Abstract
The pra2 gene encodes a pea (Pisum sativum) small GTPase belonging to the YPT/rab family, and its expression is down-regulated by light, mediated by phytochrome. We have isolated and characterized a genomic clone of this gene and constructed a fusion DNA of its 5′-upstream region in front of the gene for firefly luciferase. Using this construct in a transient assay, we determined a pra2 cis-regulatory region sufficient to direct the light down-regulation of the luciferase reporter gene. Both 5′- and internal deletion analyses revealed that the 93-bp sequence between −734 and −642 from the transcriptional start site was important for phytochrome down-regulation. Gain-of-function analysis showed that this 93-bp region could confer light down-regulation when fused to the cauliflower mosaic virus 35S promoter. Furthermore, linker-scanning analysis showed that a 12-bp sequence within the 93-bp region mediated phytochrome down-regulation. Gel-retardation analysis showed the presence of a nuclear factor that was specifically bound to the 12-bp sequence in vitro. These results indicate that this element is a cis-regulatory element involved in phytochrome down-regulated expression.
Plants use light not only as an energy source for photosynthesis but also as an environmental signal. When a plant germinates in soil and is exposed to light after elongating its stem, the stem elongation is immediately repressed, and the leaves expand, turn green, and photosynthesis is initiated. Many of these changes are caused by the regulation of gene expression mediated by photoreceptors such as phytochrome. A pea (Pisum sativum) small GTPase gene, pra2, which belongs to the YPT/rab family (Nagano et al., 1993), is one of the genes whose expression is mediated by phytochrome (Yoshida et al., 1993). The pra2 gene is mainly expressed in the growing zone of etiolated epicotyls, and its expression is repressed when the plant is illuminated (Nagano et al., 1995). An interesting character is that the expression of this gene is down-regulated by phytochrome and probably involved in the morphogenesis of etiolated seedlings after germination. Small GTPases are molecular switches that are turned on by GTP and off by the hydrolysis of GTP to GDP. Members of the YPT/rab family play important roles during intracellular transport. Thus, the pra2 protein may participate in vesicle transport occurring in stem elongation of etiolated seedlings (Nagano et al., 1995).
There are many genes up-regulated by phytochrome, such as rbcS (Sasaki et al., 1983) and Lhcb (Kehoe et al., 1994), and several cis-regulatory regions involved in light-enhanced expression have been characterized (Terzaghi and Cashmore, 1995). However, there are not so many genes down-regulated by phytochrome, compared with those up-regulated. A few of these genes have been analyzed for cis-regulatory elements. Although detailed analysis of a PHYA promoter has revealed a phytochrome-repressible element, RE1 (Bruce et al., 1991), the gene for the putative repressor RF1, has not yet been cloned. Recently, Weatherwax et al. (1998) showed that the phytochrome response of the NPR1 gene was primarily mediated by the alteration of ABA levels and identified two elements necessary for phytochrome- and ABA-mediated responses. Although other genes such as tubB1 (Tonoike et al., 1994), AS1 (Nagi et al., 1997), and Athb2 (Carabelli et al., 1996) have also been reported as light down-regulated genes, we do not have as much information about down-regulation as about up-regulation.
We have been interested in the expression of the pra2 gene. Here we have examined the cis-regulatory element contributing to light down-regulation of the pra2 gene by transient assay using a reporter gene and etiolated pea stems.
MATERIALS AND METHODS
Characterization of the pra2 Gene
The genomic clone of the pra2 gene was isolated from a pea (Pisum sativum) genomic library (Stratagene) using the pea pra2 cDNA (Nagano et al., 1993) as a probe. The plaque hybridization and DNA sequencing were conducted as described previously (Nagano et al., 1993). The nucleotide sequence reported will appear in the EMBL, GenBank, and DDBJ nucleotide sequence databases under the accession no. AB007911. Primer-extension analysis was performed by the method reported previously (Nagano et al., 1991). The synthetic oligonucleotide for primer-extension analysis was 5′-ACGGTTGTTGAATTACCGGTGTTAATAGAG-3′.
Plant Material, Particle Bombardment, and Light Treatment
Pea (cv Alaska; Snow Brand Seed, Sapporo, Japan) seeds were soaked in water and sowed in a plastic pot separately. The pot (14 mm in diameter) had a polyethylene net at its lower end and was filled with rock wool and placed in an irrigated vat. Seedlings were grown in the dark for 5 or 6 d at 25°C. A seedling was set horizontally in the bombardment device (model GIE-III, Tanaka Co. Ltd., Sapporo, Japan), which was described in detail by Takeuchi et al. (1992). To the growing zone of the etiolated stem (between 0 and 1 cm from the top of the hook), 1.5- to 3.0-μm gold particles were bombarded once. The gold particles were coated with a mixture of two kinds of plasmids, a plasmid containing one of the pra2:LUC constructs and a plasmid containing a 35S-GUS construct. Five micrograms of each plasmid was mixed with 2-mg gold particles and suspended in 200 μL of ethanol. A 4-μL aliquot of the suspension was used for each bombardment. We performed all of the steps during bombardment in a darkroom with a dim-green safety light. After bombardment, the plant was returned to the irrigated vat and incubated in continuous white light or darkness for 12 h at 25°C. White light was provided by white fluorescent tubes at an intensity of 70 μmol m−2 s−1 (measured by a quantum sensor, model LI-190SA, LI-COR, Lincoln, NE).
Equipment for red and far-red irradiation was the same as that described previously (Yoshida et al., 1993). For brief red-light irradiation, monochromatic light with a peak emission at 660 nm was supplied at an intensity of 30.5 μmol m−2 s−1 (measured by a quantum sensor, model LI-190SA, LI-COR) for 2 min. For brief far-red light irradiation, monochromatic light with a peak emission at 750 nm was supplied at an intensity of 36.5 μmol m−2 s−1 (measured by thermopiles, model MIR-100Q, Mitsubishi Oil Chemicals, Tokyo, Japan) for 5 min. A pair of modified slide projectors (Cabin III) each equipped with a 300-W halogen lamp (Philips, Eindhoven, The Netherlands) was used as a source. This light was filtered through a combination of a red-interference filter (maximum wavelength = 660 nm; DIF-BPF-2, Vacuum Optics, Tokyo, Japan) and a heat-cut filter (CF-B) to obtain red light and through a far-red interference filter (maximum wavelength = 750 nm; DIF-BPF-2) and a long-wavelength heat-cut filter (CF-A) to obtain far-red light.
Measurements of Enzyme Activities
A stem of the bombarded pea seedling (between 0 and 1 cm from the top of the hook) was ground in liquid N2 using a chilled mortar and pestle. The powder was dispensed into a microcentrifuge tube and mixed with 300 μL of the buffer consisting of 100 mm potassium phosphate, pH 7.8, 1 mm DTT, 1% Triton X-100, and 1 mm EDTA and then centrifuged at 15,000g at 4°C for 5 min. The supernatant was frozen at −80°C until the enzyme assay was conducted. LUC assays were performed as described by Miller et al. (1992) using the Pica Gene LUC assay kit (Wako, Osaka, Japan). Photon emission derived from LUC activity was counted by AUTO LUMAT LB953 (Berthold, Bad Wildbad, Germany). GUS activity was measured by the method of Jefferson et al. (1987), using 4-methylumbelliferyl β-d-glucuronide (Wako) as the substrate. Resulting 4-methylumbelliferone concentrations were determined by Fluoroskan II (Labsystems, Research Triangle Park, NC). To the reaction mixture, 10% methanol was added to enhance GUS activity (Kosugi et al., 1990). 4-Methylumbelliferone solutions dissolved in 0.2 m Na2CO3 were used as the standards. Proteins were measured using the DC Protein Assay kit (Bio-Rad). Background activities from plants bombarded with gold particles only were subtracted from each LUC and GUS value. All LUC values were normalized to the corresponding GUS values. Samples of at least four bombardments were independently assayed for each construct. The mean values were normalized to that of the full-length construct (PL1) kept in the dark.
Construction of pra2-LUC Plasmids
The plasmids were constructed by four methods as described below.
(1) The pra2 upstream regions were amplified by cloned Pfu (Pyrococcus furiosus) DNA polymerase (Stratagene) using two primers containing HindIII and NcoI sites of their 5′ ends, respectively. The NcoI site corresponds to the initiation codon of the pra2 gene. The HindIII site corresponds to the upstream region. For the creation of LS constructs, each upstream primer had 6-bp mutations corresponding to the PstI site on different positions. The amplified fragments were subcloned into the EcoRV site of the pZErO-2.1 vector (Invitrogen, San Diego, CA), and digested with HindIII and NcoI. DNA fragments were electrophoresed, purified by a DNA-extraction kit (Pharmacia Biotech), and subcloned into the HindIII-NcoI sites of the pBI221-LUC+ vector (a gift from Dr. K. Hiratsuka, NAIST Graduate School of Biological Science, Nara, Japan). HindIII-NcoI digestion of pBI221-LUC+ plasmid yielded a 35S promoter-less vector. All subcloned regions of each construct were confirmed by DNA sequencing. The constructs PL1, PL3, PL4, PL4A, and PL5, and all LS constructs, were created by this method.
(2) The constructs PL2, PL6, PL7, PL8, and PL4C were amplified by LA-Taq (Takara, Otsu, Japan)-mediated inverse PCR using PL1 as a template. The amplified fragments were blunt ended by cloned Pfu DNA polymerase and self-ligated.
(3) To create the pra2-35S90-LUC (GF) plasmid, the pra2 upstream regions were amplified by cloned Pfu DNA polymerase using two primers containing the EcoRV and PstI site, respectively. The amplified fragments were subcloned into pZErO-2.1 vector as described above and digested with EcoRV and PstI. The purified DNA fragments were subcloned into the EcoRV-PstI sites of pBI221-LUC+. EcoRV-PstI digestion of the pBI221-LUC+ plasmid yielded a 35S90-LUC vector.
(4) To create the PL4B construct, the pra2 upstream regions were amplified by cloned Pfu DNA polymerase using two primers containing the HindIII and PstI sites of their 5′ ends. The amplified fragments were subcloned into the pZErO-2.1 vector as described above and digested with HindIII and PstI. The purified DNA fragments were subcloned into the HindIII-PstI digest of LS5 carrying the LUC gene.
In all of the plasmids used in this study, we measured the luminescence of the Escherichia coli culture containing each construct and selected a representative clone for plasmid purification to exclude mutations occurring on the LUC gene. All plasmids were purified using a plasmid extraction kit (Qiagen, Chatsworth, CA).
Extraction of Protein and Immunoblotting
Total protein for immunoblotting was extracted by grinding tissue with a mortar and pestle at room temperature with sand together with 0.3 mL of buffer and three stem pieces (between 0 and 1 cm from the top of the hook). The buffer contained 125 mm Tris-HCl, pH 6.8, 6% SDS, and 20% glycerol. The mixture was heated at 100°C for 3 min and centrifuged. The supernatant protein was separated by SDS-PAGE, blotted onto a nitrocellulose membrane, probed with monoclonal IgG against the pra2 protein (Nagano et al., 1995) and goat anti-mouse IgG conjugated to peroxidase (Bio-Rad), and developed with an ECL kit (Amersham).
Preparation of Nuclear Extract
Nuclear extracts were prepared by a modification of the method described by Ishiguro and Nakamura (1992). Pea seedlings were grown in darkness for 6 d. For light-treated samples, seedlings were exposed to white light for 6 h before nuclear extraction. The upper region of pea epicotyls (1-cm section from the top of the hook, 20 g) was cut into pieces. Then they were homogenized with 250 mL of homogenization buffer containing 10 mm Pipes-KOH, pH 7.0, 1 m hexylene glycol, 10 mm MgCl2, 5 mm β-mercaptoethanol, 1 mm PMSF, 8 μm pepstatin A, and 2.4 μm leupeptin, in a homogenizer (Hitachi, Tokyo, Japan). The homogenates were filtered through Miracloth (Calbiochem). Nuclei were pelleted from the homogenate by centrifugation at 2,700g for 15 min, resuspended in 50 mL of washing buffer consisting of 50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 20% glycerol, and 5 mm β-mercaptoethanol, and then centrifuged at 5,200g for 15 min. Washing and centrifugation were repeated three times. Nuclei were resuspended in 3 mL of nuclear lysis buffer consisting of 15 mm Hepes-KOH, pH 7.5, 1.25 m KCl, 5 mm MgCl2, 2.5 mm EDTA, 1 mm DTT, 1 mm PMSF, 8 μm pepstatin A, and 2.4 μm leupeptin, with gentle stirring. Particulate material was pelleted by centrifugation at 5,200g for 15 min and then at 100,000g for 1 h. The supernatant was desalted by dialysis. The sample was then centrifuged at 12,000g for 15 min, and the supernatant was collected and stored at −80°C until the assay was performed. Proteins were measured using the DC Protein Assay kit (Bio-Rad).
Gel-Retardation Assay
The gel-retardation assay was performed by the method described by Shimizu et al. (1996) with some modification. Synthetic oligonucleotides for gel-retardation assay were as follows: 5′-GTCTGAGGATTTTACAGTAATAAAGAAACGA-3′(WT1) and 5′-TCGTTTCTTTATTACTGTAAAATCCTCAGAC-3′ (WT2). The 5′ end of a 31-bp WT1 DNA probe was labeled by incubation with [γ-32P]ATP and T4 polynucleotide kinase and then hybridized with WT2. The nuclear protein (6 μg) was mixed in binding buffer (20 μL), which consisted of 20 mm Tris-HCl, pH 8.0, 50 mm KCl, 0.5 mm EDTA, 15 mm MgCl2, 10% glycerol, 1 mm DTT, 2 μg of poly(dI-dC)-poly(dI-dC), and competitor DNA as indicated in the figure legends. The unlabeled probe was used as a wild-type competitor. For mutant competitor, the following oligonucleotides with a 3-bp substitution were prepared and hybridized: 5′-GTCTGAGGcTTTTcCcGTAATAAAGAAACGA-3′ (MT1) and 5′-TCGTTTCTTTATTACgGgAAAAgCCTCAGAC-3′ (MT2). The mutated nucleotides are indicated in lowercase. The reaction mixture was preincubated for 5 min. Then, 10,000 cpm of the labeled DNA probe was added to the reaction mixture. The binding reaction was continued for 15 min at 37°C. Samples were loaded onto a 5% polyacrylamide gel that contained 10% glycerol, 45 mm Tris-borate, and 1 mm EDTA (0.5× TBE buffer) and subjected to electrophoresis at 4°C. The gel was dried and exposed overnight to an imaging plate (Fuji Photo Film Co., Kanagawa, Japan). The image was visualized with a Bio Imaging Analyzer (model BAS2000, Fuji).
RESULTS
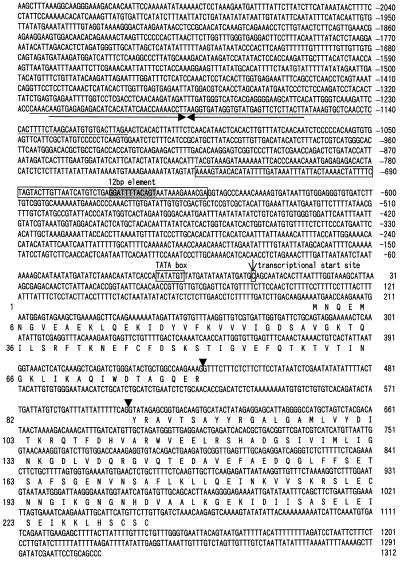
Characterization of the Genomic Clone of the pra2 Gene and Determination of Its Transcriptional Start Site
The nucleotide sequence of the genomic clone of the pra2 gene is shown in Figure 1. The gene contained two exons and one intron. The deduced amino acid sequences agreed with those of cDNA (Nagano et al., 1993), except that Gly-206 was replaced by Ala. This may be caused by the fact that the cDNA and genomic libraries were derived from different cultivars. The predicted protein sequences had the conserved domains for GTP binding (Nagano et al., 1993). Primer-extension analysis revealed that the transcriptional start site was located 196 nucleotides upstream of the translational start site of the pra2 gene (Fig. 1). The putative TATA box was located 24 bp upstream from this transcriptional start site. A characteristic feature of the upstream sequence was the presence of a 113-bp inverted repeat element (from −1226 to −1114), which displayed 88% sequence similarity with the upstream region of one of the pea lipoxygenase genes (lox1:Ps:2; Forster et al., 1994). The PCR and Southern-blot analyses showed that similar elements were dispersed throughout the genome of pea plants (data not shown). These results showed that the element is a family of interspersed DNA elements. Whereas the function of most repetitive DNAs has not been elucidated, some of them have been implicated in various cellular functions, such as the regulation of gene expression (Bureau and Wessler, 1994). However, this element is not involved in the light down-regulated expression as described below, although further study is required to elucidate the role of this element in the regulation of gene expression.
Figure 1.
Nucleotide sequence of the pea pra2 gene. Nucleotides are numbered on the right side, with the transcriptional start site designated +1. Amino acids are numbered on the left side, with the first residue of the protein designated +1. Arrowheads show exon-intron boundaries. The 113-bp inverted repeat element is underlined. Arrows show inverted repeats. The location of the 93-bp region is boxed.
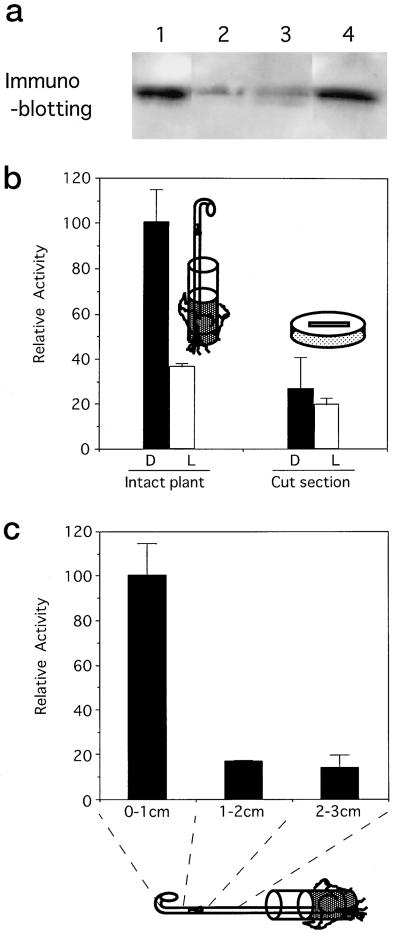
Intact Epicotyls Were Necessary for This Transient Assay
pra2 mRNA and protein are mainly present in the growing zone of etiolated pea seedlings and disappear upon exposure to light (Yoshida et al., 1993; Nagano et al., 1995). Because we wanted to introduce a reporter gene into this zone, we examined whether it would be possible to use a stem section of the growing zone. We examined whether the pra2 protein in the stem section was stable for 12 h in darkness, as it is in intact seedlings. Immunoblot analysis using a monoclonal antibody against the pra2 protein indicated that the pra2 protein of intact seedlings was found in darkness (Fig. 2a, lane 1) and decreased after 12 h of white-light irradiation (lane 2). The pra2 protein also decreased when the stem was cut and kept in darkness for 12 h (lane 3). The results indicated that cutting the stem caused some changes that affected pra2 protein expression and that the stem section could not be used instead of intact stems for our experiments. Therefore, we used intact seedlings to examine reporter-gene expression in a transient assay. Bombardment of the growing zone of intact seedlings with gold particles, which might wound plants, did not affect the expression of the pra2 protein (lane 4).
Figure 2.
Determination of the experimental conditions. a, Effect of cutting the stem on pra2 protein levels. Total proteins from the stem (1.0 cm from the top of the hook) were extracted, separated by SDS-PAGE, and probed with anti-pra2 protein IgG. The same amount of protein (30 μg) was put in each lane. Six-day-old seedlings grown in darkness (lane 1) were irradiated with white light for 12 h (lane 2). Stem sections (1.0 cm from the top of the hook) of the 6-d-old seedlings were cut and kept in darkness for 12 h on wet cotton (lane 3). The growing zone of etiolated 6-d-old seedlings were bombarded with gold particles and kept in darkness for 12 h (lane 4). b, Effect of pra2 upstream fragment on the reporter-enzyme activity in stems of intact plants and sections. The PL1 construct was introduced into the growing zone of intact etiolated stem (left) or stem section (1 cm from the top of the hook; right) by particle bombardment with the 35S-GUS construct as the internal standard. Reporter-enzyme activity was measured after 12 h of darkness (D) or 12 h of white-light irradiation (L). Relative activity was defined in “Materials and Methods,” and the average of PL1 in darkness was taken to be 100. Values are the means of at least four independently bombarded samples with error bars representing se (n ≥ 4). c, Comparison of reporter-gene expression in different parts of intact stem. PL1 construct was bombarded into the indicated parts, and the reporter-enzyme activity was measured as described.
We fused the DNA sequence extending 2325 bp upstream from the translational start site (196-bp 5′-untranslated region of mRNA and 2129-bp upstream region; Fig. 1) to the LUC reporter gene (PL1 construct; Fig. 3). Then, we introduced the PL1 construct into the growing zone of intact, etiolated stem by particle bombardment with the 35S-GUS construct as the internal standard. When the relative activity of the reporter enzyme in darkness was taken to be 100, the activity decreased to about 37 after 12 h of illumination (Fig. 2b, left). When we bombarded the PL1 construct into a cut section of growing zone, we did not observe light down-regulated expression of the reporter enzyme (Fig. 2b, right). These results indicated that the 2325-bp 5′-upstream region was sufficient to down-regulate the expression of the LUC gene in the growing zone of intact stem upon light irradiation.
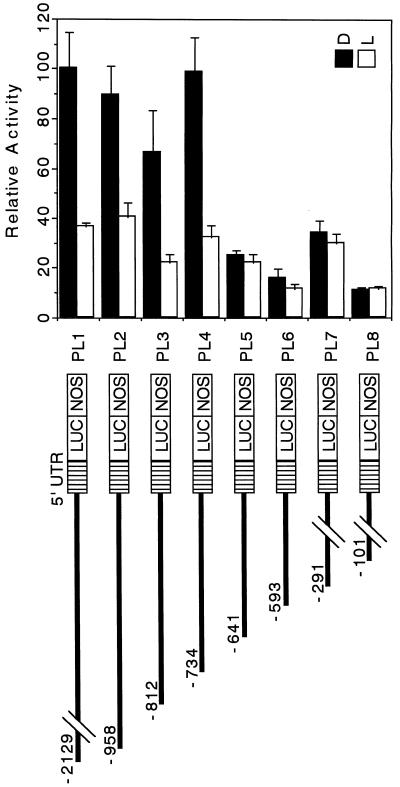
Figure 3.
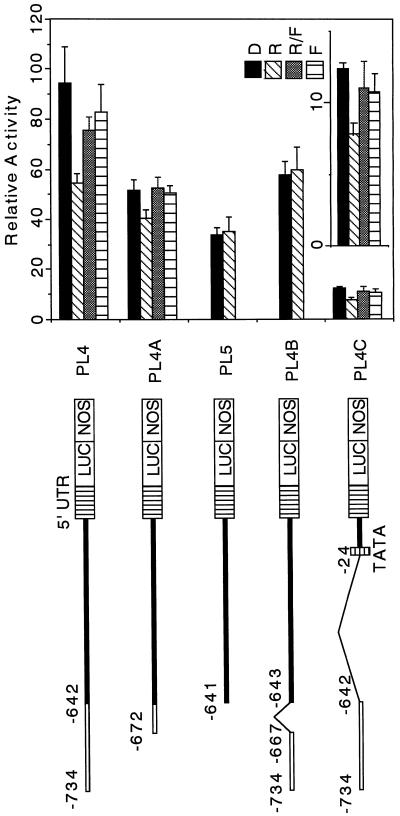
5′-Deletion analysis of the pra2 upstream region. The 5′-deletion constructs containing the pra2 upstream region were fused to the promoter-less pBI221-LUC+ plasmid. The numbers refer to the 5′ end of the pra2 upstream fragments from the transcriptional start site in Figure 1. The names of the resulting plasmids are indicated. Equivalent amounts of pBI221-LUC+ plasmid DNA fused to different pra2 upstream regions were introduced into the growing zone of etiolated pea stem by particle bombardment, with the 35S-GUS construct as the internal standard. After bombardment, samples were kept in darkness (D) or under continuous white light (L) for 12 h. Relative activity is defined in Figure 2b. Values are the means of at least four independently bombarded samples with error bars representing se (n ≥ 4). UTR, Untranslated region; NOS, nopaline synthase terminator.
We also introduced the PL1 construct into different zones of intact, etiolated pea stems (0–1, 1–2, and 2–3 cm from the top of the hook). The region of 0 to 1 cm elongated about 2-fold within 12 h, whereas 1- to 2- and 2- to 3-cm regions elongated little (data not shown). The LUC activity in the growing zone was much higher than that in the nongrowing zone (Fig. 2c). This result agreed with previous data (Nagano et al., 1995) and indicated that the 2325-bp region conferred growing-zone-specific expression on the reporter gene. It is necessary to introduce reporter-gene constructs into the growing zone to get reliable data.
5′-Deletion Analyses Showed That the pra2 Upstream Region Had a cis-Regulatory Region for Light Down-Regulated Expression
To broadly determine the light down-regulated region, we carried out the experiments using white light. Then we used red and far-red lights to precisely determine the cis element involved in phytochrome down-regulation. We constructed a series of 5′ deletions of the pra2 upstream region and fused them to the LUC reporter gene, as shown in Figure 3. The resulting plasmids were bombarded into the growing zone of etiolated, intact stems. We determined the comparative expression of the reporter gene in darkness and after 12 h of white light (Fig. 3) for each of the deletion constructs. Four constructs, PL1 to PL4, showed similar levels of activity in darkness, and this activity was decreased by light. These findings indicated that a region sufficient to confer higher expression in dark than in light was present within the −734-bp region from the transcriptional start site. However, in the PL5 to PL8 constructs the activity in darkness decreased to about one-fifth of that of PL4, although a little activity was recovered in PL7. In addition, the effect of light on expression was almost abolished. This result indicated a critical region for expression in darkness, and the response to light was at least present in the 93-bp sequence from −734 to −642. The activity increase of the PL7 construct in darkness suggested the possible presence of a repressing region from −593 to −292 and an activating region from −291 to −101.
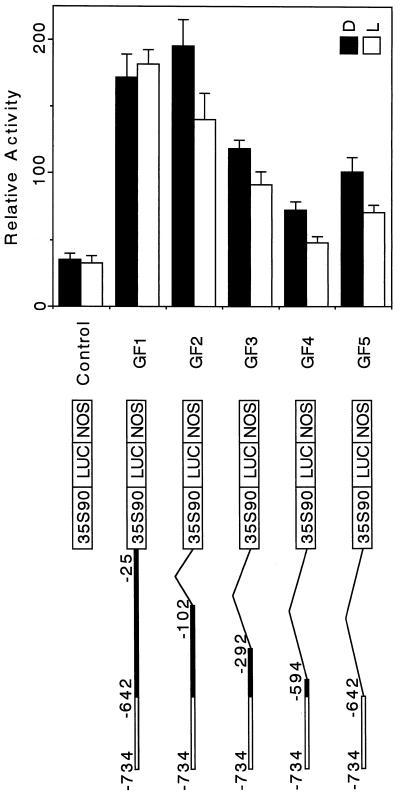
The 93-bp Region Conferred Light Down-Regulated Expression to a Heterologous Promoter
We performed gain-of-function 3′-deletion analysis to determine whether this 93-bp region was able to confer light down-regulation to a non-light-regulated heterologous promoter. We fused several 3′-deleted fragments of the pra2 upstream region to the minimal CaMV 35S promoter (−90 from the transcriptional start site, designated as 35S90) and LUC reporter gene (Fig. 4). We constructed five plasmids containing the 93-bp region, as shown in Figure 4, and compared the light responsiveness of the reporter-gene expression in each construct. In the absence of the pra2 5′ region, the activity of the reporter gene did not change upon light irradiation, and light down-regulation was not observed (Fig. 4, control). The GF1 construct, from which the sequence between the pra2's own TATA box and the translational start site (−24 to +196) was deleted, could not confer down-regulated expression of the reporter gene. However, the further deleted construct (−101 to +196), GF2, could confer expression. This suggests that the element, which is located between −101 and −25, may interact with the as-1 sequence of the CaMV 35S90 promoter. This interaction may determine the light response of the GF1 construct. The other 3′-deleted constructs, GF3, GF4, and GF5, conferred down-regulation of the reporter gene. These results indicated that the 93-bp region was sufficient to confer light down-regulated expression to a heterologous promoter, 35S90. As shown by the activity in light, the basal level decreased with the length of the fused fragment (from GF1–GF4), suggesting that the length or sequence affected expression at the basal level.
Figure 4.
Gain-of-function analysis. A, CaMV 35S promoter of 90 bp from the transcriptional start site fused to the LUC reporter gene was used as the control construct. To the control construct, several 3′-deleted fragments of the pra2 upstream region were fused. The names of the resulting plasmids are indicated on the right. White bold line, 93-bp region; thin line, deleted region. Control or GF1 to GF5 constructs were introduced into the growing zone of etiolated, intact stem by particle bombardment, with the 35S-GUS construct as the internal standard. After bombardments, the plants were kept in darkness (D) or irradiated with white light (L) for 12 h. Values are the means of at least four independently bombarded samples, with error bars representing se (n ≥ 4). Relative activity is defined in Figure 2b. NOS, Nopaline synthase terminator.
The 24-bp Region Is Involved in Phytochrome Down-Regulated Expression
pra2 expression is down-regulated by phytochrome (Yoshida et al., 1993). To investigate whether a cis element involved in the down-regulation exists in the 93-bp region, we examined the effect of a brief red-light irradiation on reporter-gene expression using the PL4 construct. After particle bombardment of the PL4 construct, the plants were irradiated with 2 min of red light and kept for 12 h in darkness, and the reporter enzyme activity was then measured. A brief red light repressed the reporter enzyme activity, and this repression was reversed by far-red irradiation provided immediately after red-light irradiation (Fig. 5, PL4). In the plants bombarded with the PL5 construct lacking the 93-bp region, red-light irradiation did not repress the reporter-enzyme activity (Fig. 5, PL5). These results suggested that the 93-bp region was necessary for phytochrome down-regulation.
Figure 5.
Red/far-red reversibility of the change in the reporter-enzyme activity. Different kinds of constructs containing the pra2 upstream region were prepared as described in Methods. The names of the resulting plasmids are indicated on the right. White bold line, 93-bp region; thin line, deleted region. Equivalent amounts of each plasmid were introduced into 5- or 6-d-old etiolated seedlings as described in Methods. After bombardment, the plants were exposed to red light for 2 min (R) or far-red light for 5 min immediately after red light for 2 min (R/FR) and then were returned to darkness for 12 h. Dark and far-red controls are indicated as D and F, respectively. Values are the means of 5 to 11 independently bombarded samples, with error bars representing se. Relative activity is defined in Figure 2b. A fine-scale figure of PL4C is shown in the inset. NOS, Nopaline synthase terminator.
To examine whether the 93-bp region was sufficient to confer phytochrome down-regulation to its own minimal promoter, we fused the 93-bp fragment to its own TATA box and 5′-untranslated region (Fig. 5, PL4C). We observed red-light-inducible and far-red-light-reversible repression, although the basal expression was reduced (Fig. 5, PL4C). These results indicated that the 93-bp region was sufficient to confer phytochrome down-regulation.
For further analysis, we deleted 62 bp of the 93-bp region in PL4 and measured its activity (Fig. 5, PL4A). We observed red-light-inducible and far-red-light-reversible repression for PL4A containing 31 bp of the 93-bp region. The reduction of the expression in PL4A implies an involvement of the 62-bp region in basal expression. The internal deletion of 24 bp (−666 to −643) in the 31-bp region abolished red-light-regulated repression (Fig. 5, PL4B). These results indicated that the 24-bp region from −666 to −643 was responsive to phytochrome down-regulated expression and that the 62-bp region from −734 to −673 affected the basal expression level. The level of PL4B activity was almost the same as that of red-light-treated plants in PL4, and the deletion of 24 bp from PL4 did not decrease the expression of basal level. However, the deletion decreased the expression in darkness (Fig. 5, PL4B). These results suggested that the 24-bp region was involved in enhanced expression in darkness.
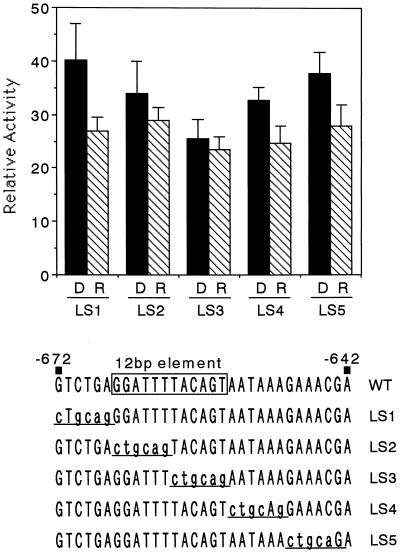
LS Revealed That the 12-bp Element Mediated Phytochrome Down-Regulated Expression
To identify the cis element necessary for phytochrome down-regulation, we conducted LS analysis. We prepared five constructs with 6-bp mutations in the 31-bp region of the PL4A construct (Fig. 5), as shown in Figure 6. The 6-bp mutations correspond to the PstI recognition sequence. We examined the effect of red light on expression using the resulting plasmids. LS2 and LS3 did not show red-light down-regulation (Fig. 6). In particular, the red-light repression was completely abolished in the LS3 construct, suggesting that the core region was located in the mutated region within LS3. The other three constructs, LS1, LS2, and LS5, retained the red-light repression. These results indicated that the phytochrome down-regulation was mediated by the element within the 12-bp region, the sequence of which was GGATTTTACAGT. This element did not show any sequence similarity with the previously reported elements involved in phytochrome- or light-regulated expression.
Figure 6.
LS analysis to define the 12-bp phytochrome down-regulated element. LS constructs in the pra2 5′-upstream region are shown. The positions of mutated nucleotides are indicated in lowercase and underlined. The WT sequence corresponds to the PL4A construct in Figure 5. The 12-bp phytochrome-responsive element is boxed. Equivalent amounts of each plasmid were introduced into 5- or 6-d-old etiolated seedlings as described. After bombardment, the plants were exposed to red light for 2 min (R) and then returned to darkness for 12 h. D, Dark control. Values are the means of at least five independently bombarded samples, with error bars representing se. Relative activity is defined in Figure 2b.
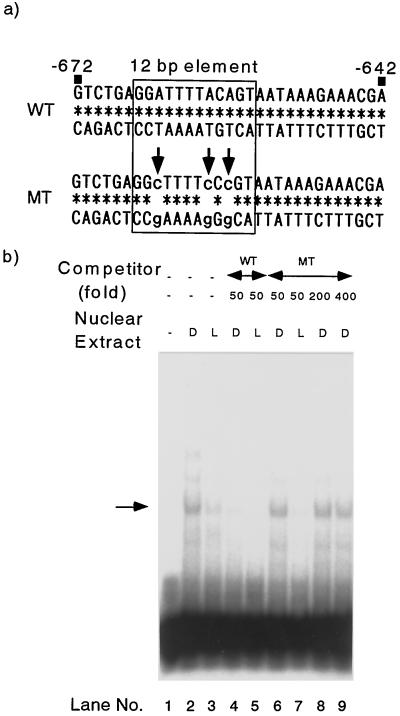
A Nuclear Factor Specifically Bound to the 12-bp Element in Vitro
To examine the presence of nuclear factors such as DNA-binding proteins bound to the 12-bp element specifically, we conducted gel-retardation assays using nuclear extracts from pea epicotyls. As a probe, we prepared a 31-bp synthetic oligonucleotide containing the sequence from −672 to −642 (WT; Fig. 7a). Addition of the nuclear extracts from both etiolated and light-irradiated (6 h) pea plants to the binding reaction mixture showed the retarded band of the DNA-protein complex (Fig. 7b, lanes 2 and 3). The signal of etiolated plants was stronger than that of light-irradiated plants (lanes 2 and 3), suggesting that the amount of this nuclear factor was higher in the dark-grown epicotyls than in the light-irradiated plants. This dark-light difference was observed in three independent extracts. To test whether the observed binding was specific to the 12-bp element, we prepared an oligonucleotide in which the adenines in the 12-bp element were changed to cytosines (MT; Fig. 7a). The band was diminished by 50-fold WT competitor (lanes 4 and 5) but not by MT competitor (lanes 6 and 7). MT competitors in 200- or 400-fold excess could not compete with labeled probe (lanes 8 and 9). These data indicated that the observed band was a complex of probe DNA and nuclear factor specifically bound to the 12-bp element.
Figure 7.
Gel-mobility shift assays with the synthetic oligonucleotides. a, Sequences of synthetic oligonucleotides used in this experiment are shown. The 12-bp sequence is boxed. Nucleotides in the MT oligonucleotide that are different from the WT oligonucleotide are indicated in lowercase with arrows. b, Binding of nuclear proteins from pea stems to the WT oligonucleotide and competition with the WT and MT oligonucleotides. The 12-bp-specific bound complex is indicated by the arrow. D and L indicate the nuclear extracts of dark-grown plants and plants exposed to light for 6 h, respectively.
DISCUSSION
We present evidence that the pra2 upstream fragment of 2325 bp from the initiation codon can direct a reporter gene to be expressed at a higher level in darkness than in light in the growing zone of etiolated pea stem. In developing a transient assay system, we found that cutting the stem drastically decreased the pra2 protein level and reporter-gene expression. The expression of the reporter gene by our transient assay was analogous to that of the pra2 gene itself (Yoshida et al., 1993; Nagano et al., 1995). This suggests that the experimental system is a reliable way to examine pra2 expression. By 5′-deletion analysis of the pra2 upstream region (Fig. 3), we showed that 93 bp from −734 to −642 could drive light down-regulated expression of the LUC gene. The 93-bp region conferred the down-regulation to a heterologous CaMV 35S90 promoter (Fig. 4). The 93-bp region was sufficient to confer phytochrome down-regulation of the reporter gene when combined with its own TATA box and 5′-untranslated region (Fig. 5). Furthermore, we discovered that a 12-bp element within the 93-bp region mediated phytochrome down-regulated expression (Fig. 6). We also discovered that nuclear factor bound to this element in a light-dependent manner (Fig. 7).
To our knowledge, 5′ regions of the genes down-regulated by phytochrome have been characterized for PHYA (Bruce et al., 1991), NPR (Okubara et al., 1993; Williams et al., 1994), AS1 (Nagi et al., 1997; Neuhaus et al., 1997), and TubB1 (Tonoike et al., 1994). The elements containing the TGGG sequence were shown to be involved in down-regulation by phytochrome for PHYA (Bruce et al., 1991) and AS1 (Neuhaus et al., 1997). However, in the case of NPR1, two elements that were necessary for both phytochrome- and ABA-mediated expression of this gene were identified, and these elements did not contain the TGGG sequence (Weatherwax et al., 1998). Rather, phytochrome regulated ABA levels, and then ABA affected the NPR1 expression. Thus, the mechanism of down-regulation of this gene is quite different from that of other genes such as PHYA and AS1. In the pra2 gene the 12-bp GGATTTTACAGT element mediates down-regulation by phytochrome. This element is similar to the −300 element that is involved in endosperm-specific expression of the high-Mr glutenin gene (Colot et al., 1987; Thomas and Flavell, 1990), but the 12-bp element is functionally different from the −300 element. Moreover, the putative core sequence of the 12-bp element TACAGT (LS3 mutated position) does not contain the same motif reported previously. The mechanism of phytochrome down-regulation of the pra2 gene is probably different from that of other genes characterized previously.
It is well known that the promoter context affects the light response when cis elements are fused to heterologous promoters (Puente et al., 1996). In the CaMV 35S90 promoter, an activator sequence, as-1, affects light response interacting with a fused fragment (Terzaghi and Cashmore, 1995). The data in Figure 4 suggest that the 93-bp region could confer light down-regulation to a 35S90 promoter. The sequence from −101 to −25 of the 5′-upstream region of pra2 did not confer the light response to the 35S90 promoter. These results suggest that the interaction between the regions from −101 to −25 and as-1 suppressed the light down-regulation.
The binding manner of nuclear protein to the 12-bp element also showed an interesting feature. Apparently, the amount of protein bound to this element decreased upon light irradiation (Fig. 7b), suggesting an involvement of this protein in light down-regulation. Here we propose one simple hypothesis: The protein bound to the 12-bp element enhances transcription of pra2 in darkness, and its dissociation from the 12-bp region upon light irradiation results in repression of the transcription. In the case of PHYA, the putative trans factor RF1 is thought to bind RE1 when irradiated and regulate gene repression. The 12-bp binding factor may be a new type of DNA-binding protein involved in down-regulation by phytochrome. Molecular cloning and characterization of this trans factor may reveal the mechanism of phytochrome signal transduction.
We also demonstrated that the pra2 5′-upstream region was involved in the growing-zone-specific expression of the reporter gene in etiolated pea stem. To our knowledge, expression of the β-tubulin TUB1 (or soybean tubB1) gene is similar to that of pra2. These two genes are specifically expressed in etiolated stems (Han et al., 1991; Nagano et al., 1995). Previously, Tonoike et al. (1994) demonstrated that a 2-kb fragment 5′ upstream of the tubB1 gene was sufficient to direct hypocotyl expression and light down-regulation. Both phytochrome A and phytochrome B regulate the TUB1 gene. However, TUB1 expression is reduced by red light but is not reversed by far-red light (Leu et al., 1995). Stem elongation is inhibited by a brief red-light irradiation, and this effect was reversed by subsequent exposure to far-red light. The pra2 gene possibly plays an important role in the morphogenesis of etiolated seedlings and the inhibition of stem elongation by light.
ACKNOWLEDGMENTS
We thank Drs. H. Mori, K. Yoshida, K. Nakamura, K. Maeo, and S. Shimizu for their technical advice. We also thank Dr. A.T. Jagendorf for discussion. The pBI221-LUC+ plasmid was a kind gift of Dr. Kazuyuki Hiratsuka.
Abbreviations:
- CaMV
cauliflower mosaic virus
- LS
linker-scanning
- LUC
luciferase
Footnotes
This work was supported by grants from the Japanese Ministry of Education, Science, Sports and Culture and from the Japan Society for the Promotion of Science (Research for the Future Program, no. JSPS-RTFT.96L006012). T.I. received Research Fellowships from the Japan Society for the Promotion of Science for Young Scientists.
The accession number for the sequence reported in this article is AB007911.
LITERATURE CITED
- Bruce WB, Deng X-W, Quail PH. A negatively acting DNA sequence element mediates phytochrome-directed repression of phyA gene transcription. EMBO J. 1991;10:3015–3024. doi: 10.1002/j.1460-2075.1991.tb07852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau TE, Wessler SR. Stowaway: a new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Plant Cell. 1994;6:907–916. doi: 10.1105/tpc.6.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli M, Morelli G, Whitelam G, Ruberti I. Twilight-zone and canopy shade induction of the Athb-2 homeobox gene in green plants. Proc Natl Acad Sci USA. 1996;93:3530–3535. doi: 10.1073/pnas.93.8.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot V, Robert LS, Kavanagh TA, Bevan MW, Thompson RD. Location of sequences in wheat endosperm protein genes which confer tissue-specific expression in tobacco. EMBO J. 1987;6:3559–3564. doi: 10.1002/j.1460-2075.1987.tb02685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster C, Knox M, Domoney C, Casey R. lox1:Ps:2, a Pisum sativum seed lipoxygenase gene. Plant Physiol. 1994;106:1227–1228. doi: 10.1104/pp.106.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han I-S, Jongewaard I, Fosket DE. Limited expression of a diverged β-tubulin gene during soybean (Glycine max [L.] Merr.) development. Plant Mol Biol. 1991;16:225–234. doi: 10.1007/BF00020554. [DOI] [PubMed] [Google Scholar]
- Ishiguro S, Nakamura K. The nuclear factor SP8BF binds to the 5′-upstream regions of three different genes coding for major proteins of sweet potato tuberous roots. Plant Mol Biol. 1992;18:997–1008. doi: 10.1007/BF00018460. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanaugh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe DM, Degenhardt J, Winicov I, Tobin EM. Two 10-bp regions are critical for phytochrome regulation of a Lemna gibba Lhcb gene promoter. Plant Cell. 1994;6:1123–1134. doi: 10.1105/tpc.6.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y, Nakajima K, Arai Y. An improved assay for β-glucuronidase in transformed cells: methanol almost completely suppresses a putative endogenous β-glucuronidase activity. Plant Sci. 1990;70:133–140. [Google Scholar]
- Leu W-M, Cao X-L, Wilson TJ, Snustad DP, Chua N-H. Phytochrome A and phytochrome B mediate the hypocotyl-specific downregulation of TUB1 by light in Arabidopsis. Plant Cell. 1995;7:2187–2196. doi: 10.1105/tpc.7.12.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AJ, Short SR, Hiratsuka K, Chua N-H, Kay SA. Firefly luciferase as a reporter of regulated gene expression in higher plants. Plant Mol Biol Rep. 1992;10:324–337. [Google Scholar]
- Nagano Y, Matsuno R, Sasaki Y. Sequence and transcriptional analysis of the gene cluster trnQ-zfpA-psaI-ORF231-petA in pea chloroplasts. Curr Genet. 1991;20:431–436. doi: 10.1007/BF00317074. [DOI] [PubMed] [Google Scholar]
- Nagano Y, Murai N, Matsuno R, Sasaki Y. Isolation and characterization of cDNAs that encode eleven small GTP-binding proteins from Pisum sativum. Plant Cell Physiol. 1993;34:447–455. [PubMed] [Google Scholar]
- Nagano Y, Okada Y, Narita H, Asaka Y, Sasaki Y. Location of light-repressible, small GTP-binding protein of the YPT/rab family in the growing zone of etiolated pea stems. Proc Natl Acad Sci USA. 1995;92:6314–6318. doi: 10.1073/pnas.92.14.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagi N, Tsai F-Y, Coruzzi G. Light-induced transcriptional repression of the pea AS1 gene: identification of cis-elements and trans-factors. Plant J. 1997;12:1021–1034. doi: 10.1046/j.1365-313x.1997.12051021.x. [DOI] [PubMed] [Google Scholar]
- Neuhaus G, Bowler C, Hiratsuka K, Yamagata H, Chua N-H. Phytochrome-regulated repression of gene expression requires calcium and cGMP. EMBO J. 1997;16:2254–2264. doi: 10.1093/emboj/16.10.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubara PA, Williams SA, Doxsee RA, Tobin EM. Analysis of genes negatively regulated by phytochrome action in Lemna gibba and identification of a promoter region required for phytochrome responsiveness. Plant Physiol. 1993;101:915–924. doi: 10.1104/pp.101.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente P, Wei N, Deng X-W. Combinatorial interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J. 1996;15:3732–3743. [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Sakihama T, Kamikubo T, Shinozaki K. Phytochrome-mediated regulation of two mRNAs, encoded by nuclei and chloroplasts of ribulose-1,5-bisphosphate carboxylase/oxygenase. Eur J Biochem. 1983;133:617–620. doi: 10.1111/j.1432-1033.1983.tb07507.x. [DOI] [PubMed] [Google Scholar]
- Shimizu S, Itoh Y, Yamazaki K. Temperature-dependent increase in the DNA-binding activity of a heat shock factor in an extract of tobacco cultured cells. Plant Mol Biol. 1996;31:13–22. doi: 10.1007/BF00020602. [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Dotson M, Keen NT. Plant transformation: a simple bombardment device based on flowing helium. Plant Mol Biol. 1992;18:835–839. doi: 10.1007/BF00020031. [DOI] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–474. [Google Scholar]
- Thomas MS, Flavell RB. Identification of an enhancer element for the endosperm-specific expression of high molecular weight glutenin. Plant Cell. 1990;2:1171–1180. doi: 10.1105/tpc.2.12.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonoike H, Han I-S, Jongewaard I, Doyle M, Guiltinan M, Fosket DE. Hypocotyl expression and light downregulation of the soybean tubulin gene, tubB1. Plant J. 1994;5:343–351. doi: 10.1111/j.1365-313x.1994.00343.x. [DOI] [PubMed] [Google Scholar]
- Weatherwax SC, Williams SA, Tingay S, Tobin EM. The phytochrome response of the Lemna gibba NPR1 gene is mediated primarily through changes in abscisic acid levels. Plant Physiol. 1998;116:1299–1305. doi: 10.1104/pp.116.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SA, Weatherwax SC, Bray EA, Tobin EM. NPR genes, which are negatively regulated by phytochrome action in Lemna gibba L. G-3, can also be positively regulated by abscisic acid. Plant Physiol. 1994;105:949–954. doi: 10.1104/pp.105.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Nagano Y, Murai N, Sasaki Y. Phytochrome-regulated expression of the genes encoding the small GTP-binding proteins in peas. Proc Natl Acad Sci USA. 1993;90:6636–6640. doi: 10.1073/pnas.90.14.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]