Abstract
Lung squamous cell carcinoma (LUSC), a type of non-small cell lung carcinoma, has a poor therapeutic response, high relapse rate and poor prognosis. The present study was designed to reveal the key long non-coding RNAs (lncRNAs) associated with the prognosis of LUSC. The lncRNA expression profiles of LUSC and adjacent samples were downloaded from The Cancer Genome Atlas database. Based on the edgeR and DEseq packages, the differentially expressed lncRNAs (DELs) between LUSC and adjacent samples were obtained and the intersecting DELs were regarded as significant DELs. Subsequently, a prognostic risk model was established using Cox regression analysis and its classification effect was detected by survival analysis. Using survival analysis, the effect of the prognostic risk model was assessed in the validation set and other types of cancer. Finally, the co-expression genes of key lncRNAs were screened using the Multi-Experiment Matrix tool and the STRING database, and their functions were predicted via enrichment analysis using the Database for Annotation, Visualization and Integrated Discovery tool. A total of 2,041 significant DELs between LUSC and adjacent samples were screened. The prognostic risk model consisting of RP5-821D11.7, APCDD1L-AS1 and RP11-277P12.9 was established, which had a good classification effect. Cox multivariate regression analysis demonstrated that risk score may serve as an independent prognostic factor. Furthermore, certain co-expression genes of RP5-821D11.7 (including proliferating cell nuclear antigen), APCDD1L-AS1 (including semaphorin 5A, semaphorin 6D, ADAMTS like 1, ADAM metallopeptidase with thrombospondin type 1 motif 6, slit guidance ligand 3, and tenascin C) and RP11-277P12.9 (including Wnt family member 2B) were identified. Additionally, ‘positive regulation of cell migration’ and ‘proteinaceous extracellular matrix’ were enriched. In conclusion, the expression levels of the lncRNAs RP5-821D11.7, APCDD1L-AS1 and RP11-277P12.9 may affect the prognosis of LUSC.
Keywords: lung squamous cell carcinoma, long non-coding RNA, prognostic risk model, survival analysis, co-expression gene
Introduction
Lung cancer, including small cell lung carcinoma and non-small cell lung carcinoma (NSCLC), is the primary cause of cancer death globally (1). NSCLC consists of lung squamous cell carcinoma (LUSC), adenocarcinoma and large cell carcinoma (2), which are responsible for 80% of lung cancer mortality (3). LUSC is associated with tobacco smoking, and more frequently occurs in men (4). It has the characteristics of poor therapeutic response, high relapse rate and poor prognosis (5), inducing ~400,000 mortalities/year worldwide (6). Therefore, revealing the genes involved in the prognosis of LUSC is of great importance.
Long non-coding RNAs (lncRNAs) are a class of non-protein-coding RNAs that mediate gene expression, which play important roles in multiple biological processes and diverse carcinomas (7). Previous studies have identified a number of lncRNAs to be associated with prognosis of LUSC. For example, Zhang et al (8) reported that lncRNA 1133 (LINC01133) is overexpressed in patients with LUSC and may shorten their survival time, thus LINC01133 is a promising biomarker for LUSC. The lncRNA metastasis associated lung adenocarcinoma transcript 1 is reported to have tumor-promoting functions and is associated with the survival time of patients with NSCLC (9,10). The lncRNA HOX transcript antisense intergenic RNA (HOTAIR) mediates the cell invasion and metastasis of NSCLC by downregulating homeobox A5, indicating that HOTAIR may serve as a prognostic marker and therapeutic target in patients with NSCLC (11,12). By inhibiting the expression of Kruppel like factor 2 (KLF2) and p21, lncRNA antisense non-coding RNA in the INK4 locus promotes cell proliferation and suppresses cellular apoptosis NSCLC (13,14). Furthermore, lncRNA associated with microvascular invasion in HCC regulates NSCLC cell proliferation and invasion, thus its overexpression may be used as a prognostic biomarker for NSCLC (15). However, the lncRNAs associated with the prognosis of LUSC have not been completely elucidated.
The present study further examined the key lncRNAs associated with the prognosis of LUSC through a series of bioinformatics methods. The lncRNA expression profiles of LUSC were downloaded from The Cancer Genome Atlas, and the differentially expressed lncRNAs (DELs) between LUSC and adjacent samples were identified. Following prognosis-associated DEL screening, a prognostic risk model was established and evaluated. Finally, the co-expression genes of important lncRNAs were obtained and their functions were predicted. The present study may contribute to predicting the prognosis of LUSC and revealing novel molecules associated with the disease.
Materials and methods
Data source
The lncRNA expression profiles of LUSC and the relevant clinical data were downloaded from The Cancer Genome Atlas (TCGA; cancergenome.nih.gov) database (downloaded on April 18, 2017). Samples without clinical data were removed and the data of 501 patients were obtained (Table I). In addition, the data of 49 tumor-adjacent normal lung tissues were obtained.
Table I.
Clinical characteristics of patients (n=501) with lung squamous cell carcinoma in the present study.
| Variable | No. patients |
|---|---|
| Age, years | |
| ≤65 | 190 |
| >65 | 311 |
| Sex | |
| Female | 139 |
| Male | 362 |
| Pathological stage | |
| I | 244 |
| II | 162 |
| III | 84 |
| IV | 7 |
| NA | 4 |
| T stage | |
| T1 | 114 |
| T2 | 293 |
| T3 | 71 |
| T4 | 23 |
| N stage | |
| N0 | 319 |
| N1 | 131 |
| N2-N3 | 45 |
| NA | 6 |
| M stage | |
| M0 | 411 |
| M1 | 7 |
| NA | 83 |
| Radiotherapy | |
| Yes | 53 |
| No | 384 |
| NA | 64 |
| Targeted molecular therapy | |
| Yes | 133 |
| No | 306 |
| NA | 62 |
| Residual tumor | |
| R0 | 398 |
| R1+R2 | 16 |
| NA | 87 |
| Neoplasm recurrence | |
| Yes | 135 |
| No | 285 |
| NA | 81 |
| Vital status | |
| Alive | 289 |
| Deceased | 212 |
| NA | 6 |
NA, data unavailable, tumor; N, node; M, metastasis.
DEL screening
Subsequent to obtaining the RNA-sequencing data of LUSC from TCGA, lncRNA annotation was performed using the GENCODE database (www.gencodegenes.org) (16). The lncRNAs with average expression values (counts/million) >0.1 were considered as sample expressing lncRNAs. The two independent R (version 3.1.0) (17) packages edgeR (version 3.8.5; www.bioconductor.org/packages/release/bioc/html/edgeR.html) (18) and DEseq (version 1.16.0; www.bioconductor.org/packages/release/bioc/html/DESeq.html) (19) were used for screening DELs between LUSC samples and adjacent samples. The adjusted P-value <0.05 and |log fold change (FC)| >1 were set as thresholds. The intersecting DELs predicted by the edgeR and DEseq packages were regarded as significant DELs.
Establishment of prognostic risk model
The DELs expressed in <10% of LUSC samples and patients with a survival time of <30 days were eliminated. The patients were randomly divided into a test set and validation set. For the test set, Cox univariate regression analysis was used to analyze the correlation between DELs and overall survival (OS). The lncRNAs with a P-value <0.05 was screened as prognosis-associated DELs. Subsequently, these prognosis-associated DELs were analyzed using Cox multivariate regression analysis to establish the prognostic risk model, with P-value <0.05 as the threshold. The prognosis risk score was calculated using the following formula (20):
Prognosis risk score=expression value of gene 1 × risk coefficient of gene 1 + expression value of gene 2 × risk coefficient of gene 2 + expression value of gene 3 × risk coefficient of gene 3.
In the test set, the expression levels of the prognosis-associated DELs in cancerous samples and adjacent samples were compared using the non-parametric Wilcoxon signed-rank test.
Detection of the classification effect of the prognostic risk model
The test set was divided into a high risk group and a low risk group, according to the median prognostic risk score. To assess the effect of prognostic risk score in determining the prognosis of patients, the difference between the survival curves of the high and low risk groups was analyzed by drawing receiver operating characteristic (ROC) curves using survivalROC package (version 1.0.3; https://cran.r-project.org/web/packages/survivalROC/index.html).
Determination of the correlation between prognostic risk score and prognosis
Using Cox univariate regression analysis, the correlation between prognostic risk score and clinical characteristics, including the OS of patients, were further analyzed. Subsequently, Cox multivariate regression analysis was applied to determinate whether the prognostic risk score was an independent prognostic factor. A P-value <0.05 was set as the threshold value. The hazard ratio and its 95% confidence intervals were used for evaluation. In order to assess the prediction accuracy of the prognostic risk model for time-dependent disease consequences, the survival ROC package (cran.r-project.org/web/packages/survivalROC/index.html) (21) in R was applied for drawing ROC curve. Furthermore, the OS differences among the patients in the high and low risk groups were evaluated by a log rank test using Kaplan Meier (KM) survival analysis (22). Based on a χ2 test (23), the correlations between DELs and clinical characteristics were detected. In addition, ROC curves were used to assess the prediction significance of the prognostic risk score following treatment, with the two-sided P-value <0.05 as the threshold.
Functional analysis of important lncRNAs
Associated genes (co-expression genes) of lncRNAs were obtained through the Human RNAseq expression data platform in the Multi-Experiment Matrix (MEM; biit.cs.ut.ee/mem/index.cgi) online tool (24). Using the STRING database (string-db.org) (25), a protein-protein interaction (PPI) network was established, with an associated score >0.4 and a number of associated nodes >3 set as the thresholds. Subsequently, the PPI network of lncRNAs and their associated genes was visualized using Cytoscape software 3.5.1 (www.cytoscape.org) (26). Additionally, Gene Ontology (GO; www.geneontology.org) (27) and Kyoto Encyclopedia of Genes and Genomes (KEGG; www.genome.ad.jp/kegg) (28) pathway enrichment analyses were performed based on the Database for Annotation, Visualization and Integrated Discovery (DAVID 6.7; david.ncifcrf.gov) (29) bioinformatics tool, with a P-value <0.05 set as the threshold.
Results
DEL screening
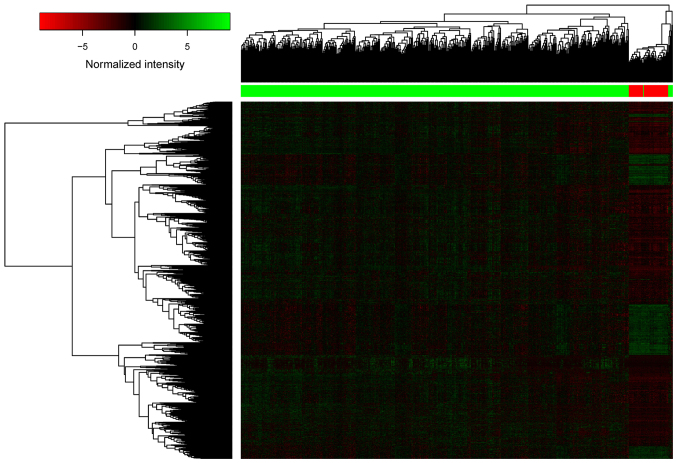
A total of 5,515 DELs were identified between LUSC samples and adjacent samples, including 2,537 DELs from the edgeR package and 2,048 DELs from the DEseq package. Finally, a total of 2,041 significant DELs were obtained by selecting the intersecting DELs predicted by the edgeR and DEseq packages. The heatmap of the 2,041 DELs is presented in Fig. 1, indicating that the lncRNA expression profiles of LUSC and adjacent samples were different.
Figure 1.
Heatmap of the 2,041 DELs. The horizontal axis and the vertical axis represent the DELs and the samples, respectively. DEL, differentially expressed long non-coding RNA.
Establishment of the prognostic risk model
A total of 489 samples and 1,468 DELs were included in the survival analysis. The patients were randomly divided into test (n=245) and validation (n=244) sets. Cox univariate regression analysis demonstrated that there were 68 prognosis-associated DELs in the test set. Subsequently, Cox multivariate regression analysis indicated that 3 prognosis-associated DELs (including RP5-821D11.7, APCDD1L-AS1 and RP11-277P12.9) had important prognostic value. The prognostic risk score was calculated using the following formula: Prognostic risk score=expression value of RP5-821D11.7 × (−0.392) + expression value of APCDD1L-AS1 × (0.101) + expression value of RP11-277P12.9 × (−0.114).
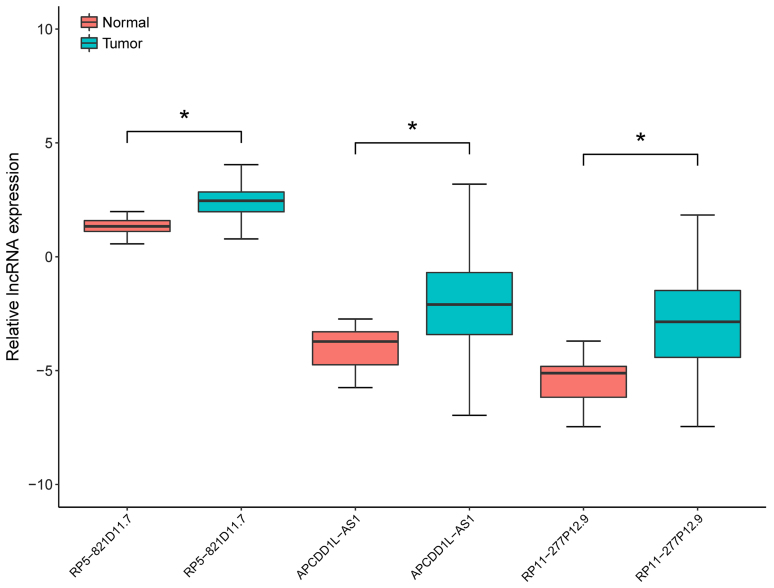
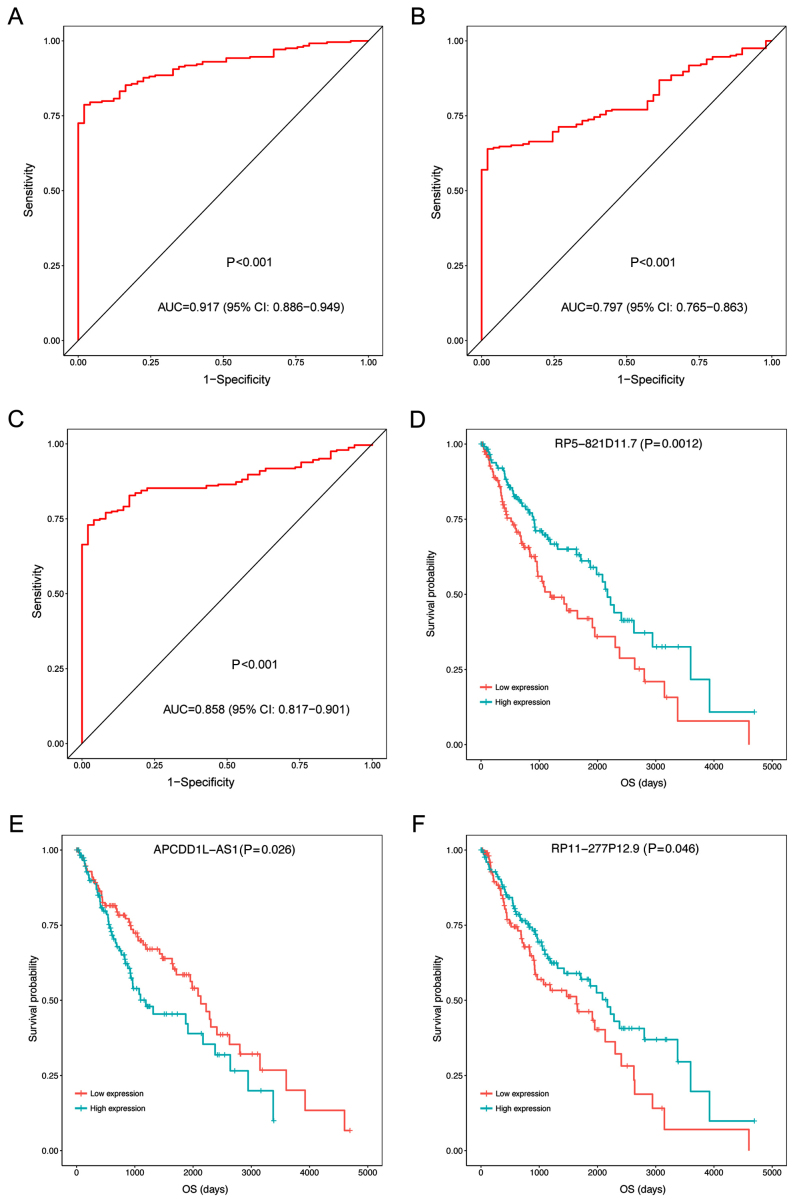
In the test set, the expression levels of the three prognosis-associated DELs in LUSC samples and adjacent samples were compared by non-parametric Wilcoxon signed-rank test. As presented in Fig. 2, the expression levels were significantly increased in LUSC samples compared with adjacent samples (P<0.05; Fig. 2). In addition, the expression levels of the three prognosis-associated DELs were able to be used to separate LUSC samples from the adjacent samples with an area under the ROC curve (AUC) >0.8 (Fig. 3A-C). Furthermore, the KM survival analysis demonstrated that the samples with high expression levels of the three prognosis-associated DELs had significantly lower OS compared with those with low expression levels (P<0.05; Fig. 3D-F).
Figure 2.
Expression levels of the three prognosis-associated differentially expressed lncRNAs in lung squamous cell carcinoma samples and adjacent samples of the test set. *P<0.05 (non-parametric Wilcoxon signed-rank test). lncRNA, long non-coding RNA.
Figure 3.
ROC curves and KM curves of the three prognosis-associated differentially expressed lncRNAs in lung squamous cell carcinoma samples and adjacent samples of the test set. (A) RP5-821D11.7 ROC curve, (B) APCDD1L-AS1 ROC curve, (C) RP11-277P12.9 ROC curve, (D) RP5-821D11.7 KM curve, (E) APCDD1L-AS1 KM curve and (F) RP11-277P12.9 KM curve. AUC, area under the curve; OS, overall survival; CI, confidence interval; ROC, receiver operating characteristic; KM, Kaplan-Meier.
Detection of the classification effect of the prognostic risk model
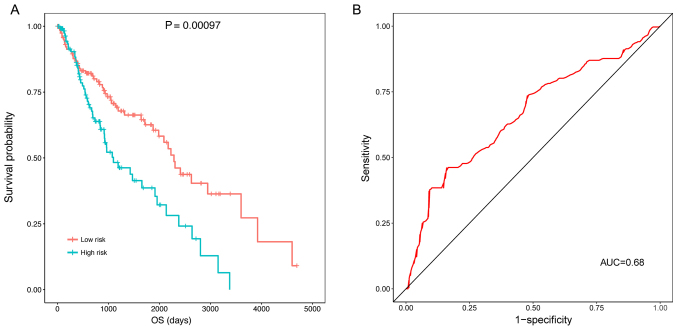
The KM survival analysis indicated that the samples in the high risk group (OS, 769.1±713.3 days) had a significantly lower OS compared with those in the low risk group (OS, 1240.0±1030.1 days) (P=0.00097) (Fig. 4A). The ROC curve demonstrated that the prognostic risk scores were able to predict the 5-year survival of patients to a certain degree (AUC, 0.68; Fig. 4B).
Figure 4.
ROC curves and KM curves of the high and low risk groups classified by the prognostic risk score in the training dataset. (A) KM curve indicating that the samples in the high risk group had a significantly higher OS, and (B) ROC curve demonstrating that prognostic risk scores were able to predict the 5-year survival of patients. AUC, area under the curve; OS, overall survival; KM, Kaplan-Meier; ROC, Receiver operating characteristic.
Evaluation of prognostic risk as an independent prognostic factor
Cox univariate regression analysis demonstrated that certain clinical characteristics were associated with the OS of patients, including risk score (P<0.001), neoplasm recurrence (P<0.001), tumor-node-metastasis (TNM) stage (P<0.001), tobacco smoking history (P=0.028), M stage (P=0.030), and T stage (P=0.038) (Table II). However, Cox multivariate regression analysis suggested that only risk score (P<0.001), and tobacco smoking history (P=0.048) had potential as independent prognostic factors (Table III).
Table II.
Cox univariate regression analysis between prognostic risk scores/clinical characteristics and overall survival of patients.
| Risk scores/clinical characteristics | Hazard ratio | Lower.95 | Upper.95 | P-value |
|---|---|---|---|---|
| Risk score | 2.718 | 1.720 | 4.297 | <0.001 |
| Neoplasm recurrence | 2.479 | 1.594 | 3.856 | <0.001 |
| TNM stage | 2.254 | 1.402 | 3.625 | <0.001 |
| Tobacco smoking history | 0.633 | 0.421 | 0.951 | 0.028 |
| M stage | 3.637 | 1.137 | 11.635 | 0.030 |
| T stage | 1.663 | 1.030 | 2.686 | 0.038 |
| Target molecular therapy | 0.689 | 0.409 | 1.160 | 0.161 |
| Residual tumor | 1.215 | 0.821 | 1.799 | 0.331 |
| Radiotherapy | 1.349 | 0.731 | 2.489 | 0.338 |
| N stage | 1.188 | 0.781 | 1.806 | 0.421 |
| Gender | 1.200 | 0.744 | 1.935 | 0.455 |
| Age | 1.016 | 0.671 | 1.538 | 0.939 |
T, tumor; N, node; M, metastasis.
Table III.
Cox multivariate regression analysis between prognostic risk scores/clinical characteristics and overall survival of patients.
| Risk scores/clinical characteristics | Hazard ratio | Lower.95 | Upper.95 | P-value |
|---|---|---|---|---|
| Risk score | 3.747 | 1.765 | 7.957 | <0.001 |
| Tobacco smoking history | 0.585 | 0.344 | 0.995 | 0.048 |
| Neoplasm recurrence | 1.486 | 0.871 | 2.535 | 0.146 |
| M stage | 3.026 | 0.550 | 16.640 | 0.203 |
| TNM stage | 1.711 | 0.658 | 4.451 | 0.271 |
| T stage | 0.838 | 0.325 | 2.161 | 0.714 |
T, tumor; N, node; M, metastasis.
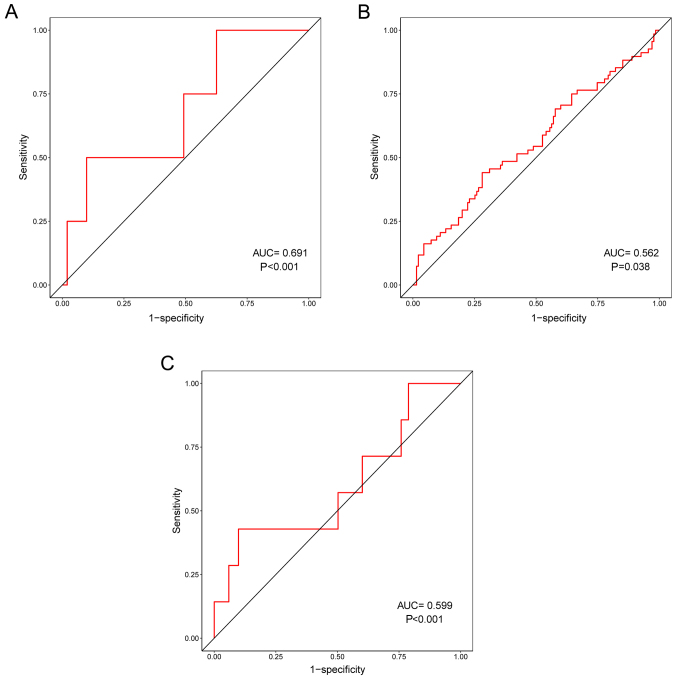
In addition, the correlation between prognostic risk scores and other clinical characteristics were evaluated. ROC curves demonstrated that prognostic risk scores were correlated with distant metastasis (pM) (P<0.001; Fig. 5A), tumor recurrence (P=0.038; Fig. 5B) and residual tumor (P=0.001; Fig. 5C). In particular, pM had the highest correlation with prognostic risk scores, indicating that the three prognosis-associated DELs may serve important roles in the distant metastasis of LUSC.
Figure 5.
Correlation between prognostic risk scores and other clinical characteristics. Receiver operating characteristic curves demonstrating that prognostic risk scores had a degree of correlation with (A) distant metastasis, (B) tumor recurrence and (C) residual tumor. AUC, area under the curve.
Assessment of the prognostic values of prognostic risk scores in the validation and universal sets
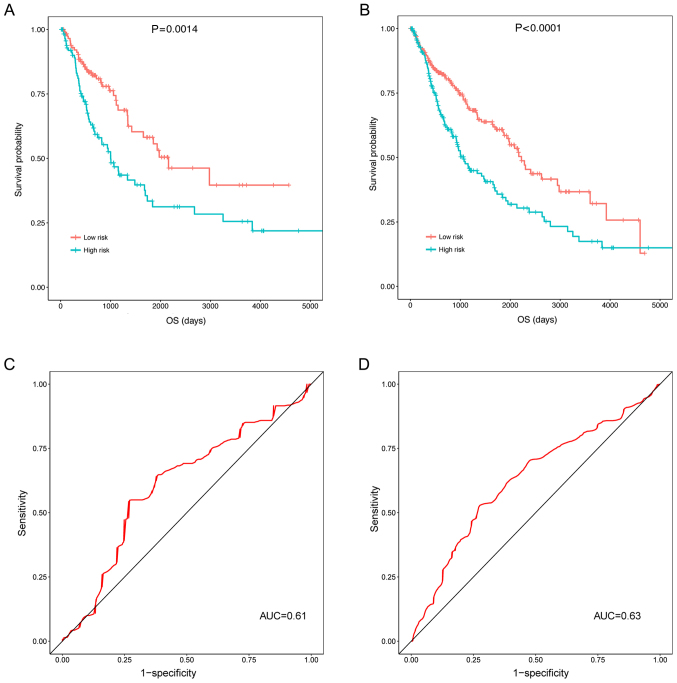
The three prognosis-associated DELs were validated in the validation and universal sets (including the test set and the validation set). In the validation set (P=0.0014; Fig. 6A) and the universal set (P<0.0001; Fig. 6B), the OS of patients in the high risk group was significantly decreased compared with the low risk group. Additionally, the ROC curves illustrated that the prognostic risk scores of the three prognosis-associated DELs were able to predict the 5-year survival of patients in the validation set (AUC, 0.61; Fig. 6C) and the universal set (AUC, 0.63; Fig. 6D). Though the AUCs of the ROCs predicting 5-year survival were small (0.61 and 0.63; Fig. 6C and D), the KM curves did illustrate an association between the three DEL-signature and survival (P=0.0014 and P<0.0001; Fig. 6A and B).
Figure 6.
Assessment of the prognostic values of prognostic risk scores in the validation and universal sets. Kaplan-Meier curves indicating that the OS of patients in the high risk group was significantly decreased for (A) the validation set and (B) the universal set, and receiver operating characteristic curves demonstrating that the three prognosis-associated differentially expressed long non-coding RNAs were able to predict the 5-year survival of patients in (C) the validation set and (D) the universal set. OS, overall survival; AUC, area under the curve.
Functional analysis of important lncRNAs
Using the MEM online tool and STRING database, co-expression genes of RP5-821D11.7 [including proliferating cell nuclear antigen (PCNA)], APCDD1L-AS1 [including semaphorin 5A (SEMA5A), semaphorin 6D (SEMA6D), ADAMTS like 1 (ADAMTSL1), ADAM metallopeptidase with thrombospondin type 1 motif 6 (ADAMTS6), slit guidance ligand 3 (SLIT3) and tenascin-C (TNC)] and RP11-277P12.9 [including Wnt family member 2B (WNT2B)] were screened. Enrichment analysis demonstrated that only the co-expression genes of APCDD1L-AS1 were enriched in certain GO terms and KEGG pathways (Table IV). Notably, ‘positive regulation of cell migration’ (P=0.048, involving SEMA5A and SEMA6D) and ‘proteinaceous extracellular matrix’ (P=0.017, involving ADAMTSL1 and ADAMTS6) were enriched.
Table IV.
The Gene Ontology terms and pathways enriched for the co-expression genes of APCDD1L-AS1.
| Category | Term | P-value | Genes |
|---|---|---|---|
| Pathway | Axon guidance | <0.001 | SEMA5A, SEMA6D, NTNG1, NFATC4, SLIT3 |
| ECM-receptor interaction | 0.013 | TNC, COL6A3, ITGA11 | |
| Olfactory transduction | 0.043 | OR4C13, OR5J2, OR1L6, OR8H2 | |
| Biological process | Cell adhesion | <0.001 | NRP2, SEMA5A, COL7A1, TNC, COL6A3, ITGA11, COL8A1, CDH6 |
| Negative chemotaxis | <0.001 | NRP2, SEMA5A, SEMA6D, SLIT3 | |
| Extracellular matrix organization | <0.001 | COL7A1, TNC, COL6A3, ITGA11, COL8A1 | |
| Semaphorin-plexin signaling pathway | 0.002 | SEMA5A, PLXNA4, SEMA6D | |
| Collagen catabolic process | 0.007 | COL7A1, COL6A3, COL8A1 | |
| Proteolysis | 0.014 | CPA4, ADAMTS6, ADAMTSL1, PAPPA2, HTRA3 | |
| Facial nerve structural organization | 0.017 | NRP2, PLXNA4 | |
| Axon extension involved in axon guidance | 0.023 | NRP2, SLIT3 | |
| Detection of chemical stimulus involved in sensory perception of smell | 0.046 | OR4C13, OR5J2, OR1L6, OR8H2 | |
| Heart development | 0.047 | NRP2, FOXL1, NFATC4 | |
| Positive regulation of cell migration | 0.048 | SEMA5A, SEMA6D, LRRC15 | |
| Negative regulation of axon extension involved in axon guidance | 0.048 | SEMA5A, SEMA6D | |
| Cellular component | Extracellular matrix | 0.003 | COL7A1, TNC, COL6A3, COL8A1, SSC5D |
| Endoplasmic reticulum lumen | 0.007 | ADAMTSL1, COL7A1, COL6A3, COL8A1 | |
| Extracellular region | 0.014 | NRP2, PTHLH, COL7A1, TNC, COL6A3, PAPPA2, HTRA3, COL8A1, SLIT3 | |
| Proteinaceous extracellular matrix | 0.017 | ADAMTS6, ADAMTSL1, COL6A3, SLIT3 | |
| Semaphorin receptor complex | 0.022 | NRP2, PLXNA4 | |
| Molecular function | Metallopeptidase activity | 0.010 | ADAMTS6, ADAMTSL1, PAPPA2 |
| Syndecan binding | 0.011 | SEMA5A, TNC | |
| Semaphorin receptor activity | 0.023 | NRP2, PLXNA4 | |
| Semaphorin receptor binding | 0.043 | SEMA5A, SEMA6D | |
| Laminin binding | 0.046 | LRRC15, SSC5D | |
| Olfactory receptor activity | 0.047 | OR4C13, OR5J2, OR1L6, OR8H2 | |
| Fibronectin binding | 0.048 | LRRC15, SSC5D | |
| Chemorepellent activity | 0.050 | SEMA5A, SEMA6D |
Discussion
In the present study, a total of 2,041 significant DELs between LUSC and adjacent samples were identified. A prognostic risk model involving three prognosis-associated DELs (including RP5-821D11.7, APCDD1L-AS1 and RP11-277P12.9) was established. The prognostic risk scores were able to predict the 5-year survival of patients to a certain degree, indicating that the classification effect of the prognostic risk model was good. Cox multivariate regression analysis suggested that only prognostic risk score and tobacco smoking history had the potential to be used as independent prognostic factors. Besides, the prognostic risk model was validated in the validation set and the universal set. In addition, certain co-expression genes of RP5-821D11.7 (including PCNA), APCDD1L-AS1 (including SEMA5A, SEMA6D, ADAMTSL1, ADAMTS6, SLIT3 and TNC) and RP11-277P12.9 (including WNT2B) were screened.
The SLIT/ROBO signaling pathway acts by regulating tumor cell metastasis, and SLIT3 serves as a promising tumor suppressor gene in lung adenocarcinoma (LAD) (30,31). The expression of TNC, S100A10 and S100A11 may predict survival in patients with LAD, indicating that these factors may be promising markers for better diagnosis and therapy in LAD (32,33). TNC expression is markedly elevated in patients with NSCLC, implying that TNC may serve as a prognostic marker for NSCLC (34,35). Semaphorins are a large number of transmembrane, glycosylphosphatidylinositol-linked and secreted proteins that are able to suppress and promote tumors (36). Downregulated SEMA5A is correlated with poor survival in patients with NSCLC, thus SEMA5A may be a prognostic marker for the disease (37). In the present study, enrichment analysis suggested that SEMA5A and SEMA6D were enriched in ‘positive regulation of cell migration’. These results indicated that APCDD1L-AS1 may affect the prognosis of LUSC by affecting the expression levels of SEMA5A, SEMA6D, SLIT3 and TNC.
Functional enrichment analysis demonstrated that ADAMTSL1 and ADAMTS6 were enriched in ‘proteinaceous extracellular matrix’. The ADAMTS enzymes are zinc metalloendopeptidases that affect the structure and function of the extracellular matrix (ECM) (38). ADAMTSL-1 belongs to the family of ADAMTS-like proteins and may serve important roles in the ECM (39). The ECM is important in mesenchymal cancer cells; in particular, its compositional and structural alterations may affect metastasis of mesenchymal lung cancer cells (40). Dysregulation of certain ADAMTS proteinases may be directly implicated in tumor development and metastasis (41). Therefore, APCDD1L-AS1 may be associated with the prognosis of LUSC through ADAMTSL1 and ADAMTS6.
A previous study reported that PCNA is specifically targeted by miR-363-3p, and may inhibit tumor growth in LAD (42). PCNA may be inhibited by the small molecule AOH1160, which suppresses the growth of SCLC cells without inducing any unacceptable side-effects and may be used as a potential anticancer therapy (43). Dysregulated Wnt signaling functions in the progression and metastasis of lung cancer, and inhibitors of WNT signaling are promising for the treatment of the disease (44,45). WNT2 may promote NSCLC cell growth by activating the Wnt/β-catenin signaling pathway, suggesting that WNT2 may be a marker for the diagnosis and prognosis of NSCLC (46). Thus, RP5-821D11.7 (co-expressed with PCNA) and RP11-277P12.9 (co-expressed with WNT2B) may be used for predicting the prognosis of LUSC.
There are certain limitations to the present study. First, these findings resulted from bioinformatics analysis and require further validation. Since the sample and experimental conditions are insufficient, it is not possible to perform verification experiments at present. Furthermore, it was identified from the Cox univariate regression analysis that only neoplasm recurrence, risk score and T stage were associated with the overall survival of patients. Therefore, only these three indicators were included in the Cox multivariate regression analysis, and only neoplasm recurrence and risk score were associated with the overall survival of patients. It was hypothesized that this may result from the relatively small sample size and its unbalanced distribution. For example, the number of patients with T3 and T4 stages was significantly lower compared with T1 and T2 stages. Similarly, the number of patients with N2-N3 stages was significantly lower compared with N0 and N1 stages. In addition, only seven patients were M1, while 411 patients were M0. However, the present prognosis risk model may be a valuable tool for further research.
In conclusion, a total of 2,041 significant DELs were identified in LUSC samples. RP5-821D11.7 (co-expressed with PCNA), APCDD1L-AS1 (co-expressed with SEMA5A, SEMA6D, ADAMTSL1, ADAMTS6, SLIT3 and TNC) and RP11-277P12.9 (co-expressed with WNT2B) may be important lncRNAs associated with the prognosis of LUSC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZW conceived and designed the study. YL and ZX designed and performed data analyses. XZ collected the data and wrote the manuscript. PZ provided critical suggestion and organized the literature. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Alberg AJ, Brock MV, Ford JG, Samet JM, Spivack SD. Epidemiology of lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143(5 Suppl):e1S–e29S. doi: 10.1378/chest.12-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ettinger DS, Akerley W, Borghaei H, Chang AC, Cheney RT, Chirieac LR, D'Amico TA, Demmy TL, Ganti AK, Govindan R, et al. Non-small cell lung cancer. J Natl Compr Canc Netw. 2012;10:1236–1271. doi: 10.6004/jnccn.2012.0130. [DOI] [PubMed] [Google Scholar]
- 3.Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, Beer DG. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 4.Kenfield SA, Wei EK, Stampfer MJ, Rosner BA, Colditz GA. Comparison of aspects of smoking among the four histological types of lung cancer. Tob Control. 2008;17:198–204. doi: 10.1136/tc.2007.022582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antal CE, Hudson AM, Kang E, Zanca C, Wirth C, Stephenson NL, Trotter EW, Gallegos LL, Miller CJ, Furnari FB, et al. Cancer-associated protein kinase C mutations reveal kinase's role as tumor suppressor. Cell. 2015;160:489–502. doi: 10.1016/j.cell.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network: Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang G, Lu X, Yuan L. LncRNA: A link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Zhu N, Chen X. A novel long noncoding RNA LINC01133 is upregulated in lung squamous cell cancer and predicts survival. Tumor Biol. 2015;36:7465–7471. doi: 10.1007/s13277-015-3460-9. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt LH, Spieker T, Koschmieder S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, Hillejan L, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol Off Pub Int Assoc Study Lung Cancer. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 10.Weber DG, Johnen G, Casjens S, Bryk O, Pesch B, Jöckel KH, Kollmeier J, Brüning T. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res Notes. 2013;6:518. doi: 10.1186/1756-0500-6-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, Wei D. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakagawa T, Endo H, Yokoyama M, Abe J, Tamai K, Tanaka N, Sato I, Takahashi S, Kondo T, Satoh K. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun. 2013;436:319–324. doi: 10.1016/j.bbrc.2013.05.101. [DOI] [PubMed] [Google Scholar]
- 13.Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R, Liu YW, Liu XH, Zhang EB, Lu KH, Shu YQ. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. 2015;14:268–277. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 14.Naemura M, Murasaki C, Inoue Y, Okamoto H, Kotake Y. Long noncoding RNA ANRIL regulates proliferation of non-small cell lung cancer and cervical cancer cells. Anticancer Res. 2015;35:5377–5382. [PubMed] [Google Scholar]
- 15.Nie FQ, Zhu Q, Xu TP, Zou YF, Xie M, Sun M, Xia R, Lu KH. Long non-coding RNA MVIH indicates a poor prognosis for non-small cell lung cancer and promotes cell proliferation and invasion. Tumour Biol. 2014;35:7587–7594. doi: 10.1007/s13277-014-2009-7. [DOI] [PubMed] [Google Scholar]
- 16.Wright JC, Mudge J, Weisser H, Barzine MP, Gonzalez JM, Brazma A, Choudhary JS, Harrow J. Improving GENCODE reference gene annotation using a high-stringency proteogenomics workflow. Nat Commun. 2016;7:11778. doi: 10.1038/ncomms11778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R Core Team. R Foundation for Statistical Computing. Vienna: 2016. R: A language and environment for statistical computing. [Google Scholar]
- 18.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anders S, Huber W. Differential expression of RNA-Seq data at the gene level-the DESeq package. EMBL. 2013 [Google Scholar]
- 20.Zeng JH, Liang L, He RQ, Tang RX, Cai XY, Chen JQ, Luo DZ, Chen G. Comprehensive investigation of a novel differentially expressed lncRNA expression profile signature to assess the survival of patients with colorectal adenocarcinoma. Oncotarget. 2017;8:16811–16828. doi: 10.18632/oncotarget.15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heagerty PJ, Thomas L, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341X.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 22.Porcher R. CORR Insights(®): Kaplan-meier survival analysis overestimates the risk of revision arthroplasty: A meta-analysis. Clin Orthopaed Relat Res. 2015;473:3431–3442. doi: 10.1007/s11999-015-4235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McHugh ML. The chi-square test of independence. Biochem Med (Zagreb) 2013;23:143–149. doi: 10.11613/BM.2013.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adler P, Kolde R, Kull M, Tkachenko A, Peterson H, Reimand J, Vilo J. Mining for coexpression across hundreds of datasets using novel rank aggregation and visualization methods. Genome Biol. 2009;10:R139. doi: 10.1186/gb-2009-10-12-r139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9. 1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T. A travel guide to Cytoscape plugins. Nat Methods. 2012;9:1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The Gene Ontology Consortium: Gene ontology consortium: Going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 30.Zhang C, Guo H, Li B, Sui C, Zhang Y, Xia X, Qin Y, Ye L, Xie F, Wang H, et al. Effects of Slit3 silencing on the invasive ability of lung carcinoma A549 cells. Oncol Rep. 2015;34:952–960. doi: 10.3892/or.2015.4031. [DOI] [PubMed] [Google Scholar]
- 31.Kong R, Yi F, Wen P, Liu J, Chen X, Ren J, Li X, Shang Y, Nie Y, Wu K, et al. Myo9b is a key player in SLIT/ROBO-mediated lung tumor suppression. J Clin Invest. 2015;125:4407–4420. doi: 10.1172/JCI81673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gocheva V, Naba A, Bhutkar A, Guardia T, Miller KM, Li CM, Dayton TL, Sanchez-Rivera FJ, Kim-Kiselak C, Jailkhani N, et al. Quantitative proteomics identify Tenascin-C as a promoter of lung cancer progression and contributor to a signature prognostic of patient survival; Proc Natl Acad Sci USA; 2017; pp. E5625–E5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang YA, Chen CH, Sun HS, Cheng CP, Tseng VS, Hsu HS, Su WC, Lai WW, Wang YC. Global Oct4 target gene analysis reveals novel downstream PTEN and TNC genes required for drug-resistance and metastasis in lung cancer. Nucleic Acids Res. 2015;43:1593–1608. doi: 10.1093/nar/gkv024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gebauer F, Gelis S, Zander H, Meyer KF, Wolters-Eisfeld G, Izbicki JR, Bockhorn M, Tachezy M. Tenascin-C serum levels and its prognostic power in non-small cell lung cancer. Oncotarget. 2016;7:20945–20952. doi: 10.18632/oncotarget.7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onion D, Isherwood M, Shridhar N, Mikalena X, Madeleine LC, Laura JD, Maria A GM, Robert GP, Alex MRS, John HS, et al. Multicomponent analysis of the tumour microenvironment reveals low CD8 T cell number, low stromal caveolin-1 and high tenascin-C and their combination as significant prognostic markers in non-small cell lung cancer. Oncotarget. 2017;9:1760–1771. doi: 10.18632/oncotarget.18880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Potiron VA, Roche J, Drabkin HA. Semaphorins and their receptors in lung cancer. Cancer Lett. 2009;273:1–14. doi: 10.1016/j.canlet.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu TP, Mong-Hsun T, Jang-Ming L, Hsu CP, Chen PC, Lin CW, Shih JY, Yang PC, Hsiao CK, Lai LC, Chuang EY. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomark Prevent. 2010;19:2590–2597. doi: 10.1158/1055-9965.EPI-10-0332. [DOI] [PubMed] [Google Scholar]
- 38.Kelwick R, Desanlis I, Wheeler GN, Edwards DR. The ADAMTS (A disintegrin and metalloproteinase with thrombospondin motifs) family. Genome Biol. 2015;16:113. doi: 10.1186/s13059-015-0676-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirohata S, Wang LW, Miyagi M, Yan L, Seldin MF, Keene DR, Crabb JW, Apte SS. Punctin, a novel ADAMTS-like molecule, ADAMTSL-1, in extracellular matrix. J Biol Chem. 2002;277:12182–12189. doi: 10.1074/jbc.M109665200. [DOI] [PubMed] [Google Scholar]
- 40.Peng DH, Ungewiss C, Tong P, Byers LA, Wang J, Canales JR, Villalobos PA, Uraoka N, Mino B, Behrens C, et al. ZEB1 induces LOXL2-mediated collagen stabilization and deposition in the extracellular matrix to drive lung cancer invasion and metastasis. Oncogene. 2017;36:1925–1938. doi: 10.1038/onc.2016.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun Y, Huang J, Yang Z. The roles of ADAMTS in angiogenesis and cancer. Tumour Biol. 2015;36:pp4039–4051. doi: 10.1007/s13277-015-3461-8. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Chen T, Huang H, Jiang Y, Yang L, Lin Z, He H, Liu T, Wu B, Chen J, et al. miR-363-3p inhibits tumor growth by targeting PCNA in lung adenocarcinoma. Oncotarget. 2017;8:20133–20144. doi: 10.18632/oncotarget.15448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu L, Hickey RJ, Reckamp KL, Malkas LH. Structural analysis identifies an orally active PCNA inhibitor that inhibits the growth of small cell lung cancer cells without causing significant toxicity to nonmalignant cells. J Thorac Oncol. 2016;11(Suppl):S22–S23. doi: 10.1016/j.jtho.2015.12.037. [DOI] [Google Scholar]
- 44.Yang J, Chen J, He J, Li J, Shi J, Cho WC, Liu X. Wnt signaling as potential therapeutic target in lung cancer. Expert Opin Ther Targets. 2016;20:999–1015. doi: 10.1517/14728222.2016.1154945. [DOI] [PubMed] [Google Scholar]
- 45.Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst. 2014;106:djt356. doi: 10.1093/jnci/djt356. [DOI] [PubMed] [Google Scholar]
- 46.Huang C, Ma R, Xu Y, Li N, Li Z, Yue J, Li H, Guo Y, Qi D. Wnt2 promotes non-small cell lung cancer progression by activating WNT/β-catenin pathway. Am J Cancer Res. 2015;5:1032–1046. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.