Abstract
The present study aimed to investigate the association between septin 7 (SEPT7) and melatonin-induced apoptosis in the human fetal osteoblastic cell line hFOB 1.19. MicroRNA (miR)-590-3p was identified by identifying overlapping miRNAs that target SEPT7, across different databases (miRDB, DIANA and Targetscan). Apoptosis was assessed via flow cytometric analysis. Small interfering RNA of SEPT7 and a miR-590-3p inhibitor were used for gene silencing and the efficiency was assessed by reverse transcription-quantitative polymerase chain reaction. Western blotting was used to measure the expression of proteins associated with pathways mediating endoplasmic reticulum stress and melatonin-induced apoptosis. The present study identified that SEPT7 was a potential target of miR-590-3p and demonstrated that SEPT7 is associated with mediating the pro-apoptotic effect of miR-590-3p in human osteoblast cell line hFOB 1.19. High concentrations of melatonin may result in the inhibition of miR-590-3p expression, leading to the upregulation of target genes that promote apoptosis.
Keywords: human osteoblastic hFOB 1.19 cells, microRNA-590-3p, melatonin-induced apoptosis, septin 7
Introduction
Previous studies have demonstrated that septins are involved in various biological processes, including cell mitosis, polarity determination, vesicle trafficking and apoptosis (1–4). It has been demonstrated that septin 7 (SEPT7) inhibits cell cycle progression in yeast, due to its high degree of homology to cell division control protein 10 (5). Furthermore, it has been revealed that SEPT7 inhibits glioma cell proliferation and invasion, and induces apoptosis, suggesting that SEPT7 inhibits the progression of glioma (6).
A previous study demonstrated that melatonin can induce cell apoposis in hFOB 1.19 human osteoblastic cells by activating the endoplasmic reticulum stress (ERS)-associated eukaryotic translation initiation factor 2α (eIF2α)-cyclic AMP-dependent transcription factor ATF-4 (ATF4) pathway and subsequently triggered the cascade effects of DNA damage-inducible transcript 3 protein, caspase-3 and mitogen-activated protein kinase 8 (7). It has also been previously demonstrated that the expression level of 78 kDa glucose-regulated protein homolog (GRP78) increases when ERS occurs (8).
Zhang et al (9) demonstrated that SEPT7 silencing inhibits the proliferation and promotes the apoptosis of human breast cancer cells, which indicated that the present of SEPT7 can promote proliferation and inhibit apoptosis. Furthermore, miR-127 is able to inhibit the proliferation of hepatocellular carcinoma cells (HCC) by suppressing the expression of SEPT7, indicating that SPET7 can promote proliferation and inhibit apoptosis (10). The aforementioned studies both demonstrated that in tumor cells, SEPT7 can promote proliferation and inhibit apoptosis, but in normal cells, especially in human osteoblasts, its function remains to be elucidated. The present study aims to elucidate the underlying mechanism of SEPT7 in melatonin-induced cell apoptosis in human osteoblast cell line hFOB 1.19.
MicroRNAs (miRNAs) are small, non-coding RNAs that are 19–22 nucleotides long and post-transcriptionally regulate protein expression by targeting protein-coding genes involved in the proliferation, differentiation, apoptosis and migration of cancer cells (11–16). Previous studies have demonstrated that miR-590-3p inhibits hepatocellular carcinoma cell growth by targeting transcriptional enhancer factor TEF-1, promotes osteogenic differentiation by suppressing expression of adenomatous polyposis coli protein and stabilizing β-catenin, and contributes to the development of radioresistance in human glioblastoma cells by directly targeting leucine-rich repeats and immunoglobulin-like domains protein 1 (17–19). Although miR-590 serves potential roles in cancer development, to the best of our knowledge, it remains unknown whether miR-590 affects normal human osteoblasts. In the present study, miR-590-3p was used following the identification of overlapping microRNAs that target SEPT7 using different databases (miRDB, DIANA and Targetscan). Experiments were performed to elucidate whether SEPT7 is a novel direct target by which miR-590-3p exerts its effect, including in vitro experiments to investigate the role of miR-590-3p in melatonin-induced apoptosis in the human fetal osteoblastic cell line hFOB 1.19.
Abnormal proliferation and apoptosis of osteoblasts may occur in patients with osteoporosis (20). Melatonin, which simulates apoptosis, may be used in future studies to establish a novel type of osteoporosis model and further investigate the mechanism underlying osteoporosis. The present study improved the understanding of the mechanism underlying ERS and apoptosis induced by melatonin in hFOB 1.19 cells, providing a theoretical basis for its subsequent applications of melatonin.
Materials and methods
In silico analysis
The miRNAs which target SEPT7 were predicted by identifying overlapping microRNAs across different databases (Targetscan, http://www.targetscan.org/; miRDB, http://mirdb.org/; and DIANA, http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=site/index). It was observed that miR-590-3p was the only microRNA which overlapped in all three databases (Fig. 1).
Figure 1.
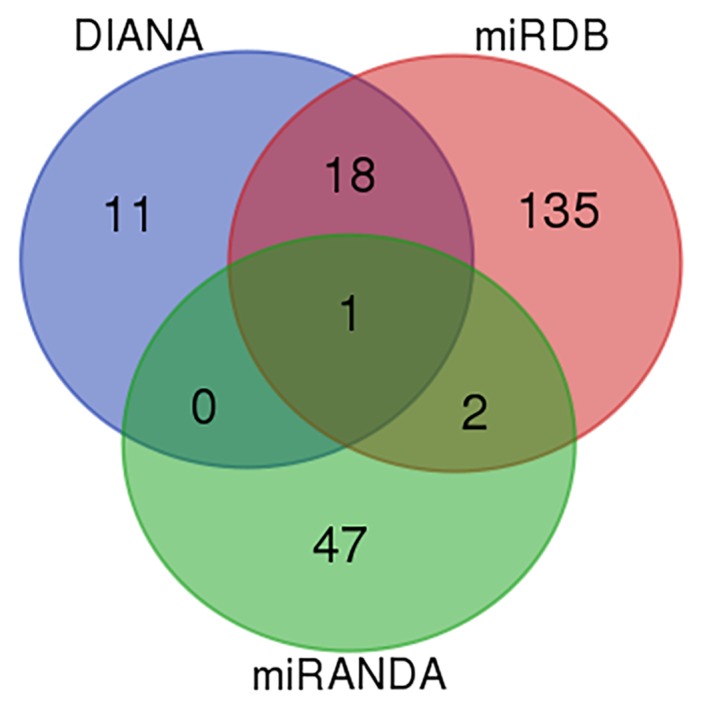
Venn diagram of the numbers of miRNAs overlapping with SEPT7 from different databases (Targetscan, miRDB and DIANA). miR, microRNA.
Cell culture and reagents
The hFOB 1.19 normal human fetal osteoblastic cell line was provided by the Department of Biochemistry and Molecular Biology, Mayo Clinic (Rochester, MN, USA) and established as previously described (21), and was maintained in a 1:1 mixture of Dulbecco's minimum essential (DME) and F12 medium without phenol red (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA), supplemented with 10% fetal bovine serum (FBS; Clark Bioscience, Richmond, VA, USA) in 5% CO2 at 37°C. The medium was replaced every other day. Cells were used for further experiments when they reached passages 8–11. Melatonin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was dissolved in 0.2% dimethyl sulfoxide; cells were subsequently treated at the indicated doses of melatonin (2, 4 and 6 mM) or vehicle media (DME and F12) containing 10% FBS for 24 h. Melatonin was obtained from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany). Primary monoclonal antibodies against proteins were purchased from Abcam (Cambridge, MA, USA). Goat anti-rabbit secondary antibodies were obtained from OriGene Technologies, Inc. (Beijing, China).
Luciferase assay
The wild-type or mutant SEPT7 3′-untranslated regions (3′-UTR; Shanghai GeneChem Co., Ltd., Shanghai, China) were cloned into the pmirGLO luciferase reporter vector (Promega Corporation, Madison, WI, USA) and co-transfected with either miR-590-3p mimic or miR-negative control (NC) (Shanghai GeneChem Co., Ltd.) into 293T cells (Scientific Center of China Medical University, Shenyang, China), using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The pRL-TK Renilla luciferase reporter vector was transfected into cells as an internal control. A total of 24 h following transfection, luciferase activity was measured using a Dual-Luciferase® Reporter assay system (Promega Corporation), following cell lysis with Passive Lysis buffer, included in the Reporter Assay System. Relative luciferase activity was calculated as the ratio of firefly luciferase activity to Renilla luciferase activity.
Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) staining and analysis using flow cytometry
An Annexin V FITC/PI staining kit (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was used to assess cell apoptosis following the manufacturer's protocol. Briefly, cells were trypsinized, washed in PBS, and resuspended in binding buffer. Cells were stained with FITC-conjugated Annexin V and PI, and analyzed using a BD LSRFortessa™ cell analyzer (BD Biosciences, Franklin Lakes, NJ, USA) and quantified using BD FACSDiva (version 6.2; BD Biosciences). Annexin V positive cells were defined as apoptotic.
Overexpression plasmid construction and rescue experiment
SEPT7 was cloned into GV230 plasmids (200 ng; Shanghai GeneChem Co., Ltd., Shanghai, China) between XhoI/KpnI sites to overexpress SEPT7 in hFOB 1.19 cells. Full-length SEPT7 gene (4377 bp; reference sequence NM_001011553) was amplified by polymerase chain reaction (PCR). The following primers were used for PCR: Forward, 5′-CTGCTCACAATAGTTGATACCCC-3′ and reverse, 5′-TGTTCACTCGTGATTCTGCATT-3′. The PrimeSTAR HS DNA polymerase, was obtained from Shanghai GeneChem Co., Ltd., and cycled for 30 cycles following initial denaturation (98°C for 5 min) with the following parameters: 98°C for 10 sec, 55°C for 10 sec, 72°C for 90 sec; and 72°C for 8 min. Following enzyme digestion (Exnase™ II, 1 µl) using ClonExpress II One Step Cloning kit (Vazyme, Piscataway, NJ, USA) and sequencing, the PCR product was cloned into the XhoI/KpnI sites of the GV230 expression vector. The recombinant GV230-SEPT7 plasmid was confirmed via endonuclease digestion and DNA sequencing (Shanghai GeneChem Co., Ltd.) prior to transfection into hFOB 1.19 cells using Lipofectamine 2000® (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The rescue experiment was preformed following transfection of the SEPT7 siRNA and the concentration of the SEPT7 overexpression plasmid was 45 nM. Time interval between transfection and subsequent experimentation was 24 h.
SEPT7 small interfering (si)RNA transfection
Cells were cultured in DME and F12 medium supplemented with 10% FBS in a humidified incubator at 37°C and 5% CO2. At 70–80% confluence, cells were transfected with 60 nM SEPT7 siRNA (forward, 5′-CGACUACAUUGAUAGUAAAUU-3′ and reverse, 5′-UUUACUAUCAAUGUAGUCGAU-3′; Shanghai Genechem Co., Ltd.) using Lipofectamine® 2000 according to the manufacturer's protocol (Invitrogen; Thermo Fisher Scientific, Inc.). There were three control groups: Blank control, transfection reagent control (to control for potentially toxic influence of Lipofectamine® 2000 and the influence of Lipofectamine 2000 on the expression of the target gene) and scramble siRNA control (5′-GAAATTTATAACGATCAGTCT-3′) (Shanghai Genechem Co., Ltd.). The time interval between transfection and subsequent experimentation was 24 h.
Western blotting
The cells were divided into 5 treatment groups (control; scramble siRNA; miR-590-3p inhibitor (Shanghai Genechem Co., Ltd., 60 nM, 37°C for 24 h) + scramble siRNA + 4 mM melatonin; miR-590-3p inhibitor + SEPT7 siRNA + 4 mM melatonin; 4 mM melatonin), 4 mM melatonin was selected as the experiment condition according to a previous study (7). Following treatment, proteins were extracted from cells using radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China), for 30 min at 4°C. The supernatant containing total protein was harvested and proteins were quantified using the bicinchoninic acid method. Aliquots containing 50 µg proteins were separated on 12% SDS-PAGE gels and transferred to polyvinylidene fluoride membranes at 60 V for 2 h at 4°C. Following transfer, membranes were immediately soaked in blocking buffer, containing 25 mg bovine serum albumin (Beyotime Institute of Biotechnology) in TBS to final volume of 0.5 l, at 4°C for 2 h. Subsequently, proteins were incubated with primary antibodies against GRP78 (cat. no. ab21685); phosphorylated (p)-eIF2α (cat. no. ab4837), SEPT7 (cat. no. ab186021) and β-actin (cat. no. ab8226; all 1:5,000 dilution) overnight at 4°C. Proteins were then incubated with goat anti-rabbit immunoglobulin G horseradish peroxidase-conjugated secondary antibody (cat. no. ab6721) at 1:10,000 dilution, for 2 h at room temperature. The DNR Imaging System (DNR Bio-Imaging Systems, Ltd., Neve Yamin, Israel) was used to visualize specific bands using the BeyoECL Plus kit (Beyotime Institute of Biotechnology) according to the manufacturer's protocol and the optical density of each band was measured using Image J software (version 1.51; National Institutes of Health, Bethesda, MD, USA).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
miRNAs were extracted from hFOB 1.19 cells using the E.Z.N.A.® Total RNA Midi kit (Omega Bio-Tek, Norcross, GA, USA) according to the manufacturer's protocol and quantified spectrophotometrically at a wavelength of 260 nm, with acceptable CCLX/280 ratios between 1.8 and 2.0. RNA quality was also verified by 1% agarose gel electrophoresis and staining with 1 µg/ml ethidium bromide. The reagent used for reverse transcription was purchased from the Promega Corporation. Reverse transcription was conducted in a total volume of 20 µl [1.0 µg total RNA, 1.0 µl forward primer, 1.0 µl reverse primer, 9.5 µl H2O, mixing at 85°C for 5 min, mixing with 2.0 µl 10 mM dNTP, 0.5 µl RNase inhibitor, 0.5 µl U6 primer, 4.0 µl 5X buffer and 0.5 µl M-MLV reverse transcriptase (200 units; cat. no. M1701) at 30°C for 10 min, 43°C for 50 min and 85°C for 10 min], the U6 primers were: Forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. Subsequently, qPCR was performed on LightCycler® 480 High-Resolution Melting Master (Roche Diagnostics, Basel, Switzerland). Specific primers for miR-590-3p (forward, 5′-TAATTTTATGTATAAGCTAGT-3′ and reverse, 5′-GCTGAGGTGCTGTGGT-3′) were obtained from Shanghai GeneChem Co., Ltd. (Shanghai, China). Amplifications were conducted in a total volume of 20 µl [10.0 µl of 2X SYBR Premix Ex Taq II (Takara Biotechnology Co., Ltd.) 0.8 µl forward primer, 0.8 µl reverse primer, 2.0 µl DNA template, 6.4 µl double distilled H2O]. The following thermocycling conditions were used for PCR: Initial denaturation at 95°C for 30 sec; 40 cycles of 95°C for 5 sec and 60°C for 30 sec. β-actin (forward, 5′-CAGGGCGTGATGGTGGGCA-3′ and reverse, 5′-CAAACATCATCTGGGTCATCTTCTC-3′) was obtained from Shanghai GeneChem Co., Ltd. (Shanghai, China) and used as an internal control. Analysis of melting curve was used to support the reliability of the results. Relative expression was calculated using the 2−ΔΔCq method (22).
Statistical analysis
SPSS software (version 20.0; IBM Corp., Armonk, NY, USA) was used for data processing. An independent samples t-test or one-way analysis of variance, followed by the Student-Newman-Keuls test, were used to evaluate the differences between groups. The results are presented as the mean ± standard error of the mean. The N-fold gene expression values in gene expression ≤0.5 and >2 were considered to be significant, compared with the values obtained for the control genes. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-590-3p expression following treatment with melatonin
The effect of melatonin treatment on miR-590-3p expression in hFOB 1.19 cells was examined using RT-qPCR. The expression of miR-590-3p was significantly decreased in the melatonin-treated groups compared with the control group (P<0.01; Fig. 2). Furthermore, it was determined that miR-590-3p expression was significantly lower in the high concentration melatonin-treated groups (4 and 6 mM), compared with the low melatonin concentration group (2 mM; P<0.01; Fig. 2).
Figure 2.
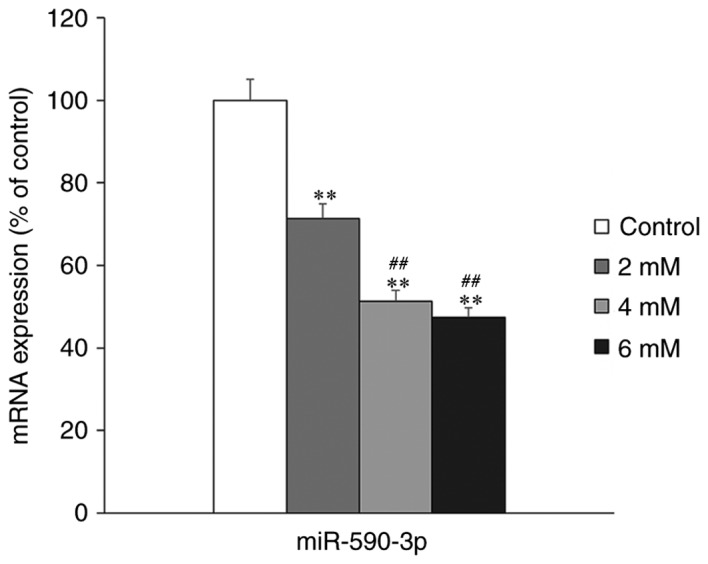
miR-590-3p expression in hFOB 1.19 cells following treatment with different concentrations of melatonin (0, 2, 4 and 6 nM) for 24 h. **P<0.01 vs. the untreated control group. ##P<0.01 vs. the 2 mM melatonin treated group. miR, microRNA.
Expression of SEPT and the association between SEPT7 and miR-590-3p
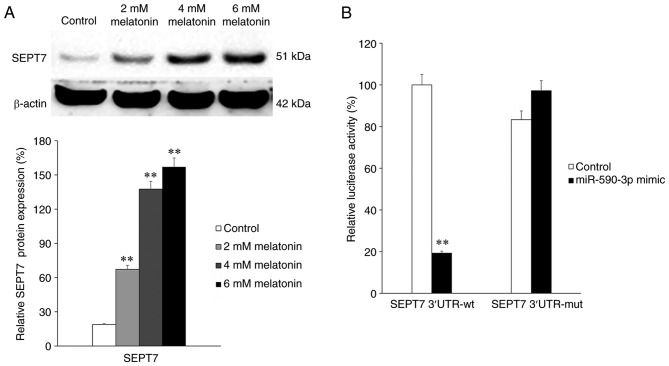
Subsequently, SEPT7 expression in groups treated with different concentrations of melatonin was determined. The results indicated that the expression of SEPT7 significantly increased following treatment with melatonin in a concentration-dependent manner (all P<0.01; Fig. 3A). This indicates that melatonin treatment is positively associated with SEPT7 expression. To elucidate the association between miR-590-3p and SEPT7, a luciferase reporter assay was performed. SEPT7 3′-UTRs containing wild-type or mutant potential target sites of miR-590-3p were constructed and co-transfected with the miR-590-3p mimic into 293T cells. The results demonstrated that the miR-590-3p mimic significantly decreased the luciferase activity of wild-type SEPT7 3′-UTR compared with the control (P<0.01), whereas the luciferase activity of the mutant SEPT7 3′-UTR was not significantly affected by the miR-590-3p mimic (Fig. 3B).
Figure 3.
SEPT7 expression and luciferase activity assay. (A) Expression of SEPT7 in hFOB 1.19 cells following treatment with different concentrations (0, 2, 4 and 6 nM) melatonin for 24 h. (B) Relative luciferase activity of wt or mut SEPT7 3′-UTR in 293T cells following co-transfection with the negative control or miR-590-3p mimic. **P<0.01 vs. control. SEPT7, septin 7; miR, microRNA; wt, wild-type; mut, mutant; UTR, untranslated region.
miR-590-3p expression and cell apoptosis in the human osteoblast cell line hFOB 1.19
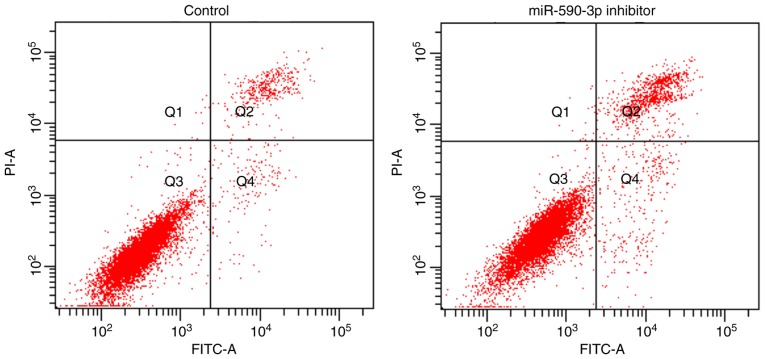
Following transfection with miR-590-3p inhibitor, the expression of miR-590-3p was significantly decreased compared with control cells that did not undergo transfection, which confirmed a successful miR-590-3p inhibition (P<0.01; Fig. 4). Furthermore, the apoptosis rate was increased following transfection with the miR-590-3p inhibitor compared with the control group (P<0.01; Fig. 5).
Figure 4.
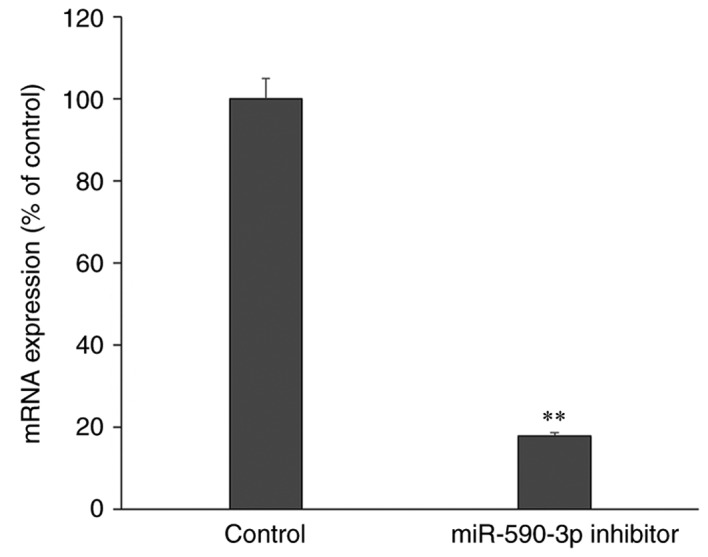
Relative expression of miR-590-3p in the human fetal osteoblast cell line hFOB 1.19 following transfection with the miR-590-3p inhibitor. **P<0.01 vs. the control. miR, microRNA.
Figure 5.
Effect on the apoptosis of the human fetal osteoblast cell line hFOB 1.19 following transfection with the miR-590-3p inhibitor. Q1, necrotic cells; Q2, late apoptotic cells; Q3, living cells; Q4, early apoptotic cells; PI, propidium iodide; miR, microRNA; FITC, fluorescein isothiocyanate.
The association between miR-590-3p and SEPT7
Transfection of the miR-590-3p inhibitor significantly increased the expression of SEPT7 in hFOB 1.19 cells compared with the control group (P<0.01; Fig. 6). However, this increased SEPT7 expression was abolished following transfection of SEPT7 small interfering (si)RNA (P<0.01).
Figure 6.
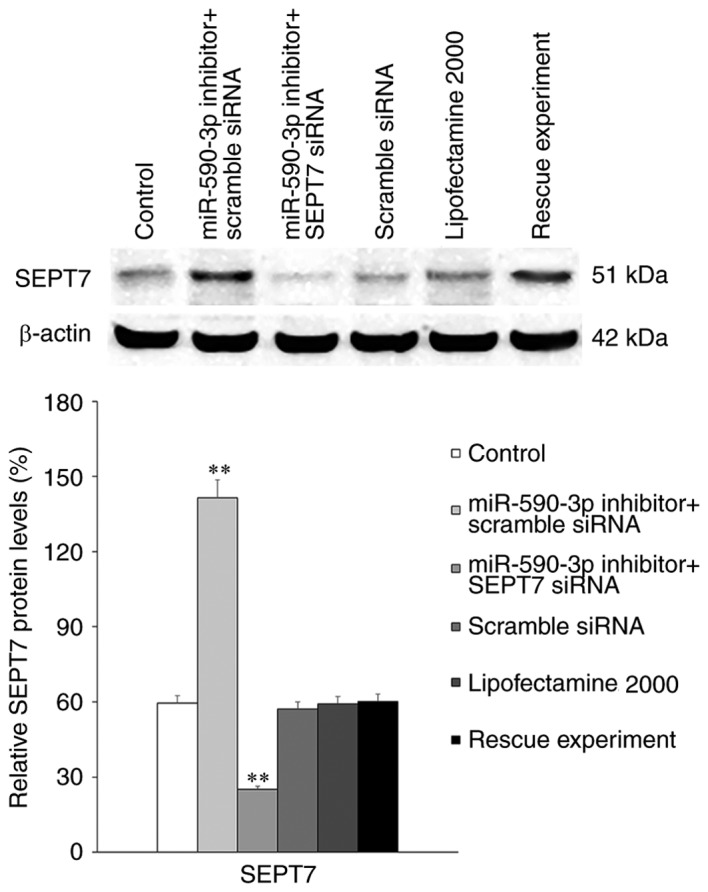
Expression of SEPT7 in cells treated with miR-590-3p inhibitor alone or in combination with SEPT7 siRNA. **P<0.01 vs. untreated control. miR, microRNA; siRN, small interfering RNA; SEPT7, septin 7.
The effects of miR-590-3p, SEPT7 and melatonin treatment on apoptosis and the expression of ERS-associated proteins
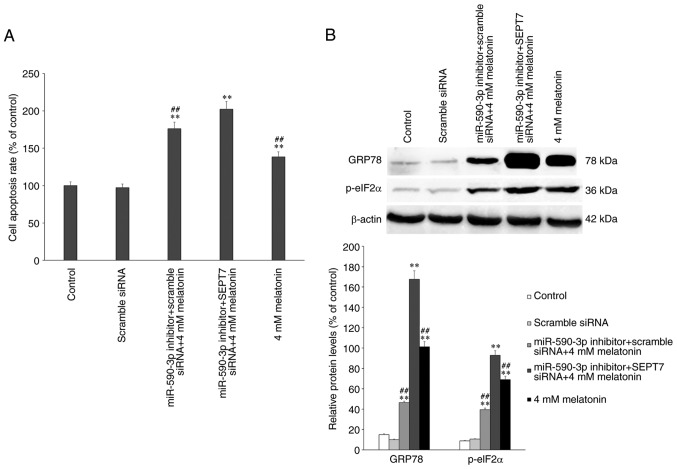
The expression of ERS- and apoptosis-associated proteins were assessed by western blotting following treatment with 4 mM melatonin alone or in combination of miR-590-3p inhibitor and/or SEPT7 siRNA. The results demonstrated that, in the 4 mM melatonin treatment group, apoptosis occurred as previously described and was significantly increased compared with the control group (P<0.01; Fig. 7A) (7), while in the miR-590-3p inhibitor treatment groups, the rate of apoptosis was significantly increased compared with the control and the melatonin-treated group (P<0.01; Fig. 7A). The apoptotic rate and ERS-associated protein expression levels increased significantly in the miR-590-3p inhibitor + SEPT7 siRNA + 4 mM melatonin treatment group, compared with the other groups (Fig. 7). The expression of ERS-associated proteins was assessed and it was demonstrated that the expression of GRP78 and p-eIF2α were significantly increased in the miR-590-3p inhibitor + SEPT7 siRNA group compared with the other groups (P<0.01) (Fig. 7B).
Figure 7.
Apoptosis rate and expression of ERS-associated proteins in hFOB 1.19 cells. (A) Cell apoptosis rate and (B) expression of ERS-associated proteins in the human fetal osteoblast cell line hFOB 1.19 following transfection with miR-590-3p inhibitor or/and SEPT7 siRNA or/and 4 mM melatonin. **P<0.01 vs. untreated control. ##P<0.01 vs. the miR-590-3p inhibitor+SEPT7 siRNA+4 mM melatonin treatment group. miR, microRNA; si, small interfering RNA; SEPT7, septin 7; GRP78, 78 kDa glucose-regulated protein homolog; p-eIF2α, phosphorylated-eukaryotic translation initiation factor 2A; ERS, endoplasmic reticulum stress.
Discussion
Melatonin serves roles in aging, reproduction, tumor growth and various biological progresses (23–25). A previous research conducted by the authors of the present study demonstrated that melatonin may cause apoptosis by inducing ERS in hFOB 1.19 cells and that during melatonin-induced apoptosis, the expression of proteins associated with the classic apoptotic pathway, including p-eIF2α, are upregulated, which is indicative of the occurrence and extent of melatonin-induced apoptosis (7).
Septins are proteins that belong to a protein superfamily. They weigh 30–65 kDa and exhibit conserved structures (26). Studies have demonstrated that septins, along with tubulin, microfilaments and intermediate fibrin, make up the cytoskeleton and serve roles in the transportation of intracellular substances, regulation of cell division and the cell cycle and physiological processes including apoptosis (27–30). The results of the present study demonstrated that miR-590-3p expression was downregulated in the human fetal osteoblastic cell line hFOB 1.19 following treatment with melatonin. This decrease occurred in a dose-dependent manner, indicating that increasing concentrations of melatonin may decrease the expression of miR-590-3p. This means that melatonin concentration-associated increases in the expression of SEPT7 may be negatively associated with miR-590-3p expression. Furthermore, the results of the luciferase reporter assay indicated that SEPT7 was a direct target of miR-590-3p. An undergoing study performed by the authors of the present study suggests that SEPT7 upregulation inhibits melatonin-induced cell apoptosis by suppressing ERS in hFOB 1.19 cells. In the present study, SEPT7 expression was significantly increased following transfection with the miR-590-3p inhibitor compared with the control group; therefore, it was hypothesized that miR-590-3p inhibition may protect hFOB 1.19 cells from melatonin-induced apoptosis. However, the results of the current study contradict this hypothesis; the inhibition of miR-590-3p expression increased the rate of apoptosis rather than decreasing it. Following treatment with 4 mM melatonin for 24 h, the miR-590-3p inhibitor and/or SEPT7 siRNA were transfected. The results demonstrated that in the 4-mM melatonin-treated group, the expression of GRP78 and p-eIF2α increased, as did the rate of apoptosis. The rate of apoptosis significantly increased in the miR-590-3p inhibitor + scramble siRNA + 4 mM melatonin-treated group and peaked in the miR-590-3p inhibitor + SEPT7 siRNA + 4 mM melatonin-treated group. Furthermore, in the miR-590-3p inhibitor + scramble siRNA + 4 mM melatonin group, the expression of the ERS-associated proteins GRP78 and p-eIF2α was the lowest of all the treatment groups, which indicated that SEPT7 may be a protective factor in melatonin-induced apoptosis. ERS expression peaked in the miR-590-3p inhibitor + SEPT7 siRNA + 4 mM melatonin-treated group compared with the 4 mM melatonin-treated group. An undergoing study suggests that, increased expression of SEPT7 may suppress ERS. Consequently, miR-590-3p inhibition may active other factors that promote apoptosis via non-ERS pathways, to oppose the inhibitory effect of SEPT7 on apoptosis.
In conclusion, the present study indicated that miR-590-3p is a potential regulator of melatonin-induced apoptosis. SEPT7, target gene of miR-590-3p, was inhibited by the presence of miR-590-3p under normal circumstances. When cells were treated with melatonin which induced apoptosis, the level of miR-590-3p decreased leading an increase of SEPT7, which can exert its protective effect on hFOB 1.19 cells. The inhibition of miR-590-3p may initially be activated by treatment with high concentrations of melatonin, upregulating the expression of SEPT7, which may induce an anti-apoptotic effect. High concentration of melatonin in human osteoblast cell line hFOB 1.19 leads to apoptosis (7). It can be hypothesized that miR-590-3p may target proapoptotic genes and SEPT7 as a target of miR-590-3p may prevent apoptosis. The results of the present study may improve understanding of the association between miRNAs and cell apoptosis in the human fetal osteoblast cell line hFOB 1.19. This may be used to identify future methods of preventing and treating osteoporosis.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China Grant (grant no. 81472044) and the Shenyang Science and Technology Program, Population and Health Special (grant no. 17-230-9-04). The authors of the present study would also like to thank Dr M. Subramaniam (Department of Biochemistry and Molecular Biology, Mayo Clinic, Rochester, MN, USA) for providing the human fetal osteoblastic cell line hFOB 1.19.
References
- 1.Beites CL, Xie H, Bowser R, Trimble WS. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat Neurosci. 1999;2:434–439. doi: 10.1038/8100. [DOI] [PubMed] [Google Scholar]
- 2.Field CM, Kellogg D. Septins: Cytoskeletal polymers or signalling GTPases? Trends Cell Biol. 1999;9:387–394. doi: 10.1016/S0962-8924(99)01632-3. [DOI] [PubMed] [Google Scholar]
- 3.Larisch S, Yi Y, Lotan R, Kerner H, Eimerl S, Tony Parks W, Gottfried Y, Birkey Reffey S, de Caestecker MP, Danielpour D, et al. A novel mitochondrial septin-like protein, ARTS, mediates apoptosis dependent on its P-loop motif. Nat Cell Biol. 2000;2:915–921. doi: 10.1038/35046566. [DOI] [PubMed] [Google Scholar]
- 4.Kartmann B, Roth D. Novel roles for mammalian septins: From vesicle trafficking to oncogenesis. J Cell Sci. 2001;114:839–844. doi: 10.1242/jcs.114.5.839. [DOI] [PubMed] [Google Scholar]
- 5.Hou M, Liu X, Cao J, Chen B. SEPT7 overexpression inhibits glioma cell migration by targeting the actin cytoskeleton pathway. Oncol Rep. 2016;35:2003–2010. doi: 10.3892/or.2016.4609. [DOI] [PubMed] [Google Scholar]
- 6.Jia ZF, Pu PY, Kang CS, Wang GX, Zhang ZY, Qiu MZ, Huang Q. Influence of SEPT7 on biological characters of glioma cell line TJ905. Zhonghua Wai Ke Za Zhi. 2007;45:1420–1423. (In Chinese) [PubMed] [Google Scholar]
- 7.Meng X, Zhu Y, Tao L, Zhao S, Qiu S. Periostin has a protective role in melatonin-induced cell apoptosis by inhibiting the eIF2α-ATF4 pathway in human osteoblasts. Int J Mol Med. 2018;41:1003–1012. doi: 10.3892/ijmm.2017.3300. [DOI] [PubMed] [Google Scholar]
- 8.Pizarro JG, Yeste-Velasco M, Esparza JL, Verdaguer E, Pallàs M, Camins A, Folch J. The antiproliferative activity of melatonin in B65 rat dopaminergic neuroblastoma cells is related to the downregulation of cell cycle-related genes. J Pineal Res. 2008;45:8–16. doi: 10.1111/j.1600-079X.2007.00548.x. [DOI] [PubMed] [Google Scholar]
- 9.Zhang NZ, Liu L, Fan N, Zhang Q, Wang W, Zheng M, Ma L, Li Y, Shi L. The requirement of SEPT2 and SEPT7 for migration and invasion in human breast cancer via MEK/ERK activation. Oncotarget. 2016;7:61587–61600. doi: 10.18632/oncotarget.11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J, Lu S, Yang S, Chen H, Shi H, Miao M, Jiao B. MicroRNA-127 post-transcriptionally downregulates Sept7 and suppresses cell growth in hepatocellular carcinoma cells. Cell Physiol Biochem. 2014;33:1537–1546. doi: 10.1159/000358717. [DOI] [PubMed] [Google Scholar]
- 11.Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: Key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 12.Iorio MV, Croce CM. MicroRNAs in cancer: Small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 14.Akao Y, Nakagawa Y, Naoe T. MicroRNA-143 and-145 in colon cancer. DNA Cell Biol. 2007;26:311–320. doi: 10.1089/dna.2006.0550. [DOI] [PubMed] [Google Scholar]
- 15.Akao Y, Nakagawa Y, Naoe T. MicroRNAs-143 and −145 are possible common onco-microRNAs in human cancers. Oncol Rep. 2006;16:845–850. [PubMed] [Google Scholar]
- 16.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge X, Gong L. MiR-590-3p suppresses hepatocellular carcinoma growth by targeting TEAD1. Tumour Biol. 2017;39:1010428317695947. doi: 10.1177/1010428317695947. [DOI] [PubMed] [Google Scholar]
- 18.Wu S, Liu W, Zhou L. MiR-590-3p regulates osteogenic differentiation of human mesenchymal stem cells by regulating APC gene. Biochem Biophys Res Commun. 2016;478:1582–1587. doi: 10.1016/j.bbrc.2016.08.160. [DOI] [PubMed] [Google Scholar]
- 19.Chen L, Wang W, Zhu S, Jin X, Wang J, Zhu J, Zhou Y. MicroRNA-590-3p enhances the radioresistance in glioblastoma cells by targeting LRIG1. Exp Ther Med. 2017;14:1818–1824. doi: 10.3892/etm.2017.4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang CX, Lv B, Wang Y. Protein phosphatase 2a mediates oxidative stress induced apoptosis in osteoblasts. Mediators Inflamm. 2015;2015:804260. doi: 10.1155/2015/804260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramaniam M, Jalal SM, Rickard DJ, Harris SA, Bolander ME, Spelsberg TC. Further characterization of human fetal osteoblastic hFOB 1.19 and hFOB/ER alpha cells: Bone formation in vivo and karyotype analysis using multicolor fluorescent in situ hybridization. J Cell Biochem. 2002;87:9–15. doi: 10.1002/jcb.10259. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Reiter RJ, Tan DX, Fuentes-Broto L. Melatonin: A multi-tasking molecule. Prog Brain Res. 2010;181:127–151. doi: 10.1016/S0079-6123(08)81008-4. [DOI] [PubMed] [Google Scholar]
- 24.Akbarzadeh M, Rahbarghazi R, Nabat E, Movassaghpour AA, Shanehbandi D, Faramarzian Azimi Maragheh B, Matluobi D, Barazvan B, Kazemi M, Samadi N, Nouri M. The impact of different extracellular matrices on melatonin effect in proliferation and stemness properties of ovarian cancer cells. Biomed Pharmacother. 2017;87:288–295. doi: 10.1016/j.biopha.2016.12.119. [DOI] [PubMed] [Google Scholar]
- 25.Bavithra S, Selvakumar K, Sundareswaran L, Arunakaran J. Neuroprotective effect of melatonin against PCBs induced behavioural, molecular and histological changes in cerebral cortex of adult male wistar rats. Neurochem Res. 2017;42:428–438. doi: 10.1007/s11064-016-2087-6. [DOI] [PubMed] [Google Scholar]
- 26.Mostowy S, Cossart P. Septins: The forth component of the cytoskeleton. Nat Rev Mol Cell Biol. 2012;13:183–194. doi: 10.1038/nrm3284. [DOI] [PubMed] [Google Scholar]
- 27.Kremer BE, Haystead T, Macara IG. Mammalian septins regulate microtubule stability through interaction with the micro tubule-binding protein MAP4. Mol Biol Cell. 2005;16:4648–4659. doi: 10.1091/mbc.E05-03-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wloka C, Nishihama R, Onishi M, Oh Y, Hanna J, Pringle JR, Krauss M, Bi E. Evidence that a septin diffusion barrier is dispensable for cytokinesis in budding yeast. Biol Chem. 2011;392:813–829. doi: 10.1515/BC.2011.083. [DOI] [PubMed] [Google Scholar]
- 29.Shehadeh L, Mitsi G, Adi N, Bishopric N, Papapetropoulos S. Expression of lewy body protein septin 4 in postmortem brain of Parkinson's disease and contro subjects. Mov Disord. 2009;24:204–210. doi: 10.1002/mds.22306. [DOI] [PubMed] [Google Scholar]
- 30.Scott M, Hyland P, McGregor G, Hillan KJ, Russell SE, Hall PA. Multimodality expression profiling shows SEPT9 to be overexpressed in a wide range of human tumours. Oncogene. 2005;24:4688–4700. doi: 10.1038/sj.onc.1208574. [DOI] [PubMed] [Google Scholar]