It is critical to conduct the imaging by at most 48 h post plating to ensure that contact inhibition or cell death does not influence the results.
Preparation of the closed-bath imaging chamber for time measurements ● TIMING 30 min
-
61|
Apply a small amount of grease to the bottom groove of the imaging chamber, and gently press one coverslip into position. Place the chamber in the imaging platform.
-
62|
Fill a 10-ml syringe with culture medium (syringe A) and another 10-ml syringe with a 1 µM solution of PARPi-FL in culture medium (syringe B). Connect the two syringes to the perfusion manifold with tubing (Supplementary Fig. 8).
-
63|
Connect the perfusion manifold to the imaging chamber, and gently fill the bath area with the culture medium through the inlet port.
-
64|
Apply a small amount of grease to the top groove of the imaging chamber, and gently press one cell-seeded coverslip into position. Remove excess grease, and seal the imaging chamber.
▲ CRITICAL STEP Make sure that the cells are oriented toward the inside of the imaging chamber.
-
65|
Connect the vacuum outlet port to the side arm of the trapping flask. Place the black rubber stopper into the top of the flask and connect it to the back end of a syringe connected by tubing to the outlet port of the imaging chamber (Supplementary Fig. 8). The vacuum trap will ensure that during perfusion of the imaging chamber the outgoing solution from the chamber outlet is dispensed into the flask, keeping the top of the imaging coverslip free of any fluorescent dye and avoiding background fluorescence signal.
▲ CRITICAL STEP Make sure that the solutions are all at a starting temperature of 37 °C. If bubbles are present within the imaging chamber, they should be removed.
-
66|
Set the heating plate at 37 °C, and fix both the plate and the imaging chamber to the microscope translation stage.
Selection of microscope parameters for live-cell imaging ● TIMING 30 min
-
67|
In the FluoView program, set the microscope laser parameters. Select two-photon-mode acquisition with near-IR femtosecond laser excitation. A typical two-photon imaging session of live cells requires between 5- and 10-mW laser power at 910 nm for PARPi-FL FAIM. For other fluorophores, the appropriate laser excitation wavelengths must be selected.
-
68|
In the ‘Image Acquisition Control’ window, press the ‘Light Settings’ button (red box 7, Supplementary Fig. 2) and, in the ‘LightPath & Dyes’ window, select the appropriate detection light paths (red box 1, Supplementary Fig. 6). Choose the right combination of PMTs (PMT 3 and PMT 4 in this protocol). Place the EF centered at 517 nm within the PFC. This EF best matches the fluorescence emission spectra of BODIPY-FL; if a different inhibitor is used, choose an appropriate EF.
-
69|
In the ‘Acquisition Setting’ window, choose a dwell time of 20 µs per pixel, with an image size of 512 × 512. Within the ‘Area’ settings, select a zoom factor equal to 3.0, and in the ‘Microscope’ settings, choose the 25× imaging objective (Olympus XLPLN25XWMP2). In the ‘Laser’ settings area, set the excitation wavelength of interest at 910 nm, and set the laser power to 10%. In the ‘Image Acquisition Control’ window, select PMT 3 and PMT 4, and set the voltages (HV) at the appropriate level previously determined during the system calibration procedure.
▲ CRITICAL STEP The laser power and the voltage settings must be determined for the experiment conducted and the quantum efficiency of the fluorophore of interest. Higher voltages enhance the detected signal level but also increase the background noise level in the image. The balance between laser exposure level and PMT gain must be determined empirically through adjustment of settings during imaging, in order to obtain the best SNR.
-
70|
Record test fluorescence images to check that the signal is well above the noise and to further optimize the microscope settings.
? TROUBLESHOOTING
-
71|
To increase the SNR, process the images for noise filtering (Step 75) or, alternatively, average images acquired over multiple acquisitions. In the ‘Image Acquisition Control’ window within the ‘Filter Mode’ group box (red box 8, Supplementary Fig. 2) select Kalman, choosing a value of 4 or 16, depending on the amount of signal recorded, and opting for ‘Line’ filtering. Alternatively, when performing measurements of dynamic events, it is recommended to perform noise filtering using the denoising options offered by the software included in the protocol.
▲ CRITICAL STEP Although averaging improves the overall SNR, it will tend to greatly increase the acquisition time (by a factor of N, where N represents the number of acquisitions). This is not recommended if following events in real time or if the dynamics are relatively fast. It is instead preferable to noise-filter the data using the software included with this protocol.
▲ CRITICAL STEP The FluoView manual erroneously reports that the Kalman accumulation algorithm is calculated according to I{n} = (I{n−1}*(n+1)+Itn)/n, where n is the number of image acquisitions, I{n} is the result of n-times Kalman accumulations and Itn is the current acquired image. For n = 1, I{1} is equal to It1. The correct formula used by the FluoView program is instead I{n} = (I{n−1}*(n−1)+Itn)/n and the result is equivalent to the average of n subsequently acquired images.
? TROUBLESHOOTING
-
72|
Take a test image in order to find the approximate imaging plane containing the cells. Use cellular autofluorescence signal to facilitate the procedure.
-
73|
Perfuse the imaging chamber with the 1 µM solution of PARPi-FL by gently pressing syringe B. Load the cells for 1 min.
-
74|
Start two-photon acquisitions while perfusing the imaging chamber with the rinsing solution from syringe A and collect a time-lapse sequence to follow drug-target engagement over time through fluorescence anisotropy images.
▲ CRITICAL STEP Make sure to maintain a constant low pressure to avoid changing the position of the imaging plane during the perfusion.
? TROUBLESHOOTING
Acquisition of fluorescence images
-
75|
Calculate and display fluorescence anisotropy images in stream processing (option A) or in postacquisition (option B) mode. Option A is recommended if you wish to have feedback in real time regarding the optimal choice of imaging parameters and to determine the best areas of interest during imaging sessions. In addition, option A is particularly recommended during time-lapse imaging sessions.
(A) Stream processing for calculation and visualization of fluorescence anisotropy images during time-lapse acquisition ● TIMING 2 h
-
Download the file OnLine.zip to the data acquisition PC or another PC with access to the directory where files are automatically saved during imaging acquisition. Unzip the file and create a new folder of your choice containing the executable file OnLine.exe. Download the file MCR_R2015a_win64_installer.exe.
▲ CRITICAL STEP If the data acquisition PC hardware resources are limited in terms of CPU speed and physical memory, it is recommended to share a temporary folder from the data acquisition PC with the data processing PC while running the OnLine software on the secondary PC (data processing PC). This will prevent the OnLine software from interfering with the FluoView program during image acquisition.
-
Execute the file MCR_R2015a_win64_installer.exe to install the MATLAB Runtime, which enables the execution of the compiled version of the OnLine.exe program. After the installation, the file can be deleted or removed from the directory.
▲ CRITICAL STEP The MATLAB Runtime installer can be downloaded for both 32- and 64-bit Windows operating systems.
Folder sharing. Connect both the data acquisition PC and the data processing PC to the same local network.
On the data acquisition PC, create a folder named: D:\FluorescenceAnisotropy\OnLine\Temporary
-
With Windows File Explorer, navigate to the newly created directory, right-click on it and on the new menu that appears, click on ‘Properties’, press the ‘Sharing’ tab, select ‘Advanced Sharing’ and tick ‘ON’ in the ‘Share this folder’ box.
▲ CRITICAL STEP Make sure that ‘Editing Permissions’ are set properly if you want to allow for writing and deleting of files.
Launch the ‘Command Prompt’ from the ‘Run Window’.
Run the command IPConfig to obtain the data acquisition PC network address.
On the data processing PC, open File Explorer and type in the data acquisition PC IP address.
Provide ‘Administrator’ and ‘Password’, if requested, and navigate to the newly shared folder.
Map a drive assigning a letter to the shared folder ‘FluorescenceAnisotropy’.
Parameter initialization. On the data acquisition PC, open the FluoView program and in the ‘Image Acquisition Control’ window, select the option ‘Hard Disk Recording ON’ in the ‘Hard Disk Recording Setting’ section (red box 9, Supplementary Fig. 2). This will force the acquisition software to save the acquired fluorescence images continuously (image by image) as they are acquired. Indicate the folder D:\FluorescenceAnisotropy\OnLine\Temporary (from now on referred to as DataFolder; red box 10, Supplementary Fig. 2) as the directory where the data should be written.
-
Press the ‘Time’ button in the ‘Image Acquisition Control’ window to activate the time-lapse acquisition option. In the ‘TimeScan’ section of the ‘Acquisition Setting’ window, select ‘FreeRun’ in the interval field, and set an appropriate value (N) for the number of frames to be acquired (typically 100). N images are then acquired and directly recorded in the folder ‘DataFolder’.
▲ CRITICAL STEP Direct recording of the data as acquired works only if the acquisition is started by selecting the ‘XY’ image acquisition scan button. The acquisition can be interrupted or paused at any point.
On the data processing PC, start the OnLine program (Supplementary Software, Fig. 13), which will open a GUI for the processing of the raw fluorescence images as acquired by the FluoView program. Each fluorescence image is directly displayed as unfiltered raw data. The program automatically adjusts the contrast of the images to fit the range of the data for the processed images. The corresponding fluorescence anisotropy image is calculated and color-mapped using a MATLAB jet color scale. The weighted fluorescence anisotropy image is also calculated and color-mapped using the weighted RGB color-mapping scheme illustrated in the Experimental design section. The user can select and define both the processing and the visualization parameters. The program will continuously read and process the data as written by the FluoView software into the ‘DataFolder’. Acquisitions can be paused, interrupted or restarted without causing any crashing of the running software.
Indicate the folder (red box i in Fig. 13) where the images are saved (DataFolder), present either on a local drive or a locally mapped disk.
If images are collected using only two PMTs, select 1 and 2 (red box ii in Fig. 13) for ‘Image A’ and ‘Image B’, respectively. If four-channel acquisition is performed, indicate which channel combination to use (1 and 2, or 3 and 4).
-
Set the threshold values previously measured, for the dark noise background subtraction of both acquisition channels (red box iii in Fig. 13).
▲ CRITICAL STEP Make sure that the dark noise background is measured using the same integration time as for the fluorescence anisotropy measurements.
-
If you require noise filtering, set parameters accordingly (red box iv in Fig. 13). Noise filtering is optional but is recommended when dealing with low-SNR images for better fluorescence anisotropy smoothing. The filters and their parameters act on the raw fluorescence images and can be also changed on-the-fly during runtime. Tick next to ‘2D Median’ and ‘2D Gauss’ to enable or disable the filters.
▲ CRITICAL STEP To avoid pixel shifting in the calculated fluorescence anisotropy images, odd values should be used for window size (filter kernel) entries.
Initiating acquisition and visualization. In the OnLine program, press the button ‘Press to Start’ (red box v in Fig. 13) to initiate visualization of the fluorescence anisotropy images (raw and weighted). If the FluoView program is not yet running or no image acquisition has been started, the OnLine program remains on hold until the first pair of fluorescence images is collected. All the fluorescence anisotropy display parameters can be changed on-the-fly during runtime.
Set the maximum (red box vi in Fig. 13) and the minimum (red box vii in Fig. 13) values for the fluorescence anisotropy color-mapping visualization scale.
-
If required, directly change the weighted fluorescence anisotropy display parameters (red box viii in Fig. 13). Different values (red box ix in Fig. 13) can be arbitrarily chosen for the minimum (min) and the maximum (Max). Three intervals (four values) are used to assign three different colors (blue—B; green—G; red—R) to the fluorescence anisotropy on a pixel-by-pixel basis. Each pixel color is statistically weighted based on the total fluorescence intensity.
▲ CRITICAL STEP Images are automatically scaled, but sometimes saturating pixels or small clusters can cause the intensity scaling to be set incorrectly.
-
Press the ‘XY’ button in the ‘Image Acquisition Control’ window of the FluoView software to start recording the measurements. Fluorescence anisotropy images are automatically calculated and displayed by the OnLine software. Time-lapse fluorescence anisotropy images of the cells during the course of the washing phase are automatically acquired and displayed (Fig. 14), allowing the researcher to follow interactions of the drug with its target.
? TROUBLESHOOTING
Figure 13.
GUI interface of the OnLine program. The program is used for real-time image processing, calculation and visualization of fluorescence anisotropy images during time-lapse acquisition (weighted and raw).
Figure 14.
Fluorescence anisotropy time-lapse imaging. (a,b) Fluorescence intensity (a) and weighted fluorescence anisotropy (b) time-lapse imaging (30 min) of HT1080 cells incubated in PARPi-FL (1 µM) for 1 min, during the course of the washing phase. Anisotropy images allow the researcher to follow target engagement in real time. An anisotropy threshold is assigned to distinguish among the different bound and unbound states. As a function of time, we observe a decrease of nonspecifically bound drug in the cytoplasmic region, whereas the bound fraction of the drug increases in the nucleoli, where PARP accumulates. The weighted fluorescence anisotropy images are color-mapped using the weighted RGB color-mapping scheme illustrated in the Experimental design section. Scale bars, 13 µm.
(B) Off-line visualization ● TIMING 30 min
Download the OffLine.zip file onto the data acquisition PC or any another PC where data previously acquired as indicated in Step 74 are present. Unzip the file and create a new folder of your choice containing the executable file OffLine.exe.
If no MATLAB Runtime has previously been installed, proceed as in Step 75A(i,ii). Otherwise, delete the file MCR_R2015a_win64_installer.exe.
Start the OffLine program (Supplementary Fig. 9), which will open a GUI equivalent to the one used for the OnLine program. The GUI can be used to postprocess previously acquired data and to save the calculated fluorescence anisotropy images (both weighted and raw) as .tif files. The user can select and define both the processing and the fluorescence anisotropy visualization parameters.
Press the ‘Load Image A’ (‘Load Image B’) button to load the fluorescence image corresponding to the first (second) acquisition channel (red boxes 1 and 2 in Supplementary Fig. 9).
The parameter initialization for the image processing of the fluorescence images (red boxes 3 and 4 in Supplementary Fig. 9) and the visualization (red boxes 5–8 in Supplementary Fig. 9) of the fluorescence anisotropy image is similar that discussed for the OnLine program (Step 75A).
Set the threshold values previously measured at Steps 22 and 23, for the dark noise background subtraction of both acquisition channels (red box 3 in Supplementary Fig. 9).
If desired, set noise-filtering parameters (red box 4 in Supplementary Fig. 9).
Set the maximum (red box 5 in Supplementary Fig. 9) and the minimum (red box 6 in Supplementary Fig. 9) values for the fluorescence anisotropy color-mapping visualization scale.
Set the weighted fluorescence anisotropy display parameters (red box 7 in Supplementary Fig. 9). Different values can be arbitrarily chosen for the minimum (min) and the maximum (Max) (red box 8 in Supplementary Fig. 9).
Press the ‘Anisotropy’ button (box 9 in Supplementary Fig. 9) to process the data and calculate the fluorescence anisotropy images (weighted and raw). If any change in the parameter values occurs after visualization, press the ‘Anisotropy’ button to update the display with the latest settings.
Tick the ‘Anisotropy.tif’ and ‘Weighted Anisotropy.tif’ checkboxes (red box 10 in Supplementary Fig. 9) of the ‘Save results’ field, and press the ‘Save’ button (red box 11 in Supplementary Fig. 9) to save the calculated fluorescence anisotropy images.
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 2.
TABLE 2.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 24 | Discrepancies between measurements | Bleaching | Reduce the power of the laser |
| Progressive misalignment because of different users | Ensure that only one person operates the imaging system | ||
| Temperature variations during night and day | Use an air conditioning unit to keep the room temperature constant at all times, not only during imaging sessions | ||
| 28 | Nonlinearity | Saturation at high values of laser power | Change the concentration of the dye |
| Power meter readings not linear across the measuring range | Calibrate power meter at low powers | ||
| 34 | Lack of reproducibility | Dark noise is changing over time | Record dark noise during the course of the experiment |
| 38 | Discrepancies between different concentrations | Solutions are not properly mixed | Work using large volumes of solution. Sonicate thoroughly |
| 46 | Ring image artifacts | Aberrations | Adjust the objective’s collar and image optical planes away from the coverslip position |
| 70 | Cells drift during imaging | Heating plate causes shift in position | Use a spacer to act as heat reservoir during imaging, switch off the heating plate |
| 71 | Anisotropy images are not reproducible for different washing experiments | Averaging times are too long | Choose the denoising option instead of averaging over long times |
| 74 | Anisotropy images are not reproducible for different washing experiments | Dark noise is changing over time | Record dark noise before each window chamber time-lapse imaging session |
| 75A(xxi) | Values of fluorescence anisotropy are erratic over time | Temperature of rinsing solution is incorrect | Make sure that the rinsing solution is at 37 °C before the washing phase |
● TIMING
The amount of time required to complete the protocol depends on the specific experiment. Some steps of the protocol are necessary only for the assembly of the different components and/or the system calibration, and these do not need to be repeated for subsequent experiments. The following guidelines provide a rough estimate of the necessary time required for the various sections and subsections of the protocol.
Steps 1–12, assembly of the polarization control unit: 3–4 h
Steps 13–18, assembly of the polarization filter cube: 1–2 h
Steps 19–31, system calibration: 4–6 h
Steps 32–34, high anisotropy measurements: 2 h
Steps 35–38, measurements of fluorescence anisotropy at increasing values of viscosity: 4 h
Steps 39–43, measurements of fluorescence anisotropy for receptor and ligand binding quantification: 4 h
Steps 44–46, imaging of phantoms: 1 h
Steps 47–55, real-time two-photon fluorescence anisotropy measurement and visualization of drug-target engagement in live cells: 30 min
Steps 56–60, preparation of adherent cells for fluorescence anisotropy imaging: 24 h
Steps 61–66, preparation of the closed-bath imaging chamber for time measurements: 30 min
Steps 67–74, selection of microscope parameters for live-cell imaging: 30 min
Step 75A, stream processing for calculation and visualization of fluorescence anisotropy images during time-lapse acquisition: 2 h
Step 75B, off-line visualization: 30 min
ANTICIPATED RESULTS
When performed correctly, this protocol allows accurate high-resolution spatial and temporal maps of fluorescence anisotropy to be obtained from live cells. Used in combination with fluorescently labeled drugs—for example, in combination with PARPi-FL—it allows engagement of small-molecules with their cognate targets to be directly quantified in intact cells and at the cellular level both in vitro and in a biological setting. In Figure 15, we show temporal evolution and quantification of fluorescently labeled olaparib, in both the cytosolic and nuclear compartments of HT1080 cells during incubation time and following drug washing. During the incubation time, the drug accumulates in the cytosolic area and enters the nucleus, where it accumulates and where its target (PARP1) mainly resides. During the washing phase, the drug present in the cytoplasm is removed, while the drug present in the nucleus remains bound to its target. The high anisotropy present in the nuclei is reduced when the fluorescently labeled drug is in the presence of unlabeled olaparib (AZD2281) as competitor at a five-fold concentration. This observation further emphasizes that the high nuclear anisotropy is due to drug-target binding and not induced by artifacts. Because imaging is performed using two-photon excitation, the modality offers extended penetration depth and reduced scattering, and is particularly indicated for in vivo studies in tissue5. In this case, the calibration and characterization procedure would be the same because it is sample-independent. However, the scattering component present within tissue must be taken into account5.
Figure 15.
Two-photon fluorescence anisotropy imaging of drug-target engagement. (a–d) Quantification of fluorescence (a,b) and anisotropy (c,d) signal in HT1080 cells during incubation time in fluorescently labeled olaparib (blue box) and following washing, in the cytoplasmic and nuclear cellular compartments. Black circles and gray squares represent values obtained in the absence and presence, respectively, of a five-fold-higher concentration of unlabeled small-molecule olaparib (AZD2281). Points on the graphs refer to a single experiment, average±s.d.; average calculated over n = 6 cells. Adapted with permission from Dubach et al.4, Nature Publishing Group.
Supplementary Material
Acknowledgments
This project was funded in part by National Institutes of Health (NIH) grants RO1-EB010011, RO1-HL122208 and P50GM107618. Investigators at Memorial Sloan Kettering Cancer Center were funded in part by NIH grant P30 CA008748 and investigators in Europe were funded by the EC Seventh Framework Programme under grant agreement no. 622182.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS All authors contributed to the writing of the paper. C.V., R.M., P.F.F. and R.W., along with others, primarily developed the technique. S. Shah and I.G. conducted one-photon experiments. C.V. and P.F.F. wrote the software. S.L. designed and machined the systems components. A.E.N., C.B., R.M. and T.R. performed the synthesis. S. Stapleton prepared cell cultures.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
References
- 1.Vinegoni C, et al. Advances in measuring single-cell pharmacokinetics and pharmacology in vivo. Drug Discov. Today. 2015;20:1087–1092. doi: 10.1016/j.drudis.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller MA, et al. Predicting therapeutic nanomedicine efficacy using a companion magnetic resonance imaging nanoparticle. Sci. Transl. Med. 2015;7:314. doi: 10.1126/scitranslmed.aac6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laughney AM, et al. Single-cell pharmacokinetic imaging reveals a therapeutic strategy to overcome drug resistance to the microtubule inhibitor eribulin. Sci. Transl. Med. 2014;6:261. doi: 10.1126/scitranslmed.3009318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubach JM, et al. In vivo imaging of specific drug-target binding at subcellular resolution. Nat. Commun. 2014;5:3946. doi: 10.1038/ncomms4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molina DM, et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 6.Savitski MM, et al. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science. 2014;346:1255784. doi: 10.1126/science.1255784. [DOI] [PubMed] [Google Scholar]
- 7.Grimwood S, Hartig PR. Target site occupancy: emerging generalizations from clinical and preclinical studies. Pharmacol. Ther. 2009;122:281–301. doi: 10.1016/j.pharmthera.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Matthews PM, Rabiner EA, Passchier J, Gunn RN. Positron emission tomography molecular imaging for drug development. Br. J. Clin. Pharmacol. 2012;73:175–186. doi: 10.1111/j.1365-2125.2011.04085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu XH, et al. Molecular imaging of drug transit through the blood–brain barrier with MALDI mass spectrometry imaging. Sci. Rep. 2013;3:2859. doi: 10.1038/srep02859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller MA, Weissleder R. Imaging the pharmacology of nanomaterials by intravital microscopy: toward understanding their biological behavior. Adv. Drug Deliv. Rev. 2016 doi: 10.1016/j.addr.2016.05.023. http://dx.doi.org/10.1016/j.addr.2016.05.023. [DOI] [PMC free article] [PubMed]
- 11.Svoboda K, Yasuda R. Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron. 2006;50:823–839. doi: 10.1016/j.neuron.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 12.Gough AH, Taylor DL. Fluorescence anisotropy imaging microscopy maps calmodulin-binding during cellular contraction and locomotion. J. Cell Biol. 1993;121:1095–1107. doi: 10.1083/jcb.121.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lakowicz JR. Principles of Fluorescence Spectroscopy. Plenum Press; 1983. [Google Scholar]
- 14.Lakowicz JR. Principles of Fluorescence Spectroscopy. 2. Kluwer Academic/Plenum Publishers; 1999. [Google Scholar]
- 15.Dix JA, Verkman AS. Mapping of fluorescence anisotropy in living cells by ratio imaging. Biophys. J. 1990;57:231–240. doi: 10.1016/S0006-3495(90)82526-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swaminathan R, Hoang CP, Verkman AS. Photobleaching recovery and anisotropy decay of green fluorescent protein GFP-S65T in solution and cells: cytoplasmic viscosity probed by green fluorescent protein translational and rotational diffusion. Biophys. J. 1997;72:1900–1907. doi: 10.1016/S0006-3495(97)78835-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster TH, Pearson BD, Mitra S, Bigelow CE. Fluorescence anisotropy imaging reveals localization of meso-tetrahydroxyphenyl chlorin in the nuclear envelope. Photochem. Photobiol. 2005;81:1544–1547. doi: 10.1562/2005-08-11-RN-646. [DOI] [PubMed] [Google Scholar]
- 18.Cao Z, Huang CC, Tan W. Nuclease resistance of telomere-like oligonucleotides monitored in live cells by fluorescence anisotropy imaging. Anal. Chem. 2006;78:1478–1484. doi: 10.1021/ac0517601. [DOI] [PubMed] [Google Scholar]
- 19.Bigelow CE, Vishwasrao HD, Frelinger JG, Foster TH. Imaging enzyme activity with polarization-sensitive confocal fluorescence microscopy. J. Microsc. 2004;215:24–33. doi: 10.1111/j.0022-2720.2004.01357.x. [DOI] [PubMed] [Google Scholar]
- 20.Li W, Wang Y, Shao H, He Y, Ma H. Probing rotation dynamics of biomolecules using polarization based fluorescence microscopy. Microsc. Res. Tech. 2007;70:390–395. doi: 10.1002/jemt.20418. [DOI] [PubMed] [Google Scholar]
- 21.Bigelow CE, Conover DL, Foster TH. Confocal fluorescence spectroscopy and anisotropy imaging system. Opt. Lett. 2003;28:695–697. doi: 10.1364/ol.28.000695. [DOI] [PubMed] [Google Scholar]
- 22.Chan FTS, Kaminski CF, Kaminski-Schierle GS. HomoFRET fluorescence anisotropy imaging as a tool to study molecular self-assembly in live cells. ChemPhysChem. 2011;12:500–509. doi: 10.1002/cphc.201000833. [DOI] [PubMed] [Google Scholar]
- 23.Hedde PN, Ranjit S, Gratton E. 3D fluorescence anisotropy imaging using selective plane illumination microscopy. Opt. Express. 2015;23:22308–22317. doi: 10.1364/OE.23.022308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yengo CM, Berger CL. Fluorescence anisotropy and resonance energy transfer: powerful tools for measuring real time protein dynamics in a physiological environment. Curr. Opin. Pharmacol. 2010;10:731–737. doi: 10.1016/j.coph.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gradinaru CC, Marushchak DO, Samim M, Krull UJ. Fluorescence anisotropy: from single molecules to live cells. Analyst. 2010;135:452–459. doi: 10.1039/b920242k. [DOI] [PubMed] [Google Scholar]
- 26.Suhling K, Levitt J, Chung PH. Time-resolved fluorescence anisotropy imaging. In: Engelborghs Y, Visser JWG, editors. Fluorescence Spectroscopy and Microscopy. Springer Science; 2014. [DOI] [PubMed] [Google Scholar]
- 27.Jameson DM, Ross JA. Fluorescence polarization/anisotropy in diagnostic and imaging. Chem. Rev. 2010;110:2685–2708. doi: 10.1021/cr900267p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matthews DR, et al. Time-lapse FRET microscopy using fluorescence anisotropy. J. Microsc. 2010;237:51–62. doi: 10.1111/j.1365-2818.2009.03301.x. [DOI] [PubMed] [Google Scholar]
- 29.Turetsky A, Kim E, Kohler RH, Miller MA, Weissleder R. Single cell imaging of Bruton’s tyrosine kinase using an irreversible inhibitor. Sci. Rep. 2014;4:4782. doi: 10.1038/srep04782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reiner T, et al. Imaging therapeutic PARP inhibition in vivo through bioorthogonally developed companion imaging agents. Neoplasia. 2012;14:169–177. doi: 10.1593/neo.12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thurber GM, et al. Single-cell and subcellular pharmacokinetic imaging allows insight into drug action in vivo. Nat. Commun. 2013;4:1504. doi: 10.1038/ncomms2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hottiger MO. Nuclear ADP-ribosylation and its role in chromatin plasticity, cell differentiation, and epigenetics. Annu. Rev. Biochem. 2015;84:227–263. doi: 10.1146/annurev-biochem-060614-034506. [DOI] [PubMed] [Google Scholar]
- 33.Barkauskaite E, Jankevicius G, Ahel I. Structures and mechanisms of enzymes employed in the synthesis and degradation of PARP-dependent protein ADP-ribosylation. Mol. Cell. 2015;58:935–946. doi: 10.1016/j.molcel.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 34.Caldecott KW. Protein ADP-ribosylation and the cellular response to DNA strand breaks. DNA Repair. 2014;19:108–113. doi: 10.1016/j.dnarep.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 35.Scott CL, Swisher EM, Kaufmann SH. Poly (ADP-ribose) polymerase inhibitors: recent advances and future development. J. Clin. Oncol. 2015;33:1397–1406. doi: 10.1200/JCO.2014.58.8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Curtin NJ, Szabo C. Therapeutic applications of PARP inhibitors: anticancer therapy and beyond. Mol. Aspects Med. 2013;34:1217–1256. doi: 10.1016/j.mam.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Connor MJ. Targeting the DNA damage response in cancer. Mol. Cell. 2015;60:547–560. doi: 10.1016/j.molcel.2015.10.040. [DOI] [PubMed] [Google Scholar]
- 38.Shen Y, Aoyagi-Scharber M, Wang B. Trapping poly (ADP-ribose) polymerase. J. Pharmacol. Exp. Ther. 2015;353:446–457. doi: 10.1124/jpet.114.222448. [DOI] [PubMed] [Google Scholar]
- 39.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mateo J, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ossovskaya V, Koo IC, Kaldjian EP, Alvares C, Sherman BM. Upregulation of poly (ADP-ribose) polymerase-1 (PARP1) in triple-negative breast cancer and other primary human tumor types. Genes Cancer. 2010;1:812–821. doi: 10.1177/1947601910383418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bièche I, Murcia GD, Lidereau R. Poly (ADP-ribose) polymerase gene expression status and genomic instability in human breast cancer. Clin. Cancer Res. 1996;2:1163–1167. [PubMed] [Google Scholar]
- 43.Rojo F, et al. Nuclear PARP-1 protein overexpression is associated with poor overall survival in early breast cancer. Ann. Oncol. 2012;23:1156–1164. doi: 10.1093/annonc/mdr361. [DOI] [PubMed] [Google Scholar]
- 44.Alanazi M, et al. Association between PARP-1 V762A polymorphism and breast cancer susceptibility in Saudi population. PLoS One. 2013;8:e855411. doi: 10.1371/journal.pone.0085541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galia A, et al. PARP-1 protein expression in glioblastoma multiforme. Eur. J. Histochem. 2012;56:e9. doi: 10.4081/ejh.2012.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barton VN, et al. PARP1 expression in pediatric central nervous system tumors. Pediatr. Blood Cancer. 2009;53:1227–1230. doi: 10.1002/pbc.22141. [DOI] [PubMed] [Google Scholar]
- 47.Staibano S, et al. Poly (adenosine diphosphate-ribose) polymerase 1 expression in malignant melanomas from photoexposed areas of the head and neck region. Hum. Pathol. 2005;36:724–731. doi: 10.1016/j.humpath.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 48.Reiner T, Earley S, Turetsky A, Weissleder R. Bioorthogonal small-molecule ligands for PARP1 imaging in living cells. Chembiochem. 2010;11:2374–2377. doi: 10.1002/cbic.201000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thurber GM, et al. Effect of small-molecule modification on single-cell pharmacokinetics of PARP inhibitors. Mol. Cancer Ther. 2014;13:986–995. doi: 10.1158/1535-7163.MCT-13-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Irwin CP, et al. PARPi-FL – a fluorescent PARP1 inhibitor for glioblastoma imaging. Neoplasia. 2014;16:432–440. doi: 10.1016/j.neo.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kossatz S, et al. Detection and delineation of oral cancer with a PARP1 targeted optical imaging agent. Sci. Rep. 2016;6:21371. doi: 10.1038/srep21371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan Y, Marriot G. Fluorescence resonance energy transfer imaging microscopy and fluorescence polarization imaging microscopy. Methods Enzymol. 2003;360:561–580. doi: 10.1016/s0076-6879(03)60128-x. [DOI] [PubMed] [Google Scholar]
- 53.Koshioka M, Sasaki K, Masuhara H. Time-dependent fluorescence depolarization analysis in three-dimensional microspectroscopy. Appl. Spectrosc. 1995;49:224–228. [Google Scholar]
- 54.Ha T, Laurence TA, Chemla DS, Weiss S. Polarization spectroscopy of single fluorescent molecules. J. Phys. Chem. B. 1999;103:6839–6850. [Google Scholar]
- 55.Becker W. In: Advanced Time-Correlated Single Photon Counting Techniques. Castleman AW, Toennis JP, Zinth W, editors. Springer Series in Chemical Physics; 2005. [Google Scholar]
- 56.Jameson DM, Weber G, Spencer RD, Mitchell G. Fluorescence polarization: measurements with a photon counting photometer. Rev. Sci. Instrum. 1978;49:510–514. doi: 10.1063/1.1135451. [DOI] [PubMed] [Google Scholar]
- 57.Benninger RKP, Ashby WJ, Ring EA, Piston DW. A single-photon counting detector for increased sensitivity in two-photon laser scanning microscopy. Opt. Lett. 2008;33:2895–2897. doi: 10.1364/ol.33.002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moon S, Kim DY. Analog single-photon counter for high-speed scanning microscopy. Opt. Express. 2008;16:13990–14003. doi: 10.1364/oe.16.013990. [DOI] [PubMed] [Google Scholar]
- 59.Hoover EE, Squier JA. Advances in multiphoton microscopy technology. Nat. Photonics. 2013;7:93–101. doi: 10.1038/nphoton.2012.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Driscoll JD, et al. Photon counting, censor corrections, and lifetime imaging for improved detection in two-photon microscopy. J. Neurophysiol. 2011;105:3106–3113. doi: 10.1152/jn.00649.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buehler C, Kim KH, Greuter U, Schlumpf N, So PTC. Single-photon counting multicolor multiphoton fluorescence microscope. J. Fluoresc. 2005;15:41–51. doi: 10.1007/s10895-005-0212-z. [DOI] [PubMed] [Google Scholar]
- 62.Dix JA, Verkman AS. Mapping of fluorescence anisotropy in living cells by ratio imaging: application to cytoplasmic viscosity. Biophys. J. 1990;57:231–240. doi: 10.1016/S0006-3495(90)82526-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson RB, Gryczynski I, Malicka J. Fluorescence polarization standards for high-throughput screening and imaging. BioTechniques. 2002;32:42. doi: 10.2144/02321bm03. [DOI] [PubMed] [Google Scholar]
- 64.van Kempen GMP, van Vliet LJ. Mean and variance of ratio estimators used in fluorescence ratio imaging. Cytometry. 2000;39:300–305. doi: 10.1002/(sici)1097-0320(20000401)39:4<300::aid-cyto8>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 65.Lidke KA, Rieger B, Lidke DS, Jovin TM. The role of photon statistics in fluorescence anisotropy imaging. IEEE Trans. Image Process. 2005;14:1237–1245. doi: 10.1109/tip.2005.852458. [DOI] [PubMed] [Google Scholar]
- 66.Siegel J, et al. Studying biological tissue with fluorescence lifetime imaging: microscopy, endoscopy, and complex decay profiles. Appl. Opt. 2003;42:2995–3004. doi: 10.1364/ao.42.002995. [DOI] [PubMed] [Google Scholar]
- 67.Suhling K, et al. Time-resolved fluorescence anisotropy imaging (TR-FAIM) applied to live cells. Opt. Lett. 2004;29:584–586. doi: 10.1364/ol.29.000584. [DOI] [PubMed] [Google Scholar]
- 68.Xu C, Webb WW. Measurement of two-photon excitation cross sections of molecular fluorophores with data from 690 to 1050 nm. J. Opt. Soc. Am. B. 1996;13:481–491. [Google Scholar]
- 69.Weber G. Dependence of polarization of the fluorescence on the concentration. Trans. Faraday Soc. 1954;50:552–555. [Google Scholar]
- 70.Yang KS, Budin G, Reiner T, Vinegoni C, Weissleder R. Bioorthogonal imaging of aurora kinase A in live cells. Angew. Chem. Int. Ed. Engl. 2012;51:6598–6603. doi: 10.1002/anie.201200994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim E, et al. Optimized near-IR fluorescent agents for in vivo imaging of Btk expression. Bioconjug. Chem. 2015;26:1513–1518. doi: 10.1021/acs.bioconjchem.5b00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fu W, et al. Epigenetic modulation of type-1 diabetes via a dual effect on pancreatic macrophages and cells. eLife. 2014;3:e04631. doi: 10.7554/eLife.04631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mikula H, Stapleton S, Kohler RH, Vinegoni C, Weissleder R. Design and development of fluorescent vemurafenib analogs for in vivo imaging. Theranostics. 2017;7:1257–1265. doi: 10.7150/thno.18238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim E, Yang K, Weissleder R. Bioorthogonal small molecule imaging agents allow single-cell imaging of MET. PLoS One. 2013;8:e81275. doi: 10.1371/journal.pone.0081275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miller MA, et al. Tumour-associated macrophages act as a slow-release reservoir of nanotherapeutic Pt(IV) pro-drug. Nat. Commun. 2015;6:8692. doi: 10.1038/ncomms9692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Earley S, et al. In vivo imaging of drug-induced mitochondrial outer membrane permeabilization at single cell resolution. Cancer Res. 2012;72:2949–2956. doi: 10.1158/0008-5472.CAN-11-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang KS, Budin G, Tassa C, Kister O, Weissleder R. Bioorthogonal approach to identify unsuspected drug targets in live cells. Angew. Chem. Int. Ed. Engl. 2013;52:10593–10597. doi: 10.1002/anie.201304096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang KS, Kohler RH, Landon M, Giedt R, Weissleder R. Single cell resolution in vivo imaging of DNA damage following PARP inhibition. Sci. Rep. 2015;5:10129. doi: 10.1038/srep10129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miller MA, Askevold B, Yang KS, Kohler RH, Weissleder R. Platinum compounds for high-resolution in vivo cancer imaging. Chem. Med. Chem. 2014;9:1131–1135. doi: 10.1002/cmdc.201300502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Song Y, Suntharalingam K, Yeung JS, Royzen M, Lippard SJ. Synthesis and characterization of Pt(IV) fluorescein conjugates to investigate Pt(IV) intracellular transformations. Bioconjug. Chem. 2013;24:1733–1740. doi: 10.1021/bc400281a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Orth JD, et al. Analysis of mitosis and antimitotic drug responses in tumors by in vivo microscopy and single-cell pharmacodynamics. Cancer Res. 2011;71:4608–4616. doi: 10.1158/0008-5472.CAN-11-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chittajallu DR, et al. In vivo cell-cycle profiling in xenograft tumors by quantitative intravital microscopy. Nat. Methods. 2015;12:577–585. doi: 10.1038/nmeth.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meimetis LG, et al. Fluorescent vinblastine probes for live cell imaging. Chem. Commun. 2016;52:9953–9956. doi: 10.1039/c6cc04129a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.