Abstract
The present study aimed to compare brown adipose-derived stem cell (BASC) and white adipose-derived stem cell (WASC) differentiation into pacemaker-like cells following T-box (TBX)18 transduction. Mouse BASCs and WASCs were induced to differentiate into pacemaker-like cells by adenovirus-TBX18 transduction in vitro. The transduction rate was determined by fluorescence microscopy and cell ultrastructural changes were observed by transmission electron microscopy at 48 h post-transduction. The mRNA and protein expression of pacemaker cell-associated markers, including TBX18, TBX3, sarcomeric α-actinin (Sr) and hyperpolarization-activated cyclic nucleotide-gated channel 4 (HCN4), were detected by reverse transcription-quantitative polymerase chain reaction, immunofluorescence staining and western blot analysis. The results demonstrated that no significant difference was observed in the transduction rate between BASCs and WASCs. The ultrastructure of BASCs was observed to be more complex than that of WASCs, indicating that BASCs may possess a better structural foundation to differentiate into pacemaker-like cells. TBX18, TBX3, Sr and HCN4 mRNA and protein expression in differentiated stem cells was significantly increased compared with the respective control groups. Furthermore, the expression levels were significantly higher in TBX18-BASCs compared with TBX18-WASCs. In conclusion, TBX18 gene transduction may facilitate the differentiation of BASCs and WASCs into pacemaker-like myocardial cells, and BASCs may have a higher capacity than WASCs for this differentiation. TBX18 gene may therefore act as an efficient candidate in cell transplantation therapy for diseases and for future research into the cardiovascular system.
Keywords: T-box 18, brown adipose-derived stem cells, white adipose-derived stem cells, myocardial cells, pacemaker-like cells
Introduction
Chronic arrhythmia is a serious threat to human health, and anti-arrhythmic medication and the fitting of electronic pacemakers are the standard treatments at present (1). A biological pacemaker based on cell and gene technology would be an improved clinical treatment (2). However, the lack of ideal seed cells hinders the development of biological pacemakers. Adipose-derived stem cells (ADSCs) are easily obtained and amplified, and exhibit weak immunogenicity and strong differentiation (3). The expression of cardiac troponin I was detected in differentiated ADSCs isolated from the myocardial cells of mice aged 1–2 weeks (4). Furthermore, it has been reported that ADSCs are able to differentiate into pacemaker-like cells in vitro (5). Therefore, it may be hypothesized that ADSCs may have the potential to become ideal seed cells for the development of biological pacemakers.
T-box (TBX) 18 is a biomarker for pacemaker progenitor cells and is associated with the development of the heart, including the formation of the venous pole (6). Gene tracing technology has demonstrated that the majority of cells in the sinoatrial node head originate from TBX18+ progenitor cells, indicating that TBX18 is an important transcription factor in the development of the embryonic sinoatrial node (7). Additionally, TBX18 is able to promote the development and differentiation of embryonic epicardial cells (8). It has also been reported that co-culture of white adipose-derived stem cells (WASCs) with newborn mouse myocardial cells transfected with TBX18 promotes WASC differentiation into pacemaker-like cells (9). Furthermore, one study also demonstrated that stem cells isolated from brown adipose tissues exhibit a high myocardial differentiation capacity and spontaneously differentiate into functional cardiomyocytes in vitro (10). Our previous study demonstrated that brown adipose-derived stem cells (BASCs) spontaneously differentiated into pacemaker cells in vitro, and TBX18 mRNA and protein expression was elevated during the differentiation process (11). Based on these findings, it was hypothesized that transduction of BASCs with the TBX18 gene may further promote their differentiation into pacemaker-like cells. WASCs were also transduced with TBX18 as they also have the ability to differentiate into myocardial cells. TBX18-induced differentiation ability was assessed in BASCs and WASCs to identify a more ideal seed cell line for the development of biological pacemakers.
Materials and methods
Materials
Dulbecco's modified Eagle's medium (DMEM) was purchased from Corning Incorporated (Corning, NY, USA) and fetal bovine serum (FBS) was purchased from Gibco (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Penicillin, streptomycin, amphotericin B were purchased from Gibco (Thermo Fisher Scientific, Inc.), and bovine serum albumin (BSA) was purchased from Sigma-Aldrich (cat. no. 0245C789; Merck KGaA, Darmstadt, Germany) and TBX18 primary antibody (rabbit; cat. no. sc-17867) was purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). TBX3 (mouse; cat. no. ab89220;), sarcomeric α-actinin (Sr; mouse; cat. no. ab9465) and hyperpolarization-activated cyclic nucleotide-gated channel 4 (HCN4; rat; cat. no. ab32675) primary antibodies were purchased from Abcam (Cambridge, MA, UK). β-actin primary antibody (WB0196; 1:1,000; Shanghai Wei Biotechnology Co., Ltd., Shanghai, China), FITC-goat anti-mouse secondary antibody (GB22301; 1:1,000; Shanghai Wei Biotechnology Co., Ltd.) was used in immunofluorescence of Sr and CY3-goat anti-rat IgG (GB; GB21302; 1:1,000; Shanghai Wei Biotechnology Co., Ltd.) was used in immunofluorescence of HCN4. DAPI-Staining-solution was purchased from Wuhan Boster Biological Technology, Ltd., Wuhan, China (cat. no. AR1176). An TRIquick total RNA extraction kit (cat. no. R1100) was purchased from Beijing Solarbio Science and Technology Co., Ltd. (Beijing, China). Adenovirus (AD)-cherry-TBX18 (1.06×1010 PFU/ml) and AD-green fluorescent protein (AD-GFP; 2.03×1010 PFU/ml) were prepared by Shanghai Genechem Co., Ltd. (Shanghai, China).
Isolation and culture of stem cells
All animal experiments were approved by the ethics committee of the Second Military Medical University (Shanghai, China). Healthy male C57BL/6 mice (n=25; weight, 30 g) were obtained from the Laboratory Animal Center of the Second Military Medical University, and they were kept at room temperature, with free access to water and food (SLACOM; cat. no. 201404001; Shanghai Pu Lu Teng Biotechnology Co., Ltd., Shanghai, China), 12-h light/dark cycle and humidity 40–50%. Brown and white adipose tissues were respectively isolated from the shoulder blade and groin of the mice at 3–4 weeks. Tissues were digested in a solution containing 0.1% BSA and 0.01% collagenase type II (Sigma-Aldrich; Merck KGaA) in a 37°C water bath for 45 min. Tissue residues were filtered by centrifugation at 800 × g for 5 min at 4°C. The supernatant was discarded and cells were resuspended in DMEM containing 15% FBS, 0.5% penicillin, 0.5%streptomycin and 0.5%amphotericin B, at 37°C with 5% CO2. The medium was replaced every two days and cells were subcultured when 80–90% confluence was reached.
Flow cytometry analysis
Third-generation WASCs and BASCs (>106/ml) were harvested, centrifuged at 230 × g for 5 min at 4°C, resuspended in PBS, transferred to 1 ml EP tubes and centrifuged again at 230 × g for 5 min at 4°C. The supernatant was discarded and WASCs and BASCs were resuspended in the blocking reagent containing 10% goat FBS and PBS for 30 min at room temperature, and then incubated with indirect labeling primary antibody CD29 (rabbit; cat. no. ab179471; 1:100), direct labeling primary antibody CD90 (cat. no. ab226; 1:100), indirect labeling primary antibody CD34 (rabbit; cat. no. ab81289; 1:100) and direct labeling primary antibody CD45 (cat no. ab33916; 1:100; all Abcam) monoclonal antibodies for 30 min at 4°C in the dark, respectively. Then the corresponding secondary antibodies, CY3-goat anti-rabbit IgG (GB21303; 1:100; Shanghai Wei Biotechnology Co., Ltd., Shanghai, China), FITC-goat anti-rabbit IgG (GB22303; 1:100; Shanghai Wei Biotechnology Co., Ltd.) were added for 30 min at 4°C in the dark. Additionally, a blank group was set up; cells were centrifuged at 230 × g for 5 min at 4°C, the supernatant was removed, and cells were washed with PBS twice and resuspended in PBS prior to analysis using a flow cytometer (BD FACSCalibur; BD Biosciences, Franklin Lakes, NJ, USA) with CellQuest Pro version 5.2.1 software.
Multipotential differentiation of BASCs and WASCs
Third-generation BASCs and WASCs under logarithmic growth phase were cultured in a complete medium (DMEM containing 15% FBS, 0.5% penicillin, 0.5% streptomycin and 0.5% amphotericin B). When cells had reached 100% confluence, the medium was discarded by careful suction. Tri-lineage differentiation medium (adipogenesis; cat. no. MUBMD-90031; osteogenesis; cat. no. MUBMX-90021; and chondrogenesis; cat. no. MUBMD-9004; Cyagen Biosciences Guangzhou, Inc., Guangzhou, China) was used to confirm mesenchymal origin. Induction medium was replaced every three days (adipogenesis, 7 days; osteogenesis, 21 days; chondrogenesis, 21 days) at 37°C with 5% CO2. Subsequently, the induced cells were fixed in 4% paraformaldehyde for 10 min, at room temperature and rinsed with PBS. Cells were stained with 0.5% Oil Red O for 10 min to detect lipids and with 1% alizarin red S (pH 7.2) for 8 min to detect calcium deposition. The accumulation of chondrocyte matrix was detected by Alcian Blue solution (anhydrous ethanol 80 ml, glacial acetic acid 20 ml, Alcian Blue powder 0.1 g) staining for 10 min (pH 2.5), all the experimental conditions were at a room temperature. All above reagents were purchased from Cyagen Biosciences Guangzhou, Inc. Cells were observed using an Olympus IX70 microscope (Olympus Corporation, Tokyo, Japan) at magnification, ×200.
Transduction of BASCs and WASCs with TBX18
Adenovirus (AD)-TBX18 (1.06×1010 PFU/ml) and AD-green fluorescent protein (AD-GFP; 2.03×1010 PFU/ml) were prepared by Shanghai Genechem Co., Ltd. (Shanghai, China). When cells had reached 95% confluence and following medium replacement, adipose stem cells were randomized into the control group (WASC/BASC), blank virus genome group (GFP-WASC/BASC) and the target gene group (TBX18-WASC/BASC). Prior to transduction with AD-TBX18 and AD-GFP, cells were cultured in 100% DMEM for 12 h, then complete medium (DMEM containing 15% FBS, 0.5% penicillin, 0.5% streptomycin and 0.5% amphotericin B), at 37°C with 5% CO2. Cell transduction efficiency was tested at a multiplicity of infection (MOI) of 10, 25, 40, 50 and 70; a MOI of 50 was subsequently selected as the transduction degree. Cells were observed and imaged (Olympus IX70; Olympus Corporation) 48 h post-transduction by fluorescence microscopy.
Transmission electron microscopy (TEM)
At 48 h post-transduction collected cell samples were washed three times with PBS and digested with 0.25% trypsin (Gibco; Thermo Fisher Scientific, Inc.). Digested cells were resuspended in a centrifuge tube containing serum medium (85% DMEM and 15% FBS) and centrifuged at 230 × g for 5 min at 4°C. The supernatant was discarded and cells were rapidly fixed in 2% glutaraldehyde acetone solution at room temperature and subjected to gradient dehydration. Epoxy resin (Epon812) was used for embedding at 37°C for 3 h, and the sections cut at 70 nm, selected, located and double-stained with uranyl acetate for 20 min and lead citrate for 5 min, both at room temperature. The ultrastructures were subsequently observed by TEM (magnification, ×10,000; Hitachi h7650; Hitachi, Ltd., Tokyo, Japan).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
TBX18, TBX3, Sr and HCN4 mRNA expression was detected by RT-qPCR. Total RNA was extracted with a TRIquick total RNA extraction kit (R1100; Beijing Solarbio Science and Technology Co., Ltd.) according to the manufacturer's protocol. A First Strand cDNA synthesis kit (K1622; Invitrogen; Thermo Fisher Scientific, Inc.) and a Quantity Nova SYBR-Green PCR kit (208052; Qiagen GmbH, Hilden, Germany) were used for qPCR, according to the manufacturer's protocols. The thermocycling conditions were as follows: Initial denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for 10 sec and 55°C for 15 sec, and a final extension at 72°C for 15 sec (12). Each sample was tested at least three times and β-actin was used as the internal reference gene. mRNA expression was quantified using the 2−ΔΔCq method (13). The primers were designed and synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) and the sequences are listed in Table I. qPCR was performed on a Roche 480II Real Time PCR System (Roche Diagnostics, Basel, Switzerland).
Table I.
Primer sequences used for reverse transcription-quantitative polymerase chain reaction.
| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| β-actin | AGCCATGTACGTAGCCATCC | GCTGTGGTGGTGAAGCTGTA |
| TBX18 | TGCTGTCCCTGCTACACATC | GCTGTAGGTCTCTGCCAAGG |
| HCN4 | TCATCTCCTCCATCCCTGTC | CTGGCCAGGTCATAGGTCAT |
| TBX3 | GCTGCTGCGAACTCTCTTCT | GAAGGTGTCGGAAACTGGAA |
| Sr | TCATCTCAGGTGAACGCTTG | AGATGTCCTGGATGGCAAAG |
TBX, T-box; HCN4, hyperpolarization-activated cyclic nucleotide-gated channel 4; Sr, sarcomeric α-actinin.
Immunofluorescence staining
At 48 h post-transduction cells (when cells had reached 60% confluence) were cultured on glass slides in a complete medium (DMEM containing 15% FBS, 0.5% penicillin, 0.5% streptomycin and 0.5% amphotericin B) for 48 h at 37°C with 5% CO2. Cells were fixed in 100% methanol for 10 min and blocked with 5% BSA (containing 0.3% Triton X-100 for permeabilization; Sigma-Aldrich; Merck KGaA) for 30 min prior to incubation with HCN4 (1:200) and Sr (1:50) primary antibodies overnight at 4°C. Cells were subsequently washed three times (5 min each time) with PBS and incubated with FITC-goat anti-mouse secondary antibody (GB22301; 1:1,000; Shanghai Wei Biotechnology Co., Ltd.) and CY3-goat anti-rabbit IgG (GB; GB21302; 1:1,000; Shanghai Wei Biotechnology Co., Ltd.) at room temperature for 1 h prior to counterstaining with DAPI for 3 min at room temperature. The prepared sample was observed under an inverted fluorescence microscope. Images were collected with a CCD digital camera (Canon, Inc., Tokyo, Japan). The obtained fluorescent images were analyzed (three fields from each sample) with Image Pro Plus version 6.0 (Media Cybernetics, Inc., Rockville, MD, USA) and Adobe Photoshop CS3 (Adobe Systems, Inc., San Jose, CA, USA).
Western blot analysis
At 48 h post-transduction, proteins were extracted from adipose stem cells by RIPA (cat. no. WB0101; Shanghai Wei Biotechnology Co., Ltd.). Protein concentration was determined with a bicinchoninic acid protein assay kit. Proteins (30 µg) were separated via SDS-PAGE on a 12 and 10% gel. The separated proteins were subsequently transferred onto a polyvinylidene difluoride membrane and blocked with 5% BSA for 2 h at room temperature. Following this, the membranes were incubated TBX18 (1:1,000), TBX3 (1:1,000), Sr (1:1,000), HCN4 (1:1,000) and β-actin (1:2,000; Shanghai Wei Biotechnology Co., Ltd.) primary antibodies overnight at 4°C. Following the addition of horseradish peroxidase-labeled goat anti-mouse secondary antibody (1:2,000; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA), goat anti-rat secondary antibody (1:2,000; WB0179; Shanghai Wei Biotechnology Co., Ltd.) and goat anti-rabbit secondary antibody (1:2,000; WB0177; Shanghai Wei Biotechnology Co., Ltd.) for 2 h at 37°C, the membrane was reacted with ECL hypersensitive chemiluminescence kit (cat. no. WB0164; Shanghai Wei Biotechnology Co., Ltd.). The X-ray film was photosensitized, developed and fixed in a dark room, and relative protein expression was calculated with Quantity One 4.6.2 software (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The gray value of the target protein band was compared with that of the β-actin band, and the obtained gray level ratio was used as the relative expression of the target protein.
Statistical analysis
Statistical analysis was performed with GraphPad Prism 5.0 statistical software (GraphPad Software, Inc., La Jolla, CA, USA). The experiments were repeated three times. Data are presented as the mean ± standard error of the mean, n=3. Data were statistically analyzed with one-way analysis of variance followed by Tukey's post-hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Characterization of BASCs and WASCs
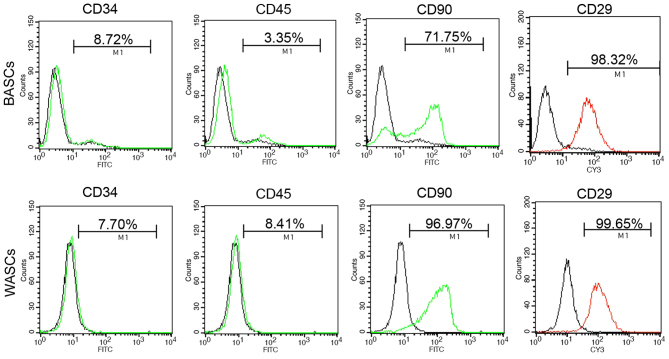
Analysis of the third generation BASC and WASC characteristics was performed using flow cytometry analysis (Fig. 1 and Table II). Flow cytometry detected the absence of hematopoietic stem cell surface antigens CD34 and CD45 expression in BASCs and WASCs (Fig. 1 and Table II). However, positive expression of mesenchymal stem cell surface antigens CD90 and CD29 was detected in both cell types (Fig. 1 and Table II).
Figure 1.
Surface marker analysis of third generation BASCs and WASCs by flow cytometry. Neither cell type expressed CD34 or CD45, but they did express CD90 and CD29. BASCs, brown adipose-derived stem cells; WASCs, white adipose-derived stem cells; FITC, fluorescein isothiocyanate; CY3, cyanine 3.
Table II.
Surface marker analysis of third generation BASCs and WASCs by flow cytometry.
| Surface marker | BASCs (%) | WASCs (%) | Positive (+) or negative (−) |
|---|---|---|---|
| CD34 | 8.39±0.3 | 7.37±0.3 | − |
| CD45 | 3.15±0.1 | 8.19±0.2 | − |
| CD90 | 70.48±1.4 | 96.6±0.6 | + |
| CD29 | 97.26±0.7 | 98.3±1.3 | + |
Values are presented as the mean ± standard error of the mean from triplicate determinations repeated in three separate experiments. BASCs, brown adipose-derived stem cells; WASCs, white adipose-derived stem cells.
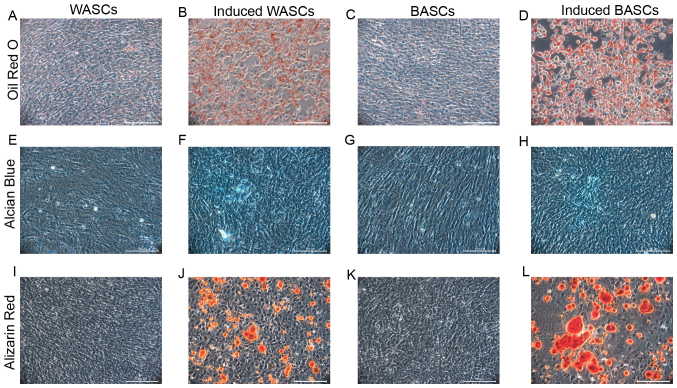
BASCs and WASCs differentiate into osteogenic, adipogenic and chondrogenic lineages
It is established that BASCs and WASCs are multipotential mesenchymal stem cells. To determine the multipotential differentiation capability of BASCs and WASCs, passage 3 cells were induced to differentiate towards osteogenic, adipogenic and chondrogenic lineages. Adipogenic induction observation in BASCs and WASCs revealed cell morphology alterations from a long, spindle shape to a quasi-spherical shape, and bright fat drops were observed intracellularly. The amount lipid droplets increased gradually with time to 80–90% of the cell volume and the nuclear volume reduced or disappeared. No volume enlargement or lipid droplet formation was observed in control cells (Fig. 2A-D). After 7 days of induction, bright red lipid droplets were observed by Oil Red O staining, while the same was not observed in cells without induction in both WASCs (Fig. 2A and B) and BASCs (Fig. 2C and D).
Figure 2.
Differentiation potential of WASCs and BASCs. Cells were cultured in adipogenic, chondrogenic or osteogenic inductive medium. (A) Control WASCs, (B) adipogenesis-induced WASCs, (C) control BASCs and (D) adipogenesis-induced BASCs were stained with Oil Red O 7 days post-adipogenic induction. (E) Control WASCs, (F) chondrogenesis-induced WASCs, (G) control BASCs and (H) chondrogenesis-induced BASCs were stained with Alcian Blue 21 days post-chondrogenic induction. (I) Control WASCs, (J) osteogenesis-induced WASCs, (K) control BASCs and (L) osteogenesis-induced BASCs were stained with alizarin red S 21 days post-osteogenic induction. Scale bar, 100 µm. WASCs, white adipose-derived stem cells; BASCs, brown adipose-derived stem cells.
Following chondrogenic differentiation induction in BASCs and WASCs, Alcian Blue staining was performed in control and induced BASCs and WASCs (Fig. 2E-H), which revealed chondrocyte aggregation into small cell masses. Prussian Blue staining demonstrated the presence of light blue-stained dense nuclei and cells around the masses grew in a radial manner in both, indicating the potential generation of sulfuric acid proteoglycan specific to the cartilage matrix in both WASCs (Fig. 2F) and BASCs (Fig. 2H). No such phenomenon was observed in the corresponding control cells (Fig. 2E and G).
After 7 days of osteogenic induction, light microscopy revealed enlarged cell bodies that were initially spindle or cork-like shaped that subsequently became polygonal and grew in multiple layers. After 14 days of induction, cell bodies had further increased in size and had the tendency to aggregate. Dense, overlapping cell growth was observed and cells gradually grew around the center of the colony in a small island-like distribution. Subsequently, small opaque calcified spots were present. These small calcified spots gradually became larger and on day 21, typical orange calcified nodules were observed following alizarin red staining (Fig. 2I-L). These observations were noted in both WASCs (Fig. 2J) and BASCs (Fig. 2L). These were not observed in the corresponding control cells (Fig. 2I and K).
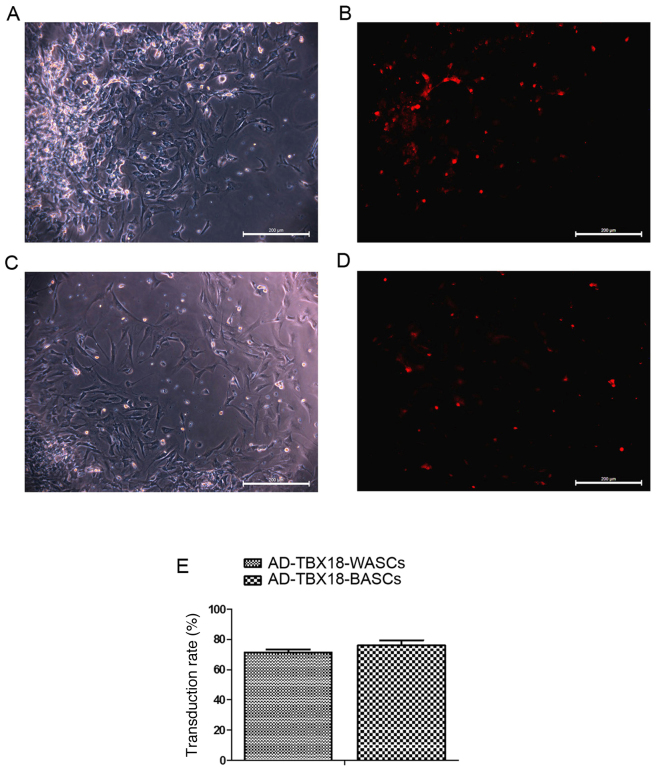
The transduction rate of BASCs and WASCs with AD-TBX18 is not significantly different
Different MOI values were tested in BASCs and WASCs for AD-TBX18 and AD-GFP transduction. A MOI value of 50 was selected as it exhibited the highest cell transduction efficiency. Following successful transduction of AD-TBX18, BASCs and WASCs emitted red fluorescence (AD-cherry-TBX18). Transduction efficiency was observed under an inverted microscope. Cells with red fluorescence were densely observed in the TBX18-BASC group (Fig. 3A and B). Fewer cells with red fluorescence were observed in the TBX18-WASC group (Fig. 3C and D). However, no significant difference was detected in the BASC and WASC TBX18 transduction rate (P>0.05; Fig. 3E), indicating that TBX18 may be stably expressed in BASCs and WASCs.
Figure 3.
Transduction of TBX18 in BASCs and WASCs. BASCs transduced with TBX18 were observed under an (A) light and (B) fluorescence microscope. WASCs transduced with TBX18 were observed under a (C) light and (D) fluorescence microscope. (E) Transduction rates in the two groups were not significantly different. Data are presented as the mean ± standard error of the mean n=3. Scale bar, 200 µm. TBX18, T-box 18; BASCs, brown adipose-derived stem cells; WASCs, white adipose-derived stem cells; AD, adenovirus.
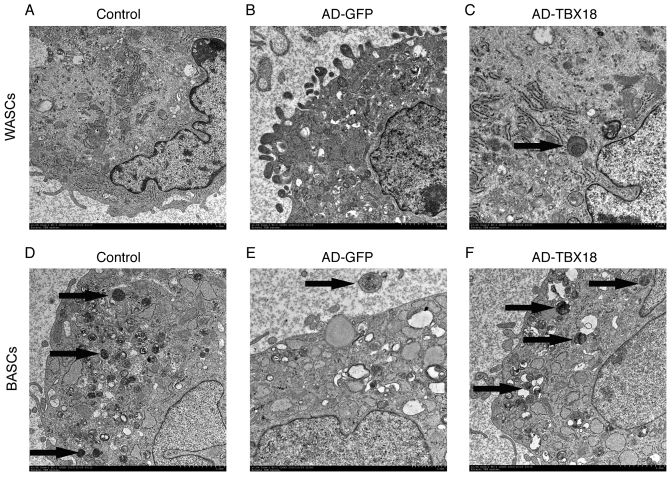
The ultrastructure of TBX18-BASCs is more similar to pacemaker-like cells than that of TBX18-WASCs
WASC, GFP-WASC, TBX18-WASC, BASC, GFP-BASC and TBX18-BASC ultrastructure was observed by TEM (Fig. 4). The results revealed marked ultrastructural differences between the WASCs and BASCs. WASCs contained only a small number of organelles (Fig. 4A-C), whereas BASCs contained more cytoplasm, organelles and mitochondrial cristae (Fig. 4D-F). Furthermore, a multilocular structure was observed in BASCs, while a monolocular structure was observed in WASCs. It was evident that the ultrastructure of BASCs was more complex compared with that of WASCs. Thus, it may be concluded that BASCs possess ultrastructural advantages for differentiation into pacemaker-like cells.
Figure 4.
Ultrastructure of BASCs and WASCs was examined by TEM (magnification, ×10,000). Ultrastructural images of (A) normal control WASCs, (B) control WASCs transduced with AD-GFP, (C) WASCs transduced with AD-TBX18, (D) normal control BASCs, (E) control BASCs transduced with AD-GFP and (F) BASCs transduced with AD-TBX18. Images were captured by TEM. The black arrows indicate the Golgi body. BASCs, brown adipose-derived stem cells; WASCs, white adipose-derived stem cells; TEM, transmission electron microscopy; AD, adenovirus; GFP, green fluorescent protein; TBX18, T-box 18.
mRNA expression of pacemaker cell-associated genes is higher in TBX18-transduced BASCs
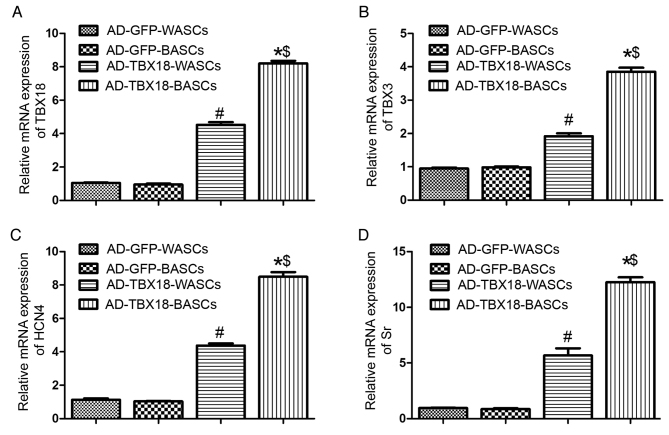
TBX18 mRNA expression was significantly higher in TBX18-BASCs compared with TBX18-WASCs (P<0.05; Fig. 5A). Additionally, TBX18 mRNA expression in TBX18-transduced BASCs and WASCs was significantly higher compared with the corresponding AD-GFP control groups (P<0.05; Fig. 5A). No significant difference was detected in TBX18 expression between the GFP-BASCs and GFP-WASCs (Fig. 5A).
Figure 5.
Reverse transcription-quantitative polymerase chain reaction analysis of pacemaker cell-associated genes in BASCs and WASCs following TBX18 transduction. The mRNA expression of (A) TBX18, (B) TBX3, (C) HCN4 and (D) Sr in WASCs and BASCs transduced with AD-GFP or AD-TBX18. Data are presented as the mean ± standard error of the mean n=3. *P<0.05 vs. AD-GFP-BASCs; #P<0.05 vs. AD-GFP-WASCs; $P<0.05 vs. AD-TBX18-WASCs. BASC, brown adipose-derived stem cells; WASCs, white adipose-derived stem cells; TBX, T-box; HCN4, hyperpolarization-activated cyclic nucleotide-gated channel 4; Sr, sarcomeric α-actinin; AD, adenovirus; GFP, green fluorescent protein.
Furthermore, TBX3 (Fig. 5B), HCN4 (Fig. 5C) and Sr (Fig. 5D) mRNA expression was significantly higher in TBX18-BASCs compared with TBX18-WASCs (P<0.05). TBX3, HCN4 and Sr mRNA expression in TBX18-BASCs and TBX18-WASCs was significantly higher compared with the corresponding GFP control groups (P<0.05; Fig. 5B-D). No difference in TBX3, HCN4 and Sr expression was observed between GFP-BASCs and GFP-WASCs (Fig. 5B-D).
Immunofluorescence analysis of Sr and HCN4 protein expression
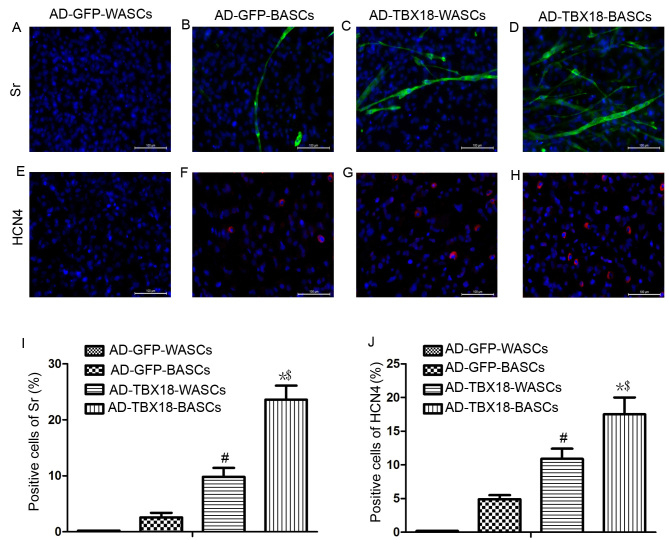
Immunofluorescence staining results in Fig. 6 revealed that the expression of Sr and HCN4 protein was significantly higher in TBX18-BASCs compared with TBX18-WASCs. Additionally, Sr and HCN4 expression in AD-TBX18-tranduced cells was significantly higher compared with WASCs and BASCs transduced with AD-GFP. It was identified that the positive cells for Sr of AD-GFP-BASCs (Fig. 6B) were lower (2.6±0.8%) compared with AD-TBX18-WASCs (9.8±1.60%; P<0.05; Fig. 6C). The positive cells for Sr of AD-TBX18-BASCs (Fig. 6D) were 23.6±2.5%, higher compared with AD-TBX18-WASCs (P<0.05; Fig. 6I). No Sr or HCN4 positive cells were identified in AD-GFP-WASCs (Fig. 6A and E). It also was found that the positive cells for HCN4 of AD-GFP-BASCs (Fig. 6F) were 4.9±0.6%, which were lower compared with AD-TBX18-WASCs (10.9±1.50%; P<0.05; Fig. 6G). The positive cells for HCN4 of AD-TBX18-BASCs (Fig. 6H) were 17.5±2.1%, higher compared with AD-TBX18-WASCs (P<0.05; Fig. 6J).
Figure 6.
Immunofluorescence staining analysis of Sr and HCN4 expression in WASCs and BASCs following TBX18 transduction. Immunofluorescence staining of Sr in (A) control WASCs transduced with AD-GFP, (B) control BASCs transduced with AD-GFP, (C) WASCs transduced with AD-TBX18 and (D) BASCs transduced with AD-TBX18. HCN4 expression was also investigated by immunofluorescence staining in (E) control WASCs transduced with AD-GFP, (F) control BASCs transduced with AD-GFP, (G) WASCs transduced with AD-TBX18 and (H) BASCs transduced with AD-TBX18. Cells were counterstained blue with DAPI to visualize the nucleus. Scale bar, 100 µm. All significant differences of positive cells between groups were presented as a graph; positive cells of (I) Sr and (J) HCN4. *P<0.05 vs. AD-GFP-BASCs; #P<0.05 vs. AD-GFP-WASCs; $P<0.05 vs. AD-TBX18-WASCs. Sr, sarcomeric α-actinin; HCN4, hyperpolarization-activated cyclic nucleotide-gated channel 4; WASCs, white adipose-derived stem cells; BASCs, brown adipose-derived stem cells; TBX18, T-box 18; AD, adenovirus; GFP, green fluorescent protein.
The expression of pacemaker cell-associated proteins is higher in TBX18-BASCs
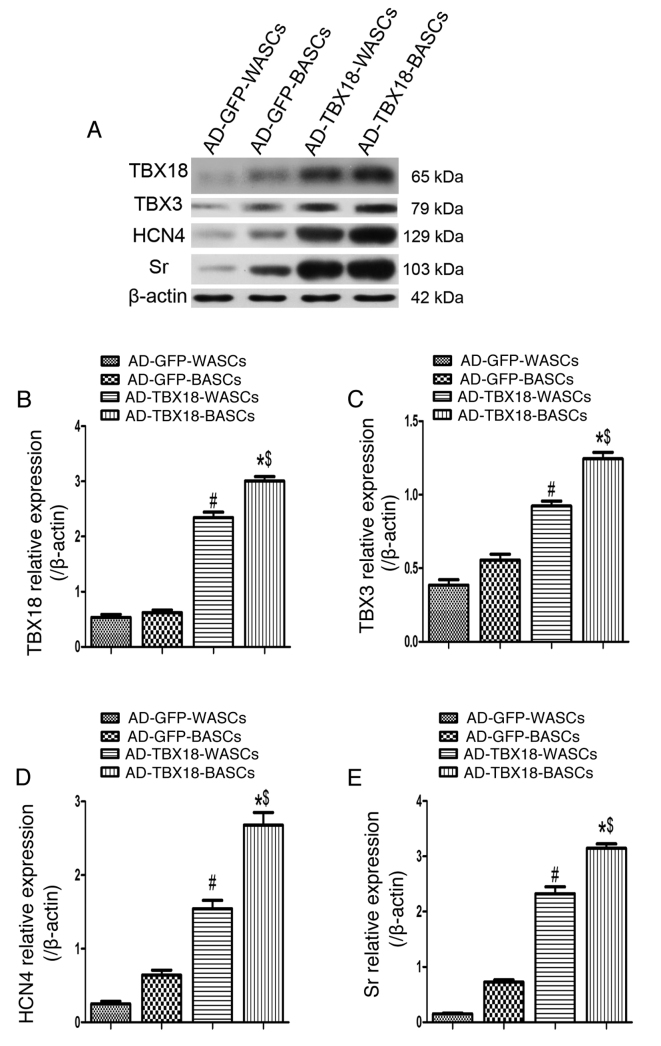
Western blot analysis was performed in TBX18-BASCs, TBX18-WASCs, GFP-BASCs and GFP-WASCs to detect the expression of TBX18, TBX3, HCN4 and Sr (Fig. 7A). Quantitative analysis revealed that TBX18 protein expression was significantly higher in TBX18-BASCs compared with TBX18-WASCs (P<0.05; Fig. 7B). Additionally, TBX18 protein expression was significantly higher in TBX18-BASCs and TBX18-WASCs, compared with the corresponding GFP control groups (P<0.05; Fig. 5B). No significant difference in TBX18 expression was detected between GFP-BASCs and GFP-WASCs (Fig. 7B).
Figure 7.
Western blot analysis of pacemaker cell-associated protein expression in BASCs and WASCs following TBX18 transduction. (A) Representative western blot bands for TBX18, TBX3, HCN4 and Sr in WASCs and BASCs transduced with either AD-TBX18 or AD-GFP. Densitometric analysis was performed to quantify the protein expression of (B) TBX18, (C) TBX3, (D) HCN4 and (E) Sr. Expression levels were normalized to β-actin. Data are presented as the mean ± standard error of the mean n=3. *P<0.05 vs. AD-GFP-BASCs; #P<0.05 vs. AD-GFP-WASCs; $P<0.05 vs. AD-TBX18-WASCs. BASCs, brown adipose-derived stem cells; WASCs, white adipose-derived stem cells; TBX, T-box; HCN4, hyperpolarization-activated cyclic nucleotide-gated channel 4; Sr, sarcomeric α-actinin; AD, adenovirus; GFP, green fluorescent protein.
Furthermore, TBX3 (Fig. 7C), HCN4 (Fig. 7D) and Sr (Fig. 7E) protein expression was significantly higher in TBX18-BASCs compared with TBX18-WASCs (P<0.05). TBX3, HCN4 and Sr protein expression was also significantly higher in TBX18-BASCs and TBX18-WASCs, compared with the corresponding GFP control groups (P<0.05; Fig. 7C-E). No significant difference in TBX3, HCN4 and Sr expression between the GFP-BASCs and GFP-WASCs was detected (Fig. 7C-E).
Discussion
The fitting of an electronic pacemaker for the treatment of heart block and sinoatrial node (SAN) dysfunction has the advantage of high reliability and low recurrence (14). However, electronic pacemakers are not sensitive to the body's humoral regulation and are susceptible to infection. These limitations have driven research into the development of biological pacemakers (15). However, the development of biological pacemakers has proven difficult due to the lack of seed cells that can be induced to differentiate into autonomic pacemaker cells. Compared with embryonic, induced pluripotent and adult stem cells, ADSCs have gradually emerged as a novel choice of target cell, owing to their easy availability, lower rejection rate and the potential of multidirectional differentiation (16).
TBX18 controls the formation of the head region in the developing SAN. In the present study, TBX18 was successfully transduced into BASCs and WASCs. The results revealed that there was no significant difference between the BASC and WASC TBX18 transduction rate, indicating that TBX18 may be stably expressed in both cell types. It has been confirmed that TBX18 controls ~75% of SAN head formation, but does not have much involvement in SAN tail formation. By contrast, TBX3 is not involved in SAN head formation, but controls SAN tail cell differentiation (6,17). No marked morphological changes were observed in the SAN of TBX3-deficient hearts (18). The present study confirmed that TBX3 mRNA and protein expression in BASCs transduced with AD-TBX18 was significantly higher compared with in WASCS transduced with AD-TBX18. Furthermore, TEM revealed that BASCs exhibited more complex ultrastructures compared with WASCs. In the present study, cells in the TBX18-BASCs group in particular had extremely similar internal structures to that of pacemaker cells (19). In the current study, fluorescence microscopy and western blot analysis demonstrated that the expression of Sr, a pacemaker cell-specific striated muscle actin, in TBX18-transduced BASCs was significantly higher compared with in TBX18-transduced WASCs. Therefore, it was concluded that BASCs may have a structural advantage over WASCs in pacemaker-like cell differentiation.
HCN4, one of the major subtypes of the hyperpolarization-activated cyclic nucleotide-gated channel family, has been reported to be highly expressed in SAN head cells and highly sensitive to cyclic AMP. HCN4 predominantly encodes pacemaker current If (20–22). If initiates the four-stage depolarization process, which means this process underlies the ability of pacemaker cells to autonomically regulate the heart and spontaneously produce current. Thus, current research has primarily focused on developing HCN4-mediated biological pacemakers (23,24). The findings of the present study confirmed that the mRNA and protein expression of HCN4 was significantly higher in TBX18-BASCs, compared with the expression in TBX18-WASCs, further indicating that BASCs may be more susceptible to pacemaker cell differentiation. As brown adipose tissue is formed during the fetal period, its amount decreases gradually with age (25), which limits the source available for autologous cell transplantation. Additionally, the origins of the brown and white adipose tissues are different, having a different embryonic source, and adults have less brown fat, which limits the clinical utilization of brown fat (26). Notably, an experiment in both mice and humans revealed that white adipose tissue may be converted into brown adipose tissue in cold environments; this may provide a basis for the clinical use of brown fat converted from white fat (27).
In summary, the present study confirmed that both BASCs and WASCs transduced with TBX18 had the potential to differentiate into pacemaker-like cells in vitro. The results also revealed that differentiated BASCs exhibited more pacemaker-like cell characteristics compared with differentiated WASCs. Therefore, BASCs may be considered for use as a carrier or platform for gene transfer therapy and may provide an effective biological intervention method for patients with SAN disease. However, investigations into the security, stability, persistent functional expression and the specific mechanisms in vivo of BASC gene transfer are required. Despite certain limitations, such as the danger of gene transduction resulting in tumorigenesis, the current study demonstrated that TBX18 gene transduction facilitated the differentiation of BASCs and WASCs into pacemaker-like myocardial cells, of which BASCs may have a higher differentiation capability.
Acknowledgements
The authors are greatly indebted to Professor Yu-Quan Li and Professor Xiang-Qun Yang, for their valuable instructions and suggestions on our thesis as well as their careful reading of the manuscript. We would like to thank colleagues for their valuable suggestions and criticisms.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 31271050 and 31170934).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YQL and XQY conceived and designed the study, and reviewed and edited the manuscript. AJS and LQ performed the experiments. CH and XZ contributed to the data analyzes and images processing. LQ, CH and XZ wrote the paper. All authors read and approved the final manuscript for publication.
Ethics approval and consent to participate
All animal experiments were approved by the ethics committee of the Second Military Medical University (Shanghai, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cho HC, Marbán E. Biological therapies for cardiac arrhythmias: Can genes and cells replace drugs and devices? Circ Res. 2010;106:674–685. doi: 10.1161/CIRCRESAHA.109.212936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapoor N, Liang W, Marbán E, Cho HC. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat Biotechnol. 2013;31:54–62. doi: 10.1038/nbt.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu Y, Liu T, Song K, Fan X, Ma X, Cui Z. Adipose-derived stem cell: A better stem cell than BMSC. Cell Biochem Funct. 2008;26:664–675. doi: 10.1002/cbf.1488. [DOI] [PubMed] [Google Scholar]
- 4.Taha MF, Hedayati V. Isolation, identification and multipotential differentiation of mouse adipose tissue-derived stem cells. Tissue Cell. 2010;42:211–216. doi: 10.1016/j.tice.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Choi YS, Dusting GJ, Stubbs S, Arunothayaraj S, Han XL, Collas P, Morrison WA, Dilley RJ. Differentiation of human adipose-derived stem cells into beating cardiomyocytes. J Cell Mol Med. 2010;14:878–889. doi: 10.1111/j.1582-4934.2010.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiese C, Grieskamp T, Airik R, Mommersteeg MT, Gardiwal A, de Gier-de Vries C, Schuster-Gossler K, Moorman AF, Kispert A, Christoffels VM. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ Res. 2009;104:388–397. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- 7.Greulich F, Rudat C, Kispert A. Mechanisms of T-box gene function in the developing heart. Cardiovasc Res. 2011;91:212–222. doi: 10.1093/cvr/cvr112. [DOI] [PubMed] [Google Scholar]
- 8.van Wijk B, van den Berg G, Abu-Issa R, Barnett P, van der Velden S, Schmidt M, Ruijter JM, Kirby ML, Moorman AF, van den Hoff MJ. Epicardium and myocardium separate from a common precursor pool by crosstalk between bone morphogenetic protein- and fibroblast growth factor-signaling pathways. Circ Res. 2009;105:431–441. doi: 10.1161/CIRCRESAHA.109.203083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang M, Zhang GG, Wang T, Wang X, Tang YH, Huang H, Barajas-Martinez H, Hu D, Huang CX. TBX18 gene induces adipose-derived stem cells to differentiate into pacemaker-like cells in the myocardial microenvironment. Int J Mol Med. 2016;38:1403–1410. doi: 10.3892/ijmm.2016.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamada Y, Wang XD, Yokoyama S, Fukuda N, Takakura N. Cardiac progenitor cells in brown adipose tissue repaired damaged myocardium. Biochem Biophys Res Commun. 2006;342:662–670. doi: 10.1016/j.bbrc.2006.01.181. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Deng ZJ, Zhou JS, Ji RJ, Zhang X, Zhang CS, Li YQ, Yang XQ. Tbx18-dependent differentiation of brown adipose tissue-derived stem cells toward cardiac pacemaker cells. Mol Cell Biochem. 2017;433:61–77. doi: 10.1007/s11010-017-3016-y. [DOI] [PubMed] [Google Scholar]
- 12.Karam JP, Bonafè F, Sindji L, Muscari C, Montero-Menei CN. Adipose-derived stem cell adhesion on laminin-coated microcarriers improves commitment toward the cardiomyogenic lineage. J Biomed Mater Res A. 2015;103:1828–1839. doi: 10.1002/jbm.a.35304. [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Potapova I, Plotnikov A, Lu Z, Danilo P, Jr, Valiunas V, Qu J, Doronin S, Zuckerman J, Shlapakova IN, Gao J, et al. Human mesenchymal stem cells as a gene delivery system to create cardiac pacemakers. Circ Res. 2004;94:952–959. doi: 10.1161/01.RES.0000123827.60210.72. [DOI] [PubMed] [Google Scholar]
- 15.Jung JJ, Husse B, Rimmbach C, Krebs S, Stieber J, Steinhoff G, Dendorfer A, Franz WM, David R. Programming and isolation of highly pure physiologically and pharmacologically functional sinus-nodal bodies from pluripotent stem cells. Stem Cell Reports. 2014;2:592–605. doi: 10.1016/j.stemcr.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizuno H, Tobita M, Uysal AC. Concise review: Adipose-derived stem cells as a novel tool for future regenerative medicine. Stem Cells. 2012;30:804–810. doi: 10.1002/stem.1076. [DOI] [PubMed] [Google Scholar]
- 17.Bakker ML, Boink GJ, Boukens BJ, Verkerk AO, van den Boogaard M, den Haan AD, Hoogaars WM, Buermans HP, de Bakker JM, Seppen J, et al. T-box transcription factor TBX3 reprogrammes mature cardiac myocytes into pacemaker-like cells. Cardiovasc Res. 2012;94:439–449. doi: 10.1093/cvr/cvs120. [DOI] [PubMed] [Google Scholar]
- 18.Hoogaars WM, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LY, Bakker ML, Clout DE, Wakker V, Barnett P, et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21:1098–1112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bleeker WK, Mackaay AJ, Masson-Pévet M, Bouman LN, Becker AE. Functional and morphological organization of the rabbit sinus node. Circ Res. 1980;46:11–22. doi: 10.1161/01.RES.46.1.11. [DOI] [PubMed] [Google Scholar]
- 20.Liang X, Wang G, Lin L, Lowe J, Zhang Q, Bu L, Chen Y, Chen J, Sun Y, Evans SM. HCN4 dynamically marks the first heart field and conduction system precursors. Circ Res. 2013;113:399–407. doi: 10.1161/CIRCRESAHA.113.301588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biel M, Schneider A, Wahl C. Cardiac HCN channels: Structure, function, and modulation. Trends Cardiovasc Med. 2002;12:206–212. doi: 10.1016/S1050-1738(02)00162-7. [DOI] [PubMed] [Google Scholar]
- 22.Plotnikov AN, Sosunov EA, Qu J, Shlapakova IN, Anyukhovsky EP, Liu L, Janse MJ, Brink PR, Cohen IS, Robinson RB, et al. Biological pacemaker implanted in canine left bundle branch provides ventricular escape rhythms that have physiologically acceptable rates. Circulation. 2004;109:506–512. doi: 10.1161/01.CIR.0000114527.10764.CC. [DOI] [PubMed] [Google Scholar]
- 23.DiFrancesco D, Tortora P. Direct activation of cardiac pacemaker channels by intracellular cyclic AMP. Nature. 1991;351:145–147. doi: 10.1038/351145a0. [DOI] [PubMed] [Google Scholar]
- 24.Wainger BJ, DeGennaro M, Santoro B, Siegelbaum SA, Tibbs GR. Molecular mechanism of cAMP modulation of HCN pacemaker channels. Nature. 2001;411:805–810. doi: 10.1038/35081088. [DOI] [PubMed] [Google Scholar]
- 25.Dinas PC, Nikaki A, Jamurtas AZ, Prassopoulos V, Efthymiadou R, Koutedakis Y, Georgoulias P, Flouris AD. Association between habitual physical activity and brown adipose tissue activity in individuals undergoing PET-CT scan. Clin Endocrinol (Oxf) 2015;82:147–154. doi: 10.1111/cen.12620. [DOI] [PubMed] [Google Scholar]
- 26.Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J, Boström P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.