Abstract
The Toll-like receptors (TLRs) of the innate immune system provide the host with the ability to detect and respond to viral infections. The present study aimed to investigate the mRNA and protein expression levels of TLR2, 3, 4 and 7 in porcine tissues upon infection with the highly virulent Shimen strain of classical swine fever virus (CSFV). Reverse transcription-quantitative polymerase chain reaction was used to detect the mRNA expression levels of CSFV and TLR, whereas western blotting was used to detect the expression levels of TLR proteins. In addition, tissues underwent histological examination and immunohistochemistry to reveal the histopathological alterations associated with highly virulent CSFV infection and to detect TLR antigens. Furthermore, porcine monocyte-derived macrophages (pMDMs) were prestimulated with peptidoglycan from Staphylococcus aureus (PGN-SA), polyinosinic-polycytidylic acid [poly (I:C)], lipopolysaccharide from Escherichia coli 055:B5 (LPS-B5) or imiquimod (R837) in order to analyze the association between TLR expression and CSFV replication. Following stimulation for 12 h (with TLR-specific ligands), cells were infected with CSFV Shimen strain. The results revealed that the expression levels of TLR2 and TLR4 were increased in the lung and kidney, but were decreased in the spleen and lymph nodes in response to CSFV. TLR3 was strongly expressed in the heart and slightly upregulated in the spleen in response to CSFV Shimen strain infection, and TLR7 was increased in all examined tissues in the presence of CSFV. Furthermore, R837 and LPS-B5 exerted inhibitory effects on CSFV replication in pMDMs, whereas PGN-SA and poly(I:C) had no significant effect. These findings highlight the potential role of TLR expression in the context of CSFV infection.
Keywords: classical swine fever virus, Toll-like receptors, tissue expression, porcine monocyte-derived macrophages, innate immunity
Introduction
Classical swine fever (CSF), which is caused by the CSF virus (CSFV), remains one of the most important swine-related diseases worldwide, which has yet to be eradicated in China, despite a nationwide vaccination program (1,2). CSFV is a single-stranded positive-sense RNA molecule ~12.3 kb that encodes a polyprotein, which belongs to the Flaviviridae family (3). The Shimen strain is the standard virulent strain of CSFV in China (2), and induces severe immunopathological symptoms associated with hemorrhagic fever, thrombocytopenia, and lymphoid organ atrophy and severe lymphopenia, which are accompanied by immune suppression (4). Research investigating the immunopathogenesis of CSFV is of great importance, since the underlying mechanisms of CSFV remain poorly understood.
Toll-like receptors (TLRs) are fundamental sensor molecules of the host innate immune system, which detect conserved viral structures and initiate innate immune responses (5,6). Among the TLR family members, some viral-envelope proteins are recognized by TLR2 and TLR4 (7); TLR7 and TLR8 are involved in the recognition of single-stranded RNA (ssRNA) sequences of RNA viruses (8); and TLR3 is able to detect double-stranded RNA (dsRNA) and ssRNA viruses (9). Our previous study investigated the gene and protein expression levels of these TLRs in response to infection with the highly virulent CSFV Shimen strain in porcine monocyte-derived macrophages (pMDMs) (10). Consequently, upregulation of TLR2, 4 and 7 was detected, whereas no effects on TLR3 were determined. At present, little is currently known regarding how the tissue expression patterns of TLRs are rapidly modulated in response to a virulent strain of CSFV, or whether the propagation of CSFV is influenced by specific TLR activation.
Accordingly, the present study aimed to determine the expression patterns of TLR2, 3, 4 and 7 in tissues (heart, liver, spleen, lung, kidney and lymph nodes) from CSFV-infected swine compared with a control group. In addition, the relevance of TLR2, 3, 4 and 7 activation to the CSFV life cycle in pMDMs was examined.
Materials and methods
Infection of animals and tissue preparation
A total of 9 Landrace pigs (castrated boars; age, 50 days; weight, 25–30 kg) provided by Professor Zhi-Zhong Jing (Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, China) were used in the present study. The animal procedures were approved and supervised by the Animal Care Commission of the College of Veterinary Medicine, Northwest A&F University (Xianyang, China). The following housing conditions were used in the present study: Temperature, 22°C; atmosphere, −40 to −80Pa, 0.08% CO2; free access to food and water. All pigs were free of CSFV, porcine reproductive and respiratory syndrome virus (PRRSV) and swine influenza virus (SIV) infection, as confirmed by the negative results from reverse transcription (RT)-polymerase chain reaction (PCR) and antibody (Ab) test kits used according to the manufacturer's instructions: IDEXX CSFV Ab test, IDEXX PRRS X3 Ab test and IDEXX Influenza A Ab test (Idexx Switzerland AG, Liebefeld, Switzerland). Subsequently, 6 pigs were injected intramuscularly with l ml CSFV Shimen strain [105 50% tissue culture infective dose (TCID50)/ml], which was obtained from the Control Institute of Veterinary Bioproducts and Pharmaceuticals (Beijing, China); the remaining pigs were used as controls. Clinical and pathomorphological data of the infected pigs were recorded daily postinfection. A total of 3 infected pigs were euthanized 24 h postinfection (hpi), whereas 4 pigs, including 3 infected pigs and 1 randomly selected pig from the control group, were euthanized at 72 hpi. Subsequently, the heart, liver, spleen, lung, kidney and lymph node tissues were collected from all euthanized pigs. The 2 remaining control pigs were used for collection of data regarding color of conjunctiva, spirit, appetite and diarrhea data, and for virus titer detection and pMDM preparation.
Cells, viruses and virus titer assays
pMDMs were differentiated from the peripheral blood mononuclear cells (PBMCs) of the 2 remaining control group pigs (all of which were negative for CSFV, PRRSV and SIV antigens and antibodies) using Ficoll-Paque density (1.077±0.001 g/ml) gradient centrifugation, as described previously (10). Prior to addition of PBMCs, the flasks were coated for 2 h with 2% sterile, endotoxin-free gelatin in 37°C and the excess gelatin was discarded. Subsequently, flasks were dried in the 5% CO2 incubator (37°C) overnight and coated for 1 h with 100% FBS (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and rinsed with 0.01 M PBS (pH 7.4). PBMCs were seeded into gelatin and fetal FBS-coated culture plate and were allowed to adhere for 2 h at 37°C in an atmosphere containing 5% CO2. Adherent monocytes were washed three times with RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) and suspended in a mixture of RPMI-1640 containing 10% FBS and 10 mM EDTA in a 1:1 dilution. Isolated monocytes were seeded into 6-well plates at a concentration of 1×106 cells/well in RPMI-1640 supplemented with 20% FBS and cultured at 37°C in a 5% CO2 incubator for 8 days to obtain pMDMs.
The CSFV Shimen strain was obtained from the Control Institute of Veterinary Bioproducts and Pharmaceuticals. The multiplicity of infection (MOI) was determined and calculated using the same method as described previously (11).
Stimulation of pMDMs and in vitro CSFV inoculation
pMDMs were seeded into 12-well plates at a concentration of 5×105 cells/well and were stimulated with TLR-specific ligands (InvivoGen, San Diego, CA, USA): Peptidoglycan from Staphylococcus aureus (PGN-SA; 10 µg/ml), polyinosinic-polycytidylic acid [poly (I:C); 10 µg/ml], lipopolysaccharide from E. coli 055:B5 (LPS-B5; 1 µg/ml) or imiquimod (R837; 5 µg/ml), which are ligands for TLR2, 3, 4 and 7, respectively (6). Following stimulation for 12 h at 37°C in an atmosphere containing 5% CO2, cells were infected with CSFV Shimen strain at an MOI of 10. At 0 (mock), 12, 24, 48 and 72 hpi, cell lysates were harvested and stored at −80°C until use.
RT-quantitative PCR (RT-qPCR)
Tissue samples (30 mg) and Buffer RLT (600 µl) (Qiagen, Inc., Valencia, CA, USA) were added to sample tubes that were prefilled with ceramic beads (MagNA Lyser Green Beads) and were homogenized using a MagNA Lyser Instrument (both from Roche Diagnostics, Indianapolis, IN, USA). Total RNA was prepared from the homogenized tissue samples and cell lysates using an RNeasy Mini kit (Qiagen, Inc.) and cDNA was synthesized with random primers using a PrimeScript™ RT reagent kit with gDNA Eraser, according to the manufacturer's instructions (Takara Biotechnology Co., Ltd., Dalian, China) in 50-µl reaction mixtures containing 2 µg total RNA. RT-qPCR was performed using a Bio-Rad iQ5 system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and SYBR Premix Ex Taq II (Takara Biotechnology Co., Ltd.) with primer pairs listed in Table I (10). qPCR was conducted according to the following thermocycling conditions: 95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec and 60°C for 30 sec. Gene expression data were normalized by assessing the mRNA accumulation of a housekeeping gene (β-actin) per sample. The 2−ΔΔCq method was used to calculate the relative quantities of mRNA (12,13). For CSFV-specific detection, a recombinant pCMV-myc plasmid containing the CSFV gene NS2 (Sangon Biotech Co., Ltd., Shanghai, China) was used to build a fluorescence quantitative standard curve, and quantification of viral RNA was performed as previously described (10).
Table I.
Primer sets used in the present study.
| Genes | Forward primer (5′-3′) | Reverse primer (5′-3′) | GenBank accession number |
|---|---|---|---|
| CSFV | GATCCTCATACTGCCCACTTAC | GTATACCCCTTCACCAGCTTG | AF092448 |
| β-actin | CAAGGACCTCTACGCCAACAC | TGGAGGCGCGATGATCTT | DQ845171.1 |
| TLR2 | GGAGCCTTAGAAGTAGAGTTTG | TGTATCCACATTACCGAGGG | AB208696.2 |
| TLR3 | TAACAACCTTCCAGGCATA | AAGAGGAGAATCAGCGAGTG | DQ266435.1 |
| TLR4 | TCTACATCAAGTGCCCCTAC | ATTCTCCCAAAACCAAC | AB188301.2 |
| TLR7 | CATGTGATCGTGGACTGC | GTGATGCTCGCTATGTGG | EF583901.1 |
CSFV, classical swine fever virus; TLR, Toll-like receptor.
Western blot analysis
For western blotting, sample tubes were prefilled with ceramic beads, tissue (100 mg) and radioimmunoprecipitation assay buffer (1 ml) with Halt™ Protease Inhibitor Cocktail (cat. no. 78410) (both from Thermo Fisher Scientific, Inc.). Subsequently, the samples were homogenized using a MagNA Lyser Instrument. The mixtures were shaken gently for 15 min on ice and were then centrifuged at 14,000 × g for 15 min to pellet the tissue debris. The supernatant was transferred to a new tube and quantified using a bicinchoninic acid kit (Thermo Fisher Scientific, Inc.). Subsequently, the proteins were boiled with SDS sample loading buffer (5X), loaded 20 µl (50 µg/ml) onto gels and separated by 12% SDS-PAGE and transferred onto polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked with TBS-0.05% Tween containing 5% skim milk at room temperature for 2 h and were then incubated with primary antibodies at 4°C overnight, as previously described (10). The following primary antibodies were used: Rabbit anti-TLR2 antibody (cat. no. LS-C116906-50; dilution 1:200), mouse anti-TLR3 antibody (cat. no. LS-C545-100; dilution 1:500) (both from LifeSpan Biosciences, Inc., Seattle, WA, USA), mouse anti-TLR4 antibody (cat. no. ab8376; dilution 1:500; Abcam, Cambridge, MA, USA), goat anti-TLR7 antibody (cat. no. sc-13207; dilution 1:200; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and mouse anti-β-actin (cat. no. GTX109639; dilution 1:1,000; GeneTex, Inc., Irvine, CA, USA). After several washes with TBST the membranes were incubated with goat anti-rabbit IgG+IgA+IgM secondary antibody horseradish peroxidase (HRP) conjugated (cat. no. LS-C347361), or goat anti-mouse IgG secondary antibody (HRP) (cat. no. LS-C60710), or rabbit anti-goat IgG Fc secondary antibodies (cat. no. LS-C69762-1000) (all 1:5,000; LifeSpan Biosciences, Inc.) for 2 h at room temperature. Unbound antibodies were removed by washing with TBST, and the signal was detected using an ChemiDoc™ XRS (Bio-Rad Laboratories, Inc.) with an exposure time ranging between 30 sec and 4 min. Image Lab™ software (5.0) (Bio-Rad Laboratories, Inc.) was used to semi-quantify the blots.
Histological examination and immunohistochemistry (IHC)
Tissues from the infected swine were immersion-fixed in cold 4% paraformaldehyde fixative (pH 7.4) for 48 h at room temperature and were then transferred to 70% ethanol prior to processing for routine paraffin embedding. Several 5-µm sections were prepared from each block and stained with hematoxylin and eosin to observe pathological alterations (14). Briefly, the paraffin sections underwent conventional dewaxing and rehydration; the cell nucleus was then stained as follows: Cells were stained with hematoxylin for 10 min, rinsed under running water for 5 min, incubated in 1% hydrochloric acid ethanol for 20 sec and rinsed under running water for 15 min, prior to rinsing with distilled water for 2 min. Cell cytoplasm was stained as follows: Cells were stained with eosin for 3–5 min, rinsed with distilled water for 30 sec, dehydrated for transparency and mounted with neutral gum; after drying, microscopic structures and pathological alterations were detected using a light microscope (Eclipse TS l00-F; Nikon, Tokyo, Japan).
IHC staining was performed to detect the presence of TLR antigens in the tissues as previously described, using the same antibodies as for western blotting above (15,16). Briefly, paraffin-embedded sections were immersed in xylene for dewaxing and rehydrated with gradient washes in pure alcohol and alcohol diluted with distilled water. The specimens were heated in a modified citrate buffer (pH 6.0; Abcam) at 94°C for 45 min to improve antigenicity. Subsequently, each section was immersed in 50 µl 3% H2O2 for 30 min at room temperature, after which sections were washed three times in 0.01 M PBS (pH 7.4) to remove any residual reagents. After blocking with 50 µl nonimmunized autologous serum (Beijing Solarbio Science and Technology Co., Ltd., Beijing, China) for 1 h at room temperature, the sections were incubated with anti-TLR2, anti-TLR3, anti-TLR4 or anti-TLR7 (1:200 dilution) at 4°C overnight. The sections were then incubated with HRP-conjugated goat anti-rabbit (or anti-mouse) IgG secondary antibody (1:5,000) for 30 min at 37°C. After incubation with 1–3 drops of peroxidase substrate (DAB; 0.6 mg/ml of 0.3% H2O2 in 0.01 M Tris-HCl buffer, pH 7.6) and subsequent washing in deionized H2O, the sections were counterstained with Gill's hematoxylin and mounted on xylene-based crystal mounts. To identify the background staining, PBS (0.01 M, pH 7.4) instead of primary antibody was used as a negative control. The light microscope used to observe IHC was the same as that for H&E.
Statistical analysis
Data are presented as the mean ± standard deviation and were analyzed by one-way analysis of variance followed by Tukey's multiple comparison test using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
In vivo inoculation of pigs with CSFV
Within 72 hpi swine from the CSFV infection group exhibited the following clinical symptoms and pathomorphological alterations: Redness of the conjunctiva with secretions, increased rectal temperature (40.8–41.4°C) and anorexia (Table II).
Table II.
Clinical and pathomorphological data of the CSFV-infected pigs.
| Clinical data | Pathomorphological data | ||||||
|---|---|---|---|---|---|---|---|
| Group | Animal ID number | Rectal temperature (°C) | Color of conjunctiva | Spirit, appetite | Diarrhea | Splenic infarction | Visceral hemorrhage |
| CSFV infection 72 h | 17 | 40.8 | Red | Depressed | – | – | Renal hemorrhage |
| 18 | 41.4 | Red with secretions | Depressed | √ | √ | Lymph node hemorrhage, renal hemorrhage | |
| 20 | 41.0 | Red with secretions | Depressed | √ | – | Lymph node hemorrhage, renal hemorrhage | |
| Control | 22 | 39.2 | – | – | – | – | – |
| 24 | 39.3 | – | – | – | / | / | |
| 25 | 39.0 | – | – | – | / | / | |
√, this symptom occurs; -, no such symptom occurs; /, no necropsy was conducted; CSFV, classical swine fever virus.
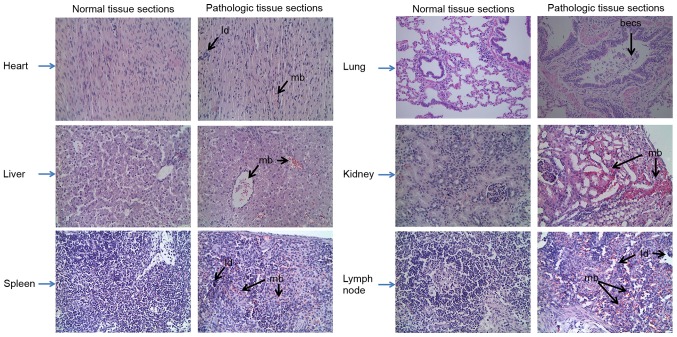
CSFV RNA was only detected in the lymph nodes at 24 hpi; however, it was detected in all organs at 72 hpi, as determined by RT-qPCR (Fig. 1). The tissues, ordered by the highest to the lowest amounts of viral RNA at 72 hpi, were as follows: Lung, spleen, lymph node, kidney, heart and liver. Furthermore, the characteristics of the various swine organs infected with CSFV Shimen strain are presented in Fig. 2; characteristics included mild bleeding, the presence of lymphocyte debris and bronchial epithelial cell shedding.
Figure 1.
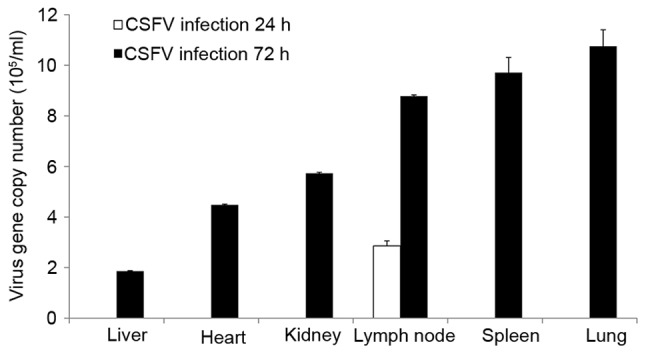
Viral loads. At 24 and 72 hpi, the copy numbers of CSFV were detected by reverse transcription-quantitative polymerase chain reaction. Data are presented as the mean ± standard deviation of three independent experiments. CSFV, classical swine fever virus.
Figure 2.
Histopathological alterations in the various swine organs infected with classical swine fever virus Shimen strain. Tissue sections were stained with hematoxylin and eosin. Original magnification, ×400. becs, bronchial epithelial cell shedding; ld, lymphocyte debris; mb, mild bleeding.
Alterations in TLR expression in response to CSFV challenge
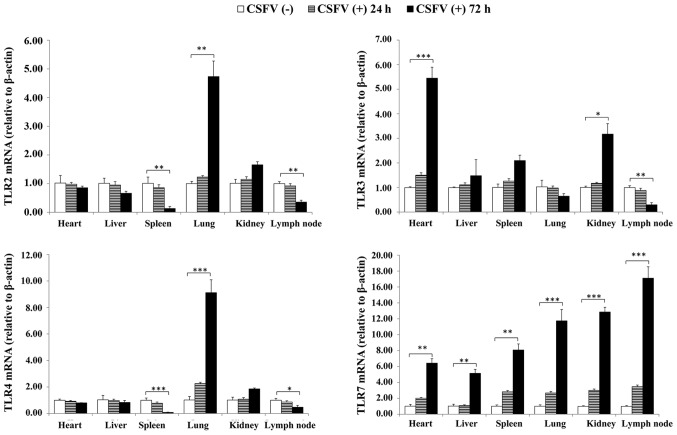
RT-qPCR was conducted to determine the expression levels of TLR2, 3, 4 and 7 in the tissues of CSFV-infected pigs at 24 and 72 hpi. As presented in Fig. 3, the expression levels of TLR2 and TLR4 were similarly upregulated in the lung and kidney, but were downregulated in the spleen and lymph nodes; TLR7 was expressed at the highest levels in all organs; and TLR3 expression was increased in the heart, spleen and kidney, but was decreased in the lymph nodes. Notably, the expression levels of TLR2, 3, 4, and 7 did not significantly differ at 24 hpi. Therefore, sample collection was performed at 72 hpi for western blotting.
Figure 3.
TLR expression levels in response to CSFV challenge of porcine tissues. Relative mRNA expression levels of TLR2, 3, 4 and 7 in porcine tissues from CSFV (+) and (−) groups were analyzed by reverse transcription-quantitative polymerase chain reaction. The housekeeping gene β-actin served as an internal control. Data are presented as the mean ± standard deviation of three independent experiments. *P<0.05, **P<0.01 and ***P<0.001 compared with the CSFV (−) group, as calculated by two-way analysis of variance. (+), infected; (−), uninfected; CSFV, classical swine fever virus; TLR, Toll-like receptor.
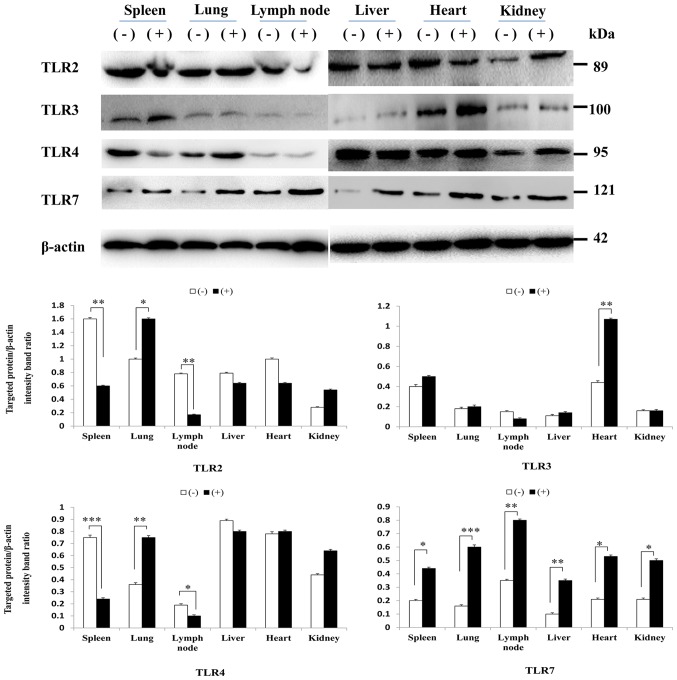
Using western blot analysis, it was indicated that the protein expression levels of TLR2, 4 and 7 in each tissue appeared to correlate with mRNA levels. However, the protein expression levels of TLR3 were not consistent with mRNA levels; there were no significant differences in TLR3 protein expression in the kidney and lymph nodes (Fig. 4).
Figure 4.
Western blotting of TLR2, 3, 4 and 7 protein expression in porcine tissues following CSFV Shimen strain infection compared with in uninfected tissues. β-actin was used to verify equal loading. Data are presented as the mean ± standard deviation of three independent experiments. *P<0.05, **P<0.01 and ***P<0.001 compared with the CSFV (−) group, as calculated by two-way analysis of variance. (+), infected; (−), uninfected; CSFV, classical swine fever virus; TLR, Toll-like receptor.
IHC staining of TLR2, 3, 4 and 7 expression in porcine tissues
TLR tissue distributions were compared by IHC staining between the CSFV-infected and uninfected groups to verify the differential expression patterns of TLR2, 3, 4 and 7. Different expression patterns for TLR2 and 4 were observed in CSFV-infected porcine tissues (Figs. 5 and 6). Compared with the uninfected group, TLR2 and 4 expression was most abundant in the lung and kidney but was decreased in the spleen and lymph nodes from the CSFV-infected group. Furthermore, analysis of TLR3-positive tissues indicated that the heart expressed the highest levels of TLR3 compared with the other tissues (Fig. 7), and TLR3 was not detected in the lymph nodes from either the CSFV-infected or uninfected groups. In addition, the intensity of TLR7 staining was much stronger in all infected tissues (heart, spleen, lung, kidney and lymph nodes) compared with in uninfected tissues (Fig. 8).
Figure 5.
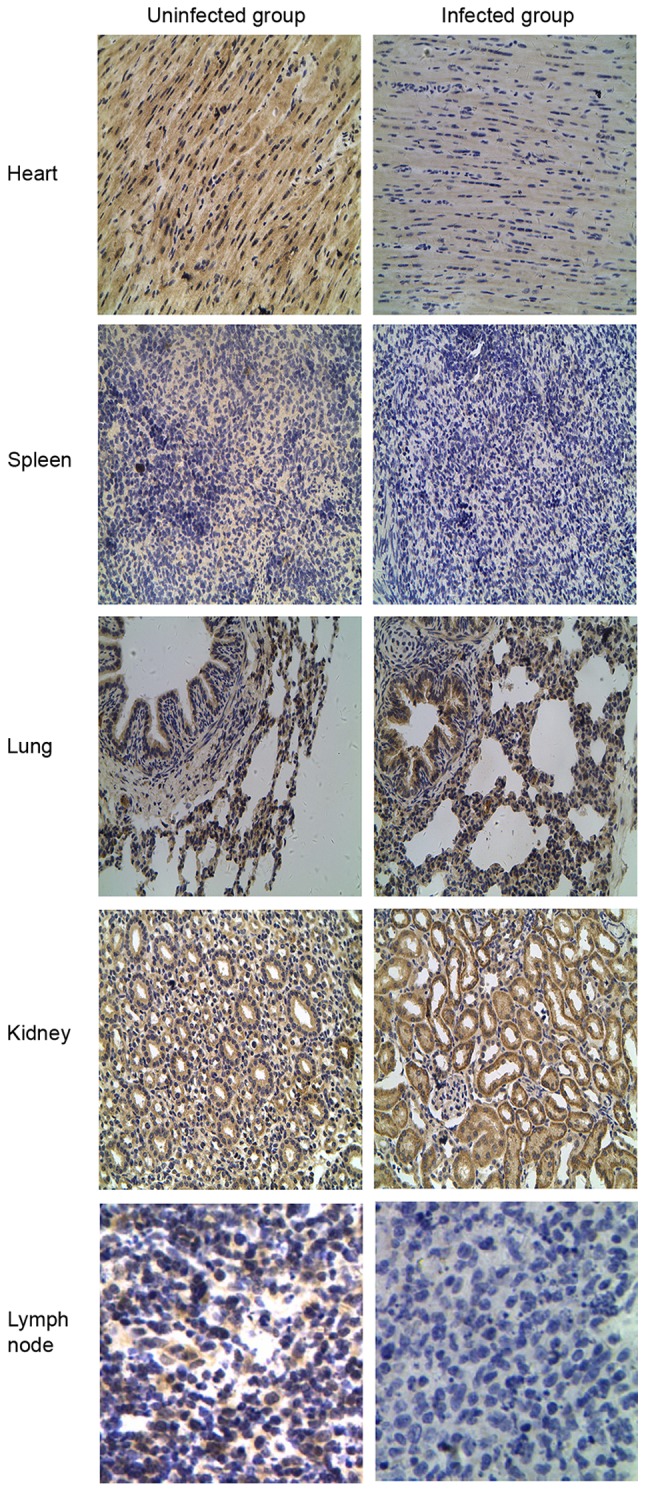
Localization of Toll-like receptor 2 protein by immunohistochemical analysis in the classical swine fever virus-infected group compared with the uninfected group. Original magnification, ×400.
Figure 6.
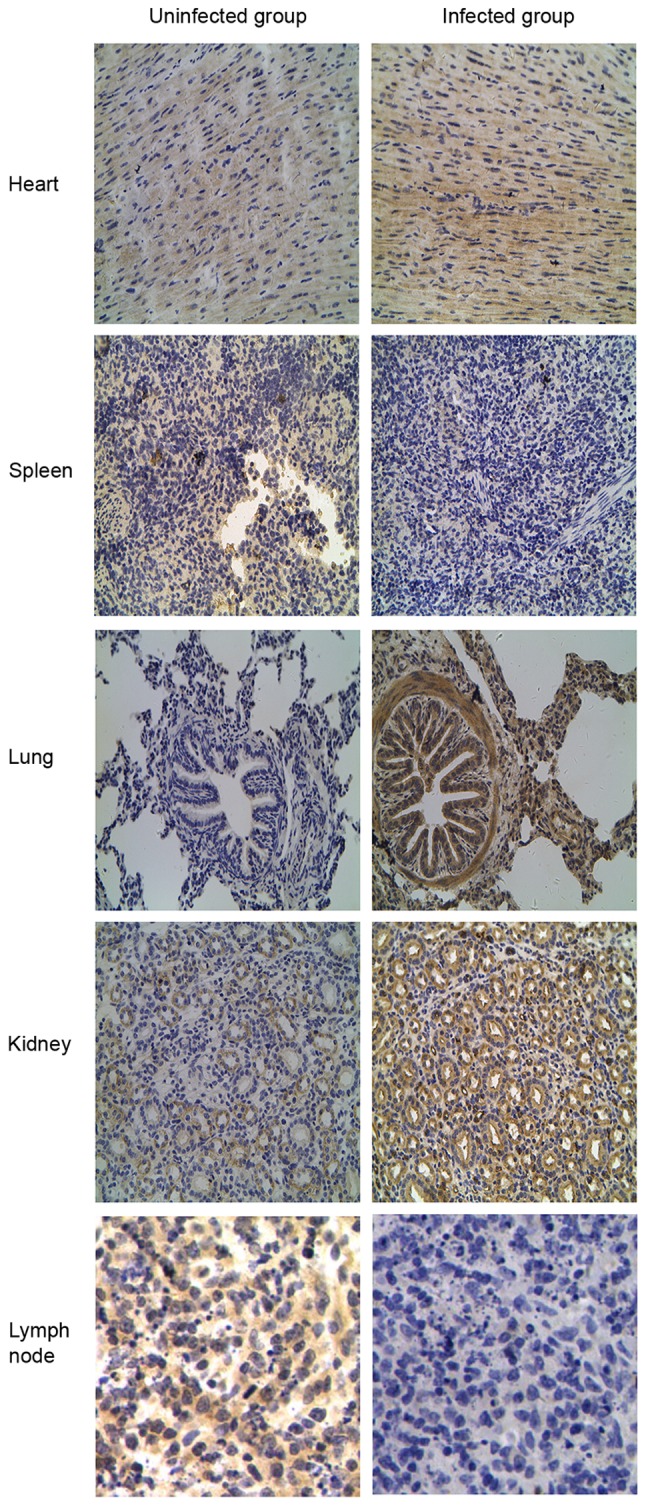
Localization of Toll-like receptor 4 protein by immunohistochemical analysis in the classical swine fever virus-infected group compared with the uninfected group. Original magnification, ×400.
Figure 7.
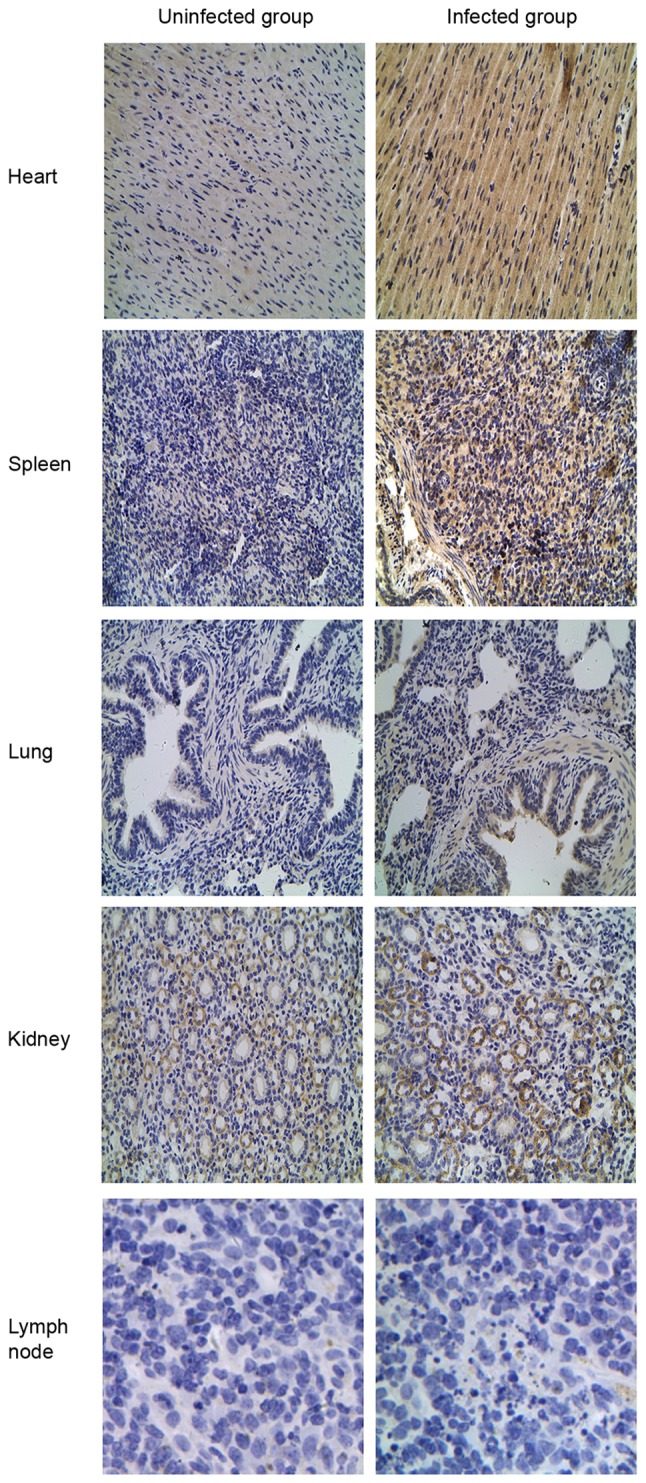
Localization of Toll-like receptor 3 by immunohistochemical analysis in the classical swine fever virus-infected group compared with the uninfected group. Original magnification, ×400.
Figure 8.
Localization of Toll-like receptor 7 protein by immunohistochemical analysis in the classical swine fever virus-infected group compared with the uninfected group. Original magnification, ×400.
Effects of TLR agonists on the growth of CSFV
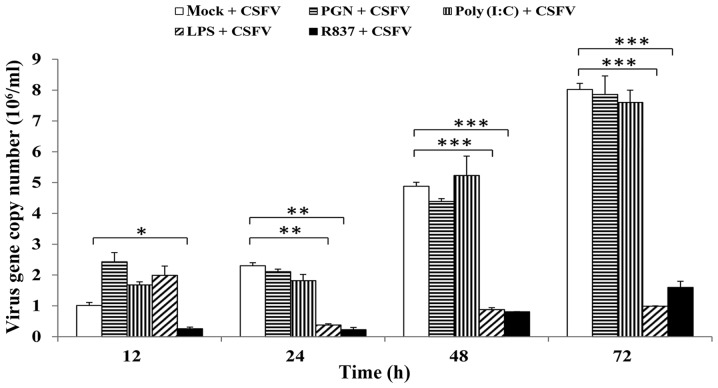
The present study aimed to reveal the relevance of TLR2, 3, 4 and 7 activation to the CSFV life cycle in pMDMs by measuring intracellular CSFV copy numbers. Therefore, following prestimulation with PGN-SA, poly(I:C), LPS-B5 or R837, the intracellular copy numbers of CSFV were detected by RT-qPCR. The results indicated that R837 was able to inhibit the proliferation of CSFV at 12, 24, 48 and 72 h.; and LPS-B5 costimulation was able to decrease CSFV propagation at 24, 48 and 72 h. Conversely, PGN-SA and poly(I:C) had no significant effect on CSFV proliferation at any time-point (Fig. 9).
Figure 9.
Effects of TLR2, 3, 4 and 7 ligands on CSFV proliferation. Porcine monocyte-derived macrophages were seeded onto 12-well plates at a concentration of 5×105 cells/well and stimulated with PGN from Staphylococcus aureus (10 µg/ml), poly (I:C) (10 µg/ml), LPS from Escherichia coli 055:B5 (1 µg/ml) or R837 (5 µg/ml). Following stimulation for 12 h, cells were infected with CSFV Shimen strain at a multiplicity of infection of 10. At 0 (mock), 12, 24, 48 and 72 h postinfection, the cells were lysed, and the copy numbers of CSFV were measured by reverse transcription-quantitative polymerase chain reaction. pCMV-myc plasmid encoding CSFV NS2 protein was used to build a fluorescence quantitative standard curve for calculating the copy numbers of CSFV in the different samples. Data are presented as the mean ± standard deviation of three independent experiments. *P<0.05, **P<0.01 and ***P<0.001 compared with the mock + CSFV group, as calculated by two-way analysis of variance. CSFV, classical swine fever virus; LPS, lipopolysaccharide; PGN, peptidoglycan; poly (I:C), polyinosinic-polycytidylic acid; R837, imiquimod.
Discussion
TLRs represent essential factors of the innate immune system and provide a crucial link to the adaptive response (17,18). It has previously been suggested that the expression patterns of TLRs may vary in different tissues and cell types, and in response to pathogen-associated molecular patterns (PAMPs) (19). The aim of the present study was to investigate TLR expression in tissues from CSFV-infected swine at early time-points.
Due to the acidification of endosomal vesicles, TLR7 is able to detect GU-rich and AU-rich ssRNA sequences of RNA viruses (20). Consistent with the role of TLR7 in immune surveillance (i.e., the recognition of viral ssRNA), the mRNA and protein expression levels of TLR7 were increased in the examined tissues in the presence of CSFV. IHC staining demonstrated that TLR7 was expressed in various tissues and was most abundant in the infected group. Furthermore, CSFV growth was significantly reduced following stimulation with R837, which is a ligand for TLR7. Taken together, these findings suggested that TLR7 signaling is important for detection of the CSFV.
TLR2 and 4 are expressed on the cell surface and primarily recognize viral glycoproteins or viral proteins released into the extracellular space (7). Notably, TLR2 and 4 expression was similarly associated with the pathogenesis of CSFV in swine, thus suggesting that their target gene expression and signaling pathways may be similar. The present study demonstrated that CSFV infection induced distinct alterations in TLR2 and 4 expression in tissues, including increased expression in the lung and kidney but decreased expression in the spleen and lymph nodes, suggesting that important roles exist for these receptors in the establishment and resolution of infections and inflammation. Furthermore, the proliferation of CSFV was inhibited following treatment with LPS, which is probably due to the activation of TLR4. However, LPS is a well-described and widely accepted antigen, which has been reported to influence a wide spectrum of immunological responses, not just TLR4.
TLR3 recognizes the dsRNA generated during viral infection as a replication intermediate for ssRNA viruses (17–21). The results of the present study indicated that TLR3 was strongly expressed in the heart and slightly upregulated in the spleen in response to CSFV Shimen strain infection. However, a further experiment demonstrated that poly(I:C) had no significant effect on the proliferation of CSFV at any time point. In a previous study, TLR3 mediated innate responses induced by poly(I:C) were inhibited in Shimen strain infected pMDMs (10). This contradiction between the expression of TLR3 in pMDMs and infected tissues requires further study.
The various expression levels of TLR2, 3, 4 and 7 detected during tissue damage in disease situations suggested that CSFV may serve as a TLR agonist or inhibitor. In addition, it remains to be determined as to whether the induction of these TLRs may have therapeutic implications. In particular, the use of TLRs themselves, PAMPs, specific cytokines and endogenous agonists, such as proteins (e.g., high mobility group box 1), lipids (e.g., oxidized phospholipids) and nucleic acids, which stimulate immune cells or nonimmune cells, may be considered targets for the development of effective vaccine adjuvants (22–25).
In conclusion, the present study is the first, to the best of our knowledge, to detect the distinct alterations in TLR2, 3, 4 and 7 expression in the presence of the highly virulent CSFV Shimen strain. Furthermore, the effects of PGN-SA, poly (I:C), LPS-B5 and R837 on CSFV replication were observed. The results highlighted the possible roles of TLR expression in the context of CSFV infection and may provide a basis for the further examination of the functions of other members of the Flaviviridae family in the innate immune system.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 31472210).
References
- 1.Edwards S, Fukusho A, Lefèvre PC, Lipowski A, Pejsak Z, Roehe P, Westergaard J. Classical swine fever: The global situation. Vet Microbiol. 2000;73:103–119. doi: 10.1016/S0378-1135(00)00138-3. [DOI] [PubMed] [Google Scholar]
- 2.Luo Y, Li S, Sun Y, Qiu HJ. Classical swine fever in China: A minireview. Vet Microbiol. 2014;172:1–6. doi: 10.1016/j.vetmic.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Paton DJ, Greiser-Wilke I. Classical swine fever-an update. Res Vet Sci. 2003;75:169–178. doi: 10.1016/S0034-5288(03)00076-6. [DOI] [PubMed] [Google Scholar]
- 4.Jamin A, Gorin S, Cariolet R, Le Potier MF, Kuntz-Simon G. Classical swine fever virus induces activation of plasmacytoid and conventional dendritic cells in tonsil, blood, and spleen of infected pigs. Vet Res. 2008;39:7. doi: 10.1051/vetres:2007045. [DOI] [PubMed] [Google Scholar]
- 5.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 6.Moresco EM, LaVine D, Beutler B. Toll-like receptors. Curr Biol. 2011;21:R488–R493. doi: 10.1016/j.cub.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 7.Lester SN, Li K. Toll-like receptors in antiviral innate immunity. J Mol Biol. 2014;426:1246–1264. doi: 10.1016/j.jmb.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 9.Guillot L, Le Goffic R, Bloch S, Escriou N, Akira S, Chignard M, Si-Tahar M. Involvement of toll-like receptor 3 in the immune response of lung evithelial cells to double-stranded RNA and influenza A virus. J Biol Chem. 2005;280:5571–5580. doi: 10.1074/jbc.M410592200. [DOI] [PubMed] [Google Scholar]
- 10.Cao Z, Guo K, Zheng M, Ning P, Li H, Kang K, Lin Z, Zhang C, Liang W, Zhang Y. A comparison of the impact of Shimen and C strains of classical swine fever virus on Toll-like receptor expression. J Gen Virol. 2015;96:1732–1745. doi: 10.1099/vir.0.000129. [DOI] [PubMed] [Google Scholar]
- 11.Ning P, Zhang Y, Guo K, Chen R, Liang W, Lin Z, Li H. Discovering up-regulated VEGF-C expression in swine umbilical vein endothelial cells by classical swine fever virus Shimen. Vet Res. 2014;45:48. doi: 10.1186/1297-9716-45-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pei J, Zhao M, Ye Z, Gou H, Wang J, Yi L, Dong X, Liu W, Luo Y, Liao M, Chen J. Autophagy enhances the replication of classical swine fever virus in vitro. Autophagy. 2014;10:93–110. doi: 10.4161/auto.26843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Fischer AH, Jacobson KA, Rose J, Zeller R. Hematoxylin and eosin staining of tissue and cell sections. CSH protocols. 2008;2008 doi: 10.1101/pdb.prot4986. pdb prot4986. [DOI] [PubMed] [Google Scholar]
- 15.Xiao SY, Zhang H, Guzman H, Tesh RB. Experimental yellow fever virus infection in the golden hamster (Mesocricetus auratus). II. pathology. J Infect Dis. 2001;183:1437–1444. doi: 10.1086/320200. [DOI] [PubMed] [Google Scholar]
- 16.Benias PC, Gopal K, Bodenheimer H, Jr, Theise ND. Hepatic expression of toll-like receptors 3, 4 and 9 in primary biliary cirrhosis and chronic hepatitis C. Clin Res Hepatol Gastroenterol. 2012;36:448–454. doi: 10.1016/j.clinre.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 19.Jiang D, Liang J, Li Y, Noble P. The role of Toll-like receptors in non-infectious lung injury. Cell Res. 2006;16:693–701. doi: 10.1038/sj.cr.7310085. [DOI] [PubMed] [Google Scholar]
- 20.Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. Recognition of single-stranded RNA viruses by Toll-like receptor 7; Proc Natl Acad Sci USA; 2004; pp. 5598–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 22.Tsung A, Sahai R, Tanaka H, Nakao A, Fink MP, Lotze MT, Yang H, Li J, Tracey KJ, Geller DA, Billiar TR. The nuclear factor HMGB1 mediates hepatic injury after murine liver ischemia-reperfusion. J Exp Med. 2005;201:1135–1143. doi: 10.1084/jem.20042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart CR, Stuart LM, Wilkinson K, van Gils JM, Deng J, Halle A, Rayner KJ, Boyer L, Zhong R, Frazier WA, et al. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat Immunol. 2010;11:155–161. doi: 10.1038/ni.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanzler H, Barrat FJ, Hessel EM, Coffman RL. Therapeutic targeting of innate immunity with Toll-like receptor agonists and antagonists. Nat Med. 2007;13:552–559. doi: 10.1038/nm1589. [DOI] [PubMed] [Google Scholar]
- 25.O'Neill LA, Bryant CE, Doyle SL. Therapeutic targeting of Toll-like receptors for infectious and inflammatory diseases and cancer. Pharmacol Rev. 2009;61:177–197. doi: 10.1124/pr.109.001073. [DOI] [PMC free article] [PubMed] [Google Scholar]