Abstract
Increasing evidence has indicated that the abnormal expressions of certain genes serve important roles in tumorigenesis, progression and metastasis. The aim of the present study was to explore the key differentially expressed genes (DEGs) between non-small cell lung cancer (NSCLC) and matched normal lung tissues by analyzing 4 different mRNA microarray datasets downloaded from the Gene Expression Omnibus (GEO) database. In improving the reliability of the bioinformatics analysis, the DEGs in each dataset that met the cut-off criteria (adjust P-value <0.05 and |log2fold-change (FC)|>1) were intersected with each other, from which 195 were identified (consisting of 57 upregulated and 138 downregulated DEGs). The GO analysis results revealed that the upregulated DEGs were significantly enriched in various biological processes (BP), including cell cycle, mitosis and cell proliferation while the downregulated DEGs were significantly enriched in angiogenesis and response to drug and cell adhesion. The hub genes, including CCNB1, CCNA2, CEP55, PBK and HMMR, were identified based on the protein-protein interaction (PPI) network. The Kaplan-Meier survival analysis indicated that the high expression level of each of these hub genes correlates with poorer overall survival in all patients with NSCLC, which indicates that they may serve important roles in the progression of NSCLC. In conclusion, the DEGs and hub genes identified in the present study may contribute to the comprehensive understanding of the molecular mechanisms of NSCLC and may be used as diagnostic and prognostic biomarkers as well as molecular targets for the treatment of NSCLC.
Keywords: non-small cell lung cancer, microarray, differentially expressed genes, bioinformatics analysis, hub genes
Introduction
Lung cancer, which accounts for more than one-tenth of all cancer cases worldwide, is one of the most common types of malignancies and perhaps, the first cause of cancer-related deaths worldwide (1). In China, lung cancer is currently the first most common cancer with more than 733,300 new cases and 610,200 deaths in 2015 (2). Based on histological sub-classification, there are non-small cell lung cancer (NSCLC) and SCLC with approximately 85% of lung cancer patients diagnosed with NSCLC (3).
To date, surgical resection remains the only treatment option for NSCLC (4,5). However, with no specific symptoms associated with the early stage of NCLSC and the lack of accurate and convenient diagnostic methods for its early detection, most patients are diagnosed at the advanced stage of this disease when surgical resection is no longer feasible (6).
With the recent advances in the molecular biology, molecule targeted therapy for NSCLC has equally made significant progress. For example, approximately 10–20% of North American and Western European patients and 30–50% of Asian patients with NSCLC were shown to have mutation of EGFR gene (7). But, with the use of EGFR tyrosine kinase inhibitors (TKIs), such as erlotinib and gefitinib, these patients were found to response well (with a response rate of 10%, and an estimated 2-month prolongation in median survival over placebo) (8,9). It was also found that 3–7% of NSCLC patients had an activated ALK gene, but treatment with crizotinib (an ALK inhibitor) elevated their response rate from 10 to 55% (compared to conventional chemotherapy), and increased their 6-month progression-free survival rate to 72% (8). Nevertheless, these situations can only represent a cohort of NSCLC patients and due to drug resistance these targeted therapy agents are often transient.
Moreover, even with the remarkable successes and withdraw that have been obtained in the targeted therapy of NSCLC, the precise molecular mechanisms of this disease are still far from being fully understood. Thus, the need to find more potential targets for more effective therapeutic strategies is very paramount.
In order to better understand the DEGs influence on molecular pathogenesis of NSCLC, in this study, we downloaded 4 NSCLC related mRNA datasets from Gene Expression Omnibus (GEO) and identified the DEGs between the NSCLC patients and healthy control samples. Subsequently, we performed Gene Ontology (GO) and protein-protein interaction (PPI) networks analysis so as to study and identify potential key DEGs for the diagnosis and treatment of NSCLC. Further studies on these identified key DEGs can aid a comprehensive understanding of the molecular development of NSCLC and be explored as potential biomarkers for its diagnosis, prognosis, and drug targets.
Materials and methods
Affymetrix microarray data
Series matrix files of the microarray data: GSE21933, GSE33532, GSE44077 and GSE74706 were downloaded from the public GEO database (http://www.ncbi.nlm.nih.gov/geo/), and the probe names based on platforms were annotated to gene symbols according to their corresponding new version of annotation files, such as GPL6254, GPL570, GPL6244 and GPL13497. In each GSE file, we only choose the NSCLC samples and their matched normal samples, and from which we obtained 21 tumor samples and 21 normal samples for GSE21933 (10), 80 tumor samples and 20 normal samples for GSE33532 (11), 65 tumor samples and 65 normal samples for GSE44077 (12) and 18 tumor samples and 18 normal samples for GSE74706 (13) to analyze downstream.
Data pre-processing and differential expression analysis
The original array data of each series were analyzed separately using R software (version 3.4.0) and R packages. In brief: Firstly, background correction and quantile normalization was performed on the raw data using the robust multi-array average (RMA) algorithm in R affy package (14). The differentially expressed proteins (DEGs) between the tumor and normal group samples were then analyzed by the paired t-test based on the limma package in R. Multiple testing was corrected by the Benjamini-Hochberg method to obtain the adjusted P-value (15). Finally, the Genes with adjusted P<0.05 and |log2fold-change (FC)|>1 were considered to be significant. To further enhance the reliability of the bioinformatics analysis, the overlapping DEGs co-existed in all 4 GSE files were identified using FunRich software version 2.1.2 (http://www.funrich.org) (16).
GO enrichment analysis of DEGs
GO analysis is widely used to identify characteristic biological processes (BP) for a large number of genes in microarray analysis (17). DAVID (Database for Annotation, Visualization and Integrated Discovery, https://david.ncifcrf.gov/), as a comprehensive set of functional annotation tools (18), was used to investigate enriched BP terms for all the overlapping DEGs. P<0.05 and Count ≥5 were considered statistically significant.
PPI network construction and hub gene analysis
STRING (Search Tool for the Retrieval of Interacting Genes, https://string-db.org/) database is a useful online tool dedicated to evaluate the PPI interaction information (19). STRING (version 10.5) was used to explore the interactive relationships of the overlapping DEGs, and only the experimentally validated interactions with a combined score >0.4 were selected as significant. Then, the PPI network was built by Cytoscape software (version 3.5.1) (20). The plug-in cytoHubba in Cytoscape was used to screen the hub genes from the PPI network and only DEGs with a degree score ≥19 was selected as hub genes.
Association of hub genes and patient prognosis
An online survival analysis tool Kaplan Meier plotter (http://kmplot.com/analysis/) (21), which includes both clinical data and gene expression data of lung, breast, gastric and ovarian cancers, was used to evaluate the prognostic significance of each hub gene in NSCLC. According to the median expression value of a gene, the patient samples were divided into high and low expression groups. In this study, the analysis was carried out under the default parameters, which is in brief, no subtypes restriction, ‘univariate’ for Cox regression and ‘exclude biased arrays’ for array quality control and in each survival plot, the log rank P-value and hazard ratio (HR, 95% confidence intervals) were calculated and displayed on the main plot.
Results
Identification of DEGs
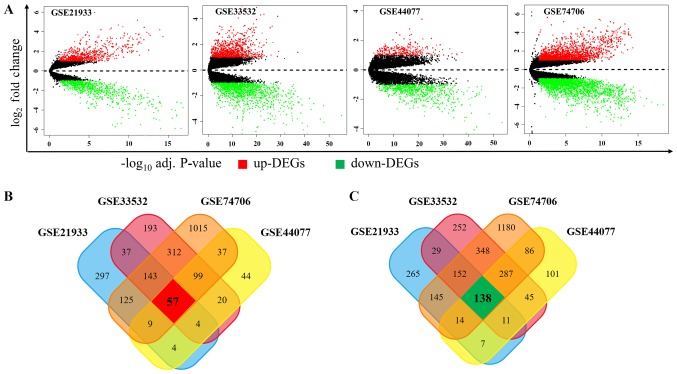
In order to identify the DEGs, we downloaded 4 public RNA-seq datasets of NSCLC from the GEO database for our analysis: GSE21933, GSE33532, GSE44077 and GSE74706. We defined DEGs between tumor samples and matched normal samples according to the threshold criterion (logFC >1 or logFC <-1 and adjusted P<0.05), and we got 1437, 2127, 963 and 4147 DEGs in GSE21933, GSE33532, GSE44077 and GSE74706, respectively. As shown in Fig. 1A, among these DEGs, 676, 865, 274 and 1797 genes were upregulated while 761, 1262, 689, 2350 ones were downregulated in GSE21933, GSE33532, GSE44077 and GSE74706, respectively. In order to obtain the most reliable DEGs, we isolated the DEGs presented in all four datasets and finally got a total of 195 DEGs, consisting of 57 upregulated (Fig. 1B) and 138 downregulated DEGs (Fig. 1C).
Figure 1.
Expression levels of expressed genes and distributions in four NSCLC datasets. (A) Volcano plots of each microarray data. Y-axis represents the log2fold-change (NSCLC vs. normal samples). Venn plots of (B) upregulated and (C) downregulated protein-coding genes were indicated. Adj. P-value, adjusted P-value; NSCLC, non-small cell lung cancer.
GO term enrichment analysis
In an attempt to find the potential biological functions associated with these 195 DEGs, the online software DAVID was used to identify the overrepresented GO categories. After GO functional enrichment analysis, as shown in Table I, the upregulated DEGs were significantly enriched in 18 biological process (BP) terms, which includes mitosis, cell cycle, protein-DNA complex assembly, microtubule-based process and cell proliferation. The downregulated DEGs were also significantly enriched in 16 BP terms, such as cellular response to hormone stimulus, angiogenesis, response to drug and cell adhesion. All these terms are closely related to the tumorigenesis and development.
Table I.
Enriched Gene Ontology terms for upregulated and downregulated differentially expressed genes.
| A, Upregulated | |||
|---|---|---|---|
| Term | Gene function | Count | P-value |
| GO:0007067 | Mitosis | 14 | 6.11×10‒13 |
| GO:0000280 | Nuclear division | 14 | 6.11×10‒13 |
| GO:0000087 | M phase of mitotic cell cycle | 14 | 7.70×10‒13 |
| GO:0048285 | Organelle fission | 14 | 1.02×10‒12 |
| GO:0000279 | M phase | 15 | 6.02×10‒12 |
| GO:0022403 | Cell cycle phase | 16 | 9.16×10‒12 |
| GO:0000278 | Mitotic cell cycle | 15 | 2.91×10‒11 |
| GO:0022402 | Cell cycle process | 16 | 7.21×10‒10 |
| GO:0007049 | Cell cycle | 16 | 5.28×10‒08 |
| GO:0051301 | Cell division | 11 | 8.09×10‒08 |
| GO:0065004 | Protein-DNA complex assembly | 5 | 3.22×10‒04 |
| GO:0007018 | Microtubule-based movement | 5 | 7.32×10‒04 |
| GO:0007017 | Microtubule-based process | 6 | 2.14×10‒03 |
| GO:0034622 | Cellular macromolecular complex assembly | 6 | 5.69×10‒03 |
| GO:0051726 | Regulation of cell cycle | 6 | 6.72×10‒03 |
| GO:0034621 | Cellular macromolecular complex subunit organization | 6 | 9.16×10‒03 |
| GO:0008283 | Cell proliferation | 6 | 2.02×10‒02 |
| B, Downregulated | |||
| Term | Gene function | Count | P-value |
| GO:0032870 | Cellular response to hormone stimulus | 6 | 1.73×10‒05 |
| GO:0001525 | Angiogenesis | 10 | 3.41×10‒05 |
| GO:0050900 | Leukocyte migration | 7 | 2.53×10‒04 |
| GO:0030336 | Negative regulation of cell migration | 6 | 6.18×10‒04 |
| GO:0042493 | Response to drug | 9 | 1.62×10‒03 |
| GO:0007165 | Signal transduction | 18 | 3.98×10‒03 |
| GO:0007155 | Cell adhesion | 10 | 5.99×10‒03 |
| GO:0016337 | Single organismal cell-cell adhesion | 5 | 6.12×10‒03 |
| GO:0002576 | Platelet degranulation | 5 | 6.56×10‒03 |
| GO:0006898 | Receptor-mediated endocytosis | 6 | 1.12×10‒02 |
| GO:0043065 | Positive regulation of apoptotic process | 7 | 2.15×10‒02 |
| GO:0018108 | Peptidyl-tyrosine phosphorylation | 5 | 2.48×10‒02 |
| GO:0045893 | Positive regulation of transcription, DNA-templated | 9 | 3.28×10‒02 |
| GO:0045087 | Innate immune response | 8 | 3.61×10‒02 |
| GO:0006810 | Transport | 7 | 4.02×10‒02 |
Count, the number of enriched genes in each term.
Hub genes screening from the PPI network
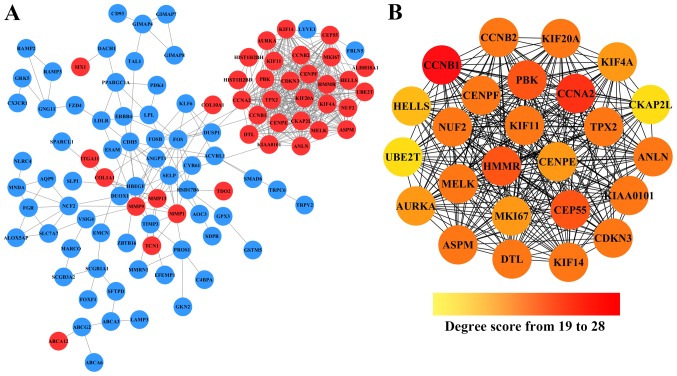
To further investigate the interaction between these 195 DEGs, the STRING database was used to analysis their PPI networks, and the resulted PPI networks were constructed by Cytoscape. As shown in Fig. 2A, after removing the effect of free protein pairs, we obtained a network with 38 upregulated DEGs, 66 downregulated DEGs and 424 edges.
Figure 2.
The constructed PPI network of DEGs. (A) A total of 195 overlapping DEGs were used as seed genes to build a PPI subnetwork based on the STRING database. Red nodes represent the upregulated DEGs, while bule nodes represent downregulated DEGs. Edges between the nodes represent their interation. (B) The screened subnetwork of hub genes. The color changes with the increase of degree score from orange to red, all nodes represent the upregulated genes.
In addition, in the PPI networks, genes that have strong interactions with many other genes are called ‘hub genes’. Due to their key positions in the PPI network, the hub genes are potential drivers of the diseases status. In order to identify the key tumorigenic genes of NSCLC, the cytoHubba plugin for Cytoscape was used to screen the hub genes among all the DEGs. As shown in Fig. 2B and Table II, we obtained 25 hub genes (the nodes with red or orange colors) that exhibited especially high degree scores (≥19). To our surprise, all these 25 hub genes were upregulated DEGs. The genes G2/mitotic-specific cyclinB1 (CCNB1), CyclinA2 (CCNA2), Centrosomal protein of 55 kDa (CEP55), lymphokine-activated killer T-cell-originated protein kinase (PBK), and hyaluronan mediated motility receptor (HMMR) were selected as the top five hub genes with high connectivity degree. CCNB1, which is also known as cyclin B1, was observed to exhibit the highest degree of connectivity as shown in Table II.
Table II.
Statistical results of connectivity degrees of the hub genes in the PPI network.
| Gene | Degree | Gene | Degree |
|---|---|---|---|
| CCNB1 | 28 | KIF11 | 24 |
| CCNA2 | 27 | KIF20A | 24 |
| CEP55 | 25 | CCNB2 | 24 |
| PBK | 25 | CENPF | 24 |
| HMMR | 25 | TPX2 | 24 |
| DTL | 24 | MKI67 | 23 |
| KIAA0101 | 24 | KIF4A | 23 |
| MELK | 24 | CENPE | 23 |
| ANLN | 24 | AURKA | 23 |
| KIF14 | 24 | HELLS | 21 |
| CDKN3 | 24 | CKAP2L | 19 |
| ASPM | 24 | UBE2T | 19 |
| NUF2 | 24 |
PPI, protein-protein interaction.
Survival analysis of each hub genes using online tool Kaplan-Meier Plotter
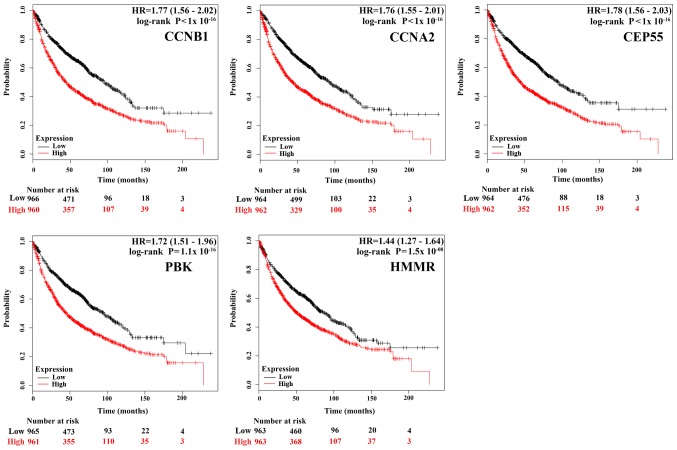
To gain insight into the associations between the hub genes and NSCLC, we did a survival analysis through an online tool Kaplan Meier Plotter, which contains a large number of microarray datasets of lung cancer. As shown in Fig. 3, the survival time of the top five hub genes was significantly separated between the high expression groups and low expression groups in NSCLC patients (P<0.05), with the low expression groups of these 5 hub genes exhibiting a good prognostic effect in NSCLC. Moreover, the remaining 20 hub genes also showed the same trend (data not shown). These imply that the expression levels of the 25 hub genes are significantly associated with clinical prognosis of NSCLC and they may play important roles in the progression of NSCLC.
Figure 3.
The prognostic value of the expression of the top five hub genes. The survival data was analyzed using Kaplan-Meier Plotter, and the survival curves were plotted based on data of all NSCLC patients (n=1,926), the red line represents patients with expression above the median, while black line represents expression below the median. HP represents hazard ratio.
Discussion
The tumorigenesis, progression and metastasis of lung cancer, like in any other cancer is considered as a very complex process as it involves aberrations of multiple genes and cellular pathways (22). Some DEGs that have multi-interactions with other DEGs are probably core functional genes in promoting the carcinogenesis (23,24). To improve diagnosis and treatment of NSCLC, it is vitally important to find these abnormal genes and understand their roles in the molecular mechanism of NSCLC. With the development of microarray and high throughput technologies, we have been able to detect the cancer etiology by examining aberrations in whole-genome level as these technologies have been widely used to predict the potential therapeutic targets for cancers (25).
In the present study, using the gene expression profiles of GSE21933, GSE33532, GSE44077 and GSE74706 to screen the co-expressed DEGs between NSCLC and normal samples, a total of 195 DEGs, consisting of 57 upregulated and 138 downregulated DEGs were obtained. The observation that, upregulated DEGs were enriched in BP terms related to mitosis, cell cycle and cell proliferation is consistent with previous knowledge that the functional deficits of cell cycle and cell proliferation regulators are the main causes of tumorigenesis and progression (26). The downregulated DEGs were enriched in BP terms such as angiogenesis and cell adhesion. Angiogenesis is an essential biological process for tumor growth and metastasis in that, its blockage in tumor tissue has been recognized as a charming strategy to inhibit tumor growth (27). According to Cavallaro and Christofori (28), changes in the expression of cell adhesion molecules could affect the adhesive repertoire of a cell, signal transduction status of cells, the interactions between cells and their environment, and play a crucial role in tumor progression, invasion and metastasis. Our results suggested that these up and downregulated DEGs are involved in these BP and may play important role in the progression of NSCLC.
Based on the PPI network, we identified a series of hub genes and among the list of the top five hub genes were CCNB1, CCNA2, CEP55, PBK and HMMR, respectively. CCNB1, as a key regulatory protein involved in mitosis, is essential for G2/M transition during the cell cycle (29). Yuan et al (30). reported that its depletion inhibited proliferation and induced apoptosis in human tumor cells. The work of Soria et al (31). established the expression of CCNB1 in a significant fraction but not all types of NSCLC. They showed that, different subtypes of NSCLC are not only biologically different but also showed variation in their CCNB1 expressions. Of all the histological subtypes examined, overexpression of CCNB1 was more frequently observed in the squamous cell carcinoma (SCC) subtypes. This overexpression was also reported to affects patient survival time and might be an adverse prognostic marker for SCC-subtype NSCLC patients (31). While our work establishes CCNB1 as a top hub gene in NSCLC in agreement with these reports, it did not specify the subtypes, which could be a possible limitation to our analysis. The second hub gene CCNA2, encoding cyclins controls both the G1/S and the G2/M transition of the cell cycle (32). Its protein expression was found to be elevated in variety of tumors, including breast, liver, prostate and lung cancers, and appears to be a prognostic marker in prediction of survival or early relapse (33). The third hub gene CEP55 (also known as c10orf3 and FLJ10540), a pivotal component of cytokinesis, is associated with the PI3K/AKT pathway activation via an interaction with the catalytic subunit of PI3K (34). CEP55 overexpression is highly correlated with carcinogenesis across multiple cancer types including lung, breast, liver, and colon cancers and has been identified as a member of several prognostic ‘gene signatures’ for cancer (35). Tao et al (36). has reported that ectopic expression of CEP55 induces tumorigenesis in nude mice, while its knockdown in gastric cancer cells suppressed cellular proliferation by inducing G2/M phase arrest, indicating its potential as an anti-tumor target. PBK also known as TOPK, was found to play an important role in tumor formation and progression. Its high expression level was detectable in the majority of lung cancer tissues and cell lines, but not in normal tissues (37–40). Overexpression of PBK was reported to promote a PI3K/AKT-dependent cell migration (5). Lei et al (40). suggested that PBK expression was positively correlated with Ki67 and p53 expression, and could be used as an independent prognostic factor in NSCLC. Another hub gene which we found to be hyper-expressed in NSCLC is HMMR, a multifunctional oncogene. Its high expression level, which was also found in breast cancer, is associated with poor disease outcome (41). Recent studies have deciphered that HMMR could regulate cell proliferation, survival, and migration via forming a complex with CD44 (a well-known cancer stem cell marker) and hyaluronan (a key component of the microenvironment of most malignant tumors) (42). In this study, HMMR showed a strong interaction with each of the top five hub genes, which indicates their joint functions in NSCLC, although further investigations are still needed to clarify underlying biological associations between HMMR and NSCLC. All these reports indicate that CCNB1, CCNA2, CEP55, PBK, and HMMR are involved in the pathogenesis of malignant tumors mainly by affecting cell cycle and mitosis. This is consistent with our findings. Furthermore, the Kaplan Meier Plotter survival analysis from our study demonstrated that mRNA expression levels of these five hub genes, as well as the remaining 20 hub genes, were significantly associated with clinical prognosis of lung cancer. While these could imply their roles in the progression of NSCLC thereby making them a potential target for its diagnosis and treatment, further experimental verification of the clinical significance of each hub gene and its subtypes is still necessary.
In summary, using the bioinformatics, we have provided information on the DEGs and hub genes that may be involved in NSCLC. This analysis is, however, limited in that it does not the capture the expression of these genes in the different NSCLC subtypes. Another limitation of this work is that, in reducing the number of false positive DEGs, we obtained co-expressed DEGs among 4 different NSCLC associated microarray datasets. By this, many likely important genes might have been lost. While our work showed the benefit and usefulness of microarray analysis in bringing out the DEGs and hub genes that can be potential diagnostic and treatment targets of NSCLC, the need for improved analysis and prospective clinical studies is still imperative. Albeit, this study can contribute to the overall understanding of the underlying molecular mechanisms of NSCLC and serve as guide to subsequent experimental studies.
Acknowledgements
Not applicable.
Funding
This study was financially supported by Fundamental Research Funds for the Central Universities (grant no. XDJK2017D143), Fundamental Research Program of Hunan University of Medicine (grant no. 2014KY03) and Science and Technology Plan Projects of Huaihua (2014–17). Funding was also received from Chongqing Science and Technology Commission (grant no. cstc2015jcyjAoo61), Chongqing Educational Ministry (grant no. Kj1500637) and The National Key Research and Development Program of China for Traditional Chinese Medicine Modernization (grant nos. 2017YFC1702600 and 2017YFC1702605).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
YBX and XGL conceived and designed the study. YBX, MF, HYR and XH performed the data analysis. YBX and HYR wrote the paper. YBX, XGL, MF, HYR and XH reviewed and edited the manuscript. All authors read and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Semenova EA, Nagel R, Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev. 2015;29:1447–1462. doi: 10.1101/gad.263145.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danesi R, Pasqualetti G, Giovannetti E, Crea F, Altavilla G, Del Tacca M, Rosell R. Pharmacogenomics in non-small-cell lung cancer chemotherapy. Adv Drug Deliv Rev. 2009;61:408–417. doi: 10.1016/j.addr.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Shih MC, Chen JY, Wu YC, Jan YH, Yang BM, Lu PJ, Cheng HC, Huang MS, Yang CJ, Hsiao M, Lai JM. TOPK/PBK promotes cell migration via modulation of the PI3K/PTEN/AKT pathway and is associated with poor prognosis in lung cancer. Oncogene. 2012;31:2389–2400. doi: 10.1038/onc.2011.419. [DOI] [PubMed] [Google Scholar]
- 6.Keith RL, Miller YE. Lung cancer chemoprevention: Current status and future prospects. Nat Rev Clin Oncol. 2013;10:334–343. doi: 10.1038/nrclinonc.2013.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oxnard GR, Lo PC, Nishino M, Dahlberg SE, Lindeman NI, Butaney M, Jackman DM, Johnson BE, Jänne PA. Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J Thorac Oncol. 2013;8:179–184. doi: 10.1097/JTO.0b013e3182779d18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerber DE, Minna JD. ALK inhibition for non-small cell lung cancer: From discovery to therapy in record time. Cancer cell. 2010;18:548–551. doi: 10.1016/j.ccr.2010.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janku F, Stewart DJ, Kurzrock R. Targeted therapy in non-small-cell lung cancer-is it becoming a reality? Nat Rev Clin Oncol. 2010;7:401–414. doi: 10.1038/nrclinonc.2010.64. [DOI] [PubMed] [Google Scholar]
- 10.Lo FY, Chang JW, Chang IS, Chen YJ, Hsu HS, Huang SF, Tsai FY, Jiang SS, Kanteti R, Nandi S, et al. The database of chromosome imbalance regions and genes resided in lung cancer from Asian and Caucasian identified by array-comparative genomic hybridization. BMC Cancer. 2012;12:235. doi: 10.1186/1471-2407-12-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meister M, Belousov A, Xu EC, Schnabel PA, Warth A, Hoffmann H, Dienemann H, Riedlinger J, Bodenmueller H, Zolg W, et al. Intra-tumor heterogeneity of gene expression profiles in early stage. Non-small cell lung cancer. 2014 [Google Scholar]
- 12.Kadara H, Fujimoto J, Yoo SY, Maki Y, Gower AC, Kabbout M, Garcia MM, Chow CW, Chu Z, Mendoza G, et al. Transcriptomic architecture of the adjacent airway field cancerization in non-small cell lung cancer. J Natl Cancer Inst. 2014;106:dju004. doi: 10.1093/jnci/dju004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marwitz S, Depner S, Dvornikov D, Merkle R, Szczygieł M, Müller-Decker K, Lucarelli P, Wäsch M, Mairbäurl H, Rabe KF, et al. Downregulation of the TGFβ Pseudoreceptor BAMBI in non-small cell lung cancer enhances TGFβ signaling and invasion. Cancer Res. 2016;76:3785–3801. doi: 10.1158/0008-5472.CAN-15-1326. [DOI] [PubMed] [Google Scholar]
- 14.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 15.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Royal Statis Soc. 1995;57:289–300. [Google Scholar]
- 16.Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim A, et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 17.Gene Ontology Consortium. The gene ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 19.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szász AM, Lánczky A, Nagy Á, Förster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 23.Saito M, Shiraishi K, Kunitoh H, Takenoshita S, Yokota J, Kohno T. Gene aberrations for precision medicine against lung adenocarcinoma. Cancer Sci. 2016;107:713–720. doi: 10.1111/cas.12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardarella S, Johnson BE. The impact of genomic changes on treatment of lung cancer. Am J Respir Crit Care Med. 2013;188:770–775. doi: 10.1164/rccm.201305-0843PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang B, Li C, Zhao J. Identification of key pathways and genes in colorectal cancer using bioinformatics analysis. Med Oncol. 2016;33:111. doi: 10.1007/s12032-016-0829-6. [DOI] [PubMed] [Google Scholar]
- 26.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 27.Feng X, Ofstad W, Hawkins D. Antiangiogenesis therapy: A new strategy for cancer treatment. US Pharm. 2010;35(Oncology Suppl):S4–S9. [Google Scholar]
- 28.Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat Rev Cancer. 2004;4:118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- 29.Petri ET, Errico A, Escobedo L, Hunt T, Basavappa R. The crystal structure of human cyclin B. Cell Cycle. 2007;6:1342–1349. doi: 10.4161/cc.6.11.4297. [DOI] [PubMed] [Google Scholar]
- 30.Yuan J, Yan R, Krämer A, Eckerdt F, Roller M, Kaufmann M, Strebhardt K. Cyclin B1 depletion inhibits proliferation and induces apoptosis in human tumor cells. Oncogene. 2004;23:5843–5852. doi: 10.1038/sj.onc.1207757. [DOI] [PubMed] [Google Scholar]
- 31.Soria JC, Jang SJ, Khuri FR, Hassan K, Liu D, Hong WK, Mao L. Overexpression of cyclin B1 in early-stage non-small cell lung cancer and its clinical implication. Cancer Res. 2000;60:4000–4004. [PubMed] [Google Scholar]
- 32.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yam CH, Fung TK, Poon RY. Cyclin A in cell cycle control and cancer. Cell Mol Life Sci. 2002;59:1317–1326. doi: 10.1007/s00018-002-8510-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen CH, Lu PJ, Chen YC, Fu SL, Wu KJ, Tsou AP, Lee YC, Lin TC, Hsu SL, Lin WJ, et al. FLJ10540-elicited cell transformation is through the activation of PI3-kinase/AKT pathway. Oncogene. 2007;26:4272–4283. doi: 10.1038/sj.onc.1210207. [DOI] [PubMed] [Google Scholar]
- 35.Jeffery J, Sinha D, Srihari S, Kalimutho M, Khanna KK. Beyond cytokinesis: The emerging roles of CEP55 in tumorigenesis. Oncogene. 2016;35:683–690. doi: 10.1038/onc.2015.128. [DOI] [PubMed] [Google Scholar]
- 36.Tao J, Zhi X, Tian Y, Li Z, Zhu Y, Wang W, Xie K, Tang J, Zhang X, Wang L, Xu Z. CEP55 contributes to human gastric carcinoma by regulating cell proliferation. Tumour Biol. 2014;35:4389–4399. doi: 10.1007/s13277-013-1578-1. [DOI] [PubMed] [Google Scholar]
- 37.Zhu F, Zykova TA, Kang BS, Wang Z, Ebeling MC, Abe Y, Ma WY, Bode AM, Dong Z. Bidirectional signals transduced by TOPK-ERK interaction increase tumorigenesis of HCT116 colorectal cancer cells. Gastroenterology. 2007;133:219–231. doi: 10.1053/j.gastro.2007.04.048. [DOI] [PubMed] [Google Scholar]
- 38.Zykova TA, Zhu F, Lu C, Higgins L, Tatsumi Y, Abe Y, Bode AM, Dong Z. Lymphokine-activated killer T-cell-originated protein kinase phosphorylation of histone H2AX prevents arsenite-induced apoptosis in RPMI7951 melanoma cells. Clin Cancer Res. 2006;12:6884–6893. doi: 10.1158/1078-0432.CCR-06-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zlobec I, Molinari F, Kovac M, Bihl MP, Altermatt HJ, Diebold J, Frick H, Germer M, Horcic M, Montani M, et al. Prognostic and predictive value of TOPK stratified by KRAS and BRAF gene alterations in sporadic, hereditary and metastatic colorectal cancer patients. Br J Cancer. 2010;102:151–161. doi: 10.1038/sj.bjc.6605452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lei B, Liu S, Qi W, Zhao Y, Li Y, Lin N, Xu X, Zhi C, Mei J, Yan Z, et al. PBK/TOPK expression in non-small-cell lung cancer: Its correlation and prognostic significance with Ki67 and p53 expression. Histopathology. 2013;63:696–703. doi: 10.1111/his.12215. [DOI] [PubMed] [Google Scholar]
- 41.Pujana MA, Han JD, Starita LM, Stevens KN, Tewari M, Ahn JS, Rennert G, Moreno V, Kirchhoff T, Gold B, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet. 2007;39:1338–1349. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- 42.Hamilton SR, Fard SF, Paiwand FF, Tolg C, Veiseh M, Wang C, McCarthy JB, Bissell MJ, Koropatnick J, Turley EA. The hyaluronan receptors CD44 and Rhamm (CD168) form complexes with ERK1,2 that sustain high basal motility in breast cancer cells. J Biol Chem. 2007;282:16667–16680. doi: 10.1074/jbc.M702078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.