Abstract
Keloids are benign skin tumors that develop in individuals who have a positive family history of keloid disorders. Keloids are characterized by a deregulated wound-healing process, atypical fibroblasts with extreme deposition of extracellular matrix components, particularly collagen, increased cell proliferation and associated failure of apoptosis. Recently ingenol-mebutate has been used as a novel agent with anti-proliferative activity on human keloids as an alternative treatment option in patients, once conventional therapies have failed. We hypothesized that microRNAs (miR/miRNA) may be involved in the balance between lesion formation and repair. A comprehensive understanding of the molecular mechanism underlying the Ingenol-mebutate response in keloid fibroblast following Ingenol-mebutate exposure has been established previously. Therefore, the present study analyzed changes in miRNAs and apoptotic gene regulation in Ingenol-mebutate treated keloid fibroblast, by reverse transcription-quantitative polymerase chain reaction and a DNA fragmentation assay. The range of upregulated miRNAs and downregulated genes encoding cell death appeared to be associated with the degree of the morphological alterations in Ingenol-mebutate treated keloids. In particular, the upregulation of miR-34a was detected in keloid fibroblasts during and following Ingenol-mebutate exposure. Keloid fibroblasts that overexpressed miR-34a showed differential expression of genes involved in the apoptotic signaling pathway such as p53. In conclusion, the Ingenol-mebutate treatment used here was effective in reducing keloid fibroblast growth in cell culture experiments and the expression of particular miRNAs modulated the pro-apoptotic gene expression following Ingenol-mebutate treatment.
Keywords: keloids, ingenol mebutate, p53, microRNA-34a, apoptosis
Introduction
Keloids are fibrotic tumors, which expand over normal skin beyond the margins of the original wound. Keloids do not revert spontaneously and are characterized by the proliferation of dermal fibroblasts, overproduction of extracellular matrix components, especially collagen, elastin, and proteoglycans, fibronectin, an increased infiltration of inflammatory cells (1).
Autosomal dominant with incomplete penetrance as well as autosomal recessive modes of inheritance have been seen among families with keloid disease. Variable gene expression patterns are also implicated in the etiology of keloid disease, but no single gene mutation has thus far been found to be responsible (2). In particular, as reported in multiple microarray studies, keloid-derived fibroblast cells exhibit dysregulated genes involved in apoptosis, mitogen-activated protein kinase, transforming growth factor-beta, interleukin-6 and plasminogen activator inhibitor-1 pathways (3).
Epigenetic alterations in keloid tissue are also involved. Actually, it has been reported that keloid fibroblast cells have altered patterns of DNA methylation, microRNAs deregulation and histone acetylation (4).
No single therapeutic modality is best for all keloids.
Lately, it has been demonstrated that ingenol-mebutate generates pro-apoptotic effects (5), even if the exact mechanism of action is unknown. Multiple apoptotic mechanisms have pleiotropic effects inducing tumor cell death or inhibiting the growth of cancer cells. Due to such outcome, topical application of ingenol mebutate has been successful for the treatment of precancerous skin; therefore, we effectively treated the lesions from a patient showing recurrent keloids with ingenol mebutate (6).
The main apoptotic effects are intermediate through the induction of specific apoptotic target genes.
Multitudes of mechanisms are operated by p53 to guarantee the induction of apoptosis in a stress signal and tissue specific manner. p53 can lead the major steps in apoptotic pathways: The intrinsic mitochondria pathways involving apoptosome realization, the extrinsic death receptor signalling and the direct caspase activation. Numerous of these effects are mediated through the activation of specific p53-target genes.
Recently, it has been shown that components of p53 pathway are direct targets of microRNAs and their over-expression lead to the induction of apoptosis, senescence and cell cycle arrest (7–9).
MicroRNAs (miRNAs) deregulation is recognized as a form of epigenetic regulation. MicroRNAs are small non-coding RNAs approximately 22 nucleotides long able to control gene expression post-transcriptionally through complementarity with a target mRNA, (10). Aberrant microRNA expression has been implicated in several cellular processes and pathogenic pathways of a number of diseases (11–13). Small non-coding RNAs termed microRNAs (miRNAs) can regulate oxidative stress and several human diseases. Recent studies suggest that microRNAs (miRNAs) might play critical roles in pharmacogenomics (14,15). MicroRNA pharmacogenomics is to study the decrease or increase the expressions of microRNAs, target genes or to change the activities of binding microRNAs following to a pharmacological treatment (16).
Since the molecular pathways underlying Ingenol mebutate mechanism of action are still unclear, in the present research, we aim to evaluate the expression levels of microRNAs and differential expression of pro-apoptotic genes expression in keloid fibroblasts following Ingenol-mebutate exposure.
Materials and methods
Cell cultures experiments and treatment
Previously, fresh tissues were obtained from skin lesions of 20 patients with keloids for establishing the primary cell cultures. Primary cell cultures were performed in a previous study (6), then used retrospectively in the present study.
Fibroblasts from keloids were isolated from samples following the same method established by Lim et al (17). All experiments used cells between the second and third passage. No morphological and biochemical differences were found with the passage. Fibroblast cells were grown to confluence in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), 10 µg/ml streptomycin, and 50 IU/ml penicillin in 5% carbon dioxide at 37°C.
From the second passage treatment time course experiments were set up for 24, 48, and 72 h without serum starvation, adding different concentrations (0.1, 0.3, 1 µM respectively) of Ingenol Mebutate (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) dissolved in water.
RNA isolation
Total RNA was isolated from cultured cells using TRIzol reagent (Sigma-Aldrich; Merck KGaA) according to the manufacturer's instructions. Quality of extracted RNA was determined according to 260/280 absorbance ratio, measured by Nano Drop spectrometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
TaqMan Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) microRNA array
TaqMan Array Human microRNA Panel Kit (ABI, Forest City, CA, USA) which includes 365 human microRNAs as well as 3 negative controls was used for microRNA expression profile.
For miR-34a analysis, reverse transcription was performed using the TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific, Inc.), and PCR products were amplified from cDNA samples using TaqMan MicroRNA Assays (Thermo Fisher Scientific, Inc.). The small nucleolar RNA human RNU48 were used as endogenous controls for normalization of miR-34a expression levels. Fold changes in microRNA expression were calculated using the ΔΔCq method.
Gene ontology analysis (GO)
We analyzed the functional distribution of miR34a target genes using the GO database: http://www.geneontology.org/.
Human apoptosis PCR array
We used the Human Apoptosis RT2 Profiler PCR Array (Qiagen, Inc., Valencia, CA, USA) for the simultaneous human pathway of 84 key genes involved in programmed cell death and the Human Cellular Stress Responses RT2 Profiler PCR Array profiles the expression of 84 genes involved in cellular stress response according to the manufacturer's instructions.
mRNA RT-qPCR analyses
For mRNA analysis, reverse transcription was performed using the High-Capacity Reverse-Transcription Kit (Thermo Fisher Scientific, Inc.). 500 ng/RNA were used in reverse-transcription reaction. RT-qPCR was performed using the SYBR Green method (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Levels of expression were determined by normalization to the housekeeping gene HPRT. PCR primers were purchased from Qiagen (RT2 qPCR Primer Assays).
Phase contrast microscopy
Phase contrast images of control and treated cells were generated with the aid of a Zeiss Axiovert 40 CFL inverted microscope equipped with a Canon Power Shot G6 digital camera.
Annexin V staining
For ANNEXIN V-PI staining, cells were washed with ice-cold PBS, trypsinized, and resuspended in 1× binding buffer [10 mm HEPES/NaOH (pH 7.4), 140 mm NaCl, and 2.5 mm CaCl2] at 1×106 cells/ml. After gentle vortex, the cells were mixed with 5 µl annexin V/fluorescein isothiocyanate (FITC) (Annexin V-FITC Apoptosis Detection kit 3, ABM Inc., USA) and 10 µl propidium iodide (PI) stock solution (50 µg/ml in PBS) followed by a 15 min incubation at room temperature in the dark for apoptosis analysis, following manufacturer's instructions.
DNA fragmentation analysis
The genomic DNA fragmentation was evaluated by agarose gel electrophoresis of DNA. For this purpose, cells were grown in the presence or absence of 3 µM Ingenol Mebutate up to 72 h. After counting and washing, cells were subjected to DNA extraction. The DNA samples were carefully resuspended in TE buffer; the nucleic acid concentration and purity were.
Measured using a NanoDrop® ND-1000 spectrophotometer (Thermo-Scientific, Wilmington, DE, USA). 2 µg of each sample was loaded onto 1.5% TAE agarose gel; DNA laddering was visualized on a UV transilluminator by ethidium bromide staining.
Caspase-3 activity assay
Caspase-3 activity was determined in keloid cells using a colorimetric kit from BioVision Inc., (Milpitas, CA, USA) in accordance with the manufacturer's instructions.
The assay is based on the spectrophotometric detection at 405 nm of the chromophore p-nitroaniline.
(pNA) after cleavage from the labeled substrate DEVDpNA by caspase-3. After Ingenol Mebutate treatment for 48 and 72 h, cells were washed with ice-cold PBS, lysed, centrifuged for 10 min at 10,000 × g. Same volume of cell lysate was added to 50 µl of reaction buffer and 5 µl of specific Caspase-3 substrate (DEVD-pNA).
Statistical analyses
All results shown are mean ± standard deviation of at least three separate experiments, measuring each parameter by triplicate (n=3). Statistical significant differences were tested by one way analysis of variance (ANOVA), and, when the F value was significant, by Student-Newman-Keul's post hoc test. Analysis was performed using UNISTAT version 10 software (Unistat Ltd., Highgate, London, England). P<0.05 was considered to indicate a statistically significant difference.
Results
MicroRNAs differential expression in keloid fibroblasts following Ingenol-mebutate treatment
We examined the microRNA profile in keloid fibroblasts following ingenol-mebutate treatment.
Apoptosis was induced in a dose-dependent manner with Ingenol Mebutate, and we showed the results at 1 microM concentration since they were significant.
Among microRNAs assembled in the list of the TaqMan MicroRNA Assays Human Panel, seven microRNAs (miR-16, −34a, −34b, −34c, -146, -147, and -205) were increased 2-fold or greater in IM-treated (1 Micromol, 72 h) keloid fibroblast cells as compared with untreated cells.
The miR-34 family, miR-34a, −34b, and -34c, were reproducibly induced over 2-fold after IM treatment, whereas four other microRNAs (miR-16, -146, -147, and -205) were not constantly induced. The expression of 17 microRNAs in IM exposed keloid fibroblast cells did not change compared with the untreated cells, representing the microRNA not being responsive to IM.
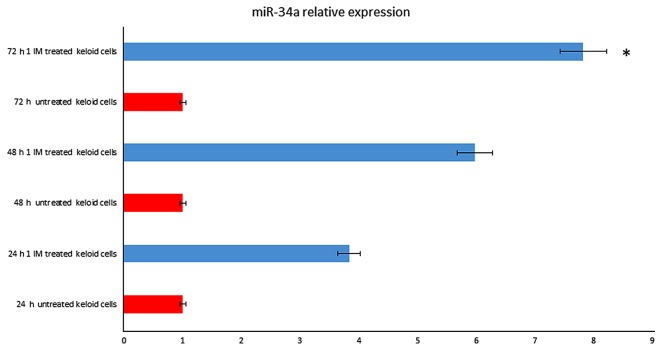
Among differentially expressed microRNA, relevant microRNAs, as miR-34a, miR-338-5p, mir-24 and let-7, were found strongly up-regulated. MicroRNAs data analysis showed that miR-34a (fold change=7.53; P=0.008) were up-regulated after 72 h treatment (Fig. 1), as well miR-24 (fold change=6.37; P=0.005) and let-7 (fold change=2.88; P=0.003 (data not shown)).
Figure 1.
miR-34a expression levels in IM treated keloid fibroblasts. The expression of miR-34a was assayed in 1 µmol IM treated keloid fibroblasts following 72 h of treatment by miRNA assay-based reverse transcription-quantitative polymerase chain reaction. RNU6B was utilized for an endogenous reference to standardize miRNA expression levels. The results were expressed as the relative expression levels following calibration with the universal reference data. *P<0.05 vs. 72 h untreated keloid cells. IM, Ingenol Mebutate; miR/miRNA, microRNA.
Functional categorization for miR-34a gene targets and Gene-expression analysis
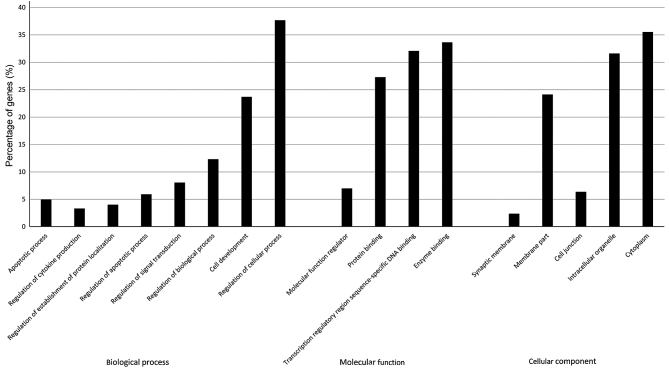
As shown in Fig. 2, the most significantly enriched GO terms identified after screening with a threshold of false discovery rate (FDR) of <0.05. Functional categorization of the GO terms demonstrated that the miR-34a gene targets were strongly associated with biological processes as regulation of biological process, apoptotic process and regulation of apoptotic process. Based on the analysis above, we assumed that the apoptosis pathway is one of the main target for mir-34a (9).
Figure 2.
Functional category enrichment analysis of the differentially expressed genes. The x-axis depicts the significantly enriched GO terms relative to microRNA-34a gene targets. GO, gene ontology.
Beside, it has reported that Ingenol Mebutate induce apoptosis in keratosis actinic cells. Therefore, we aimed to understand if similar molecular mechanism is involved in the treatment of keloids with Ingenol Mebutate. Later, in future research, we aim to investigate other molecular pathway too.
Therefore, we explored the consequences of such microRNA up-regulation in Ingenol Mebutate (1 µM after 72 h) treated keloid fibroblasts on 84 genes of the apoptosis pathway using a PCR array. miR-34a overexpression resulted related to a significant up regulation of tp53 and genes that plays a central role in regulation and activation of apoptosis such as tp53bp2, a regulator that plays a central role in regulation of apoptosis and cell growth via its interactions with TP53 (Fig. 3).
Figure 3.
Upregulated apoptotic genes in keloid fibroblasts following IM treatment. Significant upregulation of the expression of apoptotic genes, was evaluated by quantitative polymerase chain reaction in 1 µmol IM treated keloid fibroblasts following 72 h of treatment. *P<0.05 vs. untreated cells. IM, Ingenol Mebutate; BIRC3, baculoviral IAP repeat-containing protein 3; CASP, caspase; CD, cluster of differentiation; CD40LG, CD40 ligand; CIDEA, cell death inducing DFFA-like effector A; TP53, tumor protein p53; TP53BP2, TP53 binding protein 2; B2 M, β-2-microglobulin.
Among the up-regulated genes, birc3, casp1, casp10, and CIDEA were detected. Interestingly, CD40 was up-regulated, which is expressed on cultured human fibroblasts, but also shifted fibroblasts into the G0/G1 phase of the cell cycle.
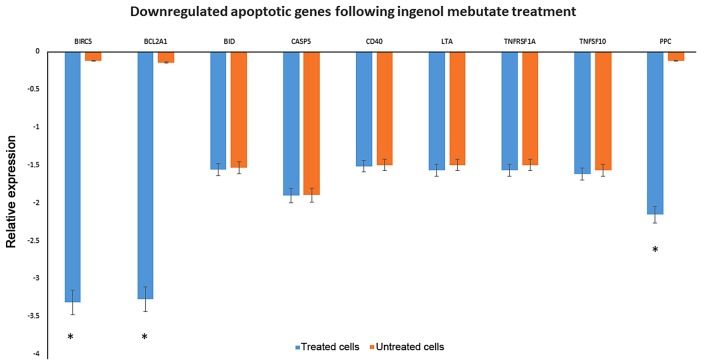
The down-regulation of Bid, casp5, cd40, cd70, and members of Tumor necrosis factor receptor superfamily as TNFRSF1A and TNFSF10, leading to signalling complexes that activate both the apoptotic caspase cascade and the NF-κB and JNK anti-apoptotic pathways, were observed among the list. Genes associated with the inhibition of apoptosis as birc5 and bcl2a1 were highly down-regulated (Fig. 4).
Figure 4.
Downregulated apoptotic genes in keloid fibroblasts following IM treatment. Significant downregulation of the expression of apoptotic genes was evaluated by quantitative polymerase chain reaction in 1 µmol IM treated keloid fibroblasts following 72 h of treatment. *P<0.05 vs. untreated cells. IM, Ingenol Mebutate; CASP, caspase; CD, cluster of differentiation; BIRC5, baculoviral IAP repeat-containing protein 5; BCL2A1, B-cell lymphoma 2 related protein A1; BID, BH3 interacting domain death agonist; LTA, lymphotoxin α; TNFRSF, tumor necrosis factor receptor superfamily member; PPC, phosphoenolpyruvate carboxylase.
miR-34a induction depends on p53 activation
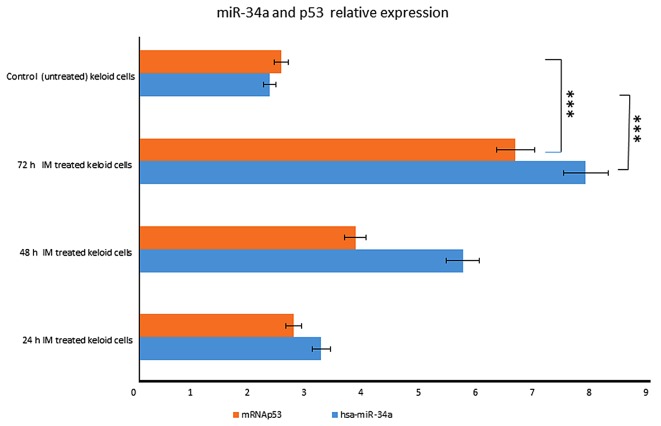
In particular, miR-34a expression was increased in a time-dependent manner after IM treatment, rising 3.2-fold at 24 h until 7 fold at 72 h (Fig. 1). We also observed that p53 started to increase at 24 h, respectively, and the increasing continued until 72 h after treatment (Fig. 5).
Figure 5.
miR-34a and p53 expression levels in IM treated keloid fibroblasts. miR-34a expression was increased in a time-dependent manner following 1 µmol IM treatment in keloid fibroblasts parallel to p53 expression levels. ***P<0.001 vs. control (untreated) keloid cells (as indicated). IM, Ingenol Mebutate; miR, microRNA.
These results indicate that miR-34a is induced in a p53-dependent manner after IM treatment.
Ingenol mebutate induces apoptosis-like phenotypes
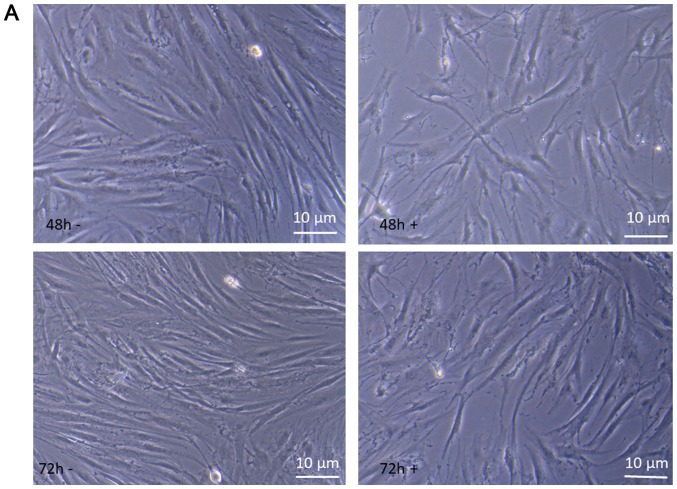
We observed that introduction of Ingenol Mebutate in keloid fibroblast cells caused apoptosis-like phenotypes showing similar morphological changes with enlarged cellular size.
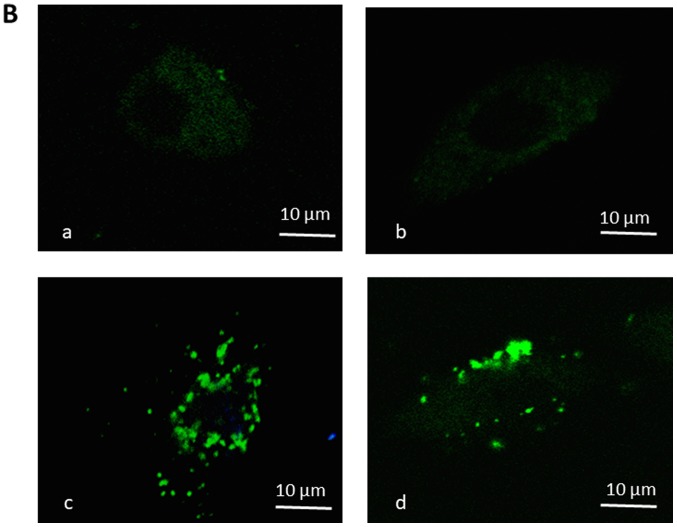
The behavior of keloid fibroblasts over the 72 h period of treatment with 1 µM Ingenol Mebutate was monitored by phase-contrast microscopy and high enlargement pictures analyzed. As shown in Fig. 6A at 48 and 72 h of incubation time, comparing treated to untreated keloid fibroblasts morphological changes are evident that strongly suggest alterations in the cytoskeleton organization, impairment of cell growing, differentiation and development. These results suggest that suppression of cell proliferation by Ingenol Mebutate could be mainly associated with the induction of apoptosis-like phenotypes. Therefore, we evaluated apoptosis with Annexin V assay. As shown in Fig. 6B, staining revealed Annexin V-positive/PI negative, apoptotic, keloid cells after 72 h ingenol mebutate treatment. Quantification of the annexin was not assayed.
Figure 6.
(A) When compared with control cells, cells treated with 1 µmol IM (48 + and 72 h +) did not grow as well and displayed heterogeneous morphological aspects with frequent cytoplasmic arborizations. (B) Annexin V staining. Images a and b are the control untreated cells, while images c and d are the keloid fibroblasts following 72 h of incubation with 1 µmol IM. Scale bars=10 µm. IM, Ingenol Mebutate.
Ingenol mebutate induces DNA fragmentation
To verify if Ingenol Mebutate could induce DNA fragmentation and thus to confirm whether apoptosis occurred, keloid fibroblasts exposed to treatment were assessed for DNA laddering by agarose gel electrophoresis (Fig. 7). We found that treated keloid fibroblasts after 48 and 72 h of incubation with 1 Micromol Ingenol mebutate, showed apoptotic DNA fragmentation profiles. On the opposite, no nucleic acid fragmentation was observed in negative controls represented by untreated cells.
Figure 7.
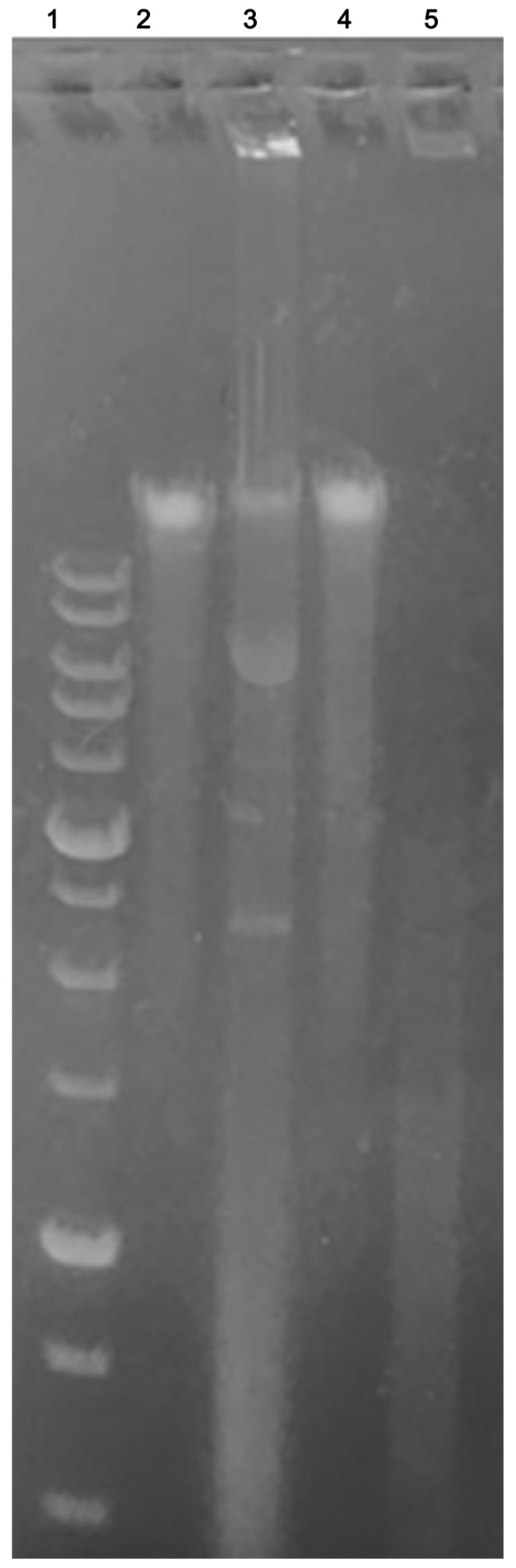
DNA fragmentation of keloid fibroblasts following 48 and 72 h of incubation with 1 µmol IM. Apoptotic DNA fragmentation was qualitatively analyzed by agarose gel electrophoresis. Lane 1, 1 kB DNA ladder marker; lanes 2 and 4, negative control (untreated cells); lanes 3 and 5, IM treated cells at 72 and 48 h, respectively. IM, Ingenol Mebutate.
Apoptotic effect mediated through caspase-3
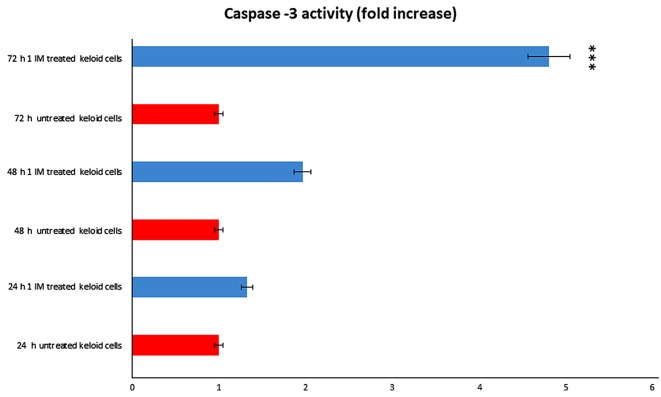
We evaluated the potential contribution of caspase-3 in Ingenol mebutate treated keloid cells. We obtained that caspase-3 was significantly activated after treatment (Fig. 8). We observed a dose and a time dependent effect, obtaining a significant result after 72 h of incubation with 1 Micromol Ingenol mebutate.
Figure 8.
IM induced apoptosis in keloid fibroblasts through caspase-3 activation. Following incubation with 1 µmol of IM for 48 and 72 h the enzymatic activity was measured by a quantitative colorimetric caspase-3 activity assay. ***P<0.001 vs. 72 h untreated keloid cells. IM, Ingenol Mebutate.
Discussion
Keloids are benign tumors that resist all clinical therapies and often recur, compromising the quality of life of many patients. Since pro-apoptotic effects of this drug have been demonstrated in several malignant cells (5), in a previous research we successfully treated a woman with a large keloid with Ingenol mebutate (6). So far, there is no molecular study on its effect on keloid fibroblasts.
Here, to explore such novel treatment modalities for keloids and to elucidate the molecular mechanism of IM treatment in keloid fibroblasts, we analyzed the microRNA and gene apoptosis expression patterns in keloid fibroblasts following exposure to Ingenol mebutate, by array strategy.
Current studies have suggested that microRNAs regulates gene expression and play important roles in mediating responses of drug molecules (18). microRNAs can regulate the expression of drug transporters and drug targets. Following Ingenol-mebutate exposure, we identified miR-34a as a major microRNA that is induced in in keloid fibroblasts.
miR-34 is a direct targets of p53 and their over-expression results in the induction of apoptosis, cell cycle arrest and senescence (7,9,19). Beside, miR-34a could partially replace p53 in inducing apoptosis or senescence. p53 directly can activate the expression of miR-34 genes, which play an important role in p53-mediated apoptotic pathway (20). Activation of miR-34a by p53 may contribute to induction of apoptosis. The miR-34a gene itself may be a tumor suppressor gene since it maps to a region on chromosome 1p36 which is commonly deleted in neuroblastoma.
The p53 tumor-suppressor protein activates a mass of genes encoding proteins with roles in apoptosis, DNA repair, senescence, and cell-cycle control. miR-34 might be key effectors of p53 tumor-suppressor function, and the inactivation might contribute to definite cancers.
Due to IM exposure in keloid fibroblasts, genotoxic stress activated p53 transcription of many genes, leading to cell cycle arrest, apoptosis or senescence (21). microRNAs, as the miR-34 family, are transcriptionally activated by p53. In turn microRNAs regulate expression of many p53-induced genes.
However, the outcome of miR-34a overexpression on p53 levels and role depends on cellular setting (7). In order to evaluate the association between the regulation of miR-34 with pro-apoptotic gene expression in Ingenol Mebutate treated keloid fibroblast, we measured expression levels of the apoptosis pathway gene was here investigated using a PCR array. We found a positive correlation between the increase of miR-34 and p53 expression.
Tp53 plays a central role in regulation and activation of apoptosis in Ingenol Mebutate treated keloid fibroblasts, since was up-regulated as well as tp53bp2, a regulator that plays a central role in regulation of apoptosis and cell growth via its interactions with TP53. Among the up-regulated genes, more apoptotic genes as birc3, casp1, casp10, and CIDEA were identified. Such genes plays a central role in the execution-phase of cell apoptosis and regulate also inflammatory signaling and immunity.
miR-34 family were shown to suppress tumor growth and metastasis by promoting processes that inhibit carcinogenesis, such as apoptosis and senescence. MiR-34 regulate these processes through downregulation of their target mRNAs. As of today, more than 77 miR-34 targets have been confirmed (22). Among miR-34 target gene, we detected birc5 and bcl2a1, genes associated with the inhibition of apoptosis, that were down-regulated.
The expressions of more miR34a target genes, such as Notch1, Delta1, Bcl2 or E2F3 in Ingenol Mebutate treated cells, will be examined in next researches.
The molecular results have been confirmed by the behavior of keloid fibroblasts over the 72 h period of treatment with 1 µM Ingenol Mebutate that was monitored by phase-contrast microscopy. Microscope observations showed an apoptosis-like phenotype such as accumulation of typical membrane blebbing, containing cytoplasm and encircled by multilayered lamellar structures, as well as fingerprint profiles and apparent mitochondrial swelling. DNA fragmentation Ingenol treated fibroblasts is also a hallmark of cell apoptosis. Apoptotic effect of Ingenol Mebutate effect on keloid cells was confirmed by Annexin-V staining and caspase-3 assay.
All together, these results indicate that Ingenol mebutate induce fibroblasts growth inhibition by the promotion of apoptosis.
Our results show, for the first time, that Ingenol Mebutate treatment induce DNA damage, which is able to generate the apoptotic response in keloid fibroblasts with higher expression of miR-34, probably because of a strong interaction between p53 and miR-34, which is a direct transactivational target of p53, inducing apoptosis, cell cycle arrest and senescence. Following Ingenol Mebutate treatment, miR-34 may modulates the pro-apoptotic genes expression promoting apoptosis in a p53-independent manner and in a p53-dependent manner.
These events all promote cell apoptosis. Our findings show that the miR-34a regulates cell-cycle progression and apoptosis together with p53 following Ingenol mebutate exposure. We put down a groundwork for further experiments in order to progress these challenges in the future.
Acknowledgements
The authors thank Professor Paolo Mondola (University of Naples ‘Federico II’, Naples, Italy), who greatly encouraged us to continue this study.
Glossary
Abbreviations
- miR/miRNA
microRNA
- p53
tumor protein p53
Funding
The present study was supported partly by a research grant from the University of Naples II, Italy (grant no. Grant L.R. N.5 2007).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
BDF performed all of the experiments and wrote the manuscript. CG and MS performed microscope observations. FM interpreted the data, corrected the English and helped draft the manuscript. MN designed the present study, critically revised the manuscript for intellectual content and gave final approval of the version to be published.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Halim AS, Emami A, Salahshourifar I, Kannan TP. Keloid scarring: Understanding the genetic basis, advances and prospects. Arch Plast Surg. 2012;3:184–189. doi: 10.5999/aps.2012.39.3.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shih B, Bayat A. Genetics of keloid scarring. Arch Dermatol Res. 2010;5:319–339. doi: 10.1007/s00403-009-1014-y. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Fu X, Sun X, Sun T, Zhao Z, Sheng Z. Analysis of differentially expressed genes in keloids and normal skin with cDNA microarray. J Surg Res. 2003;113:208–216. doi: 10.1016/S0022-4804(03)00188-4. [DOI] [PubMed] [Google Scholar]
- 4.Russell SB, Russell JD, Trupin KM, Gayden AE, Opalenik SR, Nanney LB, Broquist AH, Raju L, Williams SM. Epigenetically altered wound healing in keloid fibroblasts. J Invest Dermatol. 2010;130:2489–2496. doi: 10.1038/jid.2010.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stahlhut M, Bertelsen M, Hoyer-Hansen M, Svendsen N, Eriksson AH, Lord JM, Scheel-Toellner D, Young SP, Zibert JR. Ingenol mebutate: Induced cell death patterns in normal and cancer epithelial cells. J Drugs Dermatol. 2012;11:1181–1192. [PubMed] [Google Scholar]
- 6.De Felice B, Guida M, Boccia L, Nacca M. Ingenol mebutate treatment in keloids. BMC Res Notes. 2015;8:466. doi: 10.1186/s13104-015-1429-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Navarro F, Lieberman J. miR-34 and p53: New insights into a complex functional relationship. PLoS One. 2015;10:e0132767. doi: 10.1371/journal.pone.0132767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;3:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 9.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Fortin K, Mourelatos Z. MicroRNAs: Biogenesis and molecular functions. Brain Pathol. 2008;18:113–121. doi: 10.1111/j.1750-3639.2007.00121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: Background, mechanisms and therapeutic approaches. Physiol Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 12.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Felice B, Guida M, Guida M, Coppola C, De Mieri G, Cotrufo R. A miRNA signature in leukocytes from sporadic amyotrophic lateral sclerosis. Gene. 2012;508:35–40. doi: 10.1016/j.gene.2012.07.058. [DOI] [PubMed] [Google Scholar]
- 14.Rukov JL, Shomron N. MicroRNA pharmacogenomics: Post-transcriptional regulation of drug response. Trends Mol Med. 2011;17:412–423. doi: 10.1016/j.molmed.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Koturbash I, Tolleson WH, Guo L, Yu D, Chen S, Hong H, Mattes W, Ning B. MicroRNAs as pharmacogenomics biomarkers for drug efficacy and drug safety assessment. Biomark Med. 2015;9:1153–1176. doi: 10.2217/bmm.15.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shomron N. MicroRNAs and pharmacogenomics. Pharmaco-genomics. 2010;11:629–632. doi: 10.2217/pgs.10.26. [DOI] [PubMed] [Google Scholar]
- 17.Lim IJ, Phan TT, Song C, Tan WT, Longaker MT. Investigation of the influence of keloid-derived keratinocytes on fibroblast growth and proliferation in vitro. Plastic Reconstr Surg. 2001;107:797–808. doi: 10.1097/00006534-200103000-00022. [DOI] [PubMed] [Google Scholar]
- 18.He Y, Chevillet JR, Liu G, Kim TK, Wang K. The effects of microRNA on the absorption, distribution, metabolism and excretion of drugs. Br J Pharmacol. 2015;172:2733–2747. doi: 10.1111/bph.12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 21.Oren M. Decision making by p53: Life, death and cancer. Cell Death Differ. 2003;10:431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- 22.Rokavec M, Li H, Jiang L, Hermeking H. The p53/miR-34 axis in development and disease. J Mol Cell Biol. 2014;6:214–230. doi: 10.1093/jmcb/mju003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.