Abstract
The present study aimed to investigate key long non-coding RNAs (lncRNAs) and genes, and to obtain insights into their roles in the progression of gallbladder cancer (GBC). The gene expression profile and non-coding RNA profile of GSE62335, which included five separate GBC tissue samples and five matched adjacent gallbladder normal tissue samples, was downloaded from the Gene Expression Omnibus database. The differentially expressed lncRNAs and mRNAs in the GBC tissues were identified, following which RNA binding protein analysis was performed using starBase v2.0 and the co-expressed lncRNA-mRNA pairs were predicted. Gene Ontology enrichment analysis for mRNAs was performed using the Database for Annotation Visualization and Integrated Discovery online tool. In addition, upstream microRNAs (miRNAs) were predicted for the co-expressed lncRNAs and mRNAs. The results revealed that a total of 89 upregulated (13 lncRNAs and 76 mRNAs) and 261 downregulated transcripts (27 lncRNAs and 234 mRNAs) were identified in the GBC tissues. Only 9 lncRNAs had co-expressed mRNAs, and lncRNA forkhead box P2 (FOXP2) was co-expressed with the highest number of mRNAs, which were significant associated with the function of cell adhesion. In addition, the analysis of upstream miRNAs showed that FOXF1 adjacent non-coding developmental regulatory RNA (FENDRR) had common upstream miRNAs, including miR-18b-5p, with another 119 differentially expressed genes, and that FENDRR was co-expressed with adenomatosis polyposis coli downregulated 1 (APCDD1) and v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (KIT). Taken together, the results suggested that the lncRNAs FOXP2 and FENDRR may be crucial in promoting the progression of GBC via cell adhesion and regulating miR-18b-5p, or through interactions with KIT and APCDD1, respectively.
Keywords: gallbladder cancer, long non-coding RNAs, RNA binding protein analysis, Gene Ontology enrichment analysis, upstream microRNA analysis
Introduction
Gallbladder cancer (GBC) is a rare, aggressive biliary tract malignancy, of which only ~20% of cases diagnosed in the early stages (1). The majority of patients with GBC present at an advanced stage, which is associated with a poor prognosis and poor survival outcomes for patients, with a 5-year survival rate ranging between 20 and 40% (2). Although considerable progress has been made in elucidating the genetic mechanisms involved in GBC (3,4), the molecular pathogenesis of GBC remains to be fully elucidated. Therefore, investigating novel molecules involved in the progression of GBC is like to facilitate understanding of the molecular mechanism underlying this disease and facilitate the development of novel therapeutic strategies.
Long non-coding RNAs (lncRNAs) are >200 nucleotides in length and the extent or levels of expression of lncRNAs generally appear to be specific to the tissues (5). Increasing evidence has demonstrated that lncRNAs are important regulators of diverse cellular processes and are emerging as being important in cancer (6). They have the potential to serve as therapeutic targets and prognostic indicators. For example, Wang et al (7) showed that the expression level of lncRNA-regulator of reprogramming (ROR) was upregulated in tissues from patients with GBC, and that the overexpression of lncRNA-ROR promoted the proliferation, migration and invasion of tumor cells, which was significantly associated with a poor outcome. Another study analyzed the expression of lncRNA actin filament associated protein 1 antisense RNA1 (AFAP1-AS1) using quantitative polymerase chain reaction analysis in GBC tissues and produced survival plots, which demonstrated that lncRNA AFAP1-AS1 was correlated with a poor prognosis in patients with GBC (8). Additionally, a competitive endogenous RNA (ceRNA) hypothesis has been suggested, in which lncRNAs are involved in cancer progression via interacting with microRNAs (miRNAs) (9,10). For example, a previous study reported that lncRNA Gall bladder cancer associated suppressor of pyruvate carboxylase, a target of miRNA (miR)-17-3p, negatively regulates pyruvate carboxylase-dependent cell proliferation in GBC (11). In addition, the findings of Wang et al (12) suggested that the lncRNA H19 may regulate the expression of forkhead box M1 (FOXM1) by competitively binding endogenous miR-342-3p in GBC. Although progress has been made in understanding the roles of lncRNAs in the progression of GBC, the molecular mechanisms underlying of lncRNAs require detailed investigations.
In the present study, a set of bioinformatics approaches were used to comprehensively analyze the publicly available microarray data from the Gene Expression Omnibus (GEO) database, which included five separate GBC tissue samples and fived matched adjacent gallbladder normal tissue samples. The differentially expressed lncRNAs and mRNAs were first identified in separate GBC tissues, and compared with those of matched adjacent normal gallbladder tissues. Subsequently, co-expressed lncRNA-mRNA pairs were identified, followed by functional enrichment analysis for these mRNAs. The shared upstream miRNAs regulating the co-expressed lncRNA and mRNAs were also predicted. The present study is aimed to examine key lncRNAs potentially involved in the progression of GBC and to elucidate their molecular mechanisms in the development of GBC.
Materials and methods
Affymetrix microarray data
The microarray data of GSE62335 was downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/), which was deposited by Ma et al (13) on 15th October, 2014. In total, 10 samples were used to develop the array data, which included five separate GBC tissues and five matched adjacent normal gallbladder tissues. The raw data and annotation files were downloaded based on the platform of the GPL16686 Affymetrix Human Gene 2.0 ST Array (Affymetrix Inc., Santa Clara, CA, USA) for further analysis.
Data preprocessing and screening of differentially expressed lncRNAs and mRNAs
All the raw data were preprocessed using the robust multichip average method in the oligo package (available through Bioconductor version 3.0; http://www.bioconductor.org) (14). The comparison of differentially expressed lncRNAs and mRNAs in separate GBC tissues with normal tissues were screened using the Limma package version 3.22.7 of Bioconductor version 3.0 (http://www.bioconductor.org/packages/release/bioc/html/limma.html) (15). P<0.05 and |log2 fold-change (FC)|>0.58 were defined as the cut-off values for screening.
RNA binding protein (RBP) analysis
The starBase v2.0 database (http://starbase.sysu.edu.cn/) (16,17) is an experimentally supported database, which provides the most comprehensive protein-RNA, miRNA-mRNA and miRNA-lncRNA interactions supported by large-scale cross-linking immunoprecipitation (CLIP)-Seq (HITS-CLIP, PAR-CLIP, iCLIP and CLASH) data. The newly developed starBase v2.0 database has been used in several studies to identify between miRNAs and lncRNAs, which have also been validated by experiments (18,19). In the present study, RBPs directly interacting with differentially expressed lncRNAs and mRNAs were identified based on the information in starBase v2.0. The lncRNAs and mRNAs, which were able to bind to the same RBP, were then selected for subsequent analysis.
Co-expression network construction and functional enrichment analysis
The Pearson correlation coefficient (20,21) is widely used as a co-expression similarity measure. In the present study, the lncRNAs and mRNAs found to interact with the same RBP were selected for co-expression analysis, and the co-expressed lncRNA-mRNA pairs were screened out with the absolute value of a Pearson correlation coefficient >0.85. The co-expression network was then constructed with the significant lncRNA-mRNAs using Cytoscape software version 3.0.2 (http://www.cytoscape.org/) (22).
The Gene Ontology (GO) database (http://www. geneontology.org/) is widely used for the unification of biological functions and provides gene annotation terms for large-scale genomic or transcriptomic data (23,24). The Database for Annotation Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) (25) is used for systematically associating the functional terms with gene or protein lists. The DAVID Bioinformatics Resources consist of the DAVID Knowledgebase and a set of integrated, web-based functional annotation tool suites, which can be used to screen large gene lists in detail by comprehensively integrating several different biological angles to extract associated biological meanings (26). In the present study, the DAVID online tool was used for GO enrichment analysis of mRNAs in the co-expressed lncRNA-mRNA pairs identified. P<0.01 was considered the cut-off value.
Upstream miRNA analysis
Based on the information obtained on the miRNA-lncRNA and miRNA-mRNA pairs in starBase v2.0, the upstream miRNAs of lncRNAs and mRNAs were respectively predicted. The shared upstream miRNAs of the co-expressed lncRNAs and mRNAs were then identified according to the their expression levels.
Results
Screening of differentially expressed lncRNAs and mRNAs
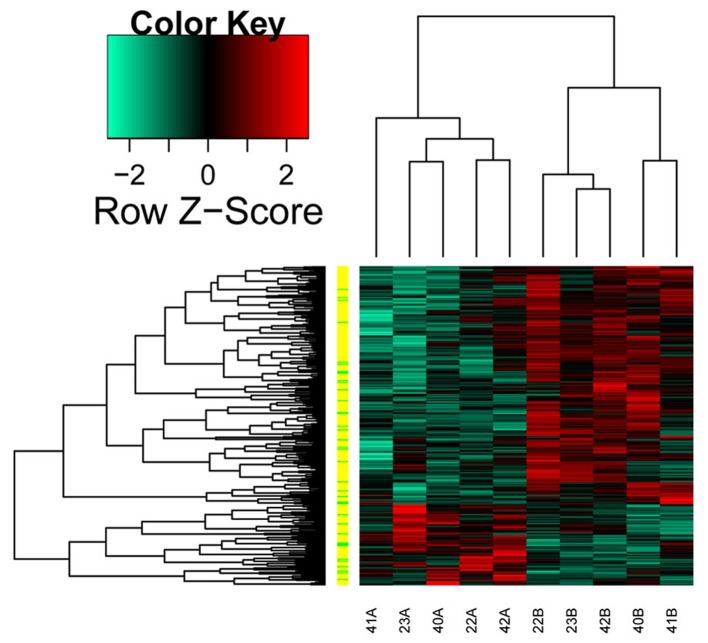
Using the Limma package with P<0.05 and |log2 FC|>0.58 as cut-off thresholds, a total of 89 upregulated transcripts (13 lncRNAs and 76 mRNAs) and 261 downregulated transcripts (27 lncRNAs and 234 mRNAs) were identified in the separate GBC tissues, compared with the normal tissues. The heatmap of the differentially expressed transcripts is shown in Fig. 1.
Figure 1.
Heatmap of differentially expressed transcripts. The left vertical strips represent the distribution of transcripts, yellow strips represent mRNAs and green strips represent long non-coding RNAs. Red indicates upregulated transcripts and green indicates downregulated transcripts.
RBP analysis
A total of 2,759 RBP-RNA pairs were initially identified according to the information of starBase v2.0, among which 1,747 were screened out as they were able to interact with not only lncRNAs, but also mRNAs.
Co-expression and functional enrichment analysis
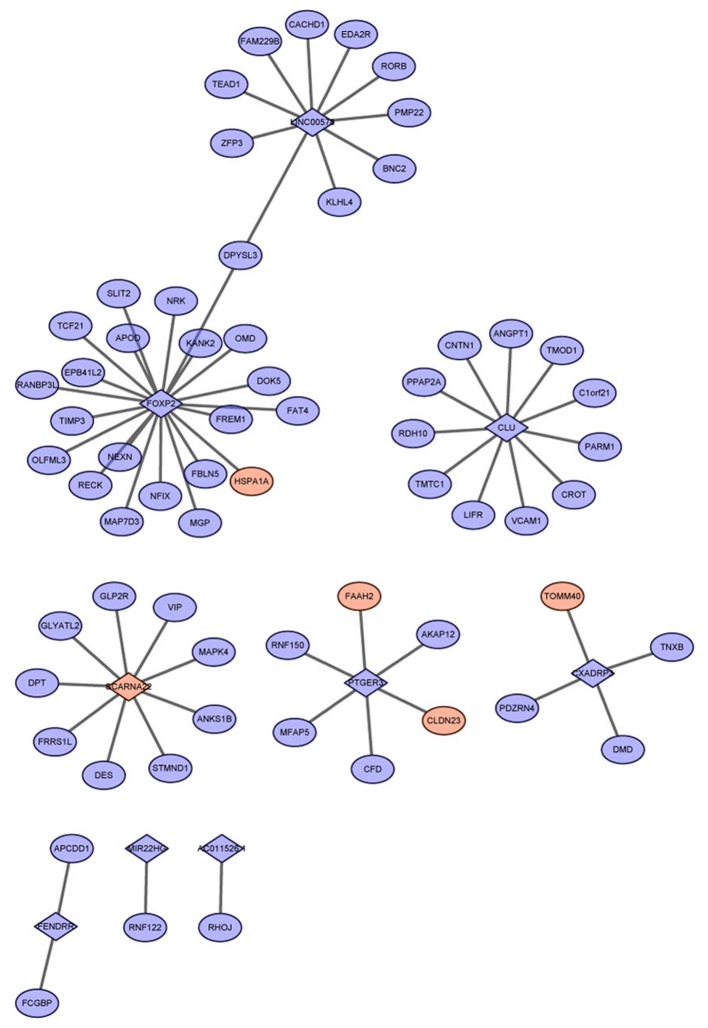
In the present study, a total of 287 co-expressed lncRNA-mRNA pairs were screened with the absolute Pearson correlation coefficient value of >0.85. Of these co-expressed lncRNA-mRNA pairs, 65 were involved in the 1,747 identified RBP-RNA pairs, from which a co-expression network was constructed (Fig. 2). Notably, all mRNAs within the 65 lncRNA-mRNA pairs were linked with only nine lncRNAs, including FOXP2, clusterin (CLU) and coxsackie virus and adenovirus receptor pseudogene 3 (CXADRP3), with co-expressed mRNAs (Table I). FOXP2 was also able to co-express with the highest number of mRNAs, followed by CLU.
Figure 2.
Co-expression network of lncRNAs and mRNAs. Red nodes represent upregulation and blue node represent downregulation. Diamond-shaped nodes represent mRNAs and circular nodes represent lncRNAs. LncRNAs, long non-coding RNAs.
Table I.
Number of co-expressed mRNAs corresponding to lncRNAs.
| LncRNA | Co-expressed mRNAs (n) |
|---|---|
| AC011526.1 | 1 |
| CLU | 11 |
| CXADRP3 | 4 |
| FENDRR | 2 |
| FOXP2 | 21 |
| LINC00578 | 10 |
| MIR22HG | 1 |
| PTGER3 | 6 |
| SCARNA22 | 9 |
CLU, clusterin; CXADRP3, coxsackie virus and adenovirus receptor pseudogene 3; LncRNAs, long non-coding RNAs; FOXP2, forkhead box P2; FENDRR, FOXF1 adjacent non-coding developmental regulatory RNA; LINC00578, long intergenic non-protein coding RNA 578; MIR22HG, MIR22 host gene; PTGER3, prostaglandin EP3 receptor; SCARNA22, small Cajal body-specific RNA 22.
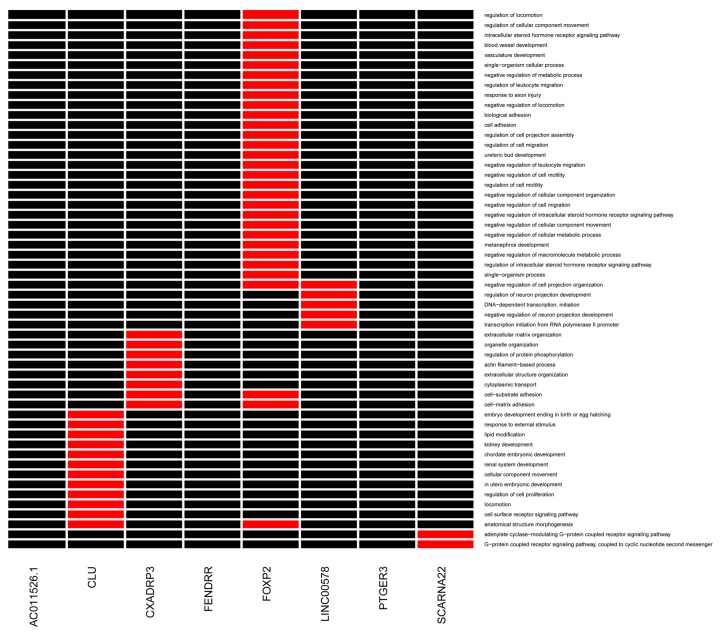
As shown in Fig. 3, the co-expressed mRNAs connected with FOXP2 and CXADRP3 were significantly enriched in several GO terms, including cell-matrix adhesion, cell-substrate adhesion and cell adhesion. The co-expressed mRNAs, which had interactions with CLU were significantly associated with the regulation of cell proliferation and cell surface receptor signaling pathway terms.
Figure 3.
GO enrichment analysis of mRNAs corresponding to lncRNAs. Red strips represent enriched GO terms. Black strips represent non-enriched GO terms. The abscissa axis above represents lncRNAs. The longitudinal axis represents enriched GO terms. GO, Gene Ontology; LncRNAs, long non-coding RNAs.
Upstream miRNA analysis
According to the information from starBase v2.0, only lncRNA FOXF1 adjacent non-coding developmental regulatory RNA (FENDRR) was able to interact with upstream miRNAs. FENDRR shared common upstream miRNAs with 119 differentially expressed mRNAs. However, based on the correlation information in the starBase version 2.0 database, not all 119 differentially expressed mRNAs exhibited a correlation with FENDRR; thus, it was not possible to construct the ceRNA interaction network with these 119 differentially expressed mRNAs, FENDRR and the shared upstream miRNAs.
However, FENDRR was revealed to be co-expressed with adenomatosis polyposis coli downregulated 1 (APCDD1; correlation coefficient, 0.905) and v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (KIT; correlation coefficient, 0.846). Notably, upstream miRNA analysis showed that miR-18b-5p, miR-28-5p and miR-4735-3p were common upstream miRNAs of FENDRR and KIT, whereas miR-18b-5p was the common upstream miRNA of FENDRR and APCDD1.
Discussion
In the present study, a comprehensive biological informatics approach was used to examine key lncRNAs involved in GBC and to elucidate their molecular mechanisms in the development of GBC. In total, 13 upregulated and 27 downregulated lncRNAs were identified in the separate GBC tissues, compared with the normal tissues. Among these lncRNAs, FOXP2 was found to be downregulated in the GBC tissues and co-expressed with the highest number of differentially expressed mRNAs, which were significantly enriched in GO terms associated with cell adhesion. FENDRR was also a downregulated lncRNA, which was found to share common upstream miRNAs with 119 differentially expressed mRNAs, including miR-18b-5p, and was co-expressed with KIT and APCDD1. These results suggested that lncRNA FOXP2 and FENDRR were likely to be important in the pathogenesis of GBC via cell adhesion and regulating miR-18b-5p, or the interaction with KIT and APCDD1, respectively.
Cell adhesion is an important event in biological processes, which determines the polarity of cells and is the morphological hallmark of malignant tumors (27). Cell-cell adhesiveness is usually reduced in several types of cancer, which is considered to be essential for cancer invasion and metastasis (27). Increasing evidence has highlighted the roles of cell adhesion molecules in perineural invasion in GBC (28). The overexpression of epithelial cell adhesion molecule antigen is an independent prognostic marker in GBC (29). In addition, E-cadherin is a transmembrane cell-adhesion protein mediating intercellular adhesion in epithelial tissues (30). It is reported that loss of the expression of E-cadherin is important in the progression from tumor formation to invasion and metastasis in several types of cancer (31). Kohya et al (32) confirmed that E-cadherin has a critical role in the proliferation, motility and invasion of GBC cells. Therefore, cell adhesion may be an important biological process involved in GBC. By contrast, lncRNAs have been shown to regulate gene expression through several mechanisms (33). LncRNAs have also been shown to allosterically modify RBPs, which regulate transcription (33). In the present study, lncRNA FOXP2 was found to be downregulated in separate GBC tissues, compared with that in matched adjacent normal gallbladder tissues, and the majority of mRNAs co-expressed with lncRNA FOXP2 were significantly associated with the function of cell adhesion. Therefore, it was hypothesized that lncRNA FOXP2 may be crucial in the progression of GBC via co-expression with important molecules in cell adhesion.
KIT is a member of the class III receptor tyrosine kinase family, which can be bound to and activated by stem cell factor (34). KIT receptor activation can cause the phosphorylation and activation of the Shc adaptor protein (35) and Ras (36). The Shc adaptor protein can link c-KIT to the Ras/mitogen-protein kinase (MAPK) pathway (37). K-ras gene mutation is frequently found in GBC cells (38). Activated Ras induces gefitinib-resistance by activating the epidermal growth factor receptor signaling pathways in human GBC cells (39). In addition, KIT be crucial in the majority of gastrointestinal stromal tumors via activation of the phosphatidyl-inositol-3-kinase (PI3-K)/AKT survival pathway. The activated MAPK and AKT pathways can lead to the uncontrolled growth of GBC epithelium, and promote the invasion and metastasis of GBC cells (40). By contrast, APCDD1 is a membrane-bound glycoprotein, which can interact in vitro with WNT3A and low-density lipoprotein receptor-related protein 5, which are two essential components of the Wnt signaling pathway (40). A previous study demonstrated that APCDD1 is the activated downstream target of the WNT/β-catenin pathway, which may enhance the proliferation of pediatric Wilms' tumor cells (41). Mutations of components of the Wnt/β-catenin pathway, including β-catenin exon 3 mutations, have been found to be associated with the progression of GBC (42). In addition, Wnt signaling can cross-talk with the E-cadherin cell adhesion system, which is involved in the morphogenesis of several types of cancer, including GBC (27,32). In the present study, FENDRR, as a downregulated lncRNA in GBC, was co-expressed with KIT and APCDD1. In a previous study, the expression profiles of lncRNAs in infantile hemangioma were assessed, and significant associations of FENDRR and its associated mRNAs were identified (43). Based on the results of the present study, and previous evidence confirming the importance of KIT and APCDD1 in the progression of GBC, FENDRR may be crucial in the progression of GBC through interaction with these two molecules by targeting several signaling pathways, including the Wnt signaling pathway.
In the present study, the analysis of upstream miRNAs also showed that miR-18b-5p was a common upstream miRNA of FENDRR, KIT and APCDD1. miR-18b-5p acts as an oncogene, and has been reported to be involved in the development and pathogenesis of human gastric cancer (44). The expression of miR-18b-5p is associated with the tumor protein 53 status, and may predict clinical outcome in patients with head and neck squamous cell carcinoma (45). Although the roles of miR-18b-5p in the development of GBC remain to be fully elucidated, the present study hypothesized that miR-18b-5p may be involved in the progression of GBC via targeting KIT and APCDD1, and lncRNA FENDRR may be involved in the development of GBC via regulating miR-18b-5p.
However, there were several limitations to the present study. Firstly, the size of samples was relatively small, therefore, further investigations with a higher number of samples are required. Secondly, statistical validation using other datasets, including meta-analysis (46,47) may be used to cross-check the results of the present study. Thirdly, due to insufficient material, experimental validation was not performed in the present study. The inclusion of these as a central component of future investigations can assist in confirming results using different approaches, including quantitative real-time polymerase chain reaction analysis.
In conclusion, lncRNA FOXP2 and FENDRR may be crucial in promoting the progression of GBC via cell adhesion and regulating miR-18b-5p or interaction with KIT and APCDD1, respectively. Therefore, these two factors may be recommended as potential targets in the management of GBC. The findings of the present study shed new light on the molecular mechanism of GBC and may provide a foundation for the development of credible therapeutic approaches. However, in the absence of experimental validation in the present study, further analysis of the microarray data and genetic experiments are required.
References
- 1.Takebe N, Yang SX. Sonic hedgehog signaling pathway and gallbladder cancer: Targeting with precision medicine approach. Chin Clin Oncol. 2016;5:1. doi: 10.3978/j.issn.2304-3865.2016.01.02. [DOI] [PubMed] [Google Scholar]
- 2.Ma M, Weng M, Zhang M, Qin Y, Gong W, Quan Z. Targeting gallbladder cancer: Hyaluronan sensitizes cancer cells to chemo-therapeutics. Int J Clin Exp Pathol. 2015;8:1822–1825. [PMC free article] [PubMed] [Google Scholar]
- 3.Nagahashi M, Ajioka Y, Lang I, Szentirmay Z, Kasler M, Nakadaira H, Yokoyama N, Watanabe G, Nishikura K, Wakai T, et al. Genetic changes of p53, K-ras, and microsatellite instability in gallbladder carcinoma in high-incidence areas of Japan and Hungary. World J Gastroenterol. 2008;14:70–75. doi: 10.3748/wjg.14.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ooi A, Suzuki S, Nakazawa K, Itakura J, Imoto I, Nakamura H, Dobashi Y. Gene amplification of Myc and its coamplification with ERBB2 and EGFR in gallbladder adenocarcinoma. Anticancer Res. 2009;29:19–26. [PubMed] [Google Scholar]
- 5.Bhat SA, Ahmad SM, Mumtaz PT, Malik AA, Dar MA, Urwat U, Shah RA, Ganai NA. Long non-coding RNAs: Mechanism of action and functional utility. Non-coding RNA Res. 2016;1:43–50. doi: 10.1016/j.ncrna.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haemmerle M, Gutschner T. Long non-coding RNAs in cancer and development: Where do we go from here? Int J Mol Sci. 2015;16:1395–1405. doi: 10.3390/ijms16011395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang SH, Zhang MD, Wu XC, Weng MZ, Zhou D, Quan ZW. Overexpression of LncRNA-ROR predicts a poor outcome in gallbladder cancer patients and promotes the tumor cells proliferation, migration, and invasion. Tumour Biol. 2016;37:12867–12875. doi: 10.1007/s13277-016-5210-z. [DOI] [PubMed] [Google Scholar]
- 8.Ma F, Wang SH, Cai Q, Zhang MD, Yang Y, Ding J. Overexpression of LncRNA AFAP1-AS1 predicts poor prognosis and promotes cells proliferation and invasion in gallbladder cancer. Biomed Pharmacother. 2016;84:1249–1255. doi: 10.1016/j.biopha.2016.10.064. [DOI] [PubMed] [Google Scholar]
- 9.Jalali S, Bhartiya D, Lalwani MK, Sivasubbu S, Scaria V. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PloS One. 2013;8:e53823. doi: 10.1371/journal.pone.0053823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juan L, Wang G, Radovich M, Schneider BP, Clare SE, Wang Y, Liu Y. Potential roles of microRNAs in regulating long intergenic noncoding RNAs. BMC Med Genomics. 2013;6(Suppl 1):S7. doi: 10.1186/1755-8794-6-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma MZ, Zhang Y, Weng MZ, Wang SH, Hu Y, Hou ZY, Qin YY, Gong W, Zhang YJ, Kong X, et al. Long noncoding RNA GCASPC, a target of miR-17-3p, negatively regulates pyruvate carboxylase-dependent cell proliferation in gallbladder cancer. Cancer Res. 2016;76:5361–5371. doi: 10.1158/0008-5472.CAN-15-3047. [DOI] [PubMed] [Google Scholar]
- 12.Wang SH, Ma F, Tang ZH, Wu XC, Cai Q, Zhang MD, Weng MZ, Zhou D, Wang JD, Quan ZW. Long non-coding RNA H19 regulates FOXM1 expression by competitively binding endogenous miR-342-3p in gallbladder cancer. J Exp Clin Cancer Res. 2016;35:160. doi: 10.1186/s13046-016-0436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma MZ, Kong X, Weng MZ, Zhang MD, Qin YY, Gong W, Zhang WJ, Quan ZW. Long non-coding RNA-LET is a positive prognostic factor and exhibits tumor-suppressive activity in gallbladder cancer. Mol Carcinog. 2015;54:1397–1406. doi: 10.1002/mc.22215. [DOI] [PubMed] [Google Scholar]
- 14.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 16.Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: A database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–D209. doi: 10.1093/nar/gkq1056. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui J, Mo J, Luo M, Yu Q, Zhou S, Li T, Zhang Y, Luo W. c-Myc-activated long non-coding RNA H19 downregulates miR-107 and promotes cell cycle progression of non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:12400–12409. [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao H, Tang K, Liu P, Chen K, Hu J, Zeng J, Xiao W, Yu G, Yao W, Zhou H, et al. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget. 2015;6:38005–38015. doi: 10.18632/oncotarget.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de la Fuente A, Bing N, Hoeschele I, Mendes P. Discovery of meaningful associations in genomic data using partial correlation coefficients. Bioinformatics. 2004;20:3565–3574. doi: 10.1093/bioinformatics/bth445. [DOI] [PubMed] [Google Scholar]
- 21.Wang J. Encyclopedia of Systems Biology. Springer; 2013. Pearson Correlation Coefficient; pp. 1671–1671. [DOI] [Google Scholar]
- 22.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Consortium GO. The gene ontology (GO) database and informatics resource. Nucleic acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID gene functional classification tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID bioinformatics resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirohashi S, Kanai Y. Cell adhesion system and human cancer morphogenesis. Cancer Sci. 2003;94:575–581. doi: 10.1111/j.1349-7006.2003.tb01485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seki H, Koyama K, Tanaka JI, Sato Y, Umezawa A. Neural cell adhesion molecule and perineural invasion in gallbladder cancer. J Surg Oncol. 1995;58:97–100. doi: 10.1002/jso.2930580205. [DOI] [PubMed] [Google Scholar]
- 29.Varga M, Obrist P, Schneeberger S, Mühlmann G, Felgel-Farnholz C, Fong D, Zitt M, Brunhuber T, Schäfer G, Gastl G, Spizzo G. Overexpression of epithelial cell adhesion molecule antigen in gallbladder carcinoma is an independent marker for poor survival. Clin Cancer Res. 2004;10:3131–3136. doi: 10.1158/1078-0432.CCR-03-0528. [DOI] [PubMed] [Google Scholar]
- 30.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 31.Katagiri A, Watanabe R, Tomita Y. E-cadherin expression in renal cell cancer and its significance in metastasis and survival. Br J Cancer. 1995;71:376. doi: 10.1038/bjc.1995.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohya N, Kitajima Y, Jiao W, Miyazaki K. Effects of E-cadherin transfection on gene expression of a gallbladder carcinoma cell line: Repression of MTS1/S100A4 gene expression. Int J Cancer. 2003;104:44–53. doi: 10.1002/ijc.10920. [DOI] [PubMed] [Google Scholar]
- 33.Su Y, Wu H, Pavlosky A, Zou LL, Deng X, Zhang ZX, Jevnikar AM. Regulatory non-coding RNA: New instruments in the orchestration of cell death. Cell Death Dis. 2016;7:e2333. doi: 10.1038/cddis.2016.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H, Chen X, Focia PJ, He X. Structural basis for stem cell factor-KIT signaling and activation of class III receptor tyrosine kinases. EMBO J. 2007;26:891–901. doi: 10.1038/sj.emboj.7601545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cutler RL, Liu L, Damen J, Krystal G. Multiple cytokines induce the tyrosine phosphorylation of Shc and its association with Grb2 in hemopoietic cells. J Biol Chem. 1993;268:21463–21465. [PubMed] [Google Scholar]
- 36.Duronio V, Welham MJ, Abraham S, Dryden P, Schrader JW. p21ras activation via hemopoietin receptors and c-kit requires tyrosine kinase activity but not tyrosine phosphorylation of p21ras GTPase-activating protein; Proc Natl Acad Sci USA; 1992; pp. 1587–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hemesath TJ, Price ER, Takemoto C, Badalian T, Fisher DE. MAP kinase links the transcription factor Microphthalmia to c-Kit signalling in melanocytes. Nature. 1998;391:298–301. doi: 10.1038/34681. [DOI] [PubMed] [Google Scholar]
- 38.Masuhara S, Kasuya K, Aoki T, Yoshimatsu A, Tsuchida A, Koyanagi Y. Relation between K-ras codon 12 mutation and p53 protein overexpression in gallbladder cancer and biliary ductal epithelia in patients with pancreaticobiliary maljunction. J Hepatobiliary Pancreat Surg. 2000;7:198–205. doi: 10.1007/s005340050176. [DOI] [PubMed] [Google Scholar]
- 39.Qin B, Ariyama H, Baba E, Tanaka R, Kusaba H, Harada M, Nakano S. Activated Src and Ras induce gefitinib resistance by activation of signaling pathways downstream of epidermal growth factor receptor in human gallbladder adenocarcinoma cells. Cancer Chemother Pharmacol. 2006;58:577–584. doi: 10.1007/s00280-006-0219-4. [DOI] [PubMed] [Google Scholar]
- 40.Horiuchi H, Kawamata H, Furihata T, Omotehara F, Hori H, Shinagawa Y, Ohkura Y, Tachibana M, Yamazaki T, Ajiki T, et al. A MEK inhibitor (U0126) markedly inhibits direct liver invasion of orthotopically inoculated human gallbladder cancer cells in nude mice. J Exp Clin Cancer Res. 2004;23:599–606. [PubMed] [Google Scholar]
- 41.Zirn B, Samans B, Wittmann S, Pietsch T, Leuschner I, Graf N, Gessler M. Target genes of the WNT/beta-catenin pathway in Wilms tumors. Genes Chromosomes Cancer. 2006;45:565–574. doi: 10.1002/gcc.20319. [DOI] [PubMed] [Google Scholar]
- 42.Yanagisawa N, Mikami T, Saegusa M, Okayasu I. More frequent beta-catenin exon 3 mutations in gallbladder adenomas than in carcinomas indicate different lineages. Cancer Res. 2001;61:19–22. [PubMed] [Google Scholar]
- 43.Liu X, Lv R, Zhang L, Xu G, Bi J, Gao F, Zhang J, Xue F, Wang F, Wu Y, et al. Long noncoding RNA expression profile of infantile hemangioma identified by microarray analysis. Tumour Biol. 2016 Oct 5; doi: 10.1007/s13277-016-5434-y. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 44.Li H, Xie S, Liu X, Wu H, Lin X, Gu J, Wang H, Duan Y. Matrine alters microRNA expression profiles in SGC-7901 human gastric cancer cells. Oncol Rep. 2014;32:2118–2126. doi: 10.3892/or_xxxxxxxx. [DOI] [PubMed] [Google Scholar]
- 45.Ganci F, Sacconi A, Ben-Moshe NB, Manciocco V, Sperduti I, Strigari L, Covello R, Benevolo M, Pescarmona E, Domany E, et al. Expression of TP53 mutation-associated microRNAs predicts clinical outcome in head and neck squamous cell carcinoma patients. Ann Oncol. 2013;24:3082–3088. doi: 10.1093/annonc/mdt380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Zhang B, Wang T, Wang H. LncRNA MALAT1 overexpression is an unfavorable prognostic factor in human cancer: Evidence from a meta-analysis. Int J Clin Exp Med. 2015;8:5499–5505. [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Ning Y, Guo X. Integrative meta-analysis of differentially expressed genes in osteoarthritis using microarray technology. Mol Med Rep. 2015;12:3439–3445. doi: 10.3892/mmr.2015.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]