Abstract
Pathogenic fungi, including Candida glabrata, develop strategies to grow and survive both in vitro and in vivo under azole stress. However, the mechanisms by which yeast cells counteract the inhibitory effects of azoles are not completely understood. In the current study, it was demonstrated that the expression of the ergosterol biosynthetic genes ERG2, ERG3, ERG4, ERG10, and ERG11 was significantly upregulated in C. glabrata following fluconazole treatment. Inhibiting ergosterol biosynthesis using fluconazole also increased the expression of the sterol influx transporter AUS1 and the sterol metabolism regulators SUT1 and UPC2 in fungal cells. The microarray study quantified 35 genes with elevated mRNA levels, including AUS1, TIR3, UPC2, and 8 ERG genes, in a C. glabrata mutant strain lacking ERG1, indicating that sterol importing activity is increased to compensate for defective sterol biosynthesis in cells. Bioinformatic analyses further revealed that those differentially expressed genes were involved in multiple cellular processes and biological functions, such as sterol biosynthesis, lipid localization, and sterol transport. Finally, to assess whether sterol uptake affects yeast susceptibility to azoles, we generated a C. glabrata aus1∆ mutant strain. It was shown that loss of Aus1p in C. glabrata sensitized the pathogen to azoles and enhanced the efficacy of drug exposure under low oxygen tension. In contrast, the presence of exogenous cholesterol or ergosterol in medium rendered the C. glabrata AUS1 wild-type strain highly resistant to fluconazole and voriconazole, suggesting that the sterol importing mechanism is augmented when ergosterol biosynthesis is suppressed in the cell, thus allowing C. glabrata to survive under azole pressure. On the basis of these results, it was concluded that sterol uptake and sterol biosynthesis may act coordinately and collaboratively to sustain growth and to mediate antifungal resistance in C. glabrata through dynamic gene expression in response to azole stress and environmental challenges.
Keywords: Candida glabrata, azole resistance, fluconazole, voriconazole, ergosterol synthesis, sterol uptake
Introduction
Candida species are ubiquitous fungi and the most common fungal pathogens affecting humans. Candida, in particular, affects high-risk patients who are either immunocompromised or critically ill. More than 100 species of Candida exist, but only a few are recognized as causing disease in humans. Candida glabrata and Candida albicans account for 70 to 80% of yeasts isolated from patients with invasive candidiasis. In recent decades, C. glabrata has become important because of its increasing world-wide incidence and because of its developing resistance to antifungals, including azoles, in the clinic.
Azole antifungal agents such as fluconazole and voriconazole are approved therapy for candidemia and invasive candidiasis caused by C. glabrata (1). Mechanisms of azole resistance include i) increased drug export (efflux pumps), ii) lost drug targets (e.g., altered drug target binding site), iii) up-regulated homeostatic stress-response pathways to deal with azole-associated damage, and iv) changed biosynthetic pathways (particularly sterol synthesis) that circumvent or attenuate the effects of azole inhibition (2).
The cellular toxicity of azole antifungals occurs primarily through their ability to affect the fungal cell membrane by inhibiting the biosynthesis of ergosterol, the principal sterol in the fungal cell membrane, leading to the depletion of ergosterol and an accumulation of possibly toxic sterol intermediates in the cytoplasmic membrane. Azole antifungal compounds (imidazoles and triazoles) inhibit cytochrome P-450 dependent sterol 14α-demethylase (Erg11p), an enzyme that catalyzes the oxidative removal of the 14α-methyl group of lanosterol in the ergosterol biosynthetic pathway (2). Fungal cells exposed to azoles must either synthesize more endogenous ergosterol or import exogenous sterol if they are to survive antimycotic treatment.
Sterol uptake appears to confer resistance to antifungal drugs since mutant strains of Candida species lacking AUS1 and TIR3, the sterol influx transporters (3), or UPC2, the transcription factor that controls AUS1 and TIR3 expression (3–5), exhibit reduced uptake of cholesterol and are hypersensitive to azoles (6,7), whereas enhanced UPC2 or AUS1 expression and cholesterol uptake have been implicated in the azole-resistant phenotype (6,7). Furthermore, several studies have even suggested that pathogenic fungi can scavenge free sterols for the cell membrane, including cholesterol, resulting in resistance to polyene and azole antifungals (8). Increased uptake of exogenous cholesterol may be associated with drug resistance in clinical isolates of bile-dependent C. glabrata cells as well as in sterol auxotrophic C. glabrata strains (9). These observations indicate that the ability to scavenge exogenous sterols, when ergosterol biosynthesis is defective or is blocked by antimycotic agents, may play an important role in increased azole resistance in pathogenic fungal species.
Besides sterol uptake, alteration in sterol biosynthesis seems to also affect fungal susceptibility to antifungals. ERG11 is one of the critical genes in the ergosterol biosynthetic pathway, and its encoding protein product Erg11p (or CYP51p depending on nomenclature) is the major target enzyme of azole antifungals. Mutations in ERG11 are the most common mechanism of azole resistance in C. albicans (10). Over-expression of ERG11 has been attributed to decreases in azole activity in C. albicans. Alterations in other enzymes of the ergosterol biosynthetic pathway, particularly ERG3 (C-5 sterol desaturase), ERG5 (C-22 sterol desaturase), ERG6 (C-24 sterol methyl-transferase), and ERG25 (C-4 sterol methyloxidase), which are up-regulated with inhibition of Erg11p, have also been documented to reduce azole susceptibility of C. albicans. In C. glabrata, depletion of the ergosterol content in a CgERG1 mutant increases the levels of susceptibility to azoles, while complementation of the CgERG1 mutation restores drug sensitivity to wild-type levels (8). In both C. albicans and C. glabrata, regulation of sterol synthesis is vital to azole susceptibility.
In this study, we assessed the role of two protective mechanisms in conferring antifungal susceptibility in C. glabrata. We showed that expression of the genes involved in both sterol uptake and sterol synthesis is up-regulated in C. glabrata when ergosterol is depleted by azole treatment and when ergosterol biosynthesis is defective. We further corroborated that pathogenic fungi can accumulate exogenous sterols from the environment as a protective strategy to survive under azole and hypoxic stress. These findings suggest that sterol uptake and sterol synthesis may act coordinately and collaboratively to sustain growth and to mediate antifungal tolerance in C. glabrata through dynamic gene expression in response to azole pressure and environmental changes.
Materials and methods
Cell culture and drug treatment
All C. glabrata strains (Table I) used in the present study were cultured on YPD agar containing 1% Bacto yeast extract (Difco Laboratories, Detroit, MI, USA), 2% Bacto peptone (Difco Laboratories), and 2% glucose (Sigma-Aldrich, St. Louis, MO, USA). NCCLS84, Cg1660, CgTn201S, and CgTn201Su/aus1 were also grown on minimal (MIN) agar containing 0.67% Yeast Nitrogen Base without amino acids (Difco Laboratories) plus 2% glucose. The ura3 mutants Cg84u, Cg1660u, and CgTn201Su were grown in MIN medium supplemented with 20 µg/ml of uracil (Sigma-Aldrich) or were selected on a MIN plus uracil agar plate containing 0.1% 5-fluoroorotic acid (FOA) (Lancaster, Pelham, NH, USA). YEPG agar was used for drug sensitivity assay, which contained 1% Bacto yeast extract (Difco Laboratories), 2% Bacto peptone (Difco Laboratories), 3% glycerol (Invitrogen/Life Technologies, Carlsbad, CA, USA), 1% ethanol (Warner-Graham, Inc., Cockeysville, MD), and 2% agar (Difco Laboratories).
Table I.
Candida glabrata strains used in this study.
| Strain | Parental strain | Genotype or description | Reference or source |
|---|---|---|---|
| NCCLS84 | Wild-type (ATCC90030)a | ATCCa | |
| Cg84u | NCCLS84 | Cgura3 | (8) |
| Cg1660 | Clinical isolate | FHCRCb | |
| Cg1660u | Cg1660 | Cgura3 | (8) |
| CgTn201S | Cg1660u | Cgura3 Cgerg1::Tn5<Cm URA3> | (8) |
| CgTn201Su | CgTn201S | Cgura3 Cgerg1::Tn5<Cm ura3> | Present study |
| CgTn201Su/aus1 | CgTn201Su | Cgura3 Cgerg1::Tn5<Cm ura3> | Present study |
| Cgaus1∆::URA3 |
American Type Culture Collection (ATCC), Manassas, VA, USA.
FHCRC, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Fluconazole (Euroasian Chemicals Private Ltd., Mumbai, India) was added to cultures of each strain at a final concentration of 200 µg/ml, followed by continued incubation with shaking for 2 h. Cell cultures without fluconazole treatment served as controls. After incubation, the cells were harvested, and the cell pellets were stored at −80°C until the subsequent isolation of RNA.
The BBL GasPak Plus gas generator envelope (Becton Dickinson Microbiology Systems, Sparks, MD, USA) or the BBL GasPak Pouch Anaerobic Systems (Becton Dickinson Microbiology Systems, Cockeysville, MD, USA) was used for cultures grown under conditions of low oxygen tension (hypoxia). To mimic the host environment of animals and humans, C. glabrata cells were also grown in the presence of 5% CO2 under low oxygen conditions using an InvivO2 400 workstation at 37°C with shaking at 200 rpm (Ruskinn Technology Ltd., Bridgend, UK). For analysis of sterol uptake and exogenous sterol utilization, 2 mg of ergosterol or cholesterol was dissolved in a mixture of 50% ethanol and 50% Tween-80 (Sigma-Aldrich) to give a 2 mg/ml stock solution, which was used to supplement the media with a final sterol concentration of 20 µg/ml. For comparison, the same final concentration of ethanol-Tween-80 without sterol was used.
Drug sensitivity assay
The susceptibility of the C. glabrata strains Cg1660, CgTn201S, and CgTn201Su/aus1 to fluconazole and voriconazole was determined on MIN and YEPG agar media using an E-test (AB Biodisk, Solna, Sweden) according to the manufacturer's instructions. In brief, logarithmic-phase cells were harvested and adjusted to the desired concentrations by counting the number of cells with a hemocytometer. From each cell suspension, 200 µl (1×106 cells/ml) was plated in duplicate on MIN and YEPG agar. All plates were incubated under conditions of low oxygen tension at 37°C for 3 days. Growth inhibition zones (minimum inhibitory concentration, MIC) to fluconazole and voriconazole were measured.
RT-qPCR analysis
Total RNA was extracted from C. glabrata (Cg84u) logarithmic-phase cultures grown in YPD broth using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's instructions. RNA was converted to cDNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) as previously described (11). Primers and probes were designed in our laboratory using the primer analysis software Primer Express 3.0 (Applied Biosystems; Thermo Fisher Scientific, Inc.). TaqMan probes were synthesized by Applied Biosystems (Thermo Fisher Scientific, Inc.) and primers were synthesized by Invitrogen (Thermo Fisher Scientific, Inc.). The sequences of TaqMan probes and forward and reverse primers, as well as the gene numbers for all genes assessed in this study, are listed in Table II. To study gene expression, the amplification was detected in real time using both SYBR Green chemistry (SYBR-Green PCR Master Mix; Applied Biosystems; Thermo Fisher Scientific, Inc.) and TaqMan chemistry (TaqMan Universal PCR Master Mix; Applied Biosystems; Thermo Fisher Scientific, Inc.) as described previously (11).
Table II.
Primers and TaqMan probes for RT-qPCR analysis of gene expression used in this study.
| Gene | Primer and probe sequence (5′→3′) | Gene numbera |
|---|---|---|
| ACT1 | ||
| F | TTGGACTCTGGTGACGGTGTTA | CAGL0K12694g |
| R | AAAATAGCGTGTGGCAAAGAGAA | |
| P | CCACGTTGTTCCAATTTACGCCGG | |
| RDN5.8 | ||
| F | CTTGGTTCTCGCATCGATGA | CAGL0L13387r |
| R | GGCGCAATGTGCGTTCA | |
| P | ACGCAGCGAAATGCGATACGTAATGTG | |
| ERG2 | ||
| F | TCCCAGGTATGACCCATCATC | CAGL0L10714g |
| R | TGCGAAGGAGTTTTGATCCAT | |
| P | ACAAAAGGGCTACGCAAAGCAATACGC | |
| ERG3 | ||
| F | TGCACTGGCCTCGTGTCTAC | CAGL0F01793g |
| R | TAACCGTCGACTGGGTGGAA | |
| P | TGGTTGGTCTGCACTCCATTCGCC | |
| ERG4 | ||
| F | CCCTCAATTAGGTGTCGTCATGT | CAGL0A00429g |
| R | GGCACGATTAATTCTTCACCCTTA | |
| P | CCACTGGCTGTACGCTAACGCTTGTG | |
| ERG10 | ||
| F | GCCAGAACCCCAATTGGTT | CAGL0L12364g |
| R | TGCAATGACACCTAGGTCAACAG | |
| P | TTCCAAGGTGCGTTGGCCTCCA | |
| ERG11 | ||
| F | TGTCTTGATGGGTGGTCAACA | CAGL0E04334g |
| R | CTGGTCTTTCAGCCAAATGCA | |
| P | CTTCCGCTGCTACCTCCGCTTGG | |
| PDR16 | ||
| F | CCTGGAGACGTGAATTTGGAAT | CAGL0J07436g |
| R | ACAGCAACCAAATCCGATGTAA | |
| P | CTCGTCACCATTTTCTTCACCCAAATGG | |
| PDR16 | ||
| F | TTGGCCTGGAGACGTGAATT | CAGL0J07436g |
| R | GTTTACCACTTTCATTCTCTACAGCAA | |
| P | TTGGGTGAAGAAAATGGTGACGAGGTTACA | |
| AUS1 | ||
| F | CCAAGCCACTGCAGGTGAA | CAGL0F01419g |
| R | GGCGTGAAACAGGGACTTGA | |
| P | CGGTGCCCCAACGTCGGGTATC | |
| SUT1 | ||
| F | GTTGATGGCATTACATGGCAAT | CAGL0I04246g |
| R | AGTAAAGGAGTTGGATGATGAGTGAA | |
| P | ACCAATTCCTATCGCCTCCAATGCCA | |
| SUT2 | ||
| F | AGGGCCTTCAAGGTATCGAAGT | CAGL0L09383g |
| R | TCGGTTTTTGGATCACACCAA | |
| P | TTGCCTCTCCAAAACAGAAACTACCCTCCC | |
| ECM22 | ||
| F | CAATTACAAGAGCATGCAAACATTG | CAGL0C01199g |
| R | GGAGTTAGCCTGACCATGAGTATTATT | |
| P | TGCATCAGAAACAGCATATCCAACGACTGT | |
| UPC2A | ||
| F | AAAATAGTACAGGAGCAACGGAGACT | CAGL0C01199g |
| R | TGGTTGCACCTGGAGATGAA | |
| P | CTGTCGCCTTCTCTGAATCTGCTTACACCC | |
| UPC2B | ||
| F | GGTCGCAAGTGCATTGTTGT | CAGL0F07865g |
| R | TCAGTCGCATTTGATGTATCTTTAGG | |
| P | CGTGGAATAATCACGATCCTCACATGCA |
Candida Genome Database (www.candidagenome.org). F, forward primer; R, reverse primer; P, TaqMan probe, labeled as 5′-FAM, 3′-TAMRA; RT-qPCR, reverse transcription-quantitative polyermase chain reaction.
Microarray hybridization and data analysis
DNA microarray analysis was used to identify genes with altered expression in the C. glabrata erg1 mutant strain CgTn201S and the wild-type strain Cg1660. Total RNA was isolated from the log phase culture of C. glabrata grown in YPD by using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) and the RNeasy MiniElute Cleanup kit (Qiagen, Valencia, CA, USA). RNA was assessed for quality using the Agilent 2100 Bioanalyzer with the RNA 6000 Nano Reagent Kit. Pin-spotted 70-mer oligonucleotide in-house arrays fabricated at the National Institute of Allergy and Infectious Diseases (NIAID) were used for analysis of the pair strains. In brief, a total of 5,908 70-mer oligonucleotides were purchased from Institut Pasteur (Paris, France) and were used for microarray printing at the NIAID Microarray Research Facility. Expression of each open reading frame (ORF) was measured by hybridization to a specific 70-mer oligonucleotide. Thirty micrograms of total RNA from the erg1 mutant strain CgTn201S and the wild-type strain Cg1660 was reverse-transcribed to cDNA to incorporate the fluorescent Cy3-dUTP and Cy5-dUTP (GE Healthcare, Piscataway, NJ, USA), respectively. The labeled cDNA of the paired mutant/wild-type strain was combined and used for microarray hybridization. Six microarrays were performed for analysis of the erg1 mutant/wild-type pair, including two with reciprocal labeling. The microarrays were prehybridized at 42°C in prehybridization buffer [5 × SSC (1 × SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1% bovine serum albumin (BSA), 0.1% SDS] for 30 to 60 min and then hybridized to the labeled cDNA in 50 µl of hybridization buffer [25% formamide, 5 × SSC, 0.2% SDS, 20 µg/ml poly (dA)40-60, 200 µg/ml Cot-1 DNA (Invitrogen; Thermo Fisher Scientific, Inc.), 80 µg/ml yeast tRNA] overnight at 42°C. The microarrays were washed three times in wash buffer A (1 × SSC, 0.05% SDS) and wash buffer B (0.1 × SSC). The in-house arrays were scanned with a GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA, USA). All microarray data archive and statistical calculations were performed on the ‘processed signal’ data by using the Web-based mAdb analysis system provided by the Bioinformatics and Molecular Analysis group (BIMAS) at the Center for Information Technology (CIT), National Institutes of Health. The data were filtered with the parameters that included genes present in four or more arrays and each array with 80% or more genes present. The data set of the paired strains was then analyzed by Student's t-test. The genes with P-values less than 0.001 and with at least 1.5-fold altered gene expression were then selected. The final data set included all genes with altered expression in the pair.
Cloning of CgAUS1
The full nucleotide sequence of the C. glabrata AUS1 homolog (CgAUS1) was obtained from the Candida Genome Database (www.candidagenome.org). A 1,087 bp partial ORF of the CgAUS1 gene was obtained by PCR with PfuUltra High-Fidelity DNA polymerase (Stratagene, La Jolla, California, USA), primer set CgAUS1S (5′-TTGAAGTTGCCTCTTGACTC-3′) and CgAUS1AS (5′-AAGGCAACAAACACAGCGGCAG-3′), and the total genomic DNA of Cg1660 was used as the template. The PCR parameters were 2 min at 95°C, followed by 30 cycles of 95°C for 30 sec, 53°C or 55°C for 30 sec, and 72°C for 1 min 30 sec, and then extended at 72°C for 10 min. The 1,087 bp PCR product, which was used as a probe for Southern blotting, was then cloned into pCR-Blunt II-TOPO (Invitrogen; Thermo Fisher Scientific, Inc.) to produce plasmid pCgAUS1.
Construction of the Candida glabrata Cgerg1 and Cgaus1∆ double mutant
The Cgerg1 mutant was generated in our laboratory as described previously (8). Gene deletion of Cgaus1 was performed as described by Vermitsky et al (12). Briefly, the primer pair CgAUS1D1S (TTGAAATTCTCGGAAAGAAACATCAAATCAAAAAATTTTAACCTTCTAAAACTTGTTCTTTTTTTGGGAAATATAAGATGTCGAAAGCTACATATAAGGA) and CgAUS1D2AS (AGTAGATTAAAGAAAAGTGTTAAATTTAAGAATAAAATGGAATTGTTATTTCATTAAAAGCTTGTAGGAGTCACTCTTATTAGTTTTGCTGGCCGCATCT) was used to generate the CgAUS1 deletion cassette using YEP24 as the template to amplify Saccharomyces cerevisiae URA3, which served as the selection marker. An additional round of PCR using the primer pair CgAUS1D3S (CCCCAACTATCAATTTTCTTTAAATCAAGGAAAATCTATTACATTCGCTATTAATCTCTACTATCTTTATCTTAGTTTTTTGAAATTCTCGGAAAGAAAC) and CgAUS1D4AS (ATTATATTTTAAATTTTGTTGTATAGCTTTTTTGCTGTAAAGGTGAAAAAACCGGGAATTTTGAGCATTAGTATTAGTAGATTAAAGAAAAGTGTTAAAT) extended the 5′ and 3′ ends with about a 160 bp homologous region on each end of the CgAUS1 deletion cassettes to increase the efficiency of homologous recombination. S. cerevisiae URA3 was transformed into CgTn201Su (Cgura3 Cgerg1::Tn5<Cm ura3>) to construct the Cgerg1/aus1 double mutant CgTn201Su/aus1 via a successful double crossover homologous recombination, which resulted in replacement of the CgAUS1 ORF with S. cerevisiae URA3 (Fig. 1A). The CgERG1 locus in the clinical isolate Cg1660 and its transposon mutant CgTn201S (Cgerg1) are shown in Fig. 1B. Transformants were selected on MIN medium, and the deletion of CgAUS1 was confirmed by Southern blot analysis of the EcoRI-digested genomic DNA (Fig. 1C). The purified 1.2-kb ScURA3 probe detected a single signal in the Cgerg1 mutant CgTn201S; two signals were detected in the two putative Cgerg1/aus1 mutants CgTn201Su/aus1-1 and CgTn201Su/aus1-2, while there was no signal in the parental strain Cg1660 (Fig. 1C, right panel). The 1087-bp CgAUS1 probe detected a single 6.3-kb signal in Cg1660 and CgTn201S, but not in the two putative Cgerg1/aus1 mutants CgTn201Su/aus1-1 and CgTn201Su/aus1-2 (Fig. 1C, left panel).
Figure 1.
Southern hybridization analysis confirms the deletion and targeted gene replacement of CgAUS1 in Candida glabrata. (A) Deletion strategy of CgAUS1 in the strain CgTn201Su (Cgerg1/ura3). The CgAUS1 ORF was replaced with ScURA3 in the mutant CgTn201Su/aus1 (Cgerg1 Cgaus1Δ). (B) The CgERG1 locus in the clinical isolate Cg1660 and its transposon mutant CgTn201S (Cgerg1). (C) Southern blot analysis. The genomic DNAs of Cg1660, CgTn201S, and the mutant CgTn201Su/aus1 were digested with EcoRI, electrophoresed, and blotted onto a nylon membrane. The membrane was hybridized with the 1087-bp CgAUS1 probe (left panel) or 1.2-kb ScURA3 probe (right panel). Lanes (from the left): 1. Cg1660, clinical isolate; 2. CgTn201S, Cgerg1 mutant; 3. and 4. CgTn201Su/aus1, Cgerg1 Cgaus1Δ double mutant. The detected signals for CgAUS1 (left panel) and ScURA3 (right panel) are indicated by arrows.
Southern blot analysis
Candida glabrata genomic DNA was isolated from cultures grown in YPD overnight by using the MasterPure Yeast Purification Kit (Epicentre, Madison, WI, USA). Purified DNA fragments were recovered using the Strataprep Gel DNA Extraction kit (Stratagene; Agilent Technologies, Inc., Santa Clara, CA, USA). Hybond-N nylon membranes (Amersham, Arlington Heights, IL, USA) were used for Southern hybridization analyses. DNA probes were labeled with [α-32P] dCTP or [α-32P] dATP (MP Biomedical, Solon, OH, USA) by using the Prime-It II kit (Stratagene; Agilent Technologies, Inc.). DNA cloning and hybridization analyses were done according to the standard protocol.
Other techniques and reagents
DNA sequencing was done using the DNA sequencing kit with a dRhodamine dye terminator (Applied Biosystems; Thermo Fisher Scientific, Inc.) and an ABI automatic DNA sequencing system (Perkin-Elmer, Foster City, CA, USA). For sequencing of PCR products, PfuUltra DNA polymerase (Stratagene; Agilent Technologies, Inc.) was used for PCR amplification to minimize the rate of PCR-introduced mutations. The PCR products were cleaned with the Strataprep PCR Purification kit (Stratagene; Agilent Technologies, Inc.) and were used as templates for DNA sequencing.
Bioinformatic and statistical analysis
The Gene Ontology (GO) enrichment analysis and the network visualization of the biological functions related to the differentially expressed genes were performed using Cytoscape software with associated plug-ins (13). To create a functional network by selecting the ClueGO: Function, GO: Biological process, all the network evidence and only the terms with various levels of significance (P<0.1-<0.0005) were taken into consideration using the plug-in ClueGO (14), which was followed by enrichment of the functional network with the plug-in CluePedia (15).
Results of RT-qPCR for each gene are presented as the fold-change over the expression in the sample not treated with fluconazole, which was set as 1. The difference in triplicate Cq values (∆Cq) was used to calculate the difference between untreated and azole-treated samples. Student's t-test was used to analyze the statistical significance of differences between untreated and fluconazole-treated C. glabrata cells, and P-values were adjusted using Holm's adjustment for multiple testing. Values of P<0.05 were taken to indicate significance.
Results
Azole stress up-regulates the genes involved in sterol uptake and biosynthesis
Ergosterol biosynthetic genes encode the enzymes in the sterol synthetic pathway leading to ergosterol biosynthesis in yeast. It is of interest to understand whether the inhibition of ergosterol biosynthesis by azoles alters endogenous sterol synthesis in C. glabrata through changes in the expression of ergosterol biosynthetic genes. We found that mRNA levels of CgERG2, CgERG3, CgERG4, CgERG10, and CgERG11 were strikingly increased in C. glabrata following fluconazole treatment. As seen in Table III, the extent of inducible up-regulation of ERG gene expression by fluconazole in these cells ranged from more than 5-fold in CgERG3 to 19-fold in CgERG10 compared with the untreated controls. Interestingly, we also showed a 3.7-fold increase in mRNA expression of CgPDR16 in fluconazole-treated C. glabrata cells (Table III); PDR16p is an ATP-binding cassette (ABC) transporter involved in the sterol biosynthetic process and phospholipid transport in S. cerevisiae (16).
Table III.
Upregulated expression of sterol biosynthetic and sterol transporter genes in Candida glabrata under fluconazole stress.
| Gene | mRNA levels |
|---|---|
| ERG2 | 11.79a |
| ERG3 | 5.04a |
| ERG4 | 11.08a |
| ERG10 | 19.07a |
| ERG11 | 5.87a |
| PDR16 | 3.73a |
| AUS1 | 2.51b |
| SUT1 | 2.60b |
| SUT2 | 0.97 |
| UPC2A | 4.71a |
| UPC2B | 1.60b |
Values indicate the fold-change in RNA transcription for each gene in 200 µg/ml fluconazole-treated C. glabrata after normalization to RDN5.8 as the reference, as compared with the level in untreated cells, which was set as 1.
P<0.05 or
P<0.01 between fluconazole-treated and untreated C. glabrata cells for all values shown in this table.
AUS1 and UPC2 (or its paralog ECM22) have been demonstrated to be responsible for sterol uptake in S. cerevisiae (17). We next addressed whether inhibition of sterol synthesis by azoles alters the expression of these genes in C. glabrata. Sequencing data have shown that C. glabrata is more closely related to Saccharomyces cerevisiae than to C. albicans, and some genes are functionally exchangeable between C. glabrata and S. cerevisiae. Therefore, to identify AUS1 and ECM22/UPC2 orthologs in C. glabrata, we selected the S. cerevisiae AUS1, ECM22, and UPC2 genes as queries to perform homology searches. A BLAST search against the C. glabrata open reading frame (ORF) nucleotide and amino acid sequences (www.candidagenome.org) predicted CAGL0F01419g (CgAUS1) to be the ortholog of AUS1. We also performed a BLAST search against C. glabrata ORF nucleotide sequence alone, which predicted CAGL0C01199g and CAGL0F07865g to be possible orthologs of ECM22 or UPC2. The amino acid sequences encoded by these two genes are particularly highly homologous to Upc2p or Ecm22p. We refer to CAGL0C01199g and CAGL0F07865g as UPC2A (ECM22) and UPC2B (ECM22), respectively. This is in concordance with the work of Nagi et al (4) and Whaley et al (18). Our results demonstrate a transcriptional up-regulation of over two-fold and over four-fold in CgAUS1 and CgUPC2A expression, respectively, in our model following fluconazole challenge (Table III). These data clearly show that inhibition of ergosterol synthesis by azoles induce the expression of sterol transporter genes and likely lead to enhanced sterol uptake in C. glabrata.
Besides its role in anaerobic sterol uptake, UPC2 has been shown to be an activator of ergosterol biosynthetic genes in yeast (4,18). As the up-regulation of CgUPC2 expression was induced by fluconazole in C. glabrata, this result prompted us to address whether azoles alter the expression of other genes encoding transactivators responsible for sterol synthesis or uptake in fungi. It has been demonstrated that SUT1 is a transcription factor involved in sterol uptake and synthesis in aerobically growing S. cerevisiae cells (19). Therefore, to identify SUT1 ortholog in C. glabrata, we selected the S. cerevisiae SUT1 gene as queries to perform homology searches. A BLAST search against the C. glabrata ORF nucleotide sequence (www.candidagenome.org) predicted CAGL0I04246g and CAGL0L09383g to be possible orthologs of SUT1. The amino acid sequences encoded by these two genes are particularly highly homologous to Sut1p. We refer to CAGL0I04246g and CAGL0L09383g as CgSUT1 and CgSUT2, respectively. In the current model, we show that the expression of CgSUT1, but not CgSUT2, was significantly increased by fluconazole in C. glabrata (Table III).
Up-regulated expression of the genes involved in sterol uptake and biosynthesis in Candida glabrata erg1 mutant
To further assess whether ergosterol depletion affects the genes involved in sterol uptake and sterol synthesis in C. glabrata, we used DNA microarray to analyze gene expression in a C. glabrata erg1 mutant with defective ergosterol biosynthesis. Transcriptional profiling of the microarrays revealed 35 genes up-regulated and 4 genes down-regulated in the C. glabrata erg1 mutant CgTn201S compared to the parental wild-type strain Cg1660. Particularly, C. glabrata erg1 mutant had 2.5-fold up-regulation of CgAUS1 mRNA, 5.6-fold up-regulation of CgTIR3 mRNA, and 3.5-fold up-regulation of CgUPC2B mRNA over the wild-type cells (Fig. 2). To a lesser extent, we also found that the up-regulation of CgERG2, CgERG3, CgERG5, CgERG6, CgERG7, CgERG10, CgERG11, and CgERG29 in the C. glabrata erg1 mutant cells ranged from 1.5-to 2.8-fold mRNA expression among these genes (Table IV), which is similar to our data from the C. glabrata cells in which ergosterol synthesis was inhibited by fluconazole (Table III). Additionally, we show a 3-fold increase in mRNA expression of HES1/KES1, which is involved in ergosterol biosynthesis, oxysterol binding, and sterol transport (Table IV). These data highlight the notion that blocking ergosterol synthetic pathway leading to ergosterol depletion causes increased sterol uptake activity and sterol biosynthesis in C. glabrata.
Figure 2.
Expression of CgAUS1, CgTIR3, and CgUPC2B is up-regulated in Candida glabrata erg1 mutant. DNA microarray was used to analyze gene expression in the C. glabrata erg1 mutant CgTn201S and its parental wild-type strain Cg1660 as described in Materials and methods. Six microarrays were performed for analysis of the Cgerg1 mutant/wild-type pair, including two with reciprocal labeling. The average mRNA expression levels were based on mean of six replicates. Transcriptional profiling of the microarrays revealed the up-regulation (fold-change) of CgAUS1, CgTIR3, and CgUPC2B in CgTn201S (Cgerg1) as compared with Cg1660 (wild-type), which was set as 1. *P<0.01 between the C. glabrata erg1 mutant CgTn201S and its parental wild-type strain Cg1660 using Student's t-test.
Table IV.
Candida glabrata genes up- and down-regulated ≥1.5-fold in response to CgERG1 disruption (Cgerg1 mutation) in Cg1660 host.
| C. glabrata designation | S. cerevisiae homologue | Description | Fold expressiona |
|---|---|---|---|
| Upregulated genes | |||
| CAGL0F01419g | AUS1 | ATP-binding cassette transporter involved in sterol uptake | 2.5006 |
| CAGL0C03872g | TIR3/YIL011w | Putative GPI-linked cell wall protein involved in sterol uptake | 5.6547 |
| CAGL0F07865g | UPC2B | Transcription factor transcriptionally regulates ergosterol biosynthetic genes and sterol transporter genes | 3.4979 |
| CAGL0L10714g | ERG2 | C-8 sterol isomerase participates in ergosterol biosynthesis | 2.0136 |
| CAGL0F01793g | ERG3 | C-5 sterol desaturase participates in ergosterol biosynthesis | 2.7363 |
| CAGL0M07656g | ERG5 | C-22 sterol desaturase participates in ergosterol biosynthesis | 2.3415 |
| CAGL0H04653g | ERG6 | C-24 sterol methyltransferase participates in ergosterol biosynthesis | 2.1176 |
| CAGL0J10824g | ERG7 | Lanosterol synthase participates in ergosterol biosynthesis | 2.31 |
| CAGL0L12364g | ERG10/POT14 | Acetyl-CoA C-acetyltransferase participates in ergosterol biosynthesis | 2.2113 |
| CAGL0E04334g | ERG11 | Lanosterol 14-a-demethylase involved in ergosterol biosynthesis | 2.8477 |
| CAGL0K03927g | ERG29 | Roles in ergosterol biosynthesis, mitochondrion organization, etc | 1.5044 |
| CAGL0J03916g | HES1/KES1 | Roles in ergosterol biosynthesis, oxysterol binding, sterol transport, etc | 2.9779 |
| CAGL0J00297g | YHR045w | Possible roles in iron, amino acid, and carbohydrate metabolisms | 1.5889 |
| CAGL0A01089g | YPL272c | Alcohol O-acetyltransferase with role in alcohol metabolic process | 4.0299 |
| CAGL0I01408g | CYC1 | Cytochrome-c isoform 1 involved in mitochondrial electron transport | 2.6164 |
| CAGL0L03828g | CYB5 | Cytochrome b5 involved in oxidation-reduction process | 1.5483 |
| CAGL0K10868g | CTA1 | Catalase A involved in cellular response to oxidative stress | 2.7464 |
| CAGL0K12100g | HEM13 | Coproporphyrinogen III oxidase involved in heme biosynthesis | 2.3296 |
| CAGL0G03905g | ISA1 | Regulation of ROS metabolic process and biotin biosynthetic process | 1.5708 |
| CAGL0H04851g | PPZ1 | Protein phosphatase Z involved in cation homeostasis and cell wall integrity | 1.633 |
| CAGL0L07480g | NRG1/NRG2 | Transcription factor activity, sequence-specific DNA binding activity | 1.613 |
| CAGL0F01485g | TIR4 | Putative GPI-linked cell wall mannoprotein of the Srp1p/Tip1p family | 5.4181 |
| CAGL0H09614g | TIR1 | Putative GPI-linked cell wall protein | 6.3371 |
| CAGL0C00110g | FLO1 | Member of the FLO family of cell wall flocculation proteins | 1.9697 |
| CAGL0M04125g | YNL320w | Roles in cell polarity, endoplasmic reticulum, mitochondrion, etc | 1.5089 |
| CAGL0E00187g | YMR317w | Putative adhesin-like protein; belongs to adhesin cluster IV | 4.3814 |
| CAGL0G04499g | SET4/YJL105w | Ortholog of S. cerevisiae: SET4 and YJL105w | 1.9778 |
| CAGL0F08965g | MSC7/YHR039c | Roles in cytosol, endoplasmic reticulum, nucleus localization, etc | 1.6906 |
| CAGL0C00209g | DAN1/YJR151c | Putative adhesin-like cell wall protein; predicted GPI-anchor | 4.6475 |
| CAGL0G10175g | DAN1/YJR151c | Adhesin-like protein; predicted GPI anchor | 4.7887 |
| CAGL0K04279g | SCM4/YGR049w | Ortholog(s) have mitochondrial outer membrane localization | 1.5542 |
| Downregulated genes | |||
| CAGL0H03971g | YCP4/PST2 | Roles in cellular response to oxidative stress, mitochondrion, etc | 1.5484 |
| CAGL0M05995g | PET10 | Roles in lipid metabolism, respiratory growth, and ATP/ADP exchange | 1.5975 |
| CAGL0G05566g | FMP45 | In mitochondria; role in ascospore formation, cellular response to drug, etc | 1.5862 |
| CAGL0L10142g | RSB1/YOR049c | Sphingolipid transporter; involved in fatty acid transport | 2.1631 |
The Candida glabrata erg1 mutant CgTn201S vs. the parental wild-type Cg1660.
Besides the genes for sterol uptake and synthesis, 23 other genes demonstrated dominant and reproducible expression in C. glabrata erg1 mutant with the average level of the six microarrays ranging from 1.5-fold to 6.3-fold greater than the wild-type strain (Table IV). These differentially expressed genes were associated with multiple different cellular metabolisms and biological processes, such as iron, amino acid, and carbohydrate metabolisms, alcohol metabolism, reactive oxygen species (ROS) metabolism, heme biosynthesis, biotin biosynthesis, mitochondrion organization, mitochondrial electron transport, oxidation-reduction process, cation homeostasis, cell wall integrity, and in-cell polarity, endoplasmic reticulum, mitochondrion, nucleus localization, and so forth (Table IV). Furthermore, mRNA expression of 4 genes was down-regulated 1.5- to 2.2-fold lower than the parental wild-type cells (Table IV). These down-regulated genes were enriched in several cellular and metabolic processes, including lipid metabolism, sphingolipid and fatty acid transport, respiratory growth, ATP/ADP exchange, and cellular response to oxidative stress (Table IV).
Potential coordination between sterol biosynthesis and sterol uptake in Candida glabrata erg1 mutant
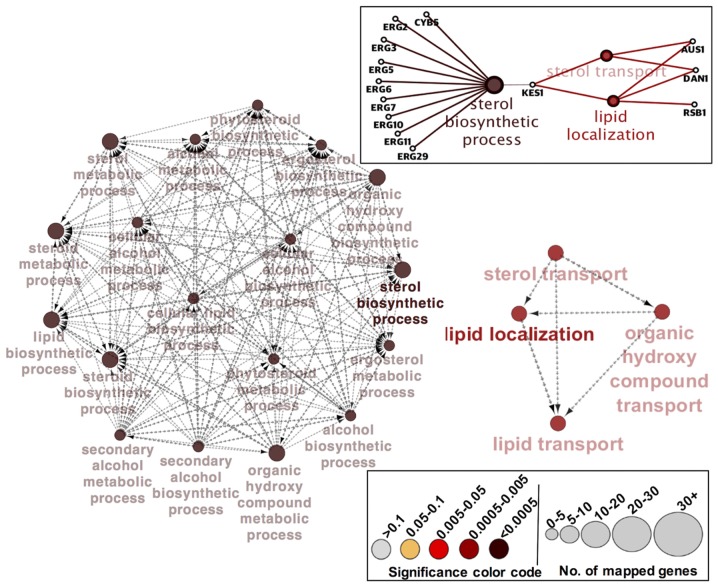
To further explore the functional themes of the above 35 differentially expressed genes in C. glabrata erg1 mutant, we used Cytoscape, a bioinformatics software for visualizing functional and molecular interaction networks. The functional interaction networks in Fig. 3 support our interpretation that the ergosterol biosynthetic process is the major biological function affected in C. glabrata mutant strain lacking ERG1. To compensate for the defective sterol biosynthetic pathway in the cell, and overcome the imbalance in cellular levels of sterol, cells increase the sterol uptake machinery and ergosterol biosynthesis processes in order to survive under azole and environmental stress. As shown in Fig. 3, nine genes (ERG2, ERG3, ERG5, ERG6, ERG7, ERG10, ERG11, ERG29, and CYB5) comprise the largest network cluster and contribute towards the enrichment of the ‘sterol biosynthetic process.’ While three genes (AUS1, DAN1, and RSB1) in the other smaller functional network cluster are enriched with ‘lipid localization’ in combination with GO-term ‘sterol transport’ (Fig. 3). KES1 and HES1 are the genes bridging the two major clusters by establishing a potential coordination between the ‘sterol biosynthetic process’ and ‘lipid localization/sterol transport’ (Fig. 3). The enriched processes identified with the Cytoscape software and GO database using the differentially expressed genes in C. glabrata erg1 mutant are shown in Table V.
Figure 3.
Two major functional networks obtained from the 35 differentially expressed genes in Candida glabrata erg1 mutant using Cytoscape software. Each node (filled circle) represents a biological process and the size and color code indicate, respectively, the number of genes mapped and the significance of the terms (lower right inset). The direction of the network is shown by arrow-head of edges; the edge-thickness is based on kappa-score level calculated automatically by ClueGO. The molecular interaction network between the ‘sterol biosynthetic process’ and ‘lipid localization’ in combination with ‘sterol transport’ is shown in the upper right inset.
Table V.
Functional enrichment with associated genes identified by using Cytoscape and GO database.
| Enriched process | Associated gene | P-value |
|---|---|---|
| Sterol biosynthetic process | CYB5, ERG2, ERG3, ERG5, ERG6, ERG7, ERG10, ERG11, ERG29 | <0.0005 |
| Lipid localization | AUS1, DAN1, RSB1 | 0.0005 to 0.005 |
GO, Gene Ontology.
Candida glabrata aus1 deletant is defective in sterol uptake and has greater susceptibility to azoles under hypoxic conditions
Lastly, we validated whether C. glabrata cells accumulate sterols from the environment through the Aus1p transporter and whether sterol uptake confers resistance to azoles in C. glabrata. To distinguish the uptake of exogenous sterol from that of endogenous ergosterol biosynthesis, we generated a C. glabrata erg1/aus1Δ double mutant strain and used the C. glabrata erg1 mutant strain CgTn201S and the wild-type strain Cg1660 as the controls. As seen in Fig. 4A and B and Tables VI and VII, in the presence of exogenous cholesterol or ergosterol, C. glabrata erg1 mutant cells showed much lower susceptibility to fluconazole and voriconazole compared to those cells in the absence of exogenous sterols, suggesting that C. glabrata erg1 mutant cells are capable of accumulating sterols from the medium through the wild-type sterol transporter CgAus1p; but not through ergosterol synthesis because the cells were defective in ergosterol synthetic pathway. In contrast, in the presence of exogenous cholesterol or ergosterol, the C. glabrata erg1/aus1Δ double mutant shows much higher susceptibility to fluconazole and voriconazole than the C. glabrata erg1 mutant, indicating that C. glabrata erg1Δaus1Δ double mutant cells are not able to uptake and synthesize sterols due to defective conditions in both the CgAus1p transporter and ergosterol biosynthesis in the cells. We also tested whether the loss of CgAus1p in C. glabrata sensitizes the pathogen to polyene amphotericin B and the echinocandins anidulafungin and caspofungin, and our E-test results reveal that C. glabrata cells lacking CgAus1p do not show altered susceptibility to non-azole antifungals in the presence or absence of exogenous sterols (data not shown). These observations confirm the findings of Zavrel et al (20) that CgAus1p is responsible for taking up exogenous cholesterol or ergosterol in C. glabrata under hypoxic stress when sterol synthesis is either absent or insufficient, and further suggest that sterol uptake plays an important role in the development of azole resistance in yeast under ergosterol starvation conditions, such as under azole drug pressure or when defective in sterol biosynthesis.
Figure 4.
CgAUS1 deletion in Candida glabrata abolishes the effect of sterol supplement on azole susceptibility under hypoxic conditions. Cg1660 (wild-type), CgTn201S (Cgerg1), or CgTn201Su/aus1 (Cgerg1/aus1Δ) were grown on (A) MIN agar medium or (B) YEPG agar medium. Plates were incubated under conditions of low oxygen tension at 37°C for 3 days. The fluconazole and voriconazole susceptibilities of Cg1660, CgTn201S, and CgTn201Su/aus1 were analyzed by E-test. The intercept of the zone of growth inhibition with the paper strip indicates the minimum inhibitory concentration (MIC). The addition of cholesterol or ergosterol is indicated by the position within each of the four groups as follows: upper left corner, MIN or YEPG; upper right corner, MIN or YEPG with ethanol-Tween 80 solvent alone; lower left corner, MIN or YEPG with cholesterol; and lower right corner, MIN or YEPG with ergosterol. The numerical data are shown in Tables VI and VII for MIN agar cultures and YEPG agar cultures, respectively.
Table VI.
Inhibitory effect of azoles in Candida glabrata wild-type, erg1 mutant, and erg1/aus1∆ double mutant on MIN agar medium with sterol supplement under hypoxic condition.
| E-test on MIN agar medium | |||||
|---|---|---|---|---|---|
| Straina | Detergent (0.5% EtOH and 0.5% Tween-80) | Cholesterol (20 µg/ml) | Ergosterol (20 µg/ml) | Fluconazole MIC (µg/ml) | Voriconazole MIC (µg/ml) |
| Cg1660 | − | − | − | >256 | 4.0 |
| Cg1660 | + | − | − | >256 | 4.0 |
| Cg1660 | + | + | − | >256 | 12 |
| Cg1660 | + | − | + | >256 | 12 |
| CgTn201S | − | − | − | 0.50 | 0.064 |
| CgTn201S | + | − | − | 0.50 | 0.064 |
| CgTn201S | + | + | − | >256 | >32 |
| CgTn201S | + | − | + | >256 | >32 |
| CgTn201Su/Cgaus1 | − | − | − | 0.50 | 0.064 |
| CgTn201Su/Cgaus1 | + | − | − | 0.50 | 0.064 |
| CgTn201Su/Cgaus1 | + | + | − | 0.50 | 0.064 |
| CgTn201Su/Cgaus1 | + | − | + | 0.50 | 0.064 |
Under hypoxic conditions (1% O2). Cg1660, C. glabrata wild-type; CgTn201S, C. glabrata erg1 mutant; CgTn201S/Cgaus1, C. glabrata erg1/aus1Δ double mutant; EtOH, ethanol; MIC, minimum inhibitory concentration.
Table VII.
Inhibitory effect of azoles in Candida glabrata wild-type, erg1 mutant, and erg1/aus1∆ double mutant on YEPG agar medium with sterol supplement under hypoxic condition.
| E-test on YEPG agar medium | |||||
|---|---|---|---|---|---|
| Straina | Detergent (0.5% EtOH and 0.5% Tween-80) | Cholesterol (20 µg/ml) | Ergosterol (20 µg/ml) | Fluconazole MIC (µg/ml) | Voriconazole MIC (µg/ml) |
| Cg1660 | − | − | − | >256 | 1.0 |
| Cg1660 | + | − | − | >256 | 1.0 |
| Cg1660 | + | + | − | >256 | 1.0 |
| Cg1660 | + | − | + | >256 | 6.0 |
| CgTn201S | − | − | − | 1.0 | 0.064 |
| CgTn201S | + | − | − | 1.0 | 0.064 |
| CgTn201S | + | + | − | >256 | >32 |
| CgTn201S | + | − | + | >256 | >32 |
| CgTn201Su/Cgaus1 | − | − | − | 1.0 | 0.064 |
| CgTn201Su/Cgaus1 | + | − | − | 1.0 | 0.064 |
| CgTn201Su/Cgaus1 | + | + | − | 1.0 | 0.064 |
| CgTn201Su/Cgaus1 | + | − | + | 1.0 | 0.064 |
Under hypoxic conditions (1% O2). Cg1660, C. glabrata wild-type; CgTn201S, C. glabrata erg1 mutant; CgTn201S/Cgaus1, C. glabrata erg1/aus1Δ double mutant; EtOH, ethanol; MIC, minimum inhibitory concentration.
Discussion
Yeast develops strategies to grow and survive in different unfavorable environments. In the present study, we observed that C. glabrata acquires two protective mechanisms to allow the pathogen to grow and survive under azole and hypoxic stress: Through increasing endogenous sterol synthesis or through importing exogenous sterols. Our data revealed that the expression of both ergosterol biosynthesis and sterol metabolism regulator genes, as well as sterol influx transporter genes, are significantly increased in C. glabrata under fluconazole stress. Likewise, the sterol influx transporter genes (CgAUS1 and CgTIR3) and the ergosterol biosynthetic genes (CgERG2, CgERG3, CgERG5, CgERG6, CgERG7, CgERG10, CgERG11, and CgERG29) are markedly up-regulated in yeast when ergosterol biosynthesis is suppressed either under hypoxia or due to defective sterol synthesis. We also confirmed that CgAUS1 in the cell is responsible for importing exogenous cholesterol and ergosterol, thus allowing C. glabrata to survive under low oxygen tension conditions or under azole pressure. The presence of both in vivo suggests an underlying mechanism for azole resistance in clinical practice.
Nakayama and colleagues show that CgAUS1 protects C. glabrata cells against azoles in the presence of serum (21). The composition of serum is complicated, and it includes many different molecules, minerals, and nutrients other than cholesterol. It is possible that, besides importing cholesterol via the sterol transporter CgAUS1, C. glabrata cells may also take up other molecules and nutrients from serum that are necessary to their survival amidst azole treatment. In our experiments, as in those of Zavrel et al (20), we did not use serum but solubilized cholesterol and ergosterol using the detergent Tween 80. This allowed for the incorporation of exogenous sterol into the fungal cell. Our results, together with the data discussed above, reinforce the understanding of how both enhanced endogenous sterol synthesis and increased exogenous sterol uptake by yeast are integral to conferring resistance to azole therapy in fungal infections. Resistance to azole treatment not only limits the usefulness of this class of drugs, it also drives the survival of intrinsically low-susceptibility C. glabrata cells that become increasingly resistant following prolonged treatment with azole therapeutic agents.
Combining our data in this study, we propose a hypothetical model in which sterol uptake and sterol biosynthesis act coordinately and collaboratively to mediate azole antifungal resistance in C. glabrata under azole and hypoxic stress as shown in Fig. 5. In this model, azole and hypoxic stresses deplete ergosterol directly or indirectly by inhibiting ergosterol biosynthesis. Hypoxia and azoles cause mitochondrial dysfunction by depriving the mitochondria of oxygen and inducing the accumulation of toxic sterol-intermediates, ultimately leading to the reduction of ergosterol biosynthesis. In response, ergosterol depletion triggers up-regulation of the genes involved in sterol transport and ergosterol biosynthesis, leading to increases in sterol levels. Because both sterol uptake and sterol biosynthesis increase sterol levels through distinct mechanisms, they both may function together to maintain sterol levels and sustain cell growth. Such a cooperative mechanism may serve to integrate the roles of sterol transport and sterol biosynthesis culminating in cell survival, which may underlie the mechanism of azole antifungal resistance in C. glabrata.
Figure 5.
A schematic diagram illustrating a hypothetical model for sterol uptake and sterol biosynthesis acting coordinately and collaboratively in mediating azole antifungal resistance in Candida glabrata under azole and hypoxic stress. See ‘Discussion’ for further elaboration.
Collectively, this study shows the up-regulation of the genes involved in ergosterol biosynthesis and sterol transport in C. glabrata cells in which sterol synthesis is defective or is abrogated by fluconazole treatment. We also corroborate that the sterol influx transporter CgAus1p imports exogenous cholesterol or ergosterol, which contributes to the development of clinical resistance to azole antifungals in C. glabrata. These findings demonstrate that sterol uptake and sterol biosynthesis may act coordinately and collaboratively to sustain growth and to mediate antifungal resistance in C. glabrata through dynamic gene expression in response to azole stress and environmental challenges.
Acknowledgements
The authors would like to thank Ms. Cindy Clark for carefully reviewing the manuscript.
Funding
This study was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
QQL conceived of the project, conducted the studies, performed drug sensitivity assay, RT-qPCR and Southern blot analysis, cloning of CgAUS1, construction of Cgerg1 and Cgaus1 double mutant, as well as other experiments, carried out the analysis and interpretation of data, wrote the manuscript, and is the primary author of this paper. HFT conceived the project, performed microarray hybridization and data analysis, and critically reviewed the manuscript. AM performed bioinformatics and statistical analysis. BAW, JAN, and YF analyzed and interpreted data and edited the manuscript. JEB participated in the design and coordination of the study, critically reviewed the manuscript, and edited the final version of the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, et al. Executive summary: Clinical practice guideline for the management of candidiasis: 2016 update by the infectious diseases society of America. Clin Infect Dis. 2016;62:409–417. doi: 10.1093/cid/civ1194. [DOI] [PubMed] [Google Scholar]
- 2.Rodrigues CF, Silva S, Henriques M. Candida glabrata: A review of its features and resistance. Eur J Clin Microbiol Infect Dis. 2014;33:673–688. doi: 10.1007/s10096-013-2009-3. [DOI] [PubMed] [Google Scholar]
- 3.Inukai T, Nagi M, Morita A, Tanabe K, Aoyama T, Miyazaki Y, Bard M, Nakayama H. The mannoprotein TIR3 (CAGL0C03872g) is required for sterol uptake in Candida glabrata. Biochim Biophys Acta. 2015;1851:141–151. doi: 10.1016/j.bbalip.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Nagi M, Nakayama H, Tanabe K, Bard M, Aoyama T, Okano M, Higashi S, Ueno K, Chibana H, Niimi M, et al. Transcription factors CgUPC2A and CgUPC2B regulate ergosterol biosynthetic genes in Candida glabrata. Genes Cells. 2011;16:80–89. doi: 10.1111/j.1365-2443.2010.01470.x. [DOI] [PubMed] [Google Scholar]
- 5.Nagi M, Tanabe K, Ueno K, Nakayama H, Aoyama T, Chibana H, Yamagoe S, Umeyama T, Oura T, Ohno H, et al. The Candida glabrata sterol scavenging mechanism, mediated by the ATP-binding cassette transporter Aus1p, is regulated by iron limitation. Mol Microbiol. 2013;88:371–381. doi: 10.1111/mmi.12189. [DOI] [PubMed] [Google Scholar]
- 6.Silver PM, Oliver BG, White TC. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot Cell. 2004;3:1391–1397. doi: 10.1128/EC.3.6.1391-1397.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacPherson S, Akache B, Weber S, De Deken X, Raymond M, Turcotte B. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob Agents Chemother. 2005;49:1745–1752. doi: 10.1128/AAC.49.5.1745-1752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai HF, Bard M, Izumikawa K, Krol AA, Sturm AM, Culbertson NT, Pierson CA, Bennett JE. Candida glabrata erg1 mutant with increased sensitivity to azoles and to low oxygen tension. Antimicrob Agents Chemother. 2004;48:2483–2489. doi: 10.1128/AAC.48.7.2483-2489.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bard M, Sturm AM, Pierson CA, Brown S, Rogers KM, Nabinger S, Eckstein J, Barbuch R, Lees ND, Howell SA, Hazen KC. Sterol uptake in Candida glabrata: Rescue of sterol auxotrophic strains. Diagn Microbiol Infect Dis. 2005;52:285–293. doi: 10.1016/j.diagmicrobio.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Flowers SA, Colón B, Whaley SG, Schuler MA, Rogers PD. Contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob Agents Chemother. 2015;59:450–460. doi: 10.1128/AAC.03470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li QQ, Skinner J, Bennett JE. Evaluation of reference genes for real-time quantitative PCR studies in Candida glabrata following azole treatment. BMC Mol Biol. 2012;13:22. doi: 10.1186/1471-2199-13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vermitsky JP, Earhart KD, Smith WL, Homayouni R, Edlind TD, Rogers PD. Pdr1 regulates multidrug resistance in Candida glabrata: Gene disruption and genome-wide expression studies. Mol Microbiol. 2006;61:704–722. doi: 10.1111/j.1365-2958.2006.05235.x. [DOI] [PubMed] [Google Scholar]
- 13.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van den Hazel HB, Pichler H, do Valle Matta MA, Leitner E, Goffeau A, Daum G. PDR16 and PDR17, two homologous genes of Saccharomyces cerevisiae, affect lipid biosynthesis and resistance to multiple drugs. J Biol Chem. 1999;274:1934–1941. doi: 10.1074/jbc.274.4.1934. [DOI] [PubMed] [Google Scholar]
- 17.Marek M, Milles S, Schreiber G, Daleke DL, Dittmar G, Herrmann A, Müller P, Pomorski TG. The yeast plasma membrane ATP binding cassette (ABC) transporter Aus1: Purification, characterization, and the effect of lipids on its activity. J Biol Chem. 2011;286:21835–21843. doi: 10.1074/jbc.M111.244525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whaley SG, Caudle KE, Vermitsky JP, Chadwick SG, Toner G, Barker KS, Gygax SE, Rogers PD. UPC2A is required for high-level azole antifungal resistance in Candida glabrata. Antimicrob Agents Chemother. 2014;58:4543–4554. doi: 10.1128/AAC.02217-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ness F, Bourot S, Régnacq M, Spagnoli R, Bergès T, Karst F. SUT1 is a putative Zn[II]2Cys6-transcription factor whose upregulation enhances both sterol uptake and synthesis in aerobically growing Saccharomyces cerevisiae cells. Eur J Biochem. 2001;268:1585–1595. doi: 10.1046/j.1432-1033.2001.02029.x. [DOI] [PubMed] [Google Scholar]
- 20.Zavrel M, Hoot SJ, White TC. Comparison of sterol import under aerobic and anaerobic conditions in three fungal species, Candida albicans, Candida glabrata, and Saccharomyces cerevisiae. Eukaryot Cell. 2013;12:725–738. doi: 10.1128/EC.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakayama H, Tanabe K, Bard M, Hodgson W, Wu S, Takemori D, Aoyama T, Kumaraswami NS, Metzler L, Takano Y, et al. The Candida glabrata putative sterol transporter gene CgAUS1 protects cells against azoles in the presence of serum. J Antimicrob Chemother. 2007;60:1264–1272. doi: 10.1093/jac/dkm321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.