Abstract
Trastuzumab, the first targeted antibody against human epidermal growth factor receptor 2 (HER2), has been used to treat gastric cancer patients with HER2 overexpression. However, trastuzumab resistance often occurs following an initial period of benefits, and the underlying mechanisms remain largely unclear. The present study revealed that collagen type IV α1 chain (COL4A1), whose expression is upregulated in gastric cancer tissues and trastuzumab-resistant gastric cancer cells, may potentially confer trastuzumab resistance in gastric cancer. By performing bioinformatics analysis of 2 microarray datasets, the present study initially identified COL4A1, overexpressed in gastric cancer tissues and trastuzumab-resistant gastric cancer cells, as a potential candidate for inducing trastuzumab resistance. The drug resistance function of COL4A1 in gastric cancer was then validated by performing protein/gene interactions and biological process annotation analyses, and further validated by analyzing the functionality of microRNAs that target COL4A1 mRNA. Collectively, these data indicated that COL4A1 may confer trastuzumab resistance in gastric cancer.
Keywords: bioinformatics analysis, gastric cancer, trastuzumab resistance, differentially expressed genes, collagen type IV α1 chain
Introduction
Gastric cancer is the third most frequently diagnosed cancer type, and has been the leading cause of cancer-related death in less-developed countries (1). Although advances in therapeutic strategies such as surgery and systemic chemotherapy, have improved the clinical outcomes for gastric cancer patients, the prognosis of these patients remains poor because of frequent cancer recurrence (2). Nevertheless, due to the encouraging development in newly targeted therapies, such a disappointing clinical condition has been improving. Trastuzumab, the first targeted antibody against human epidermal growth factor receptor 2 (HER2), has been approved for the treatment of patients with HER2-positive metastatic gastric cancer (3). However, even with trastuzumab resistance, gastric cancer almost inevitably progresses, as the tumors become resistant to trastuzumab after an initial period of clinical benefits (4). Mechanisms leading to trastuzumab resistance in breast cancer, such as cross-talk between HER2 and other intracellular kinase receptors, have been described recently (5), however, the underlying mechanisms of trastuzumab resistance in gastric cancer remain largely unknown.
Cancer cells are able to activate alternative survival pathways yielding drug resistance in response to chemotherapy, and thereby leads to the chemotherapeutic treatment failure (6). Since drug resistance is a common cause limiting the efficacy of cancer treatment, several mechanisms are elucidated to be responsible for the resistance development, such as increased rates of drug efflux, apoptosis resistance, and microRNAs (miRNAs) mediated overexpression of many drug resistance-related genes (7). Microarray technology, a high-throughput platform that analyzes gene expression, in combination with bioinformatics analysis has been widely used as a promising tool to acquire gene signature during tumorigenesis or drug resistance, and identify prognostic biomarkers in cancer patients (8–10). Abnormal expression patterns of drug resistance-related genes commonly play important roles in drug resistance (11), thus, exploring and identifying the critical drug resistance-related genes based on microarray analysis would have a significant impact.
Therefore, in the present study, we sought to identify the potential genes that promote trastuzumab resistance in gastric cancer through retrieving microarray data from public databases and comprehensive bioinformatics analysis.
Materials and methods
Microarray data
The gene expression profiles of GSE26899, GSE77346, GSE54129, and GSE65801 were obtained from the Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo). In detail, GSE26899 dataset is consisted of 96 clinical gastric tumor tissues and 12 adjacent normal tissues; GSE77346 dataset is consisted of 1 trastuzumab-sensitive cell line and 4 trastuzumab-resistant cell lines (12); GSE54129 includes 111 human gastric cancer tissues and 21 non-cancerous tissues; GSE65801 contains 32 gastric cancer tissues and 32 paired non-cancerous tissues (13).
Processing of microarray data
The raw microarray data files of the datasets downloaded from the GEO website were subsequently analyzed via using the GEO2R (www.ncbi.nlm.nih.gov/geo/geo2r/), an online tool comparing two or more groups of samples in the same experimental setting (14). False Discovery Rate (FDR) of P-value adjusted (adj. P) to 0.05 and |logFC|>1 were set as the cut-off criteria.
Functional and pathway enrichment analyses
Gene ontology (GO) analysis is a commonly used approach for functional studies with three ontologies including biological process, molecular function, and cellular component (15), while Kyoto Encyclopedia of Genes and Genomes (KEGG) is a knowledge base for the systematic study of gene functions (16). To study the functional annotations of differentially expressed genes (DEGs), we next employed Database for Annotation, Visualization and Integrated Discovery (DAVID, david.abcc.ncifcrf.gov/,) to process the GO and KEGG analyses of DEGs identified in gastric cancer samples. P<0.05 was set as the threshold.
Protein-protein interaction (PPI) network construction and module analysis
The Search Tool for the Retrieval of Interacting Genes (STRING), an online database (string-db.org) designed to evaluate PPI information, covers 9,643,763 proteins from more than 2,000 organisms, which was used to construct the PPI. To evaluate the interactive associations of DEGs identified from GSE26899, we mapped these DEGs to the STRING (version 10.5) database. Confidence score >0.4 was selected as significant. PPI networks were constructed by STRING and visualized by Cytoscape. Subsequently, the plug-in Molecular Complex Detection (MCODE) was employed to screen the modules of PPI networks in Cytoscape with the threshold set as follows: MCODE scores >10.
Survival analysis of collagen type IV α1 chain (COL4A1)
To evaluate the association between COL4A1 level and its clinical outcomes, Kaplan-Meier plotter (KM plotter; www.kmplot.com), an online survival analysis tool, was performed. KM plotter is capable of assessing the effect of 54,675 genes on overall survival via using 10,188 cancer samples including 4,142 breast, 1,648 ovarian, 2,437 lung, and 1,065 gastric cancer patients (17). Patients with gastric cancer were separated into high- and low-expression groups according to the level of COL4A1, and the overall survival was then analyzed. The hazard ratio (HR) with 95% confidence intervals and log rank P-value were calculated.
Analysis of COL4A1 by geneMANIA and coremine
GeneMANIA, an online tool (www.genemania.org/), can be used to generate hypotheses of gene function, analyze gene lists, and prioritize genes for functional assays (18). After selecting Homo sapiens from the nine optional organisms, COL4A1 was entered into the search bar and the results were then collected. Annotation of biological processes involving COL4A1 was performed by consulting the Coremine Medical online database (www.coremine.com/medical/).
Prediction of miRNAs
To predict the miRNAs targeting the mRNA of COL4A1, miRWalk (version 2.0, zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/), an online platform supplying information about predicted and experimentally validated miRNA-target interactions, was then employed (19). Herein, nine prediction programs (miRWalk, miRanda, miRDB, miRNAMap, Pictar2, PITA, RNA22, RNAhybrid and Targetscan) were selected. These predicted miRNAs were then overlapped by at least seven programs, and selected for further analysis. Pathway enrichment analysis of these miRNAs was performed by using the DIANA-mirPath web server (snf-515788.vm.okeanos.grnet.gr/index.php?r=mirpath) (20).
Statistical analysis
SPSS 22.0 software (IBM Corp., Armonk, NY, USA) was used to analyze data. Two tailed Student's t-test was used to compare the two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Module acquisition based on the DEGs identified in gastric cancer tissues
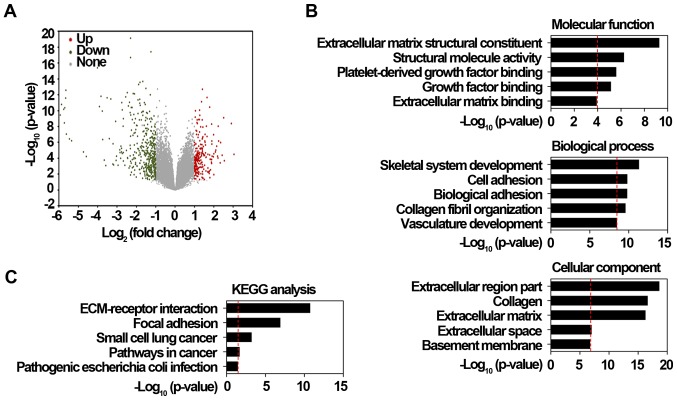
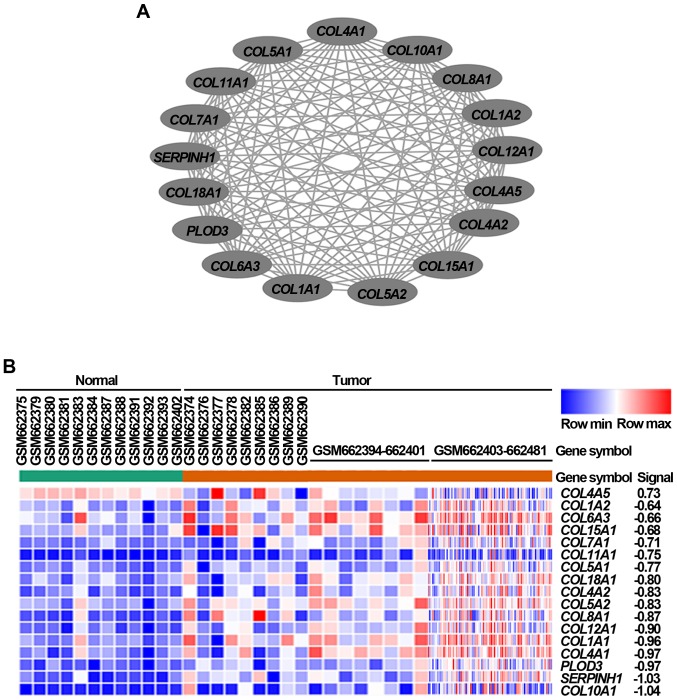
Drug resistance-related genes identified from in vitro drug-induced resistant models may simply represent transcriptional changes, thus, complementary agents targeting these genes are usually failure to translate into clinical practice (21). Because drug resistance acquisition can arise before the malignant transformation stage (22), the genes playing important roles in tumorigenesis and drug resistance is thereby more likely to be critical for resistance occurrence. Therefore, we first identified 509 DEGs in gastric cancer tissues from the GSE26899 dataset using a 2-fold-change and adj. P<0.05 as the threshold cutoff. Among these DEGs, the expression of these 172 genes was significantly upregulated, while that of 337 genes was significantly downregulated in cancer tissues (Fig. 1A). To explore the potential roles of these significantly upregulated DEGs, GO and KEGG pathway analyses, which can provide valuable insights regarding protein function, were performed (23). As shown in Fig. 1B, GO results showed that these significantly upregulated DEGs were largely associated with extracellular region part, collagen, extracellular matrix, and cell or biological adhesion processes. KEGG results also consistently revealed that these DEGs were highly enriched in extracellular matrix-receptor interaction and focal adhesion pathways (Fig. 1C). Based on further DEGs analysis with the STRING database, the PPI network of these DEGs, which contained 505 nodes and 1,207 edges, was subsequently constructed. Using the MCODE plug-in in Cytoscape, we obtained the module with the highest score (Fig. 2A), and also performed a cluster analysis of these genes in the module (Fig. 2B). The module with the highest score was selected based on these DEGs identified from gastric cancer tissues via bioinformatics methods.
Figure 1.
Identification of DEGs from GSE26899 dataset. (A) Volcano plot of DEGs between gastric cancer tissues and surrounding normal tissues. Red dots represent significantly upregulated DEGs in gastric cancer tissues; green dots represent significantly downregulated DEGs in gastric cancer tissues; gray dots represent no significant difference. P<0.05 and fold-change >2 were regarded as significant. (B) GO analysis of significantly upregulated DEGs in gastric cancer tissues. Top 5 enriched GO categories under ‘biological process’, ‘cellular component’ and ‘molecular function’ were indicated. (C) KEGG pathway enrichment analysis of significantly upregulated DEGs in gastric cancer tissues. Top 5 enriched pathways were indicated. DEGs, differentially expressed genes; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; ECM, extracellular matrix.
Figure 2.
Module with the highest score, obtained based on the DEGs identified from GSE26899 dataset. (A) Module with the highest score generated from the protein-protein interaction network of significantly upregulated DEGs in gastric cancer tissues. (B) Heat map of the 17 genes from the selected module. Red, upregulation; Blue, downregulation; DEGs, differentially expressed genes; COL, collagen; PLOD3, procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3; SERPINH1, serpin family H member 1.
Expression of COL4A1, overexpressed in gastric cancer tissues, is also upregulated in trastuzumab-resistant gastric cancer cells
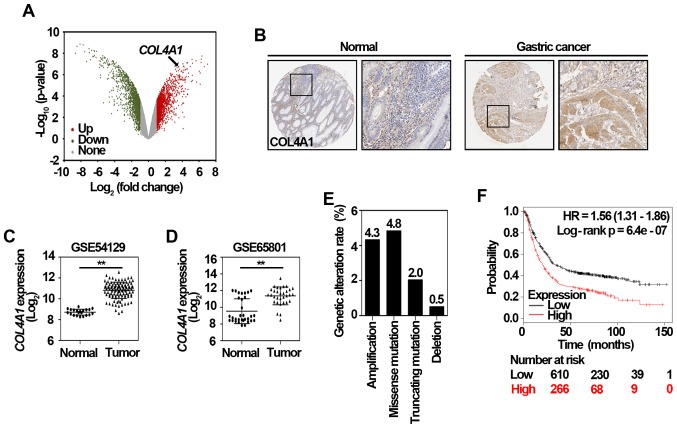
To further explore the potential genes contributing to trastuzumab resistance, we retrieved GSE77346 data, a microarray dataset consisted of one trastuzumab-sensitive gastric cancer cell line and four trastuzumab-resistant gastric cancer cell lines (12). After screening the significantly upregulated DEGs in trastuzumab-resistant cancer cells, COL4A1, one of hub genes in the selected module (Fig. 2A), was also found to be significantly upregulated in trastuzumab-resistant cells (Fig. 3A), suggesting COL4A1 might be important in both tumorigenesis and trastuzumab resistance. Since the gene expression is not always consistent with its protein amount (24), further validation of COL4A1 protein level in clinical gastric cancer tissues is quite necessary. By employing the Human Protein Atlas database, an online tool analyzing protein level from clinical specimens, we observed that COL4A1 was positively expressed in gastric cancer tissue, but negatively expressed in normal gastric tissue (Fig. 3B). Two other datasets, including GSE54129 and GSE65801 were also used to validate the mRNA level of COL4A1, which was indeed significantly upregulated in clinical gastric cancer samples (Fig. 3C and D). To investigate potential regulation mechanisms of COL4A1 in gastric cancer, the genomic alteration of COL4A1 in The Cancer Genome Atlas (TCGA) cohort was analyzed using the cBioPortal (www.cbioportal.org/) (25). As shown in Fig. 3E, the amplification, missense mutation, truncating mutation, and deletion of COL4A1 accounted for 4.3, 4.8, 2.0, and 0.5% of stomach adenocarcinoma cases, respectively. The prognostic value of COL4A1 in patients with gastric cancer was analyzed using the KM plotter according to the low and high expression of COL4A1. As shown in Fig. 3F, the high mRNA level of COL4A1 [HR 1.56 (1.31–1.86)] was associated with poor overall survival of gastric cancer patients. Altogether, these data suggest that COL4A1 is a potential gene candidate that promotes the development of gastric cancer and subsequent trastuzumab resistance.
Figure 3.
COL4A1 in the selected module is significantly upregulated in trastuzumab resistant gastric cancer cells. (A) Volcano plot of DEGs between trastuzumab-resistant and sensitive gastric cancer cells. Red dots represent significantly upregulated DEGs in trastuzumab-resistant gastric cancer cells; green dots represent significantly downregulated DEGs in trastuzumab-sensitive gastric cancer cells; and gray dots represent no significant difference. P<0.05 and fold-change >2 were regarded as significant. (B) The expression of COL4A1 in normal gastric tissue and gastric cancer tissue, respectively. Representative immunohistochemistry staining results were obtained from the Human Protein Atlas online database (magnification, ×40). The relative mRNA level of COL4A1 was validated by two other datasets including (C) GSE54129 and (D) GSE65801. The values are expressed as the mean ± standard deviation. **P<0.01, as indicated. (E) Proportion of genetic alterations of COL4A1 retrieved from the cBioportal (n=393; www.cbioportal.org/). (F) Kaplan-Meier survival analysis of COL4A1 in gastric cancer patients that was obtained from www.kmplot.com. HR, hazard ratio; DEGs, differentially expressed genes; COL4A1, collagen type IV α1.
Drug resistance function of COL4A1 is validated by protein/gene interactions and biological process annotation analyses
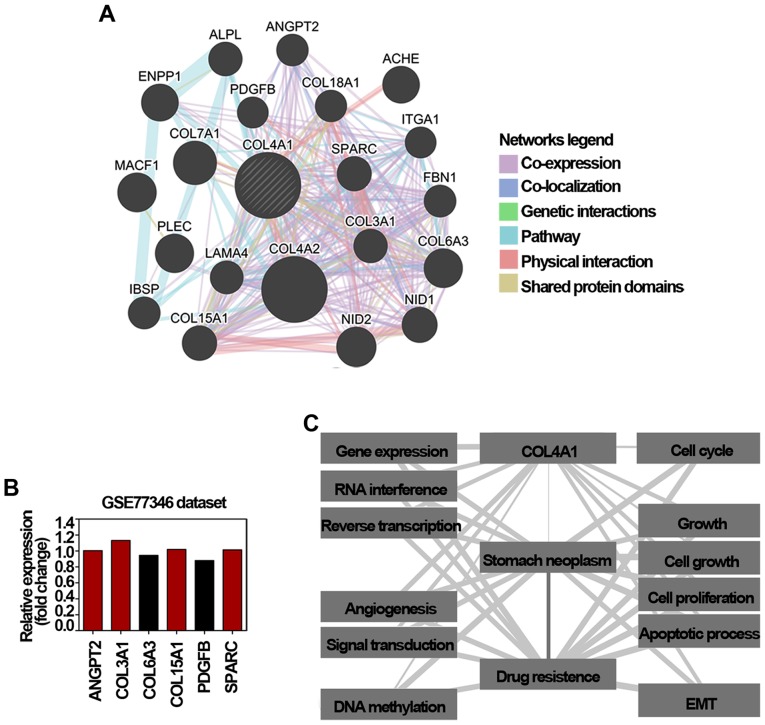
As a user-friendly web interface for functional prediction of genes, GeneMANIA has been widely used as an effective tool to predict and explore drug resistance-related genes (18). Accordingly, COL4A1 showed interactions with 20 proteins/genes; among these 6 genes were involved in conferring drug resistance in cancer (Fig. 4A). The mRNA expression of these 6 genes in gastric cancer cells and trastuzumab-resistant gastric cancer cells were also evaluated using the microarray data of GSE77346. As shown in Fig. 4B, the mRNA level of angiopoietin 2 (ANGPT2), COL3A1, COL15A1, and secreted protein acidic and cysteine rich (SPARC) were upregulated in trastuzumab-resistant gastric cancer cells. Among these genes, COL4A1 co-expressed and co-localized with ANGPT2, collagen type VI α 3 chain (COL6A3), and SPARC, respectively. It has been reported bevacizumab resistance in glioblastoma and doxorubin resistance in liver cancer are largely mediated by an enforcement of ANGPT2/TIE2 signaling (26,27). Additionally, overexpression of COL6A3, one of the most highly upregulated genes in oxaliplatin and cisplatin resistant ovarian cancer cells, has also been recognized to confer cisplatin resistance in sensitive cancer cells (28). Moreover, the accumulation of intracellular SPARC can drive imatinib resistance in chronic myelogenous leukemia cells (29), and also modulate cisplatin resistance via modulating the Let-7f-1 miRNA/HMGB1 signaling in medulloblastoma cells (30). COL3A1 and COL15A1, two types of fibrillar collagen highly expressed in chemotherapeutic drugs resistant ovarian cancer cells (31), were also exhibited to co-express, share protein domains, and co-localize with COL4A1 (Fig. 4A). Additionally, COL4A1 also was co-expressed, shared pathways, and had physical interactions with platelet derived growth factor subunit B (PDGFB). Akt/PDGF-B signaling can regulate Akt, and thus confer the hypoxia-induced cisplatin resistance in liver cancer cells (32). PDGFB may contribute to the resistant phenotype and sustain signaling through MAPK and Akt in breast cancer cells (33). The Coremine Medical database is a freely available online tool for obtaining information on health, medicine, and biology (33); thus, we used this database to annotate the biological process of COL4A1. As shown in Fig. 4C, 12 biological processes significantly associated with COL4A1, gastric cancer, and drug resistance (P<0.01) were annotated. Considering that the close relationships of COL4A1 with these processes and the close relationships of the 12 processes with gastric cancer and drug resistance, COL4A1 might be involved in the development of drug resistance in gastric cancer via its effect on these biological processes. In detail, cell growth related biological processes (including 4 cell growth, cell proliferation, and apoptotic process), gene expression regulation-related (including gene expression, RNA interference, and reverse transcription), and especially, the epithelial-mesenchymal transition (EMT) biological process were identified to be closely associated with the development of COL4A1 in the gastric cancer drug resistance. Therefore, these results suggest that COL4A1 may confer drug resistance in gastric cancer via regulating cell growth, gene expression, and in particular, epithelial mesenchymal transition (EMT) processes.
Figure 4.
Validation of drug resistance function of COL4A1. (A) Protein/gene-protein/gene interaction network of COL4A1 generated using the GeneMANIA online tool. The network legends refer to the interaction types between proteins/genes. The interaction types between proteins/genes were illustrated as indicated by the network legend. (B) Relative mRNA level of 6 genes interacting with COL4A1 based on the microarray data of GSE77346. Red column, relative expression >1; Black column, relative expression <1. (C) Annotation of biological processes of COL4A1 with gastric cancer and drug resistance using the Coremine Medical online tool. DEGs, differentially expressed genes; COL4A1, collagen type IV α1.
Drug resistance function of COL4A1 is further validated by analyzing functionality of miRNAs that target COL4A1 mRNA
Post-transcriptional gene expression regulated by miRNAs is important for multiple cellular processes during development and pathogenesis (34). Amplification and overexpression of tumor-promoting miRNAs or genetic loss of tumor-suppressing miRNAs are tightly correlated with the development of cancer and chemotherapeutic resistance (35). Therefore, studying target genes of miRNAs is a focus of interest due to the diagnostic and therapeutic relevance, and the gene functions can also be predicted according to functionality of these miRNAs targeting the gene (36). To obtain the miRNAs targeting COL4A1 mRNA, the miRNA-mRNA interaction analysis was performed by using the miRWalk. We identified 553 miRNAs predicted to transcriptionally target COL4A1 mRNA, indicating the regulation of COL4A1 by miRNAs. These miRNAs predicted by at least 8 of 9 prediction tools were then submitted to pathway enrichment using DIANA miRPath (20). As shown in Table I, the top 5 highly associated pathway were ECM-receptor interaction, TGF-β signaling pathway, viral carcinogenesis, proteoglycans in cancer, and Hippo signaling pathway, which are almost reported to be related with drug resistance (37–41). It has been established TGFβ signaling can activate autophagy process, and thereby lead to the oxaliplatin resistance in colorectal cancer (38). In addition, the activation of Hippo signaling also contributes to the drug resistance and cancer relapse (42). YAP, an effector of hippo signaling, has also been reported to alter clinical response of EGFR-tyrosine kinase inhibitors in lung cancer patients (43). Furthermore, it has also been reported that inactivation of Hippo pathway can restore gemcitabine sensitivity among a variety of cancers (44). In the present study, as shown in Table II, 9 of top 10 miRNAs that target COL4A1 mRNA were tightly associated with drug resistance in cancers (45–51). For instance, loss of intracellular miR-29b promotes cisplatin resistance in gastric cancer. Consistently, ectopic overexpression of miR-29b in cholangiocarcinoma cells can confer gemcitabine sensitivity to HuH28 cells (45). MiR-506 overexpression can confer hydroxycamptothecin resistance in colon cancer cells by inhibiting PPARα expression (50). Collectively, these data provide further supports for the drug resistance function of COL4A1 in gastric cancer.
Table I.
Top 8 enriched pathways regulated by microRNAs that target collagen type IV α1 mRNA and their associations with drug resistance in gastric cancer.
| Author, year | miRNAs (hsa-miR-) | KEGG pathway | P-value | Regulation of drug resistance in cancers | (Refs.) |
|---|---|---|---|---|---|
| Wu et al, 2017 | 29b-3p, 124-3p, 148a-3p, 29a-3p, 152-3p, 148b-3p, 506-3p, 628-5p, 29c-3p, 767-5p, 637, 33a-5p, 33b-5p, 203a, 374a-5p, 300, 106a-5p, 98-5p, 381-3p, let-7b-5p, 7c-5p, 7b-3p, 7a-5p | ECM-receptor interaction | 1.14×10‒15 | Yes | (37) |
| Sun et al, 2017 | TGF-β signaling pathway | 6.58×10‒11 | Yes | (38) | |
| – | Viral carcinogenesis | 9.72×10‒11 | None | – | |
| Lanzi et al, 2017 | Proteoglycans in cancer | 3.94×10‒10 | Yes | (39) | |
| Gujral et al, 2017 | Hippo signaling pathway | 6.66×10‒10 | Yes | (44) | |
| Lee et al, 2015 | Cell cycle | 1.01×10‒9 | Yes | (40) | |
| Wallerand et al, 2010 | Adherens junction | 2.99×10‒8 | Yes | (41) | |
| – | Pathways in cancer | 3.46×10‒8 | Cancer pathway | – |
The miRNAs predicted by at least 7 of the 9 prediction tools were submitted to DIANA-miRPath version 3.0 to perform KEGG analysis. miRNA/miR, microRNA; KEGG, Kyoto Encyclopedia of Genes and Genomes; ECM, extracellular matrix; TGF, transforming growth factor.
Table II.
Top 11 microRNAs targeting collagen type IV α1 mRNA predicted by microRNA-mRNA interactions, and their drug resistance-associated functions in cancer.
| miRNA-mRNA prediction tools | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author, year | miRNA (hsa-) | A | B | C | D | E | F | G | H | I | Drug resistance-related functions of miRNAs in cancers | (Refs.) |
| Okamoto et al, 2013 | miR-29b-3p | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Drug resistance-related | (45) |
| Liu et al, 2016 | miR-124-3p | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Drug resistance-related | (46) |
| Chen et al, 2017 | miR-148a-3p | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Drug resistance-related | (47) |
| Zhong et al, 2013 | miR-29a-3p | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Drug resistance-related | (48) |
| Chen et al, 2017 | miR-152-3p | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Drug resistance-related | (47) |
| Sui et al, 2015 | miR-148b-3p | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Drug resistance-related | (49) |
| Tong et al, 2011 | miR-506-3p | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | Drug resistance-related | (50) |
| – | miR-628-5p | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | Drug resistance-related | – |
| Zhang et al, 2013 | miR-29c-3p | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | Drug resistance-related | (51) |
| – | miR-767-5p | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Drug resistance-related | – |
| – | miR-637 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Drug resistance-related | – |
A, miRWalk; B, miRanda; C, miRDB; D, miRNAMap; E, Pictar2; F, PITA; G, RNA22; H, RNAhybrid; I, TargetScan; miRNA/miR, microRNA.
Discussion
In the present study, based on the bioinformatics analysis of two microarray datasets including GSE26899 and GSE77346, COL4A1, which was overexpressed in gastric cancer tissues and trastuzumab-resistant gastric cancer cells, was identified as a potential gene that promotes gastric cancer and trastuzumab resistance. By combining the protein/gene interactions, biological process annotation, and miRNAs-mRNA interaction analyses, we showed that COL4A1 may confer trastuzumab resistance in gastric cancer.
Traditional chemotherapy and newly targeted therapy are two important methods of cancer treatment; however, the clinical efficacy of both is largely limited due to the occurrence of subsequent drug resistance (52). Mechanisms underlying drug resistance, such as alteration of drug targets or metabolism and genetic mutation (11) have been elucidated. Similar to that in breast cancer, the HER2-positivity rate in gastric cancer can range from 20 to 42% (53). However, although the benefits of trastuzumab against HER2-positive gastric cancer have been formally established (3,53), trastuzumab resistance is almost inevitable and eventually leads to the therapeutic failure (4). Interfering with the combination of trastuzumab and HER2 (54) and constitutive dimerization of the HER2 receptor (55) have been recognized as causes of trastuzumab resistance. Recently, abnormal expression of tumorigenesis or drug resistance-related genes, such as phosphatidylinositol 3-kinase/Akt signaling (56) and insulin like growth factor 1 receptor (57), has been also identified as the important mechanisms causing trastuzumab resistance. However, a comprehensive study of specific molecular mutation underlying trastuzumab resistance is still needed. Drug resistance-related genes identified from induced drug-resistant cancer cells may simply be a result of transcriptional changes that may be irrelevant to resistance mechanisms (58). Thus, targeting these genes usually fail to translate into clinical practice (21). It has been recently shown that the ability to acquire drug resistance can arise even before the malignant transformation stage. For instance, overexpression of telomerase or inactivation of p53 can contribute to the drug-resistant phenotype in pre-tumorigenic models (22). Insulin-like growth factor signaling has also been deemed a factor that promotes both the development of various tumors and their resistance to chemotherapy (59,60). This evidence thus reminds us that the genes playing a key role in both tumor development and subsequent drug resistance likely produce the essential molecule for drug resistance. Herein, through retrieving microarray datasets and employing subsequent bioinformatics analysis, COL4A1 was validated as an important gene that drives trastuzumab resistance in gastric cancer.
The heterotrimers formed by COL4A1 and COL4A2 presents in almost all the basement membranes, which are a specialized form of the extracellular matrix. Besides that the basement membranes can mediate tissue compartmentalization and transfer environmental signals to epithelial cells (61), it is also an important structural and functional component of blood vessels. Accordingly, mutations in COL4A1 are pleiotropic and contribute to many diseases, such as myopathy, hemorrhagic stroke, and tumor progression (62). Upregulated COL4A1 produced by bladder cancer cells plays pivotal roles in tumor invasion via induction of tumor budding, while overexpression of COL4A1 also contributes to breast cancer cells proliferation, which indicates targeting COL4A1 can be an attractive approach for cancer treatment (63,64). Besides, COL4A1 has also been identified as one of biomarkers for prognosis of intrahepatic cholangiocarcinoma (65). Given that an interaction between PDGFB and HIF-1α and an interplay between SPARC, BCL-2, and caspase-8 have been recognized to augment chemotherapy-induced apoptosis, and thereby inducing resistance (32,66). Herein, our results suggest COL4A1 can drive trastuzumab resistance in gastric cancer via multiple mechanisms, such as cell proliferation and miRNAs-mediated post-transcriptional modification. Consistent with our results, a previous study also reported the sustained EMT phenotype induced by prolonged trastuzumab treatment may lead to trastuzumab resistance in gastric cancer cells (67). Small molecules that targeting drug resistance-related genes have shown promising clinical efficacy in cancer treatment. For instance, erbB3 overexpression drives paclitaxel resistance in breast cancer, MM-121/SAR256212, an erbB3-targeted antibody, was thus designed and shows augmented effect on paclitaxel resistance (68). Activation of the PI3K pathway frequently occurs in cancers and leads to drug resistance, but clinical benefits of PI3K inhibitors have been modest to date. Fortunately, LEE011, a specific CDK 4/6 inhibitor currently under clinical development, has shown promising effects against PI3K inhibition resistance (69). HER2-positive patients receiving trastuzumab may inevitably develop resistance due to excessive activation of the PI3K/AKT pathway. A phase 1 clinical trial has been performed to evaluate the efficacy of the combination of an allosteric AKT inhibitor (MK-2206) and trastuzumab in patients with HER2-positive solid tumors; interestingly, MK-2206 is safe, and reversed the trastuzumab resistance in HER2-overexpressing patients (70). Collectively, this evidence suggests that identification of drug-resistant genes through bioinformatics methods and subsequent design of small molecule drugs may have great potential.
In conclusion, our results show that COL4A1 may confer trastuzumab resistance in gastric cancer via multiple mechanisms based on bioinformatics analysis. However, further investigations elucidating the drug resistance function of COL4A1 in trastuzumab-resistant gastric cancer models are necessary.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LG conceived the project and designed the research plan. RH and WG performed the experiments and analyzed the data with input from BS who assisted in experimental design and data analyses. RH and LG wrote the manuscript, and all authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Corso S, Giordano S. How can gastric cancer molecular profiling guide future therapies? Trends Mol Med. 2016;22:534–544. doi: 10.1016/j.molmed.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 4.Aprile G, Giampieri R, Bonotto M, Bittoni A, Ongaro E, Cardellino GG, Graziano F, Giuliani F, Fasola G, Cascinu S, Scartozzi M. The challenge of targeted therapies for gastric cancer patients: The beginning of a long journey. Exp Opin Investig Drugs. 2014;23:925–942. doi: 10.1517/13543784.2014.912631. [DOI] [PubMed] [Google Scholar]
- 5.Ritter CA, Perez-Torres M, Rinehart C, Guix M, Dugger T, Engelman JA, Arteaga CL. Human breast cancer cells selected for resistance to trastuzumab in vivo overexpress epidermal growth factor receptor and ErbB ligands and remain dependent on the ErbB receptor network. Clin Cancer Res. 2007;13:4909–4919. doi: 10.1158/1078-0432.CCR-07-0701. [DOI] [PubMed] [Google Scholar]
- 6.Gottesman MM, Lavi O, Hall MD, Gillet JP. Toward a better understanding of the complexity of cancer drug resistance. Annu Rev Pharmacol Toxicol. 2016;56:85–102. doi: 10.1146/annurev-pharmtox-010715-103111. [DOI] [PubMed] [Google Scholar]
- 7.Garofalo M, Croce CM. MicroRNAs as therapeutic targets in chemoresistance. Drug Resist Updat. 2013;16:47–59. doi: 10.1016/j.drup.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu Y, Lemon W, Liu PY, Yi Y, Morrison C, Yang P, Sun Z, Szoke J, Gerald WL, Watson M, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med. 2006;3:e467. doi: 10.1371/journal.pmed.0030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen F, Xiang CX, Zhou Y, Ao XS, Zhou DQ, Peng P, Zhang HQ, Liu HD, Huang X. Gene expression profile for predicting survival of patients with meningioma. Int J Oncol. 2015;46:791–797. doi: 10.3892/ijo.2014.2779. [DOI] [PubMed] [Google Scholar]
- 10.Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5:588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 11.Yin F, Liu X, Li D, Wang Q, Zhang W, Li L. Tumor suppressor genes associated with drug resistance in ovarian cancer (Review) Oncol Rep. 2013;30:3–10. doi: 10.3892/or.2013.2446. [DOI] [PubMed] [Google Scholar]
- 12.Piro G, Carbone C, Cataldo I, Di Nicolantonio F, Giacopuzzi S, Aprile G, Simionato F, Boschi F, Zanotto M, Mina MM, et al. An FGFR3 autocrine loop sustains acquired resistance to trastuzumab in gastric cancer patients. Clin Cancer Res. 2016;22:6164–6175. doi: 10.1158/1078-0432.CCR-16-0178. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Yu B, Li J, Su L, Yan M, Zhang J, Li C, Zhu Z, Liu B. Characterization of differentially expressed genes involved in pathways associated with gastric cancer. PLoS One. 2015;10:e0125013. doi: 10.1371/journal.pone.0125013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hulsegge I, Kommadath A, Smits MA. Globaltest and GOEAST: Two different approaches for Gene Ontology analysis; BMC Proc; 2009; p. S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gyorffy B, Lánczky A, Szállási Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 18.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dweep H, Gretz N. miRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 20.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borst P, Wessels L. Do predictive signatures really predict response to cancer chemotherapy? Cell cycle (Georgetown, Tex.) 2010;9:4836–4840. doi: 10.4161/cc.9.24.14326. [DOI] [PubMed] [Google Scholar]
- 22.Yague E, Arance A, Kubitza L, O'Hare M, Jat P, Ogilvie CM, Hart IR, Higgins CF, Raguz S. Ability to acquire drug resistance arises early during the tumorigenesis process. Cancer Res. 2007;67:1130–1137. doi: 10.1158/0008-5472.CAN-06-2574. [DOI] [PubMed] [Google Scholar]
- 23.Xing Z, Chu C, Chen L, Kong X. The use of Gene Ontology terms and KEGG pathways for analysis and prediction of oncogenes. Biochim Biophys Acta. 2016;1860:2725–2734. doi: 10.1016/j.bbagen.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 24.Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 25.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Labussiere M, Cheneau C, Prahst C, Gállego Pérez-Larraya J, Farina P, Lombardi G, Mokhtari K, Rahimian A, Delattre JY, Eichmann A, Sanson M. Angiopoietin-2 May be involved in the resistance to bevacizumab in recurrent glioblastoma. Cancer Invest. 2016;34:39–44. doi: 10.3109/07357907.2015.1088948. [DOI] [PubMed] [Google Scholar]
- 27.Li T, Liu Z, Jiang K, Ruan Q. Angiopoietin2 enhances doxorubin resistance in HepG2 cells by upregulating survivin and Ref-1 via MSK1 activation. Cancer Lett. 2013;337:276–284. doi: 10.1016/j.canlet.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 28.Sherman-Baust CA, Weeraratna AT, Rangel LB, Pizer ES, Cho KR, Schwartz DR, Shock T, Morin PJ. Remodeling of the extracellular matrix through overexpression of collagen VI contributes to cisplatin resistance in ovarian cancer cells. Cancer Cell. 2003;3:377–386. doi: 10.1016/S1535-6108(03)00058-8. [DOI] [PubMed] [Google Scholar]
- 29.Fenouille N, Puissant A, Dufies M, Robert G, Jacquel A, Ohanna M, Deckert M, Pasquet JM, Mahon FX, Cassuto JP, et al. Persistent activation of the Fyn/ERK kinase signaling axis mediates imatinib resistance in chronic myelogenous leukemia cells through upregulation of intracellular SPARC. Cancer Res. 2010;70:9659–9670. doi: 10.1158/0008-5472.CAN-10-2034. [DOI] [PubMed] [Google Scholar]
- 30.Pannuru P, Dontula R, Khan AA, Herbert E, Ozer H, Chetty C, Lakka SS. miR-let-7f-1 regulates SPARC mediated cisplatin resistance in medulloblastoma cells. Cell Signal. 2014;26:2193–2201. doi: 10.1016/j.cellsig.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 31.Januchowski R, Zawierucha P, Rucinski M, Zabel M. Microarray-based detection and expression analysis of extracellular matrix proteins in drugresistant ovarian cancer cell lines. Oncol Rep. 2014;32:1981–1990. doi: 10.3892/or.2014.3468. [DOI] [PubMed] [Google Scholar]
- 32.Lau CK, Yang ZF, Ho DW, Ng MN, Yeoh GC, Poon RT, Fan ST. An Akt/hypoxia-inducible factor-1alpha/platelet-derived growth factor-BB autocrine loop mediates hypoxia-induced chemoresistance in liver cancer cells and tumorigenic hepatic progenitor cells. Clin Cancer Res. 2009;15:3462–3471. doi: 10.1158/1078-0432.CCR-08-2127. [DOI] [PubMed] [Google Scholar]
- 33.de Leeuw N, Dijkhuizen T, Hehir-Kwa JY, Carter NP, Feuk L, Firth HV, Kuhn RM, Ledbetter DH, Martin CL, van Ravenswaaij-Arts CM, et al. Diagnostic interpretation of array data using public databases and internet sources. Hum Mutat. 2012;33:930–940. doi: 10.1002/humu.22049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 35.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blenkiron C, Miska EA. miRNAs in cancer: Approaches, aetiology, diagnostics and therapy. Hum Mol Genet. 2007;16:R106–R113. doi: 10.1093/hmg/ddm056. [DOI] [PubMed] [Google Scholar]
- 37.Wu X, Wang H, Lian Y, Chen L, Gu L, Wang J, Huang Y, Deng M, Gao Z, Huang Y. GTSE1 promotes cell migration and invasion by regulating EMT in hepatocellular carcinoma and is associated with poor prognosis. Sci Rep. 2017;7:5129. doi: 10.1038/s41598-017-05311-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun C, Wang FJ, Zhang HG, Xu XZ, Jia RC, Yao L, Qiao PF. miR-34a mediates oxaliplatin resistance of colorectal cancer cells by inhibiting macroautophagy via transforming growth factor-beta/Smad4 pathway. World J Gastroenterol. 2017;23:1816–1827. doi: 10.3748/wjg.v23.i10.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lanzi C, Zaffaroni N, Cassinelli G. Targeting heparan sulfate proteoglycans and their modifying enzymes to enhance anticancer chemotherapy efficacy and overcome drug resistance. Curr Med Chem. 2017;24:2860–2886. doi: 10.2174/0929867324666170216114248. [DOI] [PubMed] [Google Scholar]
- 40.Lee B, Sandhu S, McArthur G. Cell cycle control as a promising target in melanoma. Curr Opin Oncol. 2015;27:141–150. doi: 10.1097/CCO.0000000000000159. [DOI] [PubMed] [Google Scholar]
- 41.Wallerand H, Robert G, Pasticier G, Ravaud A, Ballanger P, Reiter RE, Ferrière JM. The epithelial-mesenchymal transition-inducing factor TWIST is an attractive target in advanced and/or metastatic bladder and prostate cancers. Urol Oncol. 2010;28:473–479. doi: 10.1016/j.urolonc.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 42.Yu FX, Zhao B, Guan KL. Hippo pathway in organ size control, tissue homeostasis and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JE, Park HS, Lee D, Yoo G, Kim T, Jeon H, Yeo MK, Lee CS, Moon JY, Jung SS, et al. Hippo pathway effector YAP inhibition restores the sensitivity of EGFR-TKI in lung adenocarcinoma having primary or acquired EGFR-TKI resistance. Biochem Biophys Res Commun. 2016;474:154–160. doi: 10.1016/j.bbrc.2016.04.089. [DOI] [PubMed] [Google Scholar]
- 44.Gujral TS, Kirschner MW. Hippo pathway mediates resistance to cytotoxic drugs; Proc Natl Acad Sci USA; 2017; pp. E3729–E3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okamoto K, Miyoshi K, Murawaki Y. miR-29b, miR-205 and miR-221 enhance chemosensitivity to gemcitabine in HuH28 human cholangiocarcinoma cells. PLoS One. 2013;8:e77623. doi: 10.1371/journal.pone.0077623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu YX, Wang L, Liu WJ, Zhang HT, Xue JH, Zhang ZW, Gao CJ. MiR-124-3p/B4GALT1 axis plays an important role in SOCS3-regulated growth and chemo-sensitivity of CML. J Hematol Oncol. 2016;9:69. doi: 10.1186/s13045-016-0300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen MJ, Cheng YM, Chen CC, Chen YC, Shen CJ. MiR-148a and miR-152 reduce tamoxifen resistance in ER+ breast cancer via downregulating ALCAM. Biochem Biophys Res Commun. 2017;483:840–846. doi: 10.1016/j.bbrc.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 48.Zhong S, Li W, Chen Z, Xu J, Zhao J. MiR-222 and miR-29a contribute to the drug-resistance of breast cancer cells. Gene. 2013;531:8–14. doi: 10.1016/j.gene.2013.08.062. [DOI] [PubMed] [Google Scholar]
- 49.Sui C, Meng F, Li Y, Jiang Y. miR-148b reverses cisplatin-resistance in non-small cell cancer cells via negatively regulating DNA (cytosine-5)-methyltransferase 1 (DNMT1) expression. J Transl Med. 2015;13:132. doi: 10.1186/s12967-015-0488-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tong JL, Zhang CP, Nie F, Xu XT, Zhu MM, Xiao SD, Ran ZH. MicroRNA 506 regulates expression of PPAR alpha in hydroxycamptothecin-resistant human colon cancer cells. FEBS Lett. 2011;585:3560–3568. doi: 10.1016/j.febslet.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 51.Zhang JX, Qian D, Wang FW, Liao DZ, Wei JH, Tong ZT, Fu J, Huang XX, Liao YJ, Deng HX, et al. MicroRNA-29c enhances the sensitivities of human nasopharyngeal carcinoma to cisplatin-based chemotherapy and radiotherapy. Cancer Lett. 2013;329:91–98. doi: 10.1016/j.canlet.2012.10.033. [DOI] [PubMed] [Google Scholar]
- 52.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: An evolving paradigm. Nat Rev Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 53.Albarello L, Pecciarini L, Doglioni C. HER2 testing in gastric cancer. Adv Anat Pathol. 2011;18:53–59. doi: 10.1097/PAP.0b013e3182026d72. [DOI] [PubMed] [Google Scholar]
- 54.Price-Schiavi SA, Jepson S, Li P, Arango M, Rudland PS, Yee L, Carraway KL. Rat Muc4 (sialomucin complex) reduces binding of anti-ErbB2 antibodies to tumor cell surfaces, a potential mechanism for herceptin resistance. Int J Cancer. 2002;99:783–791. doi: 10.1002/ijc.10410. [DOI] [PubMed] [Google Scholar]
- 55.Castiglioni F, Tagliabue E, Campiglio M, Pupa SM, Balsari A, Menard S. Role of exon-16-deleted HER2 in breast carcinomas. Endocr Relat Cancer. 2006;13:221–232. doi: 10.1677/erc.1.01047. [DOI] [PubMed] [Google Scholar]
- 56.Serra V, Markman B, Scaltriti M, Eichhorn PJ, Valero V, Guzman M, Botero ML, Llonch E, Atzori F, Di Cosimo S, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008;68:8022–8030. doi: 10.1158/0008-5472.CAN-08-1385. [DOI] [PubMed] [Google Scholar]
- 57.Nahta R, Yuan LX, Zhang B, Kobayashi R, Esteva FJ. Insulin-like growth factor-I receptor/human epidermal growth factor receptor 2 heterodimerization contributes to trastuzumab resistance of breast cancer cells. Cancer Res. 2005;65:11118–11128. doi: 10.1158/0008-5472.CAN-04-3841. [DOI] [PubMed] [Google Scholar]
- 58.Boyer J, Allen WL, McLean EG, Wilson PM, McCulla A, Moore S, Longley DB, Caldas C, Johnston PG. Pharmacogenomic identification of novel determinants of response to chemotherapy in colon cancer. Cancer Res. 2006;66:2765–2777. doi: 10.1158/0008-5472.CAN-05-2693. [DOI] [PubMed] [Google Scholar]
- 59.Denduluri SK, Idowu O, Wang Z, Liao Z, Yan Z, Mohammed MK, Ye J, Wei Q, Wang J, Zhao L, Luu HH. Insulin-like growth factor (IGF) signaling in tumorigenesis and the development of cancer drug resistance. Genes Dis. 2015;2:13–25. doi: 10.1016/j.gendis.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Casa AJ, Dearth RK, Litzenburger BC, Lee AV, Cui X. The type I insulin-like growth factor receptor pathway: A key player in cancer therapeutic resistance. Front Biosci. 2008;13:3273–3287. doi: 10.2741/2925. [DOI] [PubMed] [Google Scholar]
- 61.Sado Y, Kagawa M, Kishiro Y, Sugihara K, Naito I, Seyer JM, Sugimoto M, Oohashi T, Ninomiya Y. Establishment by the rat lymph node method of epitope-defined monoclonal antibodies recognizing the six different alpha chains of human type IV collagen. Histochem Cell Biol. 1995;104:267–275. doi: 10.1007/BF01464322. [DOI] [PubMed] [Google Scholar]
- 62.Kalluri R. Basement membranes: Structure, assembly and role in tumour angiogenesis. Nat Rev Cancer. 2003;3:422–433. doi: 10.1038/nrc1094. [DOI] [PubMed] [Google Scholar]
- 63.Miyake M, Hori S, Morizawa Y, Tatsumi Y, Toritsuka M, Ohnishi S, Shimada K, Furuya H, Khadka VS, Deng Y, et al. Collagen type IV alpha 1 (COL4A1) and collagen type XIII alpha 1 (COL13A1) produced in cancer cells promote tumor budding at the invasion front in human urothelial carcinoma of the bladder. Oncotarget. 2017;8:36099–36114. doi: 10.18632/oncotarget.16432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin R, Shen J, Zhang T, Liu Q, Liao C, Ma H, Li S, Yu Z. The highly expressed COL4A1 genes contributes to the proliferation and migration of the invasive ductal carcinomas. Oncotarget. 2017;8:58172–58183. doi: 10.18632/oncotarget.17345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sulpice L, Rayar M, Desille M, Turlin B, Fautrel A, Boucher E, Llamas-Gutierrez F, Meunier B, Boudjema K, Clément B, Coulouarn C. Molecular profiling of stroma identifies osteopontin as an independent predictor of poor prognosis in intrahepatic cholangiocarcinoma. Hepatology. 2013;58:1992–2000. doi: 10.1002/hep.26577. [DOI] [PubMed] [Google Scholar]
- 66.Rahman M, Chan AP, Tang MJ, Tai IT. Correction: A peptide of SPARC interferes with the interaction between caspase8 and Bcl2 to resensitize chemoresistant tumors and enhance their regression in vivo. PLoS One. 2015;10:e0127226. doi: 10.1371/journal.pone.0127226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang Z, Guo L, Liu D, Sun L, Chen H, Deng Q, Liu Y, Yu M, Ma Y, Guo N, Shi M. Acquisition of resistance to trastuzumab in gastric cancer cells is associated with activation of IL-6/STAT3/Jagged-1/Notch positive feedback loop. Oncotarget. 2015;6:5072–5087. doi: 10.18632/oncotarget.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang S, Huang J, Lyu H, Cai B, Yang X, Li F, Tan J, Edgerton SM, Thor AD, Lee CK, Liu B. Therapeutic targeting of erbB3 with MM-121/SAR256212 enhances antitumor activity of paclitaxel against erbB2-overexpressing breast cancer. Breast Cancer Res. 2013;15:R101. doi: 10.1186/bcr3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vora SR, Juric D, Kim N, Mino-Kenudson M, Huynh T, Costa C, Lockerman EL, Pollack SF, Liu M, Li X, et al. CDK 4/6 inhibitors sensitize PIK3CA mutant breast cancer to PI3K inhibitors. Cancer Cell. 2014;26:136–149. doi: 10.1016/j.ccr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hudis C, Swanton C, Janjigian YY, Mino-Kenudson M, Huynh T, Costa C, Lockerman EL, Pollack SF, Liu M, Li X, et al. A phase 1 study evaluating the combination of an allosteric AKT inhibitor (MK-2206) and trastuzumab in patients with HER2-positive solid tumors. Breast Cancer Res. 2013;15:R110. doi: 10.1186/bcr3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.