Abstract
Hippocampal neural stem/progenitor cells (NSPCs) proliferate and differentiate to generate new neurons across the life span of most mammals, including humans. This process takes place within a characteristic local microenvironment where NSPCs interact with a variety of other cell types and encounter systemic regulatory factors. Within this microenvironment, cell intrinsic gene expression programs are modulated by cell extrinsic signals through complex interactions, in many cases involving short non-coding RNA molecules, such as miRNAs. Here we review the regulation of gene expression in NSPCs by miRNAs and its possible implications for epilepsy, which has been linked to alterations in adult hippocampal neurogenesis.
Keywords: Neural stem cell, miRNA, seizure, neurogenic niche, adult hippocampal neurogenesis
ADULT HIPPOCAMPAL NEUROGENESIS AND ITS LOCAL MICROENVIRONMENT
Throughout life new neurons are generated from neural stem cells (NSCs) in select areas of the mammalian brain, such as the olfactory bulb, subventricular zone (SVZ), and the subgranular zone (SGZ) of the Dentate Gyrus (DG). The latter is named adult hippocampal neurogenesis (AHN), a process important for several aspects of hippocampus-dependent cognitive capacity [1–5], while it has also been hypothesized to have potential therapeutic functions, responding to local loss of neurons in events of disease or pharmacological treatment [6–12]. Most details about the physiological and functional regulation of AHN come from preclinical animal studies, but it has recently been elegantly shown that adult human hippocampi possess extensive neurogenic capacity as well. In humans, about one third of all granule cells in the DG are replaced by newborn cells throughout life at a turnover rate of approximately 1.75% per year, which remarkably resembles the neurogenic capacity of the mouse hippocampus [13, 14].
AHN can be divided into several distinct stages, each of which is under specific control by cell-intrinsic gene expression programs and environmental factors, allowing tight temporal and spatial regulation of the neurogenic process within a restricted microenvironment and according to the needs of the organism [15, 16] (Fig. 1A).
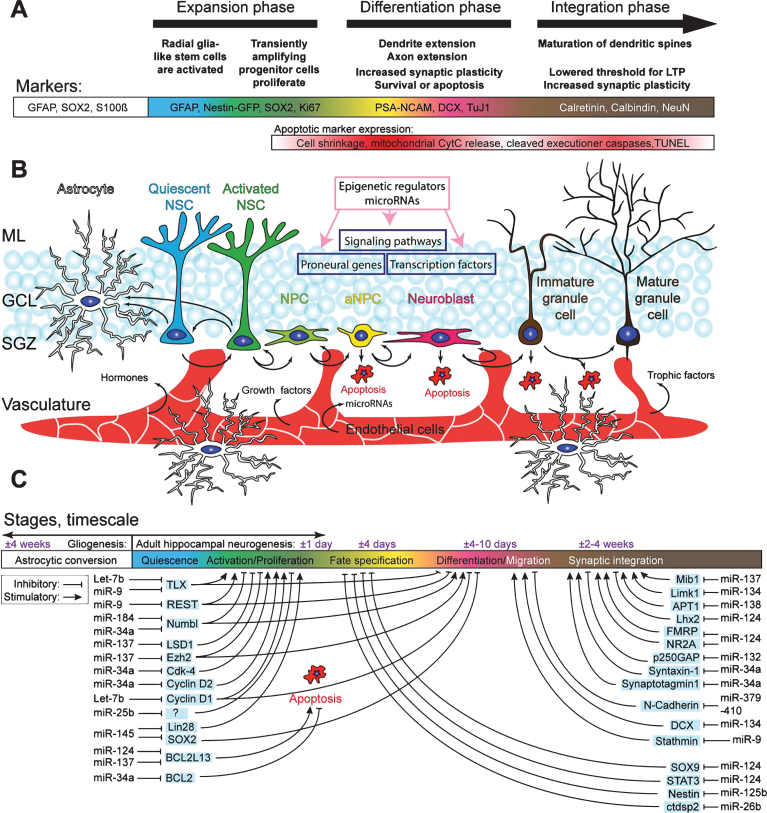
Fig.1.
Schematic overview of the hippocampal neurogenic niche, the different processes underlying AHN, and its regulation by microRNAs. A) Overview of the different stages of AHN. Each cell type can be identified by a combination of presence and absence of markers, combined with morphological cellular features. B) Overview of the neurogenic niche and the transition of a NSC into a mature neuron. The complexity of the neurogenic niche, consisting of multiple cell types in close association with the vasculature, allows for both local and distant cell communication. Distant cell communication occurs via factors released in the bloodstream, such as cell-extrinsic miRNAs, growth factors (VEGF and bFGF), hormones, and trophic factors (BDNF). Other cell-intrinsic factors, such as miRNAsm TLX signaling, notch signaling, and REST (purple boxes), and cell extrinsic factors such as HDACs, DNA methylation, and miRNAs (pink box), complete the coordinated regulated of AHN. C) MiRNAs regulate various key pathways important in AHN. Depicted are miRNAs of which a clear link with neurogenesis has been identified, together with their targets through which the miRNAs might exert their effect.
Upon stimulation, quiescent neural stem cells (NSCs) in the SGZ re-enter the cell cycle and become proliferative NSCs. These activated NSCs have the potential to both self-renew and give rise to early amplifying neural progenitors (aNPCs) maintaining the NSC pool while expanding the progenitor pool. The final fate of activated NSCs is still debatable as it has been shown that activated NSCs terminally differentiate into astrocytes after multiple rounds of asymmetric division [17], while another study suggests that NSCs can re-enter quiescence after several rounds of asymmetric division and be later activated again [18], which is thought to act as an intrinsic mechanism to maintain the NSC pool. The newly generated aNPCs undergo multiple rounds of symmetric division, expanding the neurogenic pool and giving rise to mature neurons. Soon after their birth most aNPCs are selected out by apoptosis, which may prevent excess production of new and/or unfit neurons [19, 20]. The surviving aNPCs give rise to neuroblasts, which after neuronal differentiation, migration, and maturation will integrate into the preexisting DG network, taking in total approximately 4 weeks [4, 21, 22]. The neurogenic capacity of NSCs in the adult DG is not infinite and decreases throughout life, mainly due to decreased proliferation and loss of NSCs, possibly through astrocytic conversion [17].
The complex regulation of AHN by both cell-intrinsic and cell-extrinsic factors becomes clear when studying the fate of early hippocampal NSPCs transplanted into non-neurogenic areas of the brain, where they will generate astrocytes. On the contrary, NSPCs derived from the SVZ will generate mature granule cells when transplanted into the SGZ of the DG [23, 24]. These transplant studies identified an extra layer of neurogenic regulation by the local microenvironment, or neurogenic niche, which is crucial for proper neuronal development. The hippocampal neurogenic niche is made up out of a wide array of cell types, including the NSPCs, neuroblasts and their progeny, and mature granule cells, but also astrocytes, GABAergic interneurons, microglia, and endothelial cells that connect the neurogenic niche to the vasculature (Fig. 1B) [15, 25–32]. Furthermore the local vasculature and its associated extracellular matrix allows for local cell-cell interaction, i.e. via β-Catenin regulating mitotic spindle orientation [33, 34]. Together, all these elements provide the hippocampus with a finely tuned microenvironment permissive for adult neurogenesis. Besides the crucial structural organization provided by the niche the local responsiveness of NSPCs and their progeny is tightly regulated at the molecular level, i.e. by epigenetic regulation of chromatin states allowing temporal and spatial regulation of gene transcription [35–37]. Chromatin modification is one the most studied forms of epigenetic regulation, as the chromatin state provides the first layer of epigenetic control of gene expression regulation. Histone methylation by DNA methyltransferases (DNMTs) and (de)acetylation by histone acetyl transferases (HATs) and histone deacetylases (HDACs), two of the most common forms of chromatin modification provide a first layer of molecular control over gene expression, rendering the genome (in)accessible to the transcription machinery. Another classic epigenetic regulatory mechanism active in adult neurogenesis is DNA methylation, in which cellular DNA can be covalently modified, usually, but not exclusively, at locations rich in CpG dinucleotides, through methylation of the carbon at the fifth position on the pyrimidine ring of the cytosine residue [38]. These and other levels of epigenetic control of neurogenesis have recently been extensively reviewed [39].
An extra level of epigenetic control in NSPCs comes from non-coding RNAs, such as miRNAs, [40–43]. MiRNAs are small non-coding RNA molecules composed of approximately 22 nucleotides that are transcribed from endogenous hairpin-shaped transcripts by RNA polymerase II or III. Their transcripts are called pri-miRNAs and will be cleaved into pre-miRNAs in the nucleus by the Drosha/DGCR8 complex. After exporting the pre-miRNA from the nucleus it is once again cleaved, this time by the Dicer/TRBP complex to its mature functional miRNA length [44]. Recently several other miRNA biogenesis pathways have been identified, including Drosha and Dicer independent pathways. A complete summary of miRNA biogenesis pathways has recently been published [45]. Interestingly, though outside the scope of this review, components of these miRNA biogenesis pathways, such as Drosha, Dicer and DGCR8, also have direct, miRNA-independent, effects on gene expression [46]. Furthermore, Drosha restricts the potency of adult hippocampal NSPCs in vivo by targeting NFIB through a miRNA-independent pathway [47]. miRNAs can regulate gene expression post-transcriptionally via either mRNA degradation or inhibition of mRNA translation by binding to the 3’UTR of their target mRNA. Hence, miRNAs can only exert their function when the specific substrate mRNA is available as well, introducing another layer of spatial and temporal complexity to gene expression regulation [48]. Furthermore, numerous proteins involved in the epigenetic machinery are targets of specific miRNAs and vice versa, creating complex multilayered control of gene transcription [49]. Due to several biological characteristics of miRNAs, such as their potent ability to (transiently) repress gene expression, their relative high conservation between species, and their rapid turnover, it has been hypothesized that miRNAs might be key regulators of AHN [50]. As mentioned before, local temporal control of gene expression is key in regulating the switches between different distinct stages of AHN. Down regulating non-lineage specific genes and removal of repressive marks on the chromatin of lineage-specific genes are excessively abundant forms of AHN regulation, both of which are likely candidates for miRNA regulation. Interestingly, several studies have demonstrated in other tissues that miRNAs are not only found intracellularly, but are also present in the extracellular compartment and mediate cell-to-cell communication, among others via exosomes [51–53]. In recent years, numerous miRNAs have been characterized that play crucial roles in stage transitions of AHN by regulating key epigenetic regulators, such as HDACs, Polycombgroup proteins, and REST, but also play key roles in apoptotic selection and functional maturation of immature neurons (Fig. 1C, Table 1).
Table 1.
MicroRNAs involved in regulation of AHN
| Target | miRNA function | Origin of Identification | Reference | |
| miR-184 | Numbl | Inhibits differentiation, promotes proliferation, maintenance of the neurogenic stem cell pool | Mouse NSCs | [56] |
| miR-34a | Numbl | Inhibits differentiation, promotes proliferation, maintenance of the neurogenic stem cell pool | Mouse NPCs from embryonic cortex | [57] |
| BCL2 | Promotes apoptosis | Mouse cortex/SH-SY5Y cell line | [85] | |
| Cdk-4 | Inhibits cell cycle progression | Primary keratinocytes | [83] | |
| Cyclin D2 | Inhibits cell cycle progression | Primary keratinocytes | [83] | |
| Synaptotagmin1 | Inhibits synaptic development | Mouse ES cells | [161] | |
| Syntaxin-1A | Inhibits synaptic development | Mouse ES cells | [161] | |
| miR-106b/miR-25 Cluster | IGF-signaling | Promotes NSPC proliferation, neuronal differentiation | Mouse primary NSC culture | [58] |
| miR-124 | Sox9 | Promotes neuronal differentiation | Mouse NSCs | [72] |
| STAT3 | Promotes neuronal differentiation | Mouse ESCs | [74] | |
| BCL2L13 | Inhibits apoptosis | Mouse DG | [82] | |
| Lhx2 | Promotes neurite outgrowth | Mouse hippocampus | [93] | |
| Rap2a | Promotes dendritic branching | Mouse NSCs | [162] | |
| miR-137 | BCL2L13 | Inhibits apoptosis | Mouse DG | [82] |
| Mib-1 | Inhibits dendritic growth and spine formation | Mouse hippocampus & mouse primary neuronal culture | [92] | |
| LSD1 | Promotes proliferation | Embryonic NSCs | [64] | |
| Ezh2 | Inhibits differentiation | Adult NSCs | [65] | |
| miR-9 | TLX | Promotes differentiation, inhibits differentiation | Mouse hippocampus | [63] |
| REST | Promotes neuronal differentiation | NT2 cell line | [79] | |
| Rap2a | Promotes dendritic branching | Mouse NCSs | [162] | |
| Stathmin | Inhibits migration | Human embryonic NPCs | [87] | |
| Let-7b | TLX | Inhibits proliferation, promotes differentiation | Mouse NSCs & embryonic mouse brain | [66] |
| Cyclin D1 | Inhibits cell cycle progression | Embryonic mouse brain | [66] | |
| miR-125b | Nestin | Promotes neuronal differentiation | Rat NSPCs | [70] |
| miR-145 | Sox2 | Promotes neuronal differentiation | Mouse NSC | [69] |
| miR-26b | Ctdsp2 | Promotes neuronal differentiation | Zebrafish & P19 cells | [80] |
| miR-19 | Rapgef2 | Stimulates migration | Mouse hippocampus & mouse NPCs | [88] |
| miR-379/miR410 cluster | N-Cadherin | Induces migration | Mouse embryonic cortex | [86] |
| miR-134 | Limk1 | Inhibits spinogenesis | Rat hippocampus | [95] |
| Dcx | Inhibits NPC migration | Primary NPCs, primary neurons, mouse embryonic cortex | [163] | |
| miR-138 | APT1 | Inhibits spinogenesis | Mouse hippocampus | [96] |
| miR-17/miR92 cluster | PTEN | Induces axonal outgrowth | Rat primary cortical neurons | [94] |
| miR-132 | P250GAP | Promotes spinogenesis | Mouse hippocampus | [98] |
Overview of identified microRNAs that regulate (hippocampal) neurogenesis, their targets, their role in neurogenesis, and the origin of identification.
In order to discuss the complex role miRNAs play in regulating AHN, we first review neurogenesis stage-specific regulation by miRNAs. Further, we discuss how miRNA deregulation could contribute to the development of neurological disorders, focusing on aberrant hippocampal neurogenesis and epilepsy.
miRNA-dependent control of adult neurogenesis under physiological conditions
Neural stem cell maintenance and quiescence
To preserve the neurogenic capacity throughout life it is critical that NSCs retain both their proliferative and self-renewal capacities. A fine balance between the two is needed, since each NSC has a limited number of proliferation rounds before it terminally differentiates and loses its stem cell capacities [17]. Several miRNAs have been identified that play a crucial role in the regulation of NSC maintenance.
A key pathway in maintaining neurogenic capacity is the Notch signaling pathway, which drives asymmetric division rendering both NSCs and aNPCs [54]. Activation of the Notch signaling drives proliferation, and through asymmetric expression of its repressor protein Numbl, two distinct daughter cells are derived from one single NSC [55]. Absence of Numbl in NSCs results in increased proliferation and symmetric division, exhausting the NSC pool. Both miR-184 and miR-34a regulate Numbl expression by binding to its 3’UTR, driving symmetric division [56, 57]. Similarly, high expression levels of miR-184 are associated with long-term impaired neurogenesis. miR-34a also indirectly regulates Notch signaling as it targets two downstream proteins of Notch, NeuroD1 and Mash1 [57].
Another pathway implicated in stem cell maintenance throughout ageing is the Insulin/IGF pathway. FoxO3, a component of the Insulin/IGF pathway, directly regulates the expression of the miR106b-25 cluster. The main miRNA from this cluster studied in relation to neurogenesis is miR-25. Expression of miR-25 results in increased proliferation of NSPCs, possibly depleting the NSPC pool [58].
Besides maintaining the NSC pool through controlling proliferation rates and cell division, miRNAs might also be involved in maintaining NSC quiescence. Though this phenomenon has not yet been shown in AHN and evidence from other stem cell niches is scarce, there are several interesting findings pointing towards roles for miRNAs in controlling quiescence. One study using muscle stem cells in which Dicer was conditionally knocked out showed a significant increase in proliferation rate, indicating a transition from quiescence to activation. Further analysis of miRNA expression differences between quiescent and activated stem cells yielded several candidates, from which miRNA-489 turned out to be a crucial mediator of stem cell quiescence [59].
NSPCs Proliferation versus differentiation
As mentioned before both NSCs and aNPCs can undergo several rounds of proliferation, thereby amplifying the NSPC pool before giving rise to new neurons or astrocytes. Several miRNAs and their targets tightly regulate this switch from the proliferative state towards differentiation.
One key regulator of proliferation is the nuclear receptor TLX. TLX activates the Wnt/B-catenin pathway and is crucial for NSPCs to maintain their self-renewal and proliferative capacities [60, 61]. NSPCs expressing TLX can proliferate, self-renew, and differentiate into all neuronal lineages, while NSPCs devoid of TLX fail to proliferate at all. Reintroducing TLX into NSPCs rescues their proliferative potential [62].
Expression of TLX is controlled by several miRNAs, indicating its importance in this complex regulatory system. TLX expression is regulated by miR-9, which is highly abundant in the adult brain. At the same time, TLX controls miR-9 expression through a negative feedback loop, ensuring tight control over NSPC proliferation [63]. Overexpression of miR-9 results in decreased levels of TLX and increased (premature) neuronal differentiation, while miR-9 knockdown increases NPC proliferation.
TLX forms a second regulatory loop with miR-137, which stimulates proliferation by repressing LSD1, a co-repressor of TLX, while repressing differentiation by down regulating Ezh2, a histone methyltransferase and part of the Polycombgroup proteins [64, 65]. Like miR-9, miR-137 expression levels are tightly regulated via closed regulatory loops involving TLX and its co-repressor LSD1.
A third miRNA that regulates TLX expression is Let-7b, which inhibits proliferation and drives differentiation through direct targeting of the 3’UTR of the TLX receptor [66]. Let-7b knockdown increases proliferation, while overexpression of Let-7b in NSCs decreases proliferation and increases accelerated neuronal differentiation [67]. Let-7b also regulates Cyclin D1, an effector downstream of TLX, important for cell cycle progression. Down regulation of Cyclin D1 by Let-7b inhibits progression into the S-phase of the cell cycle, lengthening the G1 –phase and thereby stimulates the transition into differentiation [68].
A fourth miRNA that indirectly also regulates TLX expression is miR-145. miR-145 regulates Sox2, a transcription factor crucial for stem cells to maintain pluripotency and self-renewal capacity, and Lin28, a well known suppressor of Let-7 biogenesis [69]. Expression of miR-145 increases during differentiation of NPCs and thus drives differentiation indirectly via TLX regulation.
These observations indicate that TLX may work as a hub in miRNA-mediated regulation of NSPC proliferation and transition into differentiation. Furthermore, the complex regulation of its expression by several miRNAs, which are themselves again regulated by TLX expression through negative feedback loops, provides the perfect mechanism for the fast and transient switch in gene expression needed to drive NPCs out of their proliferative state and into differentiation, without losing proliferative capacity on the long term.
Finally, miR-125b regulates the transition between proliferation and differentiation. miR-125b targets Nestin, an intermediate filament protein expressed in NSPCS, but not in neurons [70]. Overexpression of miR125-b results in decreased proliferation and increased differentiation of NSPCs. These effects can be rescued by introducing mutations in binding sites for miR125-b present on the Nestin mRNA 3’UTR. Besides this direct regulation of differentiation, miR-125b also targets numerous repressors of neuronal genes, mainly effectors of the ERK signaling pathway involved in the effect of retinoic acid, highlighting a role for miR-125b in neuronaldifferentiation [71].
Cell fate specification
Under normal physiological conditions most cells derived from adult hippocampal NSCs will differentiate into neurons, while only a small subset will give rise to astrocytes. This balance is tightly maintained, regulating the neurogenic capacity of the stem cell pool. DNA methylation is a main driving force behind the repression of the astrocytic fate [36], but recent studies also show methylation-independent pathways regulated by miRNAs. Most work done on cell fate specification comes from studies on miR-124, which is a key player in neuronal versus astrocytic cell fate determination. miR-124 is absent in NSPCs, while expression levels increase in neuroblasts and remain high in differentiated neurons. One of the targets of miR-124 is SOX9, a transcription factor crucial for gliogenesis present in both NSPCs and astrocytes, but not in neuroblasts and neurons [72]. Overexpression of a 3’UTR deficient SOX9 inhibits neuronal differentiation, while a knockdown of miR-124 results in increased ectopic SOX9 expression in neuroblasts. Importantly, in the subventricular zone stem cell niche, miR-124-mediated repression of SOX-9 is important for neuronal fate determination [73]. A second pathway regulated by miR-124 is the STAT3 pathway, which plays a role in terminal differentiation [74]. Upon phosphorylation STAT3 inhibits terminal neuronal differentiation, and drives cells towards a glial lineage [75, 76]. Overexpression of miR-124 reduces STAT3 phosphorylation, driving NSPCs towards the neuronal lineage [74]. Furthermore, overexpression of miR-124 in HeLa cells, a non-neuronal cell line, results in the expression of pro-neuronal genes, while silencing non-neuronal genes [77]. All together, these data indicate miR-124 is a crucial regulator of cell fate determination, stimulating neuronal lineages by suppressing glial lineage-specificgenes.
MiR-9, which controls proliferation through the TLX receptor, also targets several other genes that play crucial roles in neuronal differentiation, such as Foxg1 and Gsh2 [78]. Moreover, miR-9 also regulates several members of the REST complex, which acts as a transcriptional repressor of neuronal genes, inhibiting neuronal differentiation [79].
Another miRNA involved in cell fate determination is miR-26b, which is specifically expressed in NSPCs. miR-26b targets ctdsp2, an important component of the REST complex, which inhibits the transcription of neuronal genes [80].
Apoptotic selection of newborn neurons
A majority of the cells generated by adult hippocampal NSPCs undergo extensive apoptotic selection, thereby regulating neurogenesis levels. Apoptotic selection of NPCs is thought to serve as a quality control as well, selecting out unfit cells [17, 18]. The first indications for a role for miRNAs in controlling apoptosis comes from in vivo Dicer knockout studies, though these studies focused on SVZ neurogenesis instead of AHN. Early embryonic Dicer knockout results in increased proliferation of neural progenitors, however due to failed cell cycle progression upon differentiation most of these cells undergo apoptosis [81].
A recent study showed that miR-124 and miR-137 act cooperatively to regulate apoptosis in NSPCs, through fine-tuning expression levels of the pro-apoptotic protein Bcl2L13, upstream of Caspase3 [82]. Acting together on a common target, miR-124 and miR-137 decrease Bcl2L13 expression levels resulting in concomitant decreases of cleaved Caspase3, indicating a decrease in apoptosis. A second miRNA linked to the regulation of apoptosis is miR-34a, which targets the anti-apoptotic Bcl-2 protein, as well as several cell cycle regulators involved in cell cycle progression, such as Cdk-4 and Cyclin D2 [83–85]. Caution must be applied in interpreting these results, as these data all originate from non-neuronal cell type studies; however, both miR-34a and some of its targets are expressed in NSPCs, thereby providing a possible new regulatory mechanism for apoptotic selection mediated by miRNAs inNSPCs.
Migration
Migration of newborn neurons in the DG is crucial for establishing correct new synaptic contacts with the preexisting network. This migration is controlled by several chemoattractants, such as Reelin, and adhesion proteins. Several miRNAs have been identified that play a role in neuronal migration in the DG.
MiRNA cluster miR-379-410 regulates N-Cadherin, an adhesion protein found on cellular membranes, and is expressed in NSPCs and migrating immature neurons. Overexpression of this miRNA cluster in NSCs results in increased neuronal migration, which can be rescued by exogenous miRNA-insensitive N-Cadherin overexpression [86]. The brain-specific miR-9 is expressed in NPCs derived from human embryonic stem cells and loss of miR-9 suppressed proliferation and promoted migration of NPCs independently of precocious differentiation, possible by regulating the expression of its target, stathmin [87]. Recently, miR-19 was identified to play a critical role in neuronal migration. miR-19 is abundantly in NSPCs, while its expression decreases upon differentiation. Overexpression of miR-19 in NSPCs in the adult DG results in an increase in migration of newborn neurons [88].
Maturation and integration
After committing to their neuronal fate neuroblasts will undergo further maturation over the course of several weeks. Immature neurons migrate horizontally, start to form axonal and dendritic processes, and develop dendritic spines to allow for neuron-to-neuron communication [21]. The development of strong synaptic connections with afferent neurons is crucial as an absence of synaptic input will result in selective apoptosis [89–91] further guarding quality of newborn neurons in the DG. Several miRNAs are known to regulate these processes, including dendritic outgrowth and spine formation, crucial for maturation and integration of newborn neurons.
miR-34a and miR-137 negatively regulate neurite outgrowth and dendritic branching, limiting dendritic complexity of newborn neurons [83, 92]. miR-34a targets both synaptotagmin1 and syntaxin-1, 2 proteins crucial for functional maturation of newborn neurons. Overexpression of miR-34a results in a significant decrease of dendritic complexity, as well as a reduction in synaptic function. Inhibiting miR-34a rescues the functional deficits, while overexpression of synaptotagmin1 was able to partially rescue dendritic morphology [83]. miR-137 regulates Mib1, a protein known for its crucial role in neurodevelopment that acts on Notch signaling and induces apoptosis. Overexpression of miR-137 severely impacts dendritic outgrowth and spine formation, which can be rescued by overexpression of Mib1 [92].
While miR34a and miR-137 negatively regulate dendrite outgrowth, miR-124 positively regulated axonal and dendritic branching [93]. Knocking out Rcnr3, the primary source of miR-124, results in severe neuronal malformation and aberrant axonal sprouting. Lhx2, a primary target of miR-124, mediates these effects. Like miR-124, the miR17-92 cluster drives axonal outgrowth [94]. This cluster is specifically expressed in the distal axon of neurons, where it regulates phosphatase and tensin homolog (PTEN), a repressor of the MTOR pathway. Overexpression of this miR cluster results in increased axonal sprouting, which can be rescued by rapamycin treatment, an inhibitor of MTOR.
miR-125b, miR-134, and miR-138 are known to negatively regulate spine formation [95–97]. miR-125b has been linked to regulation of Fragile X Mental Retardation Protein (FRMP), and its overexpression results in severely altered spine morphology. Furthermore, miR-125b also regulates the NR2A subunit of the NMDA receptor, affecting synaptic plasticity of hippocampal neurons [97]. Both miR-134 and miR-138 are specifically enriched in dendritic spines, where they exert crucial roles for synaptic functioning. miR-134 regulates Limk-1, a transcription factor crucial for spinogenesis [95], while miR-138 regulates APT1, an enzyme responsible for the palmitoylation state of many synaptic proteins [96].
On the contrary, miR-132 is thought to positively regulate spine formation, in an activity-dependent manner by regulating p250GAP expression [98]. Its expression is regulated by CREB and is necessary and crucial for spine formation. miR-132 expression promotes neurite outgrowth [99] and inhibiting miR-132 results in decreased EPSC frequency and numbers of GluR1-positive spines, indicating miR-132 to play a key role in both structural and functional spinogenesis [98, 100].
Pathological alterations in AHN in the context of epilepsy
Even though AHN is tightly regulated, numerous internal and external factors can compromise the process. This becomes particularly clear when studying the neurogenic process under pathological conditions, such as epilepsy. Epileptic seizures have a strong effect on AHN, all of which have been pooled under the umbrella term “aberrant AHN” [101]. Whether these alterations causally underlie disease formation is still a matter of debate, but it has become clear that the alterations in AHN under epileptic conditions strongly affect hippocampal network excitability, thereby creating a hyperexcitable pro-epileptic environment, which possibly also underlies comorbid cognitive deficits observed in epilepsy patients [102].
Aberrant AHN and epilepsy
Epilepsy is a neurological disorder characterized by the occurrence of chronic spontaneous recurrent epileptic seizures [103]. Numerous subtypes of epilepsy exist, which can be divided by their cause (e.g. genetic versus idiopathic epilepsies) and location of origin of the epileptic insult. The largest group of patients suffers from temporal lobe epilepsy (TLE), characterized by seizures originating in many cases from the hippocampal region. Since most TLE patients also suffer from associated cognitive deficits, hippocampal functioning seems to be a key component of TLE pathology [104, 105].
Most idiopathic epilepsies share a common developmental window characterized by a primary insult (known or unknown), followed by a silent latent phase and culminating in chronic recurrent spontaneous seizures. The common conception is that the initial insult triggers multiple reactive processes in the brain tissue during the latent phase, thereby initiating epileptogenesis and finally resulting in the generation of pro-epileptic neuronal networks. These network changes eventually culminate in the occurrence of spontaneous seizures [106, 107].
AHN is particularly vulnerable to seizures. Pioneer studies showed that epileptic seizures cause a significant increase in the short-term proliferation rate of NSCs, while diminishing long-term proliferation [108]. Besides changes in proliferation rates, newborn neurons also display morphological and functional alterations, including somatic hypertrophy, presence of hilar basal dendrites [109], ectopic location of newborn neurons in the molecular layer or Hilus of the hippocampus [108], axonal sprouting towards the molecular layer forming recurrent excitatory circuits [110], and changes in dendritic spine density with concomitant alterations in neuronal excitability [111–113]. Together all these changes underlie the formation of recurrent excitatory networks within the DG, indicating a possible role for aberrant AHN in epilepsy development and the rise of chronic seizures [102]. Furthermore, long-term loss of proliferation has recently been shown to be a possible direct result of seizure activity, due to direct conversion of NSCs into astrocytes shortly after seizure onset [114]. This loss of proliferation is hypothesized to be one of the underlying causes of comorbid cognitive decline in epilepsypatients [115].
It is not yet agreed upon whether aberrant AHN plays a crucial role in epilepsy development, but multiple studies have shown that AHN is at least partially responsible for the generation of chronic epileptic seizures. Genetic induction of newborn ectopic granule cells with altered dendritic complexity using a Pten knockout mouse model results in spontaneous interictal activity and significantly increase seizure-susceptibility [116]. On the contrary, preventing aberrant AHN by completely removing AHN by either pharmacological or genetic approaches is sufficient to prevent, or at least slow down, the development of chronic epilepsy and decreases the severity of remaining chronic seizures after a primary insult known as Status Epilepticus (SE) [117, 118]. Besides its possible role in epilepsy development, aberrant AHN has already been shown to underlie comorbid cognitive deficits observed in epilepsy mouse models. Cognitive deficits that arise after SE induction can be rescued by ablating AHN, thereby preventing the occurrence of aberrant AHN[117, 119].
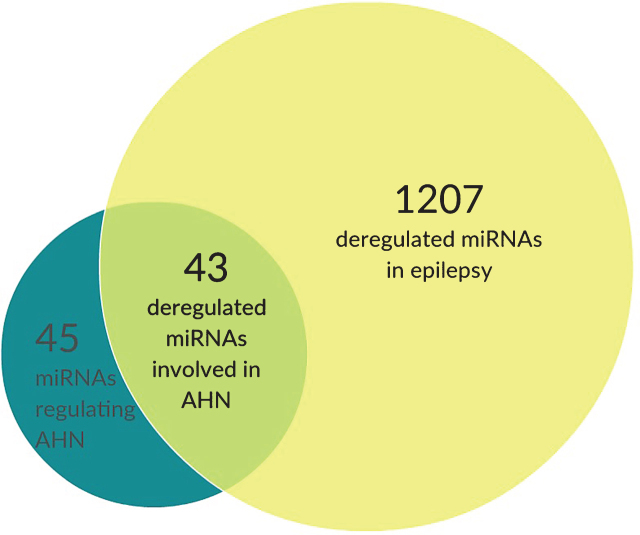
How epileptic seizures can deregulate AHN is topic of intense research, and a plethora of hypotheses have been raised. Over the recent years miRNAs have gained serious interest as it became clear that most miRNAs known to regulate AHN are also deregulated in case of seizure events or chronic epilepsy (Fig. 2). Here, we provide an overview of AHN-regulating miRNAs linked to epilepsy, based on analyses done suing the EpimiRBase developed by Mooney et al. [120]. EpimiRBase is a comprehensive manually curated database of over 2,000 miRNA-epilepsy associations, which was developed to address the need for a database to keep track of the rapidly expanding published literature on miRNAs in epilepsy. The June 2016 release of EpimiRBase lists miRNAs from 41 publications including 1207 unique miRNA (1,198 up and 870 down regulated) from three species: human [161], mouse (854) and rat (1,053). The miRNAs are categorized into four study types: expression analysis [41], functional [19], profiling-biofluid [32] and profiling-brain(1,976).
Fig.2.
AHN-regulating miRNAs are severely deregulated in epilepsy. We have identified 45 miRNAs that are established regulators of AHN (dark green). Using the EpimiRbase, which lists a total of 1207 deregulated miRNAs in epilepsy (yellow), we identified a total of 43 out of 45 AHN-regulating miRNAs to be deregulated in epilepsy (light green).
Using EpimiRBase against a list of all miRNAs known to regulate AHN provided numerous matches, again indicating a crucial role for miRNAs in the deregulation of AHN under seizure pathology (Table 2 and Fig. 2). Here we summarize all AHN-regulating miRNAs whose altered expression levels have been found in both rodent and human epilepsy studies.
Table 2.
Differentially expressed AHN-related MicroRNAs in epilepsy
| Expression during | Expression during | Predicted Biological | References | |
| acute stage | chronic stage | consequence | ||
| miR-184 | Up | Up | Increased proliferation | [121, 122] |
| miR-106b/25 cluster | Up | Up | Increased proliferation and neuronal differentiation | [123, 132, 157] |
| miR-34a | Up | Up | Increased proliferation, | [123, 124, 133] |
| Increased apoptosis | ||||
| miR-124 | Up | Up | Increased neuronal differentiation, | [82, 131] |
| Decreased apoptosis | ||||
| miR-137 | Up | Up | Increased proliferation, | [82, 129] |
| Decreased apoptosis | ||||
| miR-9 | Down | Up | Dynamic changes in proliferation and differentiation | [129, 130] |
| Let-7b | Down | Down | Increased proliferation, Decreased differentiation | [129] |
| miR-26b | Up | Up | Increased neuronal differentiation | [82, 164] |
| miR-145 | Up/Down | Up/Down | Changes in neuronal differentiation | [123, 130, 165] |
| miR-125b | Up | Up | Increased neuronal differentiation | [132, 166] |
| miR-19 | Up | Up | Increased migration | [123] |
| miR-379/410 cluster | Up | Up | Increased migration | [82, 122, 132] |
| miR-138 | Down | Down | Increased spinogenesis | [82, 132] |
| miR-134 | Up | Up | Decreased spinogenesis | [131] |
| miR-132 | Up | Up | Increased spinogenesis | [123, 132, 167] |
Overview of differentially expressed microRNAs in epilepsy known to be involved in neurogenesis, and their (predicted) biological consequence.
Stem Cell maintenance and quiescence
miR-184 and miR-34a, which play a role in stem cell maintenance through regulation of the Notch signaling pathway, as discussed before, are differentially expressed in both rodent or human profiling studies related to epilepsy. Increased miR-184 expression has been found both in chronic human epilepsy patients, as well as in the early stages of epileptogenesis in a rodent model [121, 122]. miR-34a is commonly found up regulated at different stages of disease development in rodents [123, 124]. Chronic up regulation of these miRNAs leads to depletion of the stem cell pool, one characteristic feature of epilepsy models and a potential cause of comorbid cognitive deficits observed in both rodent and human epilepsy subjects [17, 125–128].
NSPC Proliferation and differentiation
As mentioned before, TLX is involved in several negative feedback loops with different miRNAs, including miR-9, miR-137, miR-145, and Let-7b. All 3 miRNAs have also been identified in profiling studies, though the directionality of its expression differs temporally. Expression of the Let-7 family is severely altered by the initial insult. Shortly after SE-induction Let-7 expression decreases, followed by a severe increase at 24 hours post SE. Following this peak expression Let-7 is again down regulated lasting up till 50 days post SE [129]. Unlike Let-7, miR-9 is found up regulated during the chronic stage of the disease [129], while its expression decreases two hours after SE induction [130]. Finally, expression of miR-137 increases both in acute [82] and chronic [129] stages of animal SE models, while expression of miR-145 is yet inconclusive as both up and down regulation have been shown. Decreased expression of these miRNAs will result in increased expression of the TLX receptor, thereby driving proliferation over differentiation, partially explaining the increased proliferation rates found in the early stages of epileptogenesis, while chronic up regulation of these miRNAs could potentially underlie the diminished proliferation observed in chronic epileptic hippocampi.
Astrocytic conversion of NSCs
As mentioned before, it has been recently shown that hippocampal NSCs can directly convert into astrocytes under seizure conditions. Though this phenomenon has not been linked directly to miRNAs yet, it could be argued that miRNA deregulation probably plays a role. In this respect, one possible key player could be miR-124, which is a crucial regulator of NSPC fate determination, through targeting SOX9, a transcription factor critical for glionesis that is normally absent in neuroblasts and immature neurons [72]. miR-124 has been found upregulated under epileptic conditions [82, 131], providing a possible mechanistic explanation for astrocytic conversion of NSCs.
Apoptotic selection
miR-124 and miR-137, which cooperatively regulate NPC apoptosis after SE,, are both significantly up regulated during the initial stages of epileptogenesis [82]. Furthermore, both miR-124 and miR-137 are also up regulated during the chronic stage of the disease [131]. Up regulation of both miR-124 and miR-137 inhibits apoptosis of NPCs through down regulation of Bcl2L13, providing a possible explanation for the survival of unfit immature neurons commonly found in the epileptic hippocampus. On the contrary miR-34a, which down regulates anti-apoptotic Bcl2 levels, is significantly up regulated at different stages of the disease. Increased expression of miR-34a should increase apoptotic selection under epileptic conditions.
Migration of newborn neurons
The miR-379-410 cluster is also deregulated under epileptic conditions [82, 123, 132]. Interestingly, at acute stages expression of this cluster increases, down regulating N-Cadherin and making immature neurons more prone to migrate. During the latent phase expression levels decrease, indicating a clear link between the presence of epileptic seizures and aberrant migration of newborn neurons commonly observed in epileptic hippocampi.
miR-9, which promotes migration of immature neurons, is also upregulated under epileptic conditions [123]. Together, this points toward a strong effect of seizures on migration of immature neurons, providing a possible explanation of the characteristic ectopic granule cells commonly found in epileptic hippocampi.
Maturation and integration
Mir-34a and miR-134, which inhibit dendritic branching, are both up regulated in all stages of the disease [82, 123, 129, 131, 133, 134]. On the contrary miR-124, which is known to stimulate neurite outgrowth is down regulated at acute stages of epileptogenesis, but up regulated a few days after SE induction all until the chronic epileptic stage [82, 123, 131, 133]. The miR-17-92 cluster, which also promotes dendritic outgrowth, is up regulated at all stages of the disease [123, 133]. All together this points towards a complex regulation of dendritic outgrowth under seizure conditions.
A similar picture arises when looking at the expression levels of miRNAs known to regulate spinogenesis and functional integration. miR-132, which stimulates spinogenesis, has been found up regulated in all stages of the disease, indicating the occurrence of increased synaptic connectivity [123, 132, 133]. In line with these findings expression levels of miR-138, which inhibits spinogenesis, is down regulated during all stages of the disease [123, 133]. On the contrary miR-134, which also inhibits spinogenesis, is up regulated during all stages of the disease [124, 131].
miRNA-based therapeutic approaches for epilepsy
The complexity by which miRNAs regulate biological processes makes it a daunting challenge to summarize their role in the regulation of AHN. The list of miRNAs that regulate AHN keeps on growing, and more and more complex levels of miRNA regulation are being identified. Here we have summarized some examples of miRNAs and their targets that may function as hubs for the regulation of AHN. This concept is in line with recent observations that indicate functional convergence of multiple miRNAs on the same biological processes, or even on the same mRNA targets [82, 135]. By doing so, miRNAs have the potential to achieve precise temporal and spatial regulation of biological processes. Specifically, the convergence of two or more miRNAs on common targets will significantly reduce the number of potential targets because the number of common targets is lower than the list of potential individual targets in most cases. However, the converging actions of miRNAs remain a poorly understood layer of complexity in miRNA regulation, but some examples involving NSPCs have recently been described [82].
Besides the biological complexity of miRNA functioning, applied clinical RNA therapy poses multiple hurdles that still need to be overcome. First, RNA molecules are generally unstable due to the presence of hydroxyl groups. Since the discovery of siRNAs and their promised therapeutic value several strategies have been developed to stabilize small RNA molecules, which can also be applied to miRNAs. Most of these strategies involve chemical modification of the RNA, such as ribose 2’OH modification and LNA modification [137, 138] that significantly stabilize ssRNAs through multiple mechanisms. Interesting, naked small RNA molecules administered using osmotic minipumps or directly by stereotaxic injection into the brain specifically regulate intended targets in vivo, shortly after injection in some cases [139–141].
A second issue that arises in view of clinical usability is the miRNA delivery method, especially in the case of neurological disorders, since the blood-brain barrier normally excludes large polar molecules such as oligonucleotides from entering. miRNA-based treatments could be administered in several non-invasive ways, bypassing the need for intracranial administration. First, it has recently been shown that miRNAs can be successfully delivered to the brain via intranasal administration [142], though the mechanisms through which miRNAs reach their target is still unclear. Secondly, the recent discovery of exosomal trafficking of miRNAs [53] holds great therapeutic value, since exosomes can cross the blood-brain barrier and thus be administered intravenously [143]. Another possibility is to time a systemic delivery of the molecules with breaches in the integrity of the blood-brain barrier, such as may occur following brain injury or after prolonged seizures.
With all current modifications of ssRNAs and new delivery methods that will allow non-invasive miRNA administration to the brain, miRNA-based treatments for neurological disorders seem within reach, at least from a technical point of view. Though mainly in cancer research, several clinical trials are already in progress trying to establish miRNA-based therapy.
MiRNA-based therapy in epilepsy
AHN-focused miRNA-based therapy for epilepsy seem rather challenging considering the existence of multiple miRNAs targeting the same biological processes or pathways, complex regulatory feedback loops within these pathways, and bidirectional expression control in the form of feedback loops. However, several attempts to identify miRNA-based anti-epileptic therapies have been performed, though not primarily focusing on AHN. miR-34a has been the focus in two different rodent epilepsy studies. Silencing miR-34a using antagomirs during SE induction successfully reduced neuronal apoptosis in CA1 and CA3, but this was not assessed in the DG [133]. Therefore extrapolating these findings to AHN requires caution. A second study administering antagomirs against miR-34a, but now 24 hours post SE induction, did not find any neuroprotective effects, indicating a potential time-dependent treatment window [134]. More importantly, both studies also failed to show any beneficial effects of antagomir-34a administration of seizure duration and severity.
Probably the most compelling evidence for miRNA-based epilepsy therapy comes from two studies targeting miR-134 expression. As mentioned before, miR-134 is involved in the regulation of spinogenesis, and therefore is an important regulator of excitability and network formation. Administration of antagomir-134 one day before SE induction significantly decreased the proportion of animals developing pilocarpine-induced SE. Furthermore, animals that did develop SE showed a significant delay in seizure onset and a decrease in total seizure power [144]. In a second study, antagomir-134 administration one hour post SE induction reduced the occurrence of chronic spontaneous seizures by 90% [145]. Furthermore, antagomir-134 administration reduced CA3 pyramidal spine density, neuronal cell loss, and astrogliosis, which are all hallmarks of the epileptic hippocampus. Like the studies performed on miR-34a, these studies on miR-134 did not assess any AHN-related pathology.
Future perspectives
The regulation of AHN by miRNAs has been intensively studied but still needs to be further elucidated. However, possible miRNA-based therapeutic applications are already in sight, as discussed in previous sections. In order to develop strong therapeutic strategies we will first need to optimize the preclinical models used so far. For example, in the case of miRNA profiling of the epileptic brain there is no standardized procedure for tissue selection, resulting in “epilepsy” miRNA profiles from numerous different sources and timepoints. Since we here try to understand the role of miRNAs in AHN, once would prefer to have miRNA profiles from the DG specifically [82, 146]. An even more sophisticated approach that holds great promise is the recent technological advancements in single cell ‘-omics’ [147, 148]. If we could identify miRNA profiles in individual cells from all different cell types in the AHN cascade we could systematically elucidate their roles and we could be one step closer to identifying pathological miRNA expression and a potential miRNA-based therapy. Additionally, long non-coding RNAs (lnRNAs) and circular RNAs (circRNAs), which have been recently discovered as potent regulators of gene expression [149, 150], can also be profiled using single cell sequencing strategies.
Besides being a potential therapeutic target, miRNAs also hold great promise as potential biomarkers [151]. With the recent discovery that miRNAs are excreted in exosomal vesicles and can be detected in blood [53, 152–154], it has now become possible to non-invasively identify altered miRNA expression levels from pathological brain tissue [155]. Several studies have already obtained miRNA profiles from epileptic patients using blood samples [153, 156], allowing comparisons with preclinical epilepsy models [157].
Given the current limited evidence for miRNA-based anti-epileptic treatment, the future for miRNA-based therapies remains still uncertain but it is heavily investigated. Though some preclinical evidence points towards successful application of single miRNA-based anti-epileptic approaches, one might argue that the future of miRNA-based therapy comprises multi-miRNA approaches instead, based on the robust regulation of common targets or biological pathways. Fundamental research is providing more and more evidence regarding complex interplay between different miRNAs, raising questions about single-miRNA therapies.
Interestingly, recent challenging experiments have demonstrated that AHN may be associated with cognitive impairments observed in epilepsy [117]. Cognitive impairment comorbidity among epilepsy patients is high and in many cases it still progresses even when seizures have been controlled [158–160]. If these cognitive deficits indeed severely rely on the presence of aberrant AHN, and miR-based treatments can rescue cellular alterations observed under pathological conditions, one might argue that miR-based therapy against cognitive deficits associated with epilepsy, may be another promising direction for future miR-based therapeutic studies. Thus, most likely, multi-miRNA-based approaches targeting several aspects of AHN will provide promising avenues to successfully rescue disease-associated aberrant AHN, and they could pave the way towards future clinical applications of miRNA-based therapies.
REFERENCES
- [1]. Aimone JB, Li Y, Lee SW, Clemenson GD, Deng W, Gage FH. Regulation and Function of Adult Neurogenesis: From Genes to Cognition. Physiol Rev. 2014;94(4):991–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Bizon JL, Lee HJ, Gallagher M. Neurogenesis in a rat model of age-related cognitive decline. Aging Cell. 2004;3(4):227–34. [DOI] [PubMed] [Google Scholar]
- [3]. Deng W, Aimone JB, Gage FH. New neurons and new memories: How does adult hippocampal neurogenesis affect learning and memory? Nat Rev Neurosci 2010;11(5):339–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4]. Ming G-L, Song H. Adult neurogenesis in the mammalian brain: Significant answers and significant questions. Neuron. 2011;70(4):687–702 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]. Snyder JS, Kee N, Wojtowicz JM. Effects of adult neurogenesis on synaptic plasticity in the rat dentate gyrus. J Neurophysiol. 2001;85(6):2423–31. [DOI] [PubMed] [Google Scholar]
- [6]. Arvidsson A, Collin T, Kirik D, Kokaia Z, Lindvall O. Neuronal replacement from endogenous precursors in the adult brain after stroke. Nat Med. 2002;8(9):963–70. [DOI] [PubMed] [Google Scholar]
- [7]. Darsalia V, Heldmann U, Lindvall O, Kokaia Z. Stroke-induced neurogenesis in aged brain. Stroke. 2005;36(8):1790–5. [DOI] [PubMed] [Google Scholar]
- [8]. Encinas JM, Vaahtokari A, Enikolopov G. Fluoxetine targets early progenitor cells in the adult brain. Proc Natl Acad Sci U S A. 2006;103(21):8233–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9]. Jakubs K, Nanobashvili A, Bonde S, Ekdahl CT, Kokaia Z, Kokaia M, et al. Environment matters: Synaptic properties of neurons born in the epileptic adult brain develop to reduce excitability. Neuron. 2006;52(6):1047–59. [DOI] [PubMed] [Google Scholar]
- [10]. Santarelli L. Requirement of Hippocampal Neurogenesis for the Behavioral Effects of Antidepressants. Science(80-). 2003;301(5634):805–9. [DOI] [PubMed] [Google Scholar]
- [11]. Scharfman HE, McCloskey DP. Postnatal neurogenesis as a therapeutic target in temporal lobe epilepsy. Epilepsy Res. 2009;85(2-3):150–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Ziv Y, Finkelstein A, Geffen Y, Kipnis J, Smirnov I, Shpilman S, et al. A novel immune-based therapy for stroke induces neuroprotection and supports neurogenesis. Stroke. 2007;38(2 Suppl):774–82. [DOI] [PubMed] [Google Scholar]
- [13]. Knoth R, Singec I, Ditter M, Pantazis G, Capetian P, Meyer RP, et al. Murine features of neurogenesis in the human hippocampus across the lifespan from 0 to 100 years. PLoS One. 2010;5(1):e8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. Spalding KL, Bergmann O, Alkass K, Bernard S, Salehpour M, Huttner HB, et al. Dynamics of hippocampal neurogenesis in adult humans. Cell. 2013;153(6):1219–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15]. Sultan S, Li L, Moss J, Petrelli F, Cassé F, Gebara E, et al. Synaptic Integration of Adult-Born Hippocampal Neurons Is Locally Controlled by Astrocytes. Neuron. 2015;88(5):957–72. [DOI] [PubMed] [Google Scholar]
- [16]. Kempermann G. Activity Dependency and Aging in the Regulation of Adult Neurogenesis. Cold Spring Harb Perspect Biol. 2015;7(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17]. Encinas JM, Michurina T V, Peunova N, Park J-H, Tordo J, Peterson DA, et al. Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell. 2011;8(5):566–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18]. Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming G, et al. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell. 2011;145(7):1142–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19]. Kuhn HG, Biebl M, Wilhelm D, Li M, Friedlander RM, Winkler J. Increased generation of granule cells in adult Bcl-2-overexpressing mice: A role for cell death during continued hippocampal neurogenesis. Eur J Neurosci. 2005;22(8):1907–15. [DOI] [PubMed] [Google Scholar]
- [20]. Biebl M, Cooper CM, Winkler J, Kuhn HG. Analysis of neurogenesis and programmed cell death reveals a self-renewing capacity in the adult rat brain. Neurosci Lett. 2000;291(1):17–20. [DOI] [PubMed] [Google Scholar]
- [21]. Kempermann G, Jessberger S, Steiner B, Kronenberg G. Milestones of neuronal development in the adult hippocampus. Trends Neurosci. 2004;27(8):447–52. [DOI] [PubMed] [Google Scholar]
- [22]. Ming G, Song H. Adult neurogenesis in the mammalian central nervous system. Annu Rev Neurosci. 2005;28:223–50. [DOI] [PubMed] [Google Scholar]
- [23]. Seidenfaden R, Desoeuvre A, Bosio A, Virard I, Cremer H. Glial conversion of SVZ-derived committed neuronal precursors after ectopic grafting into the adult brain. Mol Cell Neurosci. 2006;32(1-2):187–98. [DOI] [PubMed] [Google Scholar]
- [24]. Shihabuddin LS, Horner PJ, Ray J, Gage FH. Adult spinal cord stem cells generate neurons after transplantation in the adult dentate gyrus. J Neurosci. 2000;20(23):8727–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25]. Goldman SA, Chen Z. Perivascular instruction of cell genesis and fate in the adult brain. Nat Neurosci. 2011;14(11):1382–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. Palmer TD, Willhoite AR, Gage FH. Vascular niche for adult hippocampal neurogenesis. J Comp Neurol. 2000;425(4):479–94. [DOI] [PubMed] [Google Scholar]
- [27]. Conover JC, Doetsch F, Garcia-Verdugo JM, Gale NW, Yancopoulos GD, Alvarez-Buylla A. Disruption of Eph/ephrin signaling affects migration and proliferation in the adult subventricular zone. Nat Neurosci. 2000;3(11):1091–7. [DOI] [PubMed] [Google Scholar]
- [28]. Doetsch F, Petreanu L, Caille I, Garcia-Verdugo JM, Alvarez-Buylla A. EGF converts transit-amplifying neurogenic precursors in the adult brain into multipotent stem cells. Neuron. 2002;36(6):1021–34. [DOI] [PubMed] [Google Scholar]
- [29]. Jin K, Zhu Y, Sun Y, Mao XO, Xie L, Greenberg DA. Vascular endothelial growth factor (VEGF) stimulates neurogenesis in vitro and in vivo. Proc Natl Acad SciU S A. 2002;99(18):11946–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Lu M, Grove EA, Miller RJ. Abnormal development of the hippocampal dentate gyrus in mice lacking the CXCR4 chemokine receptor. Proc Natl Acad Sci U S A. 2002;99(10):7090–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Nakatomi H, Kuriu T, Okabe S, Yamamoto S, Hatano O, Kawahara N, et al. Regeneration of hippocampal pyramidal neurons after ischemic brain injury by recruitment of endogenous neural progenitors. Cell. 2002;110(4):429–41. [DOI] [PubMed] [Google Scholar]
- [32]. Moss J, Gebara E, Bushong EA, Sánchez-Pascual I, O’Laoi R, El M’Ghari I, et al. Fine processes of Nestin-GFP-positive radial glia-like stem cells in the adult dentate gyrus ensheathe local synapses and vasculature. Proc Natl Acad Sci. 2016;113(18):E2536–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33]. Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297(5580):365–9. [DOI] [PubMed] [Google Scholar]
- [34]. Perez-Moreno M, Jamora C, Fuchs E. Sticky business: Orchestrating cellular signals at adherens junctions. Cell. 2003;112(4):535–48. [DOI] [PubMed] [Google Scholar]
- [35]. Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31(3):353–65. [DOI] [PubMed] [Google Scholar]
- [36]. Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, et al. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001;1(6):749–58. [DOI] [PubMed] [Google Scholar]
- [37]. Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze S-H, Chenoweth J, et al. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298(5599):1747–52. [DOI] [PubMed] [Google Scholar]
- [38]. Hsieh J, Zhao X. Genetics and Epigenetics in Adult Neurogenesis. Cold Spring Harb Perspect Biol. 2016;8(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39]. Yao B, Jin P. Unlocking epigenetic codes in neurogenesis. Genes Dev. 2014;28(12):1253–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40]. Lopez-Ramirez MA, Nicoli S. Role of miRNAs and epigenetics in neural stem cell fate determination. Epigenetics. 2014;9(1):90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. Lang M-F, Shi Y. Dynamic Roles of microRNAs in Neurogenesis. Front Neurosci. 2012;6:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42]. Jobe EM, McQuate AL, Zhao X. Crosstalk among Epigenetic Pathways Regulates Neurogenesis. Front Neurosci. 2012;6:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43]. Schouten M, Buijink MR, Lucassen PJ, Fitzsimons CP. New Neurons in Aging Brains: Molecular Control by Small Non-Coding RNAs. Front Neurosci. 2012;6:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44]. Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–34. [DOI] [PubMed] [Google Scholar]
- [45]. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–24. [DOI] [PubMed] [Google Scholar]
- [46]. Johanson TM, Lew AM, Chong MMW. MicroRNA-independent roles of the RNase III enzymes Drosha and Dicer. Open Biol. 2013;3(10):130144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47]. Rolando C, Erni A, Grison A, Beattie R, Engler A, Gokhale PJ, et al. Multipotency of Adult Hippocampal NSCs In Vivo Is Restricted by Drosha/NFIB. Cell Stem Cell. 2016;19(5):653–62. doi: 10.1016/j.stem.2016.07.003 [DOI] [PubMed] [Google Scholar]
- [48]. Kawahara H, Imai T, Okano H. MicroRNAs in Neural Stem Cells and Neurogenesis. Front Neurosci. 2012;6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49]. Sato F, Tsuchiya S, Meltzer SJ, Shimizu K. MicroRNAs and epigenetics. FEBS J. 2011;278(10):1598–609. [DOI] [PubMed] [Google Scholar]
- [50]. Ma DK, Marchetto MC, Guo JU, Ming G, Gage FH, Song H. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat Neurosci. 2010;13(11):1338–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51]. Zhu H, Fan G-C. Extracellular/circulating microRNAs and their potential role in cardiovascular disease. Am J Cardiovasc Dis. 2011;1(2):138–49. [PMC free article] [PubMed] [Google Scholar]
- [52]. Chen X, Liang H, Zhang J, Zen K, Zhang C-Y. Secreted microRNAs: A new form of intercellular communication. Trends Cell Biol. 2012;22(3):125–32. [DOI] [PubMed] [Google Scholar]
- [53]. Zhang J, Li S, Li L, Li M, Guo C, Yao J, et al. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genomics Proteomics Bioinformatics. 2015;13(1):17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54]. Liu J, Sato C, Cerletti M, Wagers A. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr Top Dev Biol. 2010;92:367–409. [DOI] [PubMed] [Google Scholar]
- [55]. Shen Q, Zhong W, Jan YN, Temple S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development. 2002;129(20):4843–53. [DOI] [PubMed] [Google Scholar]
- [56]. Liu C, Teng Z-Q, Santistevan NJ, Szulwach KE, Guo W, Jin P, et al. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell. 2010;6(5):433–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57]. Fineberg SK, Datta P, Stein CS, Davidson BL. MiR-34a represses Numbl in murine neural progenitor cells and antagonizes neuronal differentiation. PLoS One. 2012;7(6):e38562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58]. Brett JO, Renault VM, Rafalski VA, Webb AE, Brunet A. The microRNA cluster miR-106b 25 regulates adult neural stem/progenitor cell proliferation and neuronal differentiation. Aging (Albany NY). 2011;3(2):108–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59]. Cheung TH, Quach NL, Charville GW, Liu L, Park L, Edalati A, et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482(7386):524–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60]. Lie D-C, Colamarino SA, Song H-J, Désiré L, Mira H, Consiglio A, et al. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437(7063):1370–5. [DOI] [PubMed] [Google Scholar]
- [61]. Qu Q, Sun G, Li W, Yang S, Ye P, Zhao C, et al. Orphan nuclear receptor TLX activates Wnt/beta-catenin signalling to stimulate neural stem cell proliferation and self-renewal. Nat Cell Biol. 2010;12(1):31–40; sup pp 1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62]. Shi Y, Chichung Lie D, Taupin P, Nakashima K, Ray J, Yu RT, et al. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature. 2004;427(6969):78–83. [DOI] [PubMed] [Google Scholar]
- [63]. Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16(4):365–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64]. Sun G, Ye P, Murai K, Lang M-F, Li S, Zhang H, et al. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat Commun. 2011;2:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65]. Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, et al. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189(1):127–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66]. Zhao C, Sun G, Li S, Lang M-F, Yang S, Li W, et al. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci U S A. 2010;107(5):1876–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67]. Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 2009;136(5):913–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68]. Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5(3):320–31. [DOI] [PubMed] [Google Scholar]
- [69]. Morgado AL, Rodrigues CMP, Solá S. MicroRNA-145 Regulates Neural Stem Cell Differentiation Through the Sox2-Lin28/let-7 Signaling Pathway. Stem Cells. 2016;34(5):1386–95. [DOI] [PubMed] [Google Scholar]
- [70]. Cui Y, Xiao Z, Han J, Sun J, Ding W, Zhao Y, et al. MiR-125b orchestrates cell proliferation, differentiation and migration in neural stem/progenitor cells by targeting Nestin. BMC Neurosci. 2012;13:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71]. Le MTN, Xie H, Zhou B, Chia PH, Rizk P, Um M, et al. MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol Cell Biol. 2009;29(19):5290–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72]. Stolt CC, Lommes P, Sock E, Chaboissier M-C, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17(13):1677–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73]. Cheng L-C, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12(4):399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74]. Krichevsky AM, Sonntag K-C, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24(4):857–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75]. Gu F, Hata R, Ma Y-J, Tanaka J, Mitsuda N, Kumon Y, et al. Suppression of Stat3 promotes neurogenesis in cultured neural stem cells. J Neurosci Res. 2005;81(2):163–71. [DOI] [PubMed] [Google Scholar]
- [76]. Moon C, Yoo J-Y, Matarazzo V, Sung YK, Kim EJ, Ronnett G V. Leukemia inhibitory factor inhibits neuronal terminal differentiation through STAT3 activation. Proc Natl Acad Sci U S A. 2002;99(13):9015–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77]. Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–73. [DOI] [PubMed] [Google Scholar]
- [78]. Conaco C, Otto S, Han J-J, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103(7):2422–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79]. Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci. 2008;28(53):14341–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80]. Dill H, Linder B, Fehr A, Fischer U. Intronic miR-26b controls neuronal differentiation by repressing its host transcript, ctdsp2. Genes Dev. 2012;26(1):25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81]. De Pietri Tonelli D, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135(23):3911–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82]. Schouten M, Fratantoni SA, Hubens CJ, Piersma SR, Pham T V, Bielefeld P, et al. MicroRNA-124 and -137 cooperativity controls caspase-3 activity through BCL2L13 in hippocampal neural stem cells. Sci Rep. 2015;5:12448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83]. Antonini D, Russo MT, De Rosa L, Gorrese M, Del Vecchio L, Missero C. Transcriptional repression of miR-34 family contributes to p63-mediated cell cycle progression in epidermal cells. J Invest Dermatol. 2010;130(5):1249–57. [DOI] [PubMed] [Google Scholar]
- [84]. Aranha MM, Santos DM, Xavier JM, Low WC, Steer CJ, Solá S, et al. Apoptosis-associated microRNAs are modulated in mouse, rat and human neural differentiation. BMC Genomics. 2010;11:514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85]. Wang X, Liu P, Zhu H, Xu Y, Ma C, Dai X, et al. miR-34a, a microRNA up-regulated in a double transgenic mouse model of Alzheimer’s disease, inhibits bcl2 translation. Brain Res Bull. 2009;80(4-5):268–73. [DOI] [PubMed] [Google Scholar]
- [86]. Rago L, Beattie R, Taylor V, Winter J. miR379-410 cluster miRNAs regulate neurogenesis and neuronal migration by fine-tuning N-cadherin. EMBO J. 2014;33(8):906–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87]. Delaloy C, Liu L, Lee J-A, Su H, Shen F, Yang G-Y, et al. MicroRNA-9 Coordinates Proliferation and Migration of Human Embryonic Stem Cell-Derived Neural Progenitors. Cell Stem Cell. 2010;6(4):323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88]. Han J, Kim HJ, Schafer ST, Paquola A, Clemenson GD, Toda T, et al. Functional Implications of miR-19 in the Migration of Newborn Neurons in the Adult Brain. Neuron. 2016;91(1):79–89. [DOI] [PubMed] [Google Scholar]
- [89]. Kim WR, Sun W. Programmed cell death during postnatal development of the rodent nervous system. Dev Growth Differ. 2011;53(2):225–35. [DOI] [PubMed] [Google Scholar]
- [90]. Buss RR, Sun W, Oppenheim RW. Adaptive roles of programmed cell death during nervous system development. Annu Rev Neurosci. 2006;29:1–35. [DOI] [PubMed] [Google Scholar]
- [91]. Davies AM. Regulation of neuronal survival and death by extracellular signals during development. EMBO J. 2003;22(11):2537–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92]. Smrt RD, Szulwach KE, Pfeiffer RL, Li X, Guo W, Pathania M, et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28(6):1060–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93]. Sanuki R, Onishi A, Koike C, Muramatsu R, Watanabe S, Muranishi Y, et al. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat Neurosci. 2011;14(9):1125–34. [DOI] [PubMed] [Google Scholar]
- [94]. Zhang Y, Ueno Y, Liu XS, Buller B, Wang X, Chopp M, et al. The MicroRNA-17-92 cluster enhances axonal outgrowth in embryonic cortical neurons. J Neurosci. 2013;33(16):6885–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95]. Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439(7074):283–9. [DOI] [PubMed] [Google Scholar]
- [96]. Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11(6):705–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97]. Edbauer D, Neilson JR, Foster KA, Wang C-F, Seeburg DP, Batterton MN, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65(3):373–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98]. Impey S, Davare M, Lesiak A, Lasiek A, Fortin D, Ando H, et al. An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol Cell Neurosci. 2010;43(1):146–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99]. Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102(45):16426–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100]. Luikart BW, Bensen AL, Washburn EK, Perederiy JV, Su KG, Li Y, et al. miR-132 mediates the integration of newborn neurons into the adult dentate gyrus. PLoS One. 2011;6(5):e19077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101]. Jessberger S, Zhao C, Toni N, Clemenson GD, Li Y, Gage FH. Seizure-associated, aberrant neurogenesis in adult rats characterized with retrovirus-mediated cell labeling. J Neurosci. 2007;27(35):9400–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102]. Bielefeld P, van Vliet EA, Gorter JA, Lucassen PJ, Fitzsimons CP. Different subsets of newborn granule cells: A possible role in epileptogenesis? Eur J Neurosci 2014;39(1):1–11. [DOI] [PubMed] [Google Scholar]
- [103]. Engel J. Etiology as a risk factor for medically refractory epilepsy: A case for early surgical intervention. Neurology. 1998;51(5):1243–4. [DOI] [PubMed] [Google Scholar]
- [104]. Helmstaedter C, Kurthen M, Lux S, Reuber M, Elger CE. Chronic epilepsy and cognition: A longitudinal study in temporal lobe epilepsy. Ann Neurol. 2003;54(4):425–32. [DOI] [PubMed] [Google Scholar]
- [105]. Elger CE, Helmstaedter C, Kurthen M. Chronic epilepsy and cognition. Lancet Neurol. 2004;3(11):663–72. [DOI] [PubMed] [Google Scholar]
- [106]. Pitkänen A, Immonen RJ, Gröhn OHJ, Kharatishvili I. From traumatic brain injury to posttraumatic epilepsy: What animal models tell us about the process and treatment options. Epilepsia. 2009;50(Suppl 2):21–9. [DOI] [PubMed] [Google Scholar]
- [107]. Pitkänen A, Kharatishvili I, Karhunen H, Lukasiuk K, Immonen R, Nairismägi J, et al. Epileptogenesis in experimental models. Epilepsia. 2007;48(Suppl 2):13–20. [DOI] [PubMed] [Google Scholar]
- [108]. Parent JM, Yu TW, Leibowitz RT, Geschwind DH, Sloviter RS, Lowenstein DH. Dentate granule cell neurogenesis is increased by seizures and contributes to aberrant network reorganization in the adult rat hippocampus. J Neurosci. 1997;17(10):3727–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109]. Murphy BL, Hofacer RD, Faulkner CN, Loepke AW, Danzer SC. Abnormalities of granule cell dendritic structure are a prominent feature of the intrahippocampal kainic acid model of epilepsy despite reduced postinjury neurogenesis. Epilepsia. 2012;53(5):908–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110]. Scharfman HE, Goodman JH, Sollas AL. Granule-like neurons at the hilar/CA3 border after status epilepticus and their synchrony with area CA3 pyramidal cells: Functional implications of seizure-induced neurogenesis. J Neurosci. 2000;20(16):6144–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111]. Dashtipour K, Tran PH, Okazaki MM, Nadler JV, Ribak CE. Ultrastructural features and synaptic connections of hilar ectopic granule cells in the rat dentate gyrus are different from those of granule cells in the granule cell layer. Brain Res. 2001;890(2):261–71. [DOI] [PubMed] [Google Scholar]
- [112]. Wood JC, Jackson JS, Jakubs K, Chapman KZ, Ekdahl CT, Kokaia Z, et al. Functional integration of new hippocampal neurons following insults to the adult brain is determined by characteristics of pathological environment. Exp Neurol. 2011;229(2):484–93. [DOI] [PubMed] [Google Scholar]
- [113]. Murphy BL, Pun RYK, Yin H, Faulkner CR, Loepke AW, Danzer SC. Heterogeneous integration of adult-generated granule cells into the epileptic brain. J Neurosci. 2011;31(1):105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114]. Sierra A, Martín-Suárez S, Valcárcel-Martín R, Pascual-Brazo J, Aelvoet S-A, Abiega O, et al. Neuronal hyperactivity accelerates depletion of neural stem cells and impairs hippocampal neurogenesis. Cell Stem Cell. 2015;16(5):488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115]. Hattiangady B, Shetty AK. Implications of decreased hippocampal neurogenesis in chronic temporal lobe epilepsy. Epilepsia. 2008;49(Suppl 5)(0 5):26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116]. Pun RYK, Rolle IJ, Lasarge CL, Hosford BE, Rosen JM, Uhl JD, et al. Excessive activation of mTOR in postnatally generated granule cells is sufficient to cause epilepsy. Neuron. 2012;75(6):1022–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117]. Cho K-O, Lybrand ZR, Ito N, Brulet R, Tafacory F, Zhang L, et al. Aberrant hippocampal neurogenesis contributes to epilepsy and associated cognitive decline. Nat Commun. 2015;6:6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118]. Jung K-H, Chu K, Kim M, Jeong S-W, Song Y-M, Lee S-T, et al. Continuous cytosine-b-D-arabinofuranoside infusion reduces ectopic granule cells in adult rat hippocampus with attenuation of spontaneous recurrent seizures following pilocarpine-induced status epilepticus. Eur J Neurosci. 2004;19(12):3219–26. [DOI] [PubMed] [Google Scholar]
- [119]. Jessberger S, Nakashima K, Clemenson GD, Mejia E, Mathews E, Ure K, et al. Epigenetic modulation of seizure-induced neurogenesis and cognitive decline. J Neurosci. 2007;27(22):5967–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120]. Mooney C, Becker BA, Raoof R, Henshall DC. EpimiRBase: A comprehensive database of microRNA-epilepsy associations. Bioinformatics. 2016;32(9):1436–8. [DOI] [PubMed] [Google Scholar]
- [121]. Haenisch S, Zhao Y, Chhibber A, Kaiboriboon K, Do LV, Vogelgesang S, et al. SOX11 identified by target gene evaluation of miRNAs differentially expressed in focal and non-focal brain tissue of therapy-resistant epilepsy patients. Neurobiol Dis. 2015;77:127–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122]. McKiernan RC, Jimenez-Mateos EM, Sano T, Bray I, Stallings RL, Simon RP, et al. Expression profiling the microRNA response to epileptic preconditioning identifies miR-184 as a modulator of seizure-induced neuronal death. Exp Neurol. 2012;237(2):346–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123]. Gorter JA, Iyer A, White I, Colzi A, van Vliet EA, Sisodiya S, et al. Hippocampal subregion-specific microRNA expression during epileptogenesis in experimental temporal lobe epilepsy. Neurobiol Dis. 2014;62:508–20. [DOI] [PubMed] [Google Scholar]
- [124]. Jimenez-Mateos EM, Henshall DC. Epilepsy and microRNA. Neuroscience. 2013;238:218–29. [DOI] [PubMed] [Google Scholar]
- [125]. Kempermann G. The neurogenic reserve hypothesis: What is adult hippocampal neurogenesis good for? Trends Neurosci 2008;31(4):163–9. [DOI] [PubMed] [Google Scholar]
- [126]. Bishop NA, Lu T, Yankner BA. Neural mechanisms of ageing and cognitive decline. Nature. 2010;464(7288):529–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127]. Bizon JL, Gallagher M. Production of new cells in the rat dentate gyrus over the lifespan: Relation to cognitive decline. Eur J Neurosci. 2003;18(1):215–9. [DOI] [PubMed] [Google Scholar]
- [128]. van Praag H, Shubert T, Zhao C, Gage FH. Exercise enhances learning and hippocampal neurogenesis in aged mice. J Neurosci. 2005;25(38):8680–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129]. Song Y-J, Tian X-B, Zhang S, Zhang Y-X, Li X, Li D, et al. Temporal lobe epilepsy induces differential expression of hippocampal miRNAs including let-7e and miR-23a/b. Brain Res. 2011;1387:134–40. [DOI] [PubMed] [Google Scholar]
- [130]. Pichardo-Casas I, Goff LA, Swerdel MR, Athie A, Davila J, Ramos-Brossier M, et al. Expression profiling of synaptic microRNAs from the adult rat brain identifies regional differences and seizure-induced dynamic modulation. Brain Res. 1436:20–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131]. Peng J, Omran A, Ashhab MU, Kong H, Gan N, He F, et al. Expression patterns of miR-124, miR-134, miR-132, and miR-21 in an immature rat model and children with mesial temporal lobe epilepsy. J Mol Neurosci. 2013;50(2):291–7. [DOI] [PubMed] [Google Scholar]
- [132]. Kretschmann A, Danis B, Andonovic L, Abnaof K, van Rikxoort M, Siegel F, et al. Different microRNA profiles in chronic epilepsy versus acute seizure mouse models. J Mol Neurosci. 2015;55(2):466–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133]. Hu K, Xie Y-Y, Zhang C, Ouyang D-S, Long H-Y, Sun D-N, et al. MicroRNA expression profile of the hippocampus in a rat model of temporal lobe epilepsy and miR-34a-targeted neuroprotection against hippocampal neurone cell apoptosis post-status epilepticus. BMC Neurosci. 2012;13:115. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [134]. Sano T, Reynolds JP, Jimenez-Mateos EM, Matsushima S, Taki W, Henshall DC. MicroRNA-34a upregulation during seizure-induced neuronal death. Cell Death Dis. 2012;3:e287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135]. Barca-Mayo O, De Pietri Tonelli D. Convergent microRNA actions coordinate neocortical development. Cell Mol Life Sci. 2014;71(16):2975–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136]. Allerson CR, Sioufi N, Jarres R, Prakash TP, Naik N, Berdeja A, et al. Fully 2‘-Modified Oligonucleotide Duplexes with Improved in Vitro Potency and Stability Compared to Unmodified Small Interfering RNA. J Med Chem. 2005;48(4):901–4. [DOI] [PubMed] [Google Scholar]
- [137]. Elmen J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33(1):439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138]. Mook OR, Baas F, Wissel MB de, Fluiter K. Evaluation of locked nucleic acid-modified small interfering RNA in vitro and in vivo. Am Assoc Cancer Res. 2007;6(3):833–43. [DOI] [PubMed] [Google Scholar]
- [139]. Dykxhoorn DM, Palliser D, Lieberman J. The silent treatment: SiRNAs as small molecule drugs. Gene Ther. 2006;13(6):541–52. [DOI] [PubMed] [Google Scholar]
- [140]. Heidel JD, Hu S, Liu XF, Triche TJ, Davis ME. Lack of interferon response in animals to naked siRNAs. Nat Biotechnol. 2004;22(12):1579–82. [DOI] [PubMed] [Google Scholar]
- [141]. DiFiglia M, Sena-Esteves M, Chase K, Sapp E, Pfister E, Sass M, et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci U S A. 2007;104(43):17204–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142]. Lee S-T, Chu K, Jung K-H, Kim JH, Huh J-Y, Yoon H, et al. miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann Neurol. 2012;72(2):269–77. [DOI] [PubMed] [Google Scholar]
- [143]. Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJA. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–5. [DOI] [PubMed] [Google Scholar]
- [144]. Jimenez-Mateos EM, Engel T, Merino-Serrais P, Fernaud-Espinosa I, Rodriguez-Alvarez N, Reynolds J, et al. Antagomirs targeting microRNA-134 increase hippocampal pyramidal neuron spine volume in vivo and protect against pilocarpine-induced status epilepticus. Brain Struct Funct. 2015;220(4):2387–99. [DOI] [PubMed] [Google Scholar]
- [145]. Jimenez-Mateos EM, Engel T, Merino-Serrais P, McKiernan RC, Tanaka K, Mouri G, et al. Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat Med. 2012;18(7):1087–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146]. Schouten M, Bielefeld P, Fratantoni SA, Hubens CJ, Piersma SR, Pham TV, et al. Multi-omics profile of the mouse dentate gyrus after kainic acid-induced status epilepticus. Sci data. 2016;3:160068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147]. Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, et al. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell. 2015;17(3):360–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148]. Schwartzman O, Tanay A. Single-cell epigenomics: Techniques and emerging applications. Nat Rev Genet. 2015;16(12):716–26. [DOI] [PubMed] [Google Scholar]
- [149]. Ng S-Y, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. 2013;51(3):349–59. [DOI] [PubMed] [Google Scholar]
- [150]. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–8. [DOI] [PubMed] [Google Scholar]
- [151]. Henshall DC. MicroRNA and epilepsy: Profiling, functions and potential clinical applications. Curr Opin Neurol. 2014;27(2):199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152]. Hu G, Drescher KM, Chen X-M. Exosomal miRNAs: Biological Properties and Therapeutic Potential. Front Genet. 2012;3:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153]. Wang J, Tan L, Tan L, Tian Y, Ma J, Tan C-C, et al. Circulating microRNAs are promising novel biomarkers for drug-resistant epilepsy. Sci Re. 2015;5:10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154]. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–9. [DOI] [PubMed] [Google Scholar]
- [155]. Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. [DOI] [PubMed] [Google Scholar]
- [156]. Wang J, Yu J-T, Tan L, Tian Y, Ma J, Tan C-C, et al. Genome-wide circulating microRNA expression profiling indicates biomarkers for epilepsy. Sci Re. 2015;5:9522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157]. Roncon P, Soukupová M, Binaschi A, Falcicchia C, Zucchini S, Ferracin M, et al. MicroRNA profiles in hippocampal granule cells and plasma of rats with pilocarpine-induced epilepsy–comparison with human epileptic samples. Sci Re. 2015;5:14143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158]. Lin JJ, Mula M, Hermann BP. Uncovering the neurobehavioural comorbidities of epilepsy over the lifespan. Lancet. 2012;380(9848):1180–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159]. Hermann B, Seidenberg M, Jones J. The neurobehavioural comorbidities of epilepsy: Can a natural history be developed? Lancet Neurol 2008;7(2):151–60. [DOI] [PubMed] [Google Scholar]
- [160]. Brooks-Kayal AR, Bath KG, Berg AT, Galanopoulou AS, Holmes GL, Jensen FE, et al. Issues related to symptomatic and disease-modifying treatments affecting cognitive and neuropsychiatric comorbidities of epilepsy. Epilepsia. 2013;54(s4):44–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161]. Agostini M, Tucci P, Steinert JR, Shalom-Feuerstein R, Rouleau M, Aberdam D, et al. microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc Natl Acad Sci U S A. 2011. Dec 27;108(52):21099–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162]. Xue Q, Yu C, Wang Y, Liu L, Zhang K, Fang C, et al. miR-9 and miR-124 synergistically affect regulation of dendritic branching via the AKT/GSK3β pathway by targeting Rap2a. Sci Re. 2016;6:26781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163]. Gaughwin P, Ciesla M, Yang H, Lim B, Brundin P. Stage-specific modulation of cortical neuronal development by Mmu-miR-134. Cereb Cortex. 2011;21(8):1857–69. [DOI] [PubMed] [Google Scholar]
- [164]. Kan AA, van Erp S, Derijck AAHA, de Wit M, Hessel EVS, O’Duibhir E, et al. Genome-wide microRNA profiling of human temporal lobe epilepsy identifies modulators of the immune response. Cell Mol Life Sci. 2012;69(18):3127–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165]. Li M-M, Jiang T, Sun Z, Zhang Q, Tan C-C, Yu J-T, et al. Genome-wide microRNA expression profiles in hippocampus of rats with chronic temporal lobe epilepsy. Sci Re. 2014;4:4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166]. Alsharafi W, Xiao B. Dynamic Expression of MicroRNAs (183, 135a, 125b, 128, 30c and 27a) in the Rat Pilocarpine Model and Temporal Lobe Epilepsy Patients. CNS Neurol Disord Drug Targets. 2015;14(8):1096–102. [DOI] [PubMed] [Google Scholar]
- [167]. Jimenez-Mateos EM, Bray I, Sanz-Rodriguez A, Engel T, McKiernan RC, Mouri G, et al. miRNA Expression profile after status epilepticus and hippocampal neuroprotection by targeting miR-132. Am J Pathol. 2011;179(5):2519–32. [DOI] [PMC free article] [PubMed] [Google Scholar]