Abstract
The present study aimed to screen potential biomarkers for uterine leiomyomas disease, particularly target genes associated with the mediator of RNA polymerase II transcription subunit 12 (MED12) mutation. The microarray data of GSE30673, including 10 MED12 wild-type myometrium, 8 MED12 mutation leiomyoma and 2 MED12 wild-type leiomyoma samples, were downloaded from the Gene Expression Omnibus database. Compared with myometrium samples, differently-expressed genes (DEGs) in the MED12 mutation and wild-type leiomyoma samples were identified using the Limma package. The two sets of DEGs obtained were intersected to screen common DEGs. The DEGs in the MED12 mutation and wild-type leiomyoma samples, and common DEGs were defined as group A, B and C. Gene Ontology (GO) and pathway enrichment analyses were performed using the Database for Annotation, Visualization and Integrated Discovery online tool. Based on the Kyoto Encyclopedia of Genes and Genomes database, pathway relation networks were constructed. DEGs in GO terms and pathways were intersected to screen important DEGs. Subsequently, a gene co-expression network was constructed and visualized using Cytoscape software. Reverse transcription-quantitative polymerase chain reaction was used to detect the expression levels of important DEGs. A total of 1,258 DEGs in group A were screened, and enriched for extracellular matrix (ECM) organization and ECM-receptor interaction. In addition, a total of 1,571 DEGs in group B were enriched for cell adhesion. Furthermore, 391 DEGs were involved in extracellular matrix organization. Pathway relation networks of group A, B and C were constructed with nodes of 48, 39, and 28, respectively. Finally, 135 important DEGs were obtained, including Acyl-CoA synthetase medium-chain family member 3, protein S (α) (PROS1) and F11 receptor. A gene co-expression network with 68 nodes was constructed. The expression of caspase 1 (CASP1) and aldehyde dehydrogenase 1 family member A1 (ALDH1A1) was significant higher in SK-UT-1 compared with that in PHM1-31 cells, while the expression of PROS1 was significant lower in SK-UT-1 cells. These results that CASP1, ALDH1A1 and PROS1 may be potential biomarkers for uterine leiomyomas. Furthermore, hematopoietic prostaglandin D synthase and carbonyl reductase 3 (CBR3) may be particular genes associated with the MED12 mutation in this disease.
Keywords: uterine leiomyomas, differentially-expressed genes, pathway relation network, gene co-expression network
Introduction
Uterine leiomyomas, also known as uterine fibroids, is a benign smooth muscle tumor with symptoms including abdominal discomfort or bloating, abnormal uterine bleeding, back ache and urinary frequency, and always found during the middle and later reproductive years (1). In addition, it is one of the common reasons for surgical removal of the uterus (2). As we all know, fibroids run is closely related to estrogen, progesterone and reproductive years (3). Specific mutations of the mediator of RNA polymerase II transcription subunit 12 (MED12) protein have been found in 70% percent of this disease (4). However, the exact cause of uterine leiomyomas has not been stated clearly. Therefore, explore the exact cause of uterine fibroids and effective diagnosis methods are urgent.
Recently, several genes were found to participate into the molecular pathogenesis of uterine fibroids. For example, fos proto-oncogene, AP-1 transcription factor subunit (c-FOS) was found downregulated in uterine fibroids samples and also participated into the pathway of serum response factor-Fos proto-oncogene, AP-1 transcription factor subunit-JunB proto-oncogene, AP-1 transcription factor subunit (SRF-FOS-JUNB) pathway (5). In addition, Shen et al (6)found that c-FOS also involved in the pathways of estrogen receptor 1 (ERa) and transforming growth factor β 1 (TGF-β) signaling, and both of these pathways could control the uterine fibroids cells proliferation. Besides, overexpression of high-mobility group I (HMGI) proteins was verified to be important in the pathogenesis of this disease (7). Furthermore, a drug trial of Chuang et al (8) showed that berberine could decrease the expression of cyclooxygenase 2 (COX2) and pituitary tumor-transforming gene 1 (PTTG1), and further inhibit the cell proliferation and induce apoptosis of uterine fibroids cells. These genes proved to play an important role in the pathogenesis of uterine fibroid by previous studies. However, few genes and their functions and involved pathways were verified for diagnosis and treatment of this disease.
In order to find more target genes and reveal their potential molecular mechanisms, comprehensive bioinformatics approach was applied to screen differentially-expressed genes (DEGs) in MED12 mutation leiomyoma samples and MED12 wild-type leiomyoma samples compared with MED12 wild-type myometrium samples. Functional and pathway enrichment analysis, pathway relation network and gene co-expression network analysis were performed. The aims of this study were to screening potential biomarkers of uterine leiomyomas disease, especially target genes related to MED12 mutation leiomyoma samples, and further study the molecular mechanism of this disease.
Materials and methods
Gene expression data and preprocessing
One gene expression profiling of GSE30673, including 10 MED12 wild-type myometrium samples, 8 MED12 mutation leiomyoma samples and 2 MED12 wild-type leiomyoma samples, was downloaded from Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) (4). This profiling was deposited on the platform of Affymetrix Human Genome U133 Plus 2.0 Array (HG-U133_Plus_2; Affymetrix, Santa Clara, CA, USA).
The raw data was downloaded and read by Affy package of R-based software. Robust Multi-chip Average (RMA) method was used to calculate the expression value of probes, which included three steps of background correction, normalization and collection (9).
Identification of DEGs
Compared with MED12 wild-type myometrium samples, the DEGs in MED12 mutation leiomyoma samples and MED12 wild-type leiomyoma samples were separately identified by Limma package. Thereinto, Beniamini-Hochberg (BH) method was used for multiple testing corrections and T-test in limma package was used to screen the DEGs (10). Threshold for the DEGs were set as P<0.05 and log2 fold-change (FC)|≥2. The obtained two sets of DEGs were intersected to screen the common DEGs based on the information of gene symbols. The DEGs in MED12 mutation leiomyoma samples, DEGs in MED12 wild-type leiomyoma samples and the common DEGs were defined as group A, B and C.
Functional enrichment analysis and pathway enrichment analysis
Gene Ontology (GO, http://geneontology.org/) is a useful model of biology, which defines classes of gene functions (molecular function, cellular component and biological process). In addition, Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/) is a database for analysis of high-level functions and utilities of the biological system. The GO and KEGG pathway enrichment analysis of DEGs in group A, B and C were separately performed by Database for Annotation Visualization and Integrated Discovery (DAVID) online tool (11). P-value <0.05 was chosen as the threshold.
Construction of pathway relation network
The interactions in KEGG database are used to construct the interaction network between pathways. Based on the information of KEGG database, pathway relation network of three groups were constructed and visualized by Cytoscape software (12). The correlation coefficient score >1 was shown in the network.
Construction of gene co-expression network
In group C, DEGs which enriched in GO terms and pathways, were intersected to screen the more important DEGs based on the information of gene symbols. Then, based on the expression value of these obtained DEGs, gene co-expression network was constructed and visualized by Cytoscape software (12). The correlation coefficient score >1 was shown in the network.
Cells and culture conditions
Uterine smooth muscle cell line PHM1-31 and human leiomyosarcoma cell line SK-UT-1 was obtained from the cell bank of our laboratory. Both cell lines were cultured in high-glucose DMEM (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) (10% fetal calf serum and 1 mg/ml geneticin, 100 U/ml penicillin and 50 µg/ml streptomycin) in the atmosphere of 5% CO2 at 37°C.
Total RNA extraction and primer design
Total RNA extraction from uterine smooth muscle cell line PHM1-31 and human leiomyosarcoma cell line SK-UT-1 were processed as described (13). Based on human CASP1, ALDH1A, PROS1 and GADPH gene and their transcript variant in gene bank, the primers was designed as follows: CASP1 upstream, 5′-CGCAGATGCCCACCACT-3′ and downstream primer, 5′-TGCCCACAGACATTCATACAG-3′; ALDH1A upstream, 5′-GCCAGGTAGAAGAAGGAGATAAGG-3′ and downstream primer, 5′-GTGGAGAGCAGTGAGAGGAGTTT-3′; PROS1 upstream, 5′-CAACATGCTAAAAGTCTTGG-3′ and downstream primer, 5′-GAAACATAAGTATAATTACAC-3′.GAPDH upstream 5′-TGATGACATCAAGAAGGTGGTGAAG-3′ and downstream primer, 5′-TCCTTGGAGGCCATGTGGGCCAT-3′.
Measurement of CASP1, ALDH1A, PROS1 and GADPH by qRT-PCR
All amplifcations were performed on the ABI 7900 real-time PCR instrument. According to the manufacturer' guidelines, cDNA synthesis were processed using the PrimeScript™ RT Master Mix (Takara Bio, Inc., Otsu, Japan). Each gene and sample was repeated for 3 times. The reaction condition was 1 cycle of 95°C for 10 min, 40 cycles of 95°C for 15 sec, and 60°C for 1 min. SYBR Premix Ex Taq™ (Takara Bio, Inc.) was used the following PCR procedure.
Results
Screening of DEGs
Based on the threshold of P<0.05 and |log2 FC| >2, a total of 1,258 DEGs in MED12 mutation leiomyoma samples compared with MED12 wild-type myometrium samples were screened, including 646 upregulated and 612 downregulated DEGs. Besides, compared with MED12 wild-type myometrium samples, total 1,571 DEGs in MED12 wild-type leiomyoma samples were also identified, including 726 upregulated and 845 downregulated DEGs. Taking the intersection of DEGs in group A and B, a total of 391 DEGs were obtained, such as F11 receptor (F11R), relaxin/insulin-like family peptide receptor 1 (RXFP1) and ferredoxin reductase (FDXR).
GO functional and KEGG pathway enrichment analysis
After enrichment analysis, the 1,258 DEGs of group A were significant enriched into various functions including extracellular matrix organization, cell adhesion and extracellular matrix disassembly (P<0.05). These DEGs also enriched into pathways of extracellular matrix (ECM)-receptor interaction, focal adhesion and P13K-Akt signaling pathway (P<0.05, Table I). In addition, the 1,571 DEGs of group B were mainly related to cell adhesion, extracellular matrix organization and blood coagulation (P<0.05, Table II). Meanwhile, these genes also enriched into pathways of P13K-Akt signaling pathway, pathways in cancer and focal adhesion (P<0.05, Table II). As shown in Table III, the common DEGs were significantly enriched in GO terms of extracellular matrix organization, collagen catabolic process and positive regulation of apoptotic process, and pathways of cytokine-cytokine receptor interaction, ECM-receptor interaction and amoebiasis (P<0.05, Table III).
Table I.
The top 5 enriched GO terms and pathways for DEGs in group A.
| GO ID | GO name | Diff gene counts in GO | Enrichment score | P-value | Gene symbols |
|---|---|---|---|---|---|
| GO:0030198 | Extracellular matrix organization | 49 | 13.62996005 | 2.04E-40 | COL16A1, COL4A2, COL23A1, COL4A1, COL21A1, MMP11, COL5A1, FN1, MMP13, CR61, SMOC2, POSTN, COL6A3, CTSG, HSPG2, MMP16, TPSAB1, RXFP1, B4GALT1, COL4A4, ADAMTS2, COL9A2, COL1A1, TGFBI, TNFRSF11B, COL3A1, COL4A3, TGFB2, COL1A2, COL11A1, EFEMP1, EGFL6, COL22A1, COL4A6, ECM2, COL6A6, ITGA5, LGALS3, MFAP2, COL2A1, FBN2, FBLN1, COL5A2, TGFB3, LEPREL1, OLFML2B, COL12A1, PXDN, MMP7 |
| GO:0007155 | Cell adhesion | 49 | 6.304607073 | 3.24E-24 | B4GALT1, CCL2, CDH2, NEGR1, FN1, OMD, FLRT2, CCL4, TGFBI, ADAM23, WISP2, LAMC3, DPT, PERP, COMP, HAPLN1, SUSD5, POSTN, ITGA9, THBS4, CDH11, DPP4, NLGN4X, CD36, COL5A1, EFNB2, BCAN, F11R, ITGA5, COL4A6, VCAN, COL16A1, CD97, COL6A3, GPNMB, ADAM12, NRP2, CLDN10, NCAM1, COL6A6, SELE, COL12A1, CNTN1, COL4A3, MMRN1, LOXL2, TNC, SRPX, EGFL6 |
| GO:0022617 | Extracellular matrix disassembly | 24 | 17.74606011 | 2.62E-23 | COL3A1, TPSAB1, COL16A1, COL4A1, MMP11, COL5A1, COL2A1, COL4A2, MMP16, MMP13, COL1A2, COL9A2, COL6A6, COL1A1, COL4A3, COL12A1, COL5A2, CTSG, COL23A1, COL4A6, MMP7, COL11A1, COL6A3, COL4A4 |
| GO:0030574 | Collagen catabolic process | 23 | 18.66006436 | 5.96E-23 | MMP11, COL4A3, COL23A1, COL2A1, COL5A1, COL11A1, COL4A1, COL16A1, COL4A4, MMP16, MMP13, MMP7, COL1A1, ADAMTS2, COL6A3, COL6A6, COL12A1, COL3A1, COL9A2, COL4A6, COL4A2, COL1A2, COL5A2 |
| GO:0007165 | Signal transduction | 69 | 3.913178545 | 1.45E-21 | CCL2, PDE2A, MET, HPGDS, PDE10A, IL15, TENM4, GRIA2, TNFRSF11B, LRP12, ANXA1, TLR3, CASP1, WISP2, CHRNA1, PDE11A, ERBB3, RORA, BST2, RND2, CRABP2, IRS1, INPP4B, PRKD1, RGS1, SMC3, CDC42EP3, CCL4, FGF13, RGS5, CD48, MAPK10, NDP, PPP1R1A, GUCY1A2, NR4A2, C3, TNFSF10, ADORA3, TEK, DTNA, EDNRA, EPAS1, GABRB3, ARHGAP15, WISP3, NR4A1, FCGR1A, SMOC2, PDE4D, PSD, FGF9, PRKAA2, UNC5D, IGFBP6, PPARG, IL7R, APOL3, CXCL14, PRKCH, ECM1, PIK3R1, GNG11, PRKCB, IGFBP5, XCL1, CYTL1, NR3C2, NDRG2 |
| Pathway ID | Pathway name | Diff gene counts in pathway | Enrichment score | P-value | Gene symbols |
| 4512 | ECM-receptor | 25 | 16.78566509 | 1.48E-23 | COL6A6, COMP, COL6A3, COL4A4, COL4A6, COL5A1, HSPG2, ITGA5, FN1, THBS4, COL11A1, COL5A2, COL4A1, TNC, LAMC3, COL1A1, COL4A3, COL1A2, ITGA9, CD36, TNN, SDC1, COL4A2, COL2A1, COL3A1 |
| 4510 | Focal adhesion interaction | 34 | 9.641164531 | 4.65E-23 | COL4A3, THBS4, COL4A6, CCND1, COL4A4, MYLPF, FYN, COL4A2, COL4A1, ITGA5, TNN, COL1A2, FN1, SHC3, MAPK10, ITGA9, COL11A1, MET, COL5A2, COL2A1, COL3A1, PRKCB, COL5A1, BIRC3, JUN, TNC, COMP, FIGF, PDGFC, COL1A1, LAMC3, COL6A3, PIK3R1, COL6A6 |
| 4151 | PI3K-Akt signaling pathway | 42 | 7.070296281 | 8.27E-23 | MYC, MET, COL3A1, PRKAA2, COL2A1, COMP, COL4A1, FGF13, COL1A2, F2R, GNG11, COL1A1, MCL1, FN1, COL11A1, FGF9, COL6A6, TLR4, ITGA9, COL4A6, COL4A2, PIK3R1, COL4A4, IL7R, TNN, LAMC3, THBS4, COL4A3, FIGF, COL6A3, CCND1, IRS1, TEK, TNC, ITGA5, SGK1, COL5A2, PDGFC, COL5A1, PRL, LPAR6, NR4A1 |
| 5146 | Amoebiasis | 23 | 12.32591407 | 1.95E-18 | COL11A1, CTSG, COL5A2, LAMC3, IL1R1, COL4A6, SERPINB1, TGFB3, COL4A4, COL1A1, TLR4, COL5A1, COL2A1, TGFB2, FN1, PIK3R1, COL4A1, COL3A1, PRKCB, COL4A2, PLCB4, COL1A2, COL4A3 |
| 4974 | Protein digestion and absorption | 20 | 13.27593512 | 7.53E-17 | COL2A1, COL4A2, COL3A1, COL11A1, COL4A3, COL12A1, COL6A6, COL1A1, COL5A1, COL1A2, COL5A2, COL4A1, COL4A6, DPP4, COL9A2, COL21A1, CPA3, COL22A1, COL4A4, COL6A3 |
GO, Gene Ontology; DEGs, differently-expressed genes.
Table II.
The top 5 enriched GO terms and pathways for DEGs in group B.
| GO ID | GO name | Diff gene counts in GO | Enrichment score | P-value | Gene symbols |
|---|---|---|---|---|---|
| GO:0007155 | Cell adhesion | 57 | 5.737272852 | 4.76E-26 | CCL4, VCL, F11R, PRKCA, PERP, CD36, NEGR1, OMD, PECAM1, ADAM23, NCAM1, PCDHA1, CHL1, ROBO2, CLDN10, CXCR7, CDH3, CD58, COL16A1, COL6A6, CLDN1, COL5A1, PCDHAC2, MMRN1, EMILIN1, ADAM22, SVEP1, PCDHB15, DPP4, BOC, LSAMP, TGFBI, DPT, FLRT2, FLRT1, SRPX, CCL2, CNTN3, ITGA6, COL4A3, FLRT3, CDH2, LOXL2, DSG2, SELE, CDON, CNTN1, GPNMB, COL4A6, MSLN, CTNNA2, ITGBL1, CD H17, NRP2, WISP2, SPP1, EDIL3 |
| GO:0030198 | Extracellular matrix organization | 38 | 8.268958333 | 2.18E-23 | COL6A6, CTSK, CCDC80, SOX9, COL4A3, TGFBI, MMP7, LOXL1, COL1A2, COL4A6, COL16A1, COL11A1, MFAP5, LEPREL1, EFEMP1, MMP16, LGALS3, FBN2, COL4A5, TNFRSF11B, COL4A4, COL27A1, ADAMTS2, MMP10, CYR61, SERPINH1, ECM2, COL11A2, COL4A2, PXDN, RXFP1, EMILIN1, COL5A2, COL5A1, COL1A1, TGFB2, ITGA6, COL4A1 |
| GO:0007596 | Blood coagulation | 47 | 4.618823925 | 9.15E-18 | COL1A2, VCL, IGF2, PROS1, COL1A1, DGKB, CD58, CDK5, PRKCH, LRP8, ITGA4, PECAM1, RASGRP1, PRKCA, ATP2B2, LRRC16A, SERPINA1, HBB, PDE3B, GUCY1A3, TGFB2, ZFPM2, ENPP4, F2RL2, CD36, TEK, PDE5A, PRKCB, PLA2G4A, THBD, TFPI2, KCNMA1, DOCK11, SPARC, F11R, GATA5, SRGN, F2RL1, F3, CFD, TFPI, MAFF, CXADR, LEFTY2, ITGA6, MMRN1, HGF, |
| GO:0001525 | Angiogenesis | 31 | 7.047776741 | 3.38E-17 | PRKCA, FGFR1, HTATIP2, CCL2, ANG, TEK, TSPAN12, HOXB3, TGFBI, ARHGAP22, SOX17, ENPEP, EPHB1, TGFB2, PDE3B, PTGS2, KDR, NRP2, FGF9, THSD7A, SHB, SCG2, SAT1, COL4A2, KLF5, ID1, CXCR7, MEOX2, CALCRL, ESM1, COL4A1 |
| GO:0007165 | Signal transduction | 70 | 3.105612864 | 1.91E-16 | ERBB3, SPARC, CASP1, BST2, LRP8, PLCB1, CCL4, PRKCA, CD83, EDNRA, SPOCK3, EPS8, EPO, GNG11, NDP, FGF9, TEK, PDE3B, CRABP2, ADRA1A, ARHGAP15, PRKCH, ASB2, PECAM1, SKAP2, NR2F2, SOX9, FGF13, RASD1, VLDLR, C3, PPP1R1A, TSPAN8, CXCL10, TENM4, GPRC5A, PDE5A, ARHGEF7, ARHGAP22, RASGRP1, SHB, TNFSF10, ANXA1, PRKCB, CHN1, DAPK1, IGFBP6, RASSF9, CXCL14, AKT3, CCL2, CRHBP, GRIA2, XCL1, CDC42EP3, SIGIRR, ERG, CLIC2, IL11RA, TNFRSF11B, CHL1, PRKAA2, TENM2, WISP2, SH3GL2, IL15, RASSF2, RGS1, DTNA, DAPP1 |
| Pathway ID | Pathway name | Diff gene counts in pathway | Enrichment score | P-value | Gene symbols |
| 4151 | PI3K-Akt signaling pathway | 40 | 5.267651297 | 3.01E-17 | FGF9, EPO, COL4A1, ITGA6, SPP1, PRKCA, CDKN1A, COL4A5, COL11A1, CCND1, PRLR, ITGA4, CDK6, AKT3, FGF8, COL4A4, COL1A2, KDR, COL4A3, PRKAA2, COL6A6, CCNE2, COL5A2, COL1A1, MCL1, HGF, LPAR4, SGK1, PRL, COL4A2, COL4A6, COL11A2, MDM2, COL27A1, GNG11, TEK, FGF13, FGFR1, CCND2, COL5A1 |
| 5200 | Pathways in cancer | 35 | 4.891102829 | 3.04E-14 | TGFB2, FOS, PRKCA, CDH1, CCND1, PRKCB, CCNE2, MDM2, COL4A5, HGF, WNT2, COL4A4, FGFR1, CKS2, FZD2, WNT4, ITGA6, FGF9, COL4A3, COL4A2, FGF13, WNT2B, CDKN2B, COL4A1, BIRC3, MECOM, FGF8, PTCH1, PTGS2, CDK6, DAPK1, CTNNA2, CDKN1A, AKT3, COL4A6 |
| 4510 | Focal adhesion | 28 | 6.211225728 | 3.33E-14 | COL11A1, COL6A6, BIRC3, SPP1, KDR, MYLK, COL4A4, ITGA6, COL5A1, CCND1, COL4A2, PRKCA, ITGA4, COL11A2, PRKCB, COL5A2, COL1A2, COL4A6, COL4A3, MYL12A, CCND2, COL4A1, COL27A1, COL4A5, COL1A1, AKT3, HGF, VCL |
| 4512 | ECM-receptor interaction | 19 | 9.979777299 | 8.39E-14 | COL1A1, COL5A1, COL4A4, COL4A3, COL11A2, COL4A6, CD36, COL4A2, COL11A1, COL4A1, COL4A5, COL5A2, SPP1, COL27A1, COL6A6, COL1A2, SDC1, ITGA6, ITGA4 |
| 4514 | Cell adhesion molecules (CAMs) | 23 | 7.198822774 | 3.02E-13 | SELE, CDH1, CDH3, CLDN1, CDH2, PTPRC, SDC1, F11R, CLDN10, CLDN11, NCAM1, LRRC4C, CD58, VTCN1, ITGA4, HLA-E, ITGA6, PECAM1, CD8A, CNTN1, NEGR1, CLDN3, LRRC4B |
GO, Gene Ontology; DEGs, differently-expressed genes.
Table III.
The top 5 enriched GO terms and pathways for DEGs in group C.
| GO ID | GO name | Diff gene counts in GO | Enrichment score | P-value | Gene symbols |
|---|---|---|---|---|---|
| GO:0030198 | Extracellular matrix organization | 26 | 14.18120104 | 6.85E-22 | COL4A1, EFEMP1, COL11A1, COL4A3, LEPREL1, ADAMTS2, MMP16, COL5A1, COL4A4, TGFB2, COL1A2, TNFRSF11B, COL5A2, CYR61, RXFP1, TGFBI, PXDN, COL4A2, MMP7, COL1A1, FBN2, ECM2, LGALS3, COL16A1, COL6A6, COL4A6, |
| GO:0030574 | Collagen catabolic process | 15 | 23.86259791 | 1.46E-16 | COL5A1, COL4A1, COL1A1, COL11A1, COL4A4, COL4A3, COL4A6, COL6A6, COL4A2, ADAMTS2, COL5A2, MMP7, COL1A2, MMP16, COL16A1 |
| GO:0043065 | Positive regulation of apoptotic process | 21 | 12.20989781 | 1.63E-16 | HOXA13, HOXA5, DUSP1, MAP3K5, TGFB2, TNFSF10, CNR1, KCNMA1, SRPX, ID3, CSRNP3, PTPRC, PTGS2, ALDH1A2, G0S2, SFRP1, GZMA, GRIN2A, CYR61, SFRP4, MSX1 |
| GO:0007155 | Cell adhesion | 28 | 7.064169954 | 2.01E-15 | NRP2, CDH2, OMD, COL16A1, COL4A6, MMRN1, LOXL2, DPP4, SELE, GPNMB, NCAM1, SRPX, F11R, FLRT2, COL4A3, CCL2, ADAM23, WISP2, CNTN1, DPT, PERP, NEGR1, CLDN10, CD36, CCL4, TGFBI, COL5A1, COL6A6 |
| GO:0022617 | Extracellular matrix disassembly | 14 | 20.29831113 | 1.76E-14 | MMP7, COL4A1, COL1A2, COL16A1, COL5A1, COL1A1, COL4A3, COL11A1, COL4A4, COL6A6, MMP16, COL4A6, COL5A2, COL4A2 |
| Pathway ID | Pathway name | Diff gene counts in pathway | Enrichment score | P-value | Gene symbols |
| 4060 | Cytokine-cytokine receptor interaction | 20 | 8.579810485 | 7.63E-13 | IL1R1, CCL5, IL20RA, LIFR, XCL1, INHBA, CXCL2, TNFRSF11B, LEPR, IL15, PRL, CCL2, IL6ST, TNFSF10, IL17B, CCL4, CXCL14, CCL14, TGFB2, BMPR1B |
| 4512 | ECM-receptor interaction | 13 | 17.11524264 | 1.60E-12 | COL4A3, COL4A1, COL6A6, COL1A2, COL4A2, COL11A1, COL5A1, SDC1, COL4A4, CD36, COL4A6, COL5A2, COL1A1 |
| 5146 | Amoebiasis | 14 | 14.71162 | 1.79E-12 | COL4A6, PLCB4, COL4A3, COL11A1, COL1A1, PRKCB, COL4A2, COL5A1, IL1R1, COL4A1, TGFB2, COL1A2, COL5A2, COL4A4 |
| 4974 | Protein digestion and absorption | 12 | 15.619155 | 3.74E-11 | COL5A1, COL5A2, COL4A6, COL11A1, COL4A1, COL4A3, COL4A2, COL6A6, COL4A4, COL1A1, DPP4, COL1A2 |
| 4151 | PI3K-Akt signaling pathway | 20 | 6.601756194 | 9.08E-11 | FGF9, FGF13, GNG11, COL11A1, PRKAA2, COL6A6, SGK1, COL1A1, COL1A2, COL4A6, COL5A1, TEK, PRL, MCL1, COL4A3, COL5A2, COL4A4, CCND1, COL4A2, COL4A1 |
GO, Gene Ontology; DEGs, differently-expressed genes.
Construction of pathway relation network
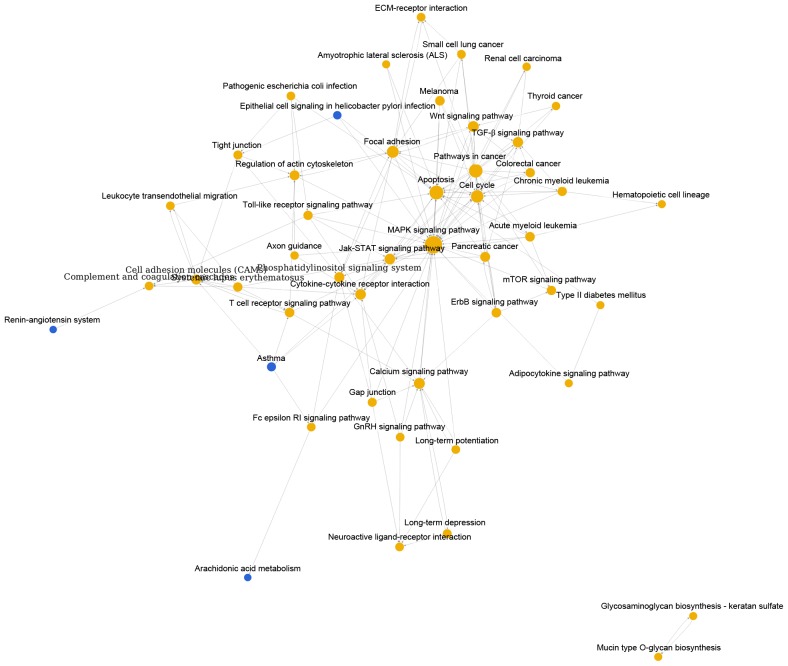
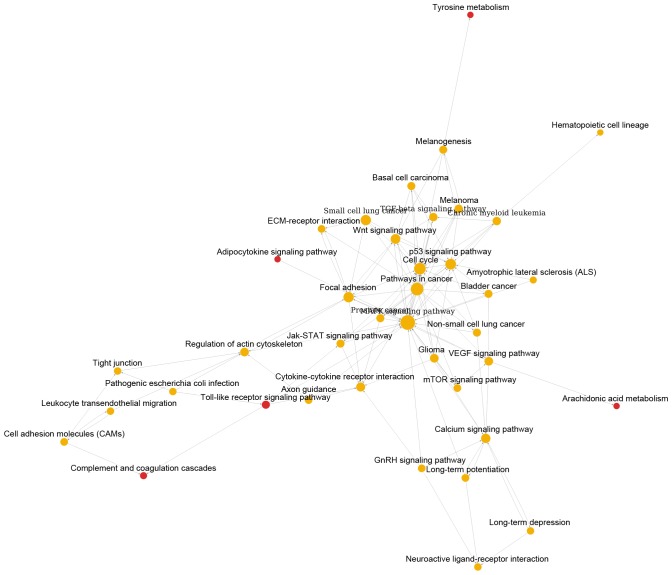
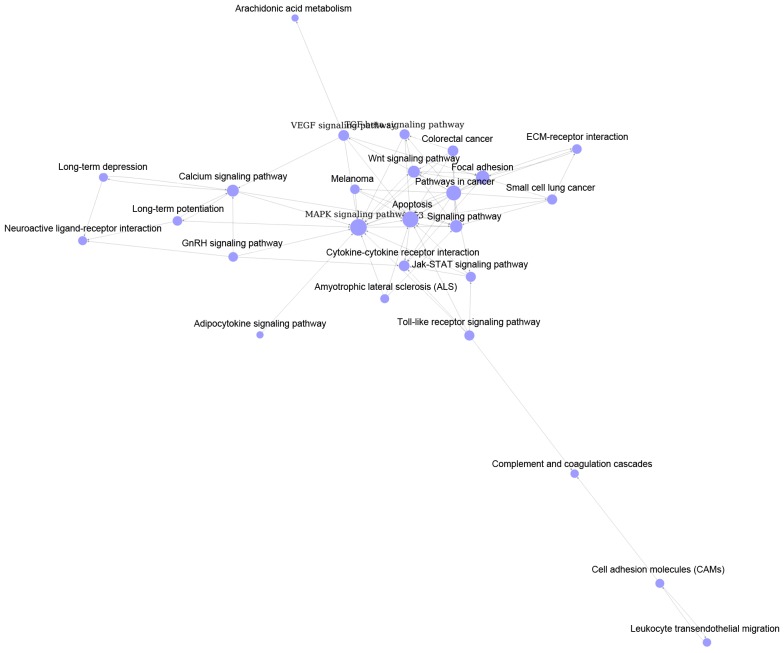
As shown in Fig. 1, pathway relation network of group A with 48 nodes and 157 edges was constructed. The top 3 significant pathways with higher degrees were mitogen-activated protein kinase (MAPK) signaling pathway (degree = 31), apoptosis (degree = 20) and pathways in cancer (degree = 19). Interestingly, there were four downregulated pathways including epithelial cell signaling in helicobacter pylori infection, asthma, renin-angiotensin system and arachidonic acid metabolism. Similarly, Fig. 2 showed the pathway relation network of group B (39 nodes and 119 edges). In addition, this network was with 5 upregulated pathways, such as toll-like receptor signaling pathway and complement and coagulation cascades. Besides, the pathway relation network of group C with 28 nodes and 76 degrees were shown in Fig. 3. Top 3 significant pathways with higher degrees were MAPK signaling pathway (degree = 17), apoptosis (degree = 15) and pathways in cancer (degree = 13).
Figure 1.
Pathway relation network of DEGs in MED12 mutation leiomyoma samples compared with MED12 wild-type myometrium samples. The red nodes represent pathways involved upregulated DEGs and the blue nodes represent pathways involved downregulated DEGs. DEGs, differentially-expressed genes; MED12, mediator of RNA polymerase II transcription subunit 12.
Figure 2.
Pathway relation network of DEGs in MED12 wild-type leiomyoma samples compared with MED12 wild-type myometrium samples. The red nodes represent pathways involved upregulated DEGs. The yellow nodes represent pathways involved both up- and downregulated DEGs. DEGs, differentially-expressed genes; MED12, mediator of RNA polymerase II transcription subunit 12.
Figure 3.
Pathway relation network of common DEGs. The violet nodes represent pathways of various groups of DEGs. DEGs, differentially-expressed genes.
Construction of gene co-expression network
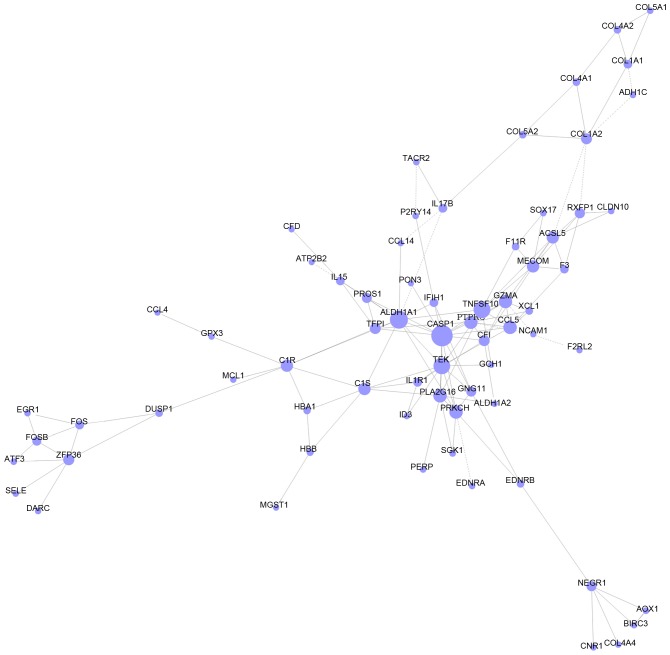
DEGs, which enriched in GO terms and pathways in group C were intersected to screen the more important DEGs based on the information of gene symbols. As a result, a total of 135 important DEGs were obtained, including Acyl-CoA synthetase medium-chain family member 3 (ACSM3), protein S (α) (PROS1) and F11 receptor (F11R) (Table IV). As shown in Fig. 4, gene co-expression network with 68 nodes and 133 edges was constructed. Thereinto, the hub nodes included caspase 1 (CASP1, degree = 14), aldehyde dehydrogenase 1 family member A1 (ALDH1A1, degree = 12), tumor necrosis factor superfamily member 10 (TNFSF10, degree = 11) and TEK receptor tyrosine kinase (TEK, degree = 11).
Table IV.
The top 10 intersected important DEGs of GO terms and pathways in group C.
| Gene symbol | Accession no. | Gene description | Biotype | GO ID | Pathway ID |
|---|---|---|---|---|---|
| ACSM3 | NM_005622 | Acyl-CoA synthetase medium-chain family member 3 | Coding | GO:0008217 | 01100 |
| PROS1 | NM_000313 | Protein S (α) | Coding | GO:0007596, GO:0030168, GO:0006508, GO:0045087, GO:0030449, GO:0002576, GO:0050900 | 04610 |
| F11R | NM_016946 | F11 receptor | Coding | GO:0007155, GO:0007596, GO:0006954, GO:0050900, GO:0007179, GO:0019048, GO:0045216 | 04514, 04670 |
| RXFP1 | NM_001253727 | Relaxin/insulin-like family peptide receptor 1 | Coding | GO:0030198, GO:0030154 | 04080 |
| CXCL14 | NM_004887 | Chemokine (C-X-C motif) ligand 14 | Coding | GO:0006955, GO:0007267, GO:0007165, GO:0006935 | 04060, 04062 |
| RIMS2 | NM_001100117 | Regulating synaptic membrane exocytosis 2 | Coding | GO:0042391, GO:0030073 | 04911 |
| EDNRA | NM_001166055 | Endothelin receptor type A | Coding | GO:0008217, GO:0032496, GO:0007165, GO:0007507, GO:0007568, GO:0043066, GO:0008283, GO:0019233, GO:0008284, GO:0001934, GO:0007186, GO:0050678, GO:0007204, GO:0001666, GO:0071260, GO:0051281, GO:0043278, GO:0060137, GO:0003094 | 04080, 04020, 04270 |
| ZFP36 | NM_003407 | ZFP36 ring finger protein | Coding | GO:0000122, GO:0006950 | 05166 |
| EDNRB | NM_000115 | Endothelin receptor type B | Coding | GO:0008217, GO:0007166, GO:0007568, GO:0000122, GO:0043066, GO:0019233, GO:0008284, GO:0001934, GO:0050678, GO:0007204, GO:0007399, GO:0048246, GO:0048265, GO:0019934 | 04080, 04020 |
| SOXs17 | NM_022454 | SRY (sex determining region Y)-box 17 | Coding | GO:0001525, GO:0000122, GO:0045944, GO:0030308, GO:0001706, GO:0060070, GO:0031648, GO:0090090, GO:0045893, GO:0072001 | 04310 |
GO, Gene Ontology; DEGs, differentially-expressed genes.
Figure 4.
Gene co-expression network of important DEGs. The nodes represent the important DEGs and the edges represent the relationship between two DEGs. DEGs, differentially-expressed genes.
The expression of CASP1, ALDH1A, PROS1 and GADPH in uterine leiomyomas and control samples
The expression of CASP1, ALDH1A, PROS1 and GADPH were measured by qRT-PCR. As shown in Fig. 5, the expression of CASP1 and ALDH1A was significant higher in leiomyosarcoma cell line SK-UT-1 than that in uterine smooth muscle cell line PHM1-31. However, the expression of PRPS1 was significant lower in leiomyosarcoma cell line SK-UT-1. These results were consistent with above bioinformatics analysis.
Figure 5.
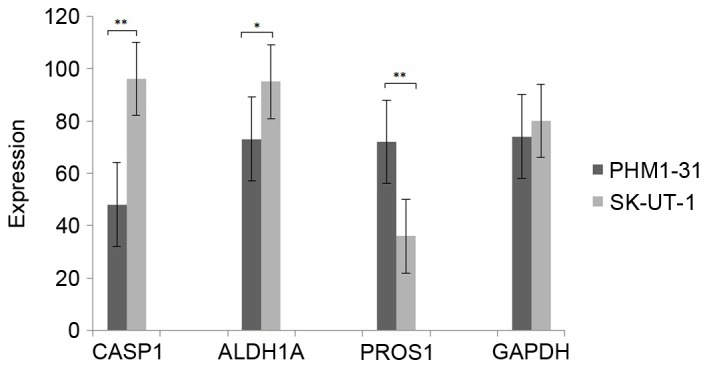
The expression of CASP1, ALDH1A, PROS1 and GADPH in uterine leiomyomas and control samples. The black and gray column represent the expression level of CASP1, ALDH1A, PROS1 and GADPH in PHM1-31 and SK-UT-1, respectively (*P<0.05; **P<0.01). CASP1, caspase 1; ALDH1A1, aldehyde dehydrogenase 1 family member A1; PROS1, protein S (α).
Discussion
Uterine leiomyomas disease is one of the most common benign smooth muscle tumors of uterus (14). In order to research more effective treatment methods, numerous studies have revealed various particular genes and pathways which are closely related with the development of uterine leiomyomas (15,16). Recently, the bioinformatics methods were performed to research the molecular mechanism of uterine leiomyomas disease. In this study, CASP1, ALDH1A1, PROS1, hematopoietic prostaglandin D synthase (HPGDS) and carbonyl reductase 3 (CBR3) were found differentially expressed in uterine leiomyomas disease samples.
CASP1 is an evolutionarily conserved enzyme, which plays important role in inflammatory immune response. It is highly expressed in various immune organs including spleen, kidney, liver and blood. The expression value of CASP1 is positively related to the inflammatory response and also with many other functions, including proteolytic cleavage, pyroptosis response and inducing necrosis. In addition, an increased inflammatory environment may cause leiomyomas, and further induce negatively impact (17). In psoriasis patients, Thirupaithi et al (18) undertook western blot experiments and found that CASP1 could be suppressed by Th-1 response for methotrexate and involved in immunopathogenesis. Gloria-Bottini et al (19) also found that the pathogenesis of uterine leiomyomas was closely related with the chronically inflammatory state. Except the pathway of innate immune response, CASP1 was also found to be enriched in signal transduction and apoptotic process and with higher expression level in uterine leiomyomas samples in this study. Likewise, Christman et al (20) showed that characters of uterine leiomyomas were decreased apoptosis and alterations in various signaling pathways, such as Notch signaling pathway. Therefore, we infer that CASP1 may be an attractive target of uterine leiomyomas by participating pathways including immune response, signal transduction and apoptotic process.
ALDH1A1 encoded the protein which a member of aldehyde dehydrogenases family, and participated the oxidative pathway of alcohol metabolism. In many studies, the expression of ALDH1A1 was confirmed to be higher in myoma samples than in the myometrium cells (21). However, the mechanism of this gene for uterine leiomyomas has not been identified. Through an experimental study of myoma and myometrium, Shveiky and Rojansky (22) found that acetaldehyde was with the inhibitory effect on myoma by participating in the pathway of cell growth. In the present study, ALDH1A1 was a key gene in two pathways of small molecule metabolic process and positive regulation of Ras GTPase activity and with higher expression level in uterine leiomyomas samples. In similarly to our results, Luo and Chegini (23) found that Ras GTPase activating protein was associated with cell differentiation, hypertrophy and apoptosis, which could further influence the growth and regression of leiomyoma. The results of western blot and RT-PCR in previous study showed that ALDH isozymes affected cell growth and motility (24). Besides, the risk of fibroids was verified to be statistically positively associated with small molecule metabolic process, such as metabolic process of triglyceride (25). Thus, we speculate that ALDH1A1 may be associated with small molecule metabolic process and positive regulation of Ras GTPase activity, and could be a key target for uterine leiomyomas treatment.
PROS1 encoded S-protein which is a vitamin K-dependent plasma glycoprotein synthesized in the liver. It always combined with protein C and further participated into the pathway of coagulation (26). Previous study processed western blot and confirmed that the nonsense mutation of PROS1 could induce anticoagulation protein S deficiency (27). In addition, this protein could also bind to negatively charged phospholipids to form a bridging which enhanced the phagocytosis of the apoptotic cell and reduced the inflammation occurring. Furthermore, Cunin et al (28) found that numbers of apoptotic cells might induce an autoimmune response. In this study, PROS1 participated into pathways including innate immune response, platelet activation and leukocyte migration and expressed lower in uterine leiomyomas samples. All of these pathways were closely related to immune process. Above all, PROS2 might be a potential target for uterine leiomyomas treatment by participating into the immune process.
Besides, there were several special pathways in MED12 mutation leiomyoma samples and 2 MED12 wild-type leiomyoma samples. For example, the pathway of arachidonic acid metabolism was downregulated expressed in MED12 mutation leiomyoma samples but upregulated expressed in MED12 wild-type leiomyoma samples. In MED12 mutation leiomyoma samples, this pathway was enriched by HPGDS, phospholipase A2 group XVI (PLA2G16), glutathione peroxidase 3 (GPX3), PTGS2 and phospholipase A2 group IVA (PLA2G4A). Similarly, in MED12 wild-type leiomyoma samples, this pathway involved genes including CBR3, PTGS2, PLA2G16, PLA2G4A and GPX3. By far, there's no studies showed the relationship between MED12 mutation and arachidonic acid metabolism. Based on the results of this study, we inferred that HPGDS and CBR3 might be the special genes related with MED12 mutation in uterine leiomyomas disease.
In conclusions, CASP1, ALDH1A and PROS1 might be the potential biomarkers for uterine leiomyomas diagnose and treatment by participating into the pathways of immune response and small molecule metabolic process. In addition, HPGDS and CBR3 might be the special genes related with MED12 mutation in uterine leiomyomas disease. The present findings offer us a new perspective for further clinical diagnosis.
References
- 1.Moroni RM, Vieira CS, Ferriani RA, Candido-dos-Reis FJ, Brito LGO. Pharmacological treatment of uterine fibroids. Ann Med Health Sci Res. 2014;4(Suppl 3):S185–S192. doi: 10.4103/2141-9248.141955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Razavi MK, Hwang G, Jahed A, Modanlou S, Chen B. Abdominal myomectomy versus uterine fibroid embolization in the treatment of symptomatic uterine leiomyomas. AJR Am J Roentgenol. 2003;180:1571–1575. doi: 10.2214/ajr.180.6.1801571. [DOI] [PubMed] [Google Scholar]
- 3.He X. Estrogen receptor and progesterone receptor expressions at the edge of the targeted region of uterus fibroids ablated by acoustic power. J Chongqing Med Univ. 2011;36:5–7. [Google Scholar]
- 4.Mäkinen N, Mehine M, Tolvanen J, Kaasinen E, Li Y, Lehtonen HJ, Gentile M, Yan J, Enge M, Taipale M, et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science. 2011;334:252–255. doi: 10.1126/science.1208930. [DOI] [PubMed] [Google Scholar]
- 5.Raimundo N, Vanharanta S, Aaltonen LA, Hovatta I, Suomalainen A. Downregulation of SRF-FOS-JUNB pathway in fumarate hydratase deficiency and in uterine leiomyomas. Oncogene. 2009;28:1261–1273. doi: 10.1038/onc.2008.472. [DOI] [PubMed] [Google Scholar]
- 6.Shen Y, Wu Y, Lu Q, Zhang P, Ren M. Transforming growth factor-β signaling pathway cross-talking with ERα signaling pathway on regulating the growth of uterine leiomyoma activated by phenolic environmental estrogens in vitro. Tumour Biol. 2016;37:455–462. doi: 10.1007/s13277-015-3813-4. [DOI] [PubMed] [Google Scholar]
- 7.Klotzbücher M, Wasserfall A, Fuhrmann U. Misexpression of wild-type and truncated isoforms of the high-mobility group I proteins HMGI-C and HMGI(Y) in uterine leiomyomas. Am J Pathol. 1999;155:1535–1542. doi: 10.1016/S0002-9440(10)65469-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chuang TY, Min J, Wu HL, McCrary C, Layman LC, Diamond MP, Azziz R, Al-Hendy A, Chen YH. Berberine inhibits uterine leiomyoma cell proliferation via downregulation of cyclooxygenase 2 and pituitary tumor-transforming gene 1. Reprod Sci. 2017;24:1005–1013. doi: 10.1177/1933719116675055. [DOI] [PubMed] [Google Scholar]
- 9.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 10.Huan JL, Gao X, Xing L, Qin XJ, Qian HX, Zhou Q, Zhu L. Screening for key genes associated with invasive ductal carcinoma of the breast via microarray data analysis. Genet Mol Res. 2014;13:7919–7925. doi: 10.4238/2014.September.29.5. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Chu C, Lu J, Kong X, Huang T, Cai YD. Gene ontology and KEGG pathway enrichment analysis of a drug target-based classification system. PLoS One. 2015;10:e0126492. doi: 10.1371/journal.pone.0126492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Southby J, Murphy LM, Martin TJ, Gillespie MT. Cell-specific and regulator-induced promoter usage and messenger ribonucleic acid splicing for parathyroid hormone-related protein. Endocrinology. 1996;137:1349–1357. doi: 10.1210/endo.137.4.8625910. [DOI] [PubMed] [Google Scholar]
- 14.Commandeur AE, Styer AK, Teixeira JM. Epidemiological and genetic clues for molecular mechanisms involved in uterine leiomyoma development and growth. Hum Reprod Update. 2015;21:593–615. doi: 10.1093/humupd/dmv030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kessel PdAHMGv, Pauwels PAA, Schoenmakers EFPM. Molecular genetic and functional characterization of signalling networks that govern the development of human uterine leiomyomas; IEEE International Symposium on Multiple-valued Logic; 1998.p. 233.
- 16.Commandeur AE, Styer AK, Teixeira JM. Epidemiological and genetic clues for molecular mechanisms involved in uterine leiomyoma development and growth. Hum Reprod Update. 2015;21:593–615. doi: 10.1093/humupd/dmv030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rice KE, Secrist JR, Woodrow EL, Hallock LM, Neal JL. Etiology, diagnosis, and management of uterine leiomyomas. J Midwifery Womens Health. 2012;57:241–247. doi: 10.1111/j.1542-2011.2012.00176.x. [DOI] [PubMed] [Google Scholar]
- 18.Thirupathi A, Elango T, Subramanian S, Gnanaraj P. Methotrexate regulates Th-1 response by suppressing caspase-1 and cytokines in psoriasis patients. Clin Chim Acta. 2016;453:164–169. doi: 10.1016/j.cca.2015.12.022. [DOI] [PubMed] [Google Scholar]
- 19.Gloria-Bottini F, Pietropolli A, Ammendola M, Saccucci P, Bottini E. PTPN22 and uterine leiomyomas. Eur J Obstet Gynecol Reprod Biol. 2015;185:96–98. doi: 10.1016/j.ejogrb.2014.11.043. [DOI] [PubMed] [Google Scholar]
- 20.Christman GM, Tang H, Ahmad I, Stribley JM. Differential expression of the Notch signal transduction pathway: Ligands, receptors and Numb in uterine leiomyomas vs. myometrium. Fertil Steril. 2007;88(Suppl 1):S72. doi: 10.1016/j.fertnstert.2007.07.242. [DOI] [Google Scholar]
- 21.Zaitseva M, Vollenhoven BJ, Rogers PAW. 443. The fibroblast-smooth muscle cell relationship is altered in uterine leiomyoma. Reprod Fertil Dev. 2008;20:123. doi: 10.1071/SRB08Abs443. [DOI] [Google Scholar]
- 22.Shveiky D, Rojansky N. 255: Alcohol has a permissive effect on the growth of uterine leiomyomata cells in tissue cultures. J Minim Invasive Gynecol. 2007;14(Suppl):S92–S93. doi: 10.1016/j.jmig.2007.08.197. [DOI] [Google Scholar]
- 23.Luo X, Chegini N. The expression and potential regulatory function of microRNAs in the pathogenesis of leiomyoma. Semin Reprod Med. 2008;26:500–514. doi: 10.1055/s-0028-1096130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreb JS, Baker HV, Chang LJ, Amaya M, Lopez MC, Ostmark B, Chou W. ALDH isozymes downregulation affects cell growth, cell motility and gene expression in lung cancer cells. Mol Cancer. 2008;7:87. doi: 10.1186/1476-4598-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeda T, Sakata M, Isobe A, Miyake A, Nishimoto F, Ota Y, Kamiura S, Kimura T. Relationship between metabolic syndrome and uterine leiomyomas: A case-control study. Gynecol Obstet Invest. 2008;66:14–17. doi: 10.1159/000114250. [DOI] [PubMed] [Google Scholar]
- 26.Castoldi E, Hackeng TM. Regulation of coagulation by protein S. Curr Opin Hematol. 2008;15:529–536. doi: 10.1097/MOH.0b013e328309ec97. [DOI] [PubMed] [Google Scholar]
- 27.Fang BQ, Zhang JW. Molecular mechanisms of protein S deficiency caused by a novel nonsense mutation. Chin J Vasc Surg. 2015;2:32–35. [Google Scholar]
- 28.Cunin P, Beauvillain C, Miot C, Augusto JF, Preisser L, Blanchard S, Pignon P, Scotet M, Garo E, Fremaux I, et al. Clusterin facilitates apoptotic cell clearance and prevents apoptotic cell-induced autoimmune responses. Cell Death Dis. 2016;7:e2215. doi: 10.1038/cddis.2016.113. [DOI] [PMC free article] [PubMed] [Google Scholar]