Abstract.
The gram-negative, pleomorphic endosymbiont Wolbachia is known to infect a large number of insects and other arthropods naturally. This bacterium modifies the host biology, mainly causing reproductive alterations including feminization, death of male, parthenogenesis, and importantly cytoplasmic incompatibility. Wolbachia-induced cytoplasmic incompatibility results in nonviable offspring and vector population suppression. In addition, this bacterium rapidly spreads and propagates within the host population. This study is the first report on Wolbachia detection and characterization from Culex quinquefasciatus collected from Lahore, Pakistan. For this purpose, mosquito adults were collected from different localities of Lahore and identified at the species level. A total of 145 pairs of ovaries were individually subjected to DNA isolation, and polymerase chain reaction amplification of three (wsp, 16S rRNA, and ftsZ) genes were investigated. In all, 128 females were found positive, representing 82.3% infection rate. The phylogenetic analysis indicated that the detected endosymbiont had 100% homology with Wolbachia pipientis wPip strain and supergroup B. The detection of the local strain of Wolbachia (wPip) will be useful in investigating its potential for the control of dengue vector (Aedes aegypti) and reducing dengue transmission in Pakistan.
INTRODUCTION
Wolbachia is a gram-negative endosymbiotic bacterium, and it belongs to the phylum α-proteobacteria, order Rickettsiales,1 and family Anaplasmataceae.2 Wolbachia was first detected as Rickettsia-like organisms from the common house mosquito Culex pipiens.3 Since then, Wolbachia has been reported in a large number of arthropods and filarial nematodes. It is abundantly found in more than 65% of insects in nature.4,5 In Cx. pipiens, 76–98% of total microbiota comprises Wolbachia.6
Wolbachia is considered as a reproductive parasite and causes various reproductive modifications in its hosts including parthenogenesis, feminization of males, death of males, and cytoplasmic incompatibility.7 Cytoplasmic incompatibility is one of the most important characteristics of Wolbachia that results in reduction of the viable progeny. Cytoplasmic incompatibility can be either unidirectional in which Wolbachia-infected sperm fertilizes uninfected egg,8 or bidirectional where sperm and egg both are infected but with different strains of Wolbachia, resulting in failure of egg hatching and leading to suppression of host population.9
There is a large variety of Wolbachia strains present in arthropods. Multiple Wolbachia strains and various changes in Wolbachia sequences within the same host have been reported.10 A huge variety of Wolbachia strains have been reported in the recent decades. Wolbachia has incorporated a wide variety of branches in phylogenetic hierarchy, and 14 supergroups have been described so far.11 Supergroups A and B, being the most abundant, are found in insects.9 Molecular approach targeting different genes for detection and identification of Wolbachia is considered as a reliable method. The most widely used detection method is carried out by amplifying specific regions of the Wolbachia surface protein (wsp) gene by polymerase chain reaction (PCR). Other different molecular markers include the 16S rRNA gene, dnaA gene which is crucial for the initiation of DNA replication process, the ftsZ (filamenting temperature sensitive) protein-encoding gene, the groE operon encoding two important heat shock proteins of bacteria, hcpA, gltA, fbpA, gatB, and coxA.12
Many classified strains of Wolbachia (wMelPop and wMelPop-CLA) are known to inhibit transmission of parasites or pathogens through infected mosquito vectors and to decrease the rate of disease transmission.13,14 It has been suggested that introduction of specific Wolbachia strains in mosquito vectors and release of these infected vectors into a local population of vectors could prevent the transmission of arboviruses and other human parasites or pathogens such as dengue viruses and Plasmodium.15,16 The use of Wolbachia as a biological tool for mosquito vector control, detection, and identification of its strains in local insects and other organisms is of prime importance. It is being well documented that Wolbachia is abundantly found in reproductive tissues of various insects. Objectives of the current study are the molecular detection of Wolbachia in the reproductive tissues of Culex mosquitoes collected from various sites of Lahore and the assessment of the phylogenetic status of detected Wolbachia on the basis of wsp, 16S rRNA, and ftsZ genes.
METHODS
Culex quinquefasciatus collection and identification.
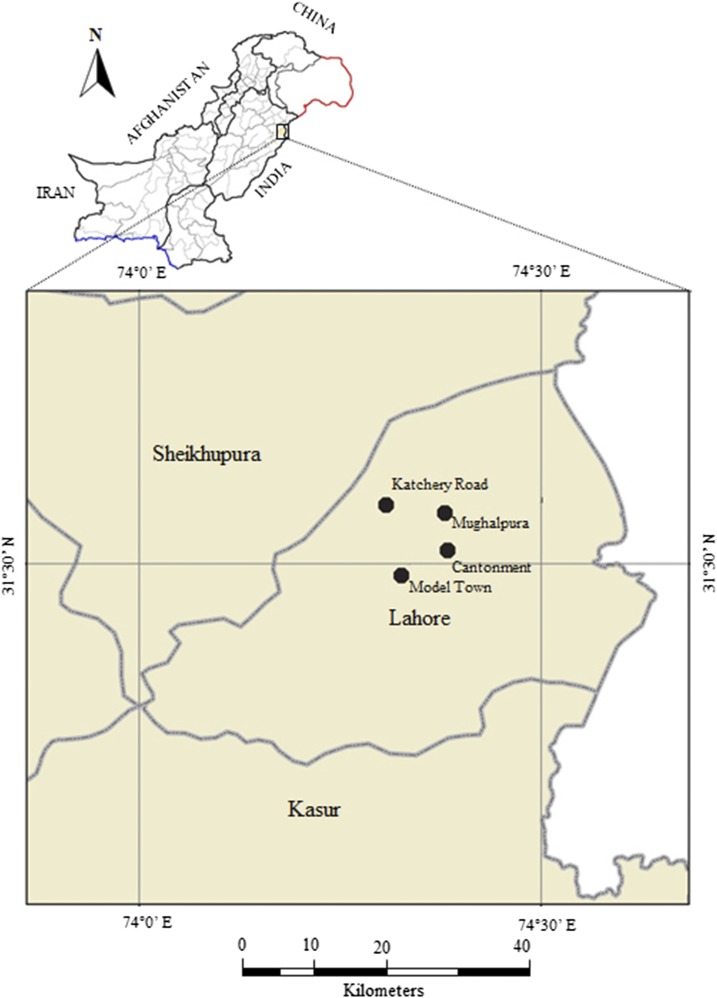
Surveillance of Cx. quinquefasciatus mosquitoes was conducted for a period of 4 months from July to October 2014. Adults were collected from four different (indoor and outdoor) locations (Cantonment, Katchery Road, Model Town, and Mughalpura) of Lahore, Pakistan (Figure 1), using a backpack aspirator (John W. Hock Company, Gainesville, FL). Geographical coordinates were collected using GPS Garmin GPSMAP®; 76CSx. The collected mosquito samples were reared in the insectary, GC University Lahore, Pakistan, under standard conditions (temperature: 27 ± 2°C, relative humidity: 80% ± 5%, and light dark cycle: 12:12 hours). The identification of adult Culex mosquitoes was carried out up to the species level using standard morphological keys.17,18
Figure 1.
Map of Lahore, Punjab, Pakistan showing sampling sites of Culex quinquefasciatus for the screening of Wolbachia. This figure appears in color at www.ajtmh.org.
Dissections and separation of ovaries.
The females were isolated from males by examining mouth parts, antennae, and the last portion of the abdomen. The selected females were used in the screening process and were paralyzed at 4°C for 5–10 minutes, using the freeze shock method. The mosquitoes were dipped 3× in sterile water, followed by rinsing in 70% ethanol for 5 minutes, and then transferred to successive sterile water baths 5× and once in sterilized 0.8% NaCl.19 Each individual female was transferred to a clean glass slide containing a drop of phosphate buffer saline (PBS). The ovaries were dissected with the help of sterile dissecting needles under dissecting microscope. A single pair of ovaries was considered as one sample.
PCR amplification of target genes.
Whole genomic DNA was extracted from each sample as described previously.20 Exponential amplification of the extracted DNA (template) was carried out with a PCR thermal cycler (Techne Progene, Cambridge, United Kingdom) in a final volume of 50 µL containing 1× Taq buffer (without MgCl2), 1.5 mM MgCl2, 0.2 mM dNTPs, 0.4 mM each primer, 1 U Taq polymerase, and 2 µL whole genomic DNA. Genomic DNA from Aedes aegypti and Aedes albopictus was used as negative and positive control, respectively. The reagents were mixed in 0.5-mL PCR tubes placed on ice in a safety cabinet. The primer sequences targeting three genes (wsp, 16S rRNA, and ftsZ) along with the respective annealing temperature used in PCR are mentioned in Table 1. The following thermal profiles were used: initial denaturation at 94°C for 2 minutes followed by 35 cycles (denaturation at 94°C for 1 minute, annealing optimized at 51°C for 16S rRNA and wsp, and 55°C for ftsZ for 1 minute, and extension at 72°C for 1 minute), and final extension at 72°C for 10 minutes.
Table 1.
The details of gene primers used for the detection and characterization of Wolbachia from Cx. quinquefasciatus collected from Lahore Pakistan
| Primers pair 5′-3′ | Annealing | Target gene | Estimated product size | GenBank accession number |
|---|---|---|---|---|
| wsp_F | 55°C | wsp | 510 bp | KX650071 |
| AAGGAACCGAAGTTCATG | ||||
| wsp_R | ||||
| AAAAATTAAACGCTACTCCA | ||||
| 16S rRNA_F | 51°C | 16S rRNA | 890 bp | KX611381 |
| TTGTAGCCTGCTATGGTATAACT | ||||
| 16S rRNA _R | ||||
| GAATAGGTATGATTTTCATGT | ||||
| ftsZ_F | 55°C | ftsZ | 570 bp | KY753917 |
| TACTGACTGTTGGAGTTGTAACTAAGCCGT | ||||
| ftsZ_R | ||||
| TGCCAGTTGCAAGAACAGAAACTCTAACTC |
Agarose gel electrophoresis.
The PCR products were subjected to 2% agarose gel electrophoresis using 0.5 μg/mL ethidium bromide and 1× TAE buffer. Four microliters of PCR products along with 2 μL of 3× loading dye (bromophenol blue) was loaded with 1-kb ladder as a standard to determine the size of the PCR products. The amplicons were visualized and photographed by using a UV transilluminator and BioDoc-It® TS imaging system (Ultra-Violet Products Limited, Cambridge, United Kingdom). The DNA samples were stored at −20°C until DNA sequencing.
Sequencing of amplicons and data analysis.
The selected amplified PCR products (two samples from each locality) were purified using gel extraction method followed by DNA sequencing from First BASE Laboratories Sdn Bhd, Kembangan, Malaysia. Briefly, the PCR products underwent Sanger sequencing in both directions using their respective (forward and reverse) primers using a DNA sequencer (96-capillary 3730xl DNA Analyzer; Applied Biosystems, Foster City, CA) and an ABI BigDye Terminator v3.1 cycle sequencing kit.
The quality of the obtained results (chromatogram) was observed using Chromas Lite software (version 2.5.1), and then FASTA format of the forward and reverse partial gene sequences was aligned using the MEGA6 software. Ambiguous sequences were omitted from the results. The partial DNA sequences of selected gene were subjected to BLAST search against the National Center for Biotechnology Information (NCBI) public sequence database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The nucleotide sequences generated in the present study were deposited in GenBank, and accession numbers were obtained. The phylogenetic tree of each strain was constructed with other taxa of related Wolbachia strains using the Maximum Likelihood method rooted on the Tamura-Nei model with the help of MEGA6 software. Maximum composite likelihood approach was used, and all possible codon frames were considered in the analysis after eliminating gaps.
RESULTS
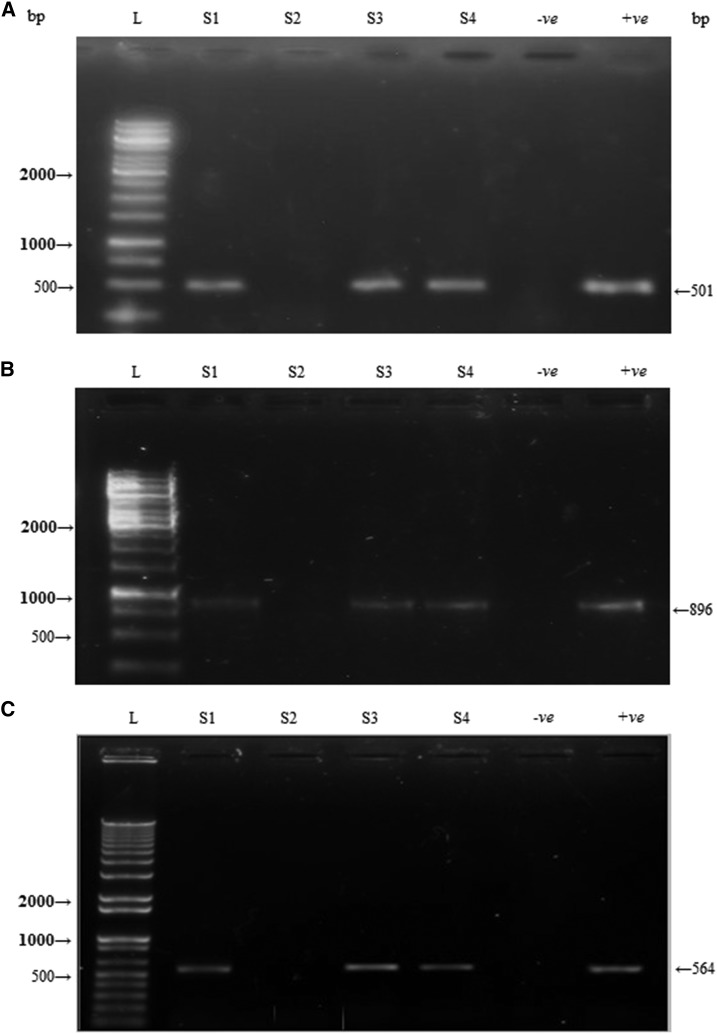
The present study involves detection and characterization of Wolbachia from local Cx. quinquefasciatus adults collected from the selected localities of Lahore, Pakistan, using the PCR method. Wolbachia infection was calculated as 82.3% (128/145) in collected Cx. quinquefasciatus females using Wolbachia-specific wsp gene primers in PCR. Partially amplified DNA fragments of wsp, 16S rRNA, and ftsZ genes revealed 510, 890, and 570 bp bands of selected four samples on agarose gel (Figure 2). DNA sequencing results of all genes revealed the presence of a single strain of Wolbachia in Culex mosquitoes collected from selected localities.
Figure 2.
Gel electrophoresis analysis of polymerase chain reaction products using wsp (A), 16S rRNA (B), and ftsZ (C) gene primers targeting genomic DNA of Culex quinquefasciatus collected from different areas of Punjab, Pakistan.
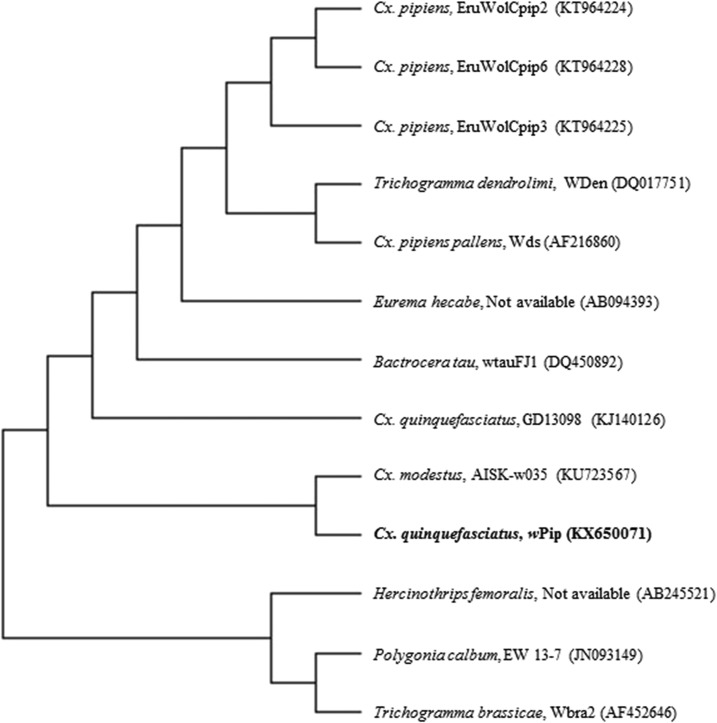
Wsp gene sequence was submitted to GenBank with Accession no. KX650071 and its NCBI BLAST results (Table 2) showed 100% homology (Max score 926) with Wolbachia endosymbiont of Cx. quinquefasciatus (Accession no. KJ140126), Culex modestus from China (Accession no. KU723567), and Cx. pipiens from Turkey (Accession no. KT964225). The wsp-gene based phylogenetic tree with the highest log likelihood (−4,762.2634) is given in Figure 3. The final dataset involved 13 nucleotide sequences with 501 positions. The evolutionary tree inferred closeness with Cx. modestus (Accession no. KJ140126) Cx. quinquefasciatus, followed by EruWolCpip2 of Cx. pipiens (Accession no. KT964225).
Table 2.
Details of arthropods having identical gene sequences with wsp partial cds (Accession KX650071) of wPip Wolbachia in Cx. quinquefasciatus collected from Lahore Pakistan
| Sr. No. | Strain/isolate | Host | Order, family | Country/origin | Total score | Query coverage (%) | E value | Ident (%) | Accession |
|---|---|---|---|---|---|---|---|---|---|
| 1 | GD13098 | Culex quinquefasciatus | Diptera, Culicidae | China | 926 | 100 | 0.0 | 100 | KJ140126 |
| 2 | AlSK-w035 | Culex modestus | Diptera, Culicidae | China | 926 | 100 | 0.0 | 100 | KU723567 |
| 3 | EruWolCpip3 | Culex pipiens | Diptera, Culicidae | Turkey | 926 | 100 | 0.0 | 100 | KT964225 |
| 4 | wTde-HEB | Trichogramma dendrolimi | Hymenoptera, Trichogrammatidae | China | 926 | 100 | 0.0 | 100 | JX027991 |
| 5 | EW 13-7 | Polygonia calbum | Lepidoptera, Nymphalidae | Not Available | 926 | 100 | 0.0 | 100 | JN093149 |
| 6 | Not available | Hercinothrips femoralis | Thysanoptera, Thripidae | Japan | 926 | 100 | 0.0 | 100 | AB245521 |
| 7 | Wbra2 | Trichogramma brassicae | Hymenoptera, Trichogrammatidae | China | 926 | 100 | 0.0 | 100 | AF452646 |
| 8 | WDen Qiqihae | T. dendrolimi | Hymenoptera, Trichogrammatidae | China | 926 | 100 | 0.0 | 100 | DQ017751 |
| 9 | WHecCI1 | Eurema hecabe | Lepidoptera, Pieridae | Japan | 926 | 100 | 0.0 | 100 | AB094393 |
| 10 | WBra | Mamestra brassicae | Lepidoptera, Noctuidae | Japan | 926 | 100 | 0.0 | 100 | AB094375 |
| 11 | Wds | Cx. pipiens pallens | Diptera, Culicidae | China | 926 | 100 | 0.0 | 100 | AF216860 |
| 12 | EruWolCpip2 | Cx. pipiens | Diptera, Culicidae | Turkey | 924 | 99 | 0.0 | 100 | KT964224 |
| 13 | wPip | Cx. quinquefasciatus | Diptera, Culicidae | Not available | 922 | 99 | 0.0 | 100 | AM999887 |
| 14 | EruWolCpip6 | Cx. pipiens | Diptera, Culicidae | Turkey | 920 | 100 | 0.0 | 99 | KT964228 |
| 15 | wtauFJ1 | Bactrocera tau | Diptera, Tephritidae | China | 920 | 100 | 0.0 | 99 | DQ450892 |
| 16 | WDen | E. hecabe | Lepidoptera, Pieridae | China | 920 | 100 | 0.0 | 99 | DQ017750 |
Figure 3.
wsp gene-based phylogenetic tree of Wolbachia pipientis wPip strain found in Culex quinquefasciatus using maximum likelihood method (Accession no. KX650071).
The 16S rRNA gene sequence of Wolbachia pipientis was submitted to GenBank as Accession no. KX611381. NCBI BLAST results of the obtained 16S rRNA sequence showed 99% homology with the wPip strain of Wolbachia endosymbiont found in Cx. quinquefasciatus Pel, indicating that the strain obtained in this study belongs to subgroup wPip. The evolutionary tree was traced on 13 nucleotide sequences with 895 positions in the final dataset including the highest log likelihood (−7,883.4825). The phylogenetic tree shows that the query sequence is closely related to Wolbachia of Ectropis grisescens (Lepidoptera, Geometridae) (Accession no. KT862198) and wPip strain of Cx. quinquefasciatus (Accession no. AM999887) (Figure 4).
Figure 4.
16Sr RNA gene-based phylogenetic tree of Wolbachia pipientis wPip strain found in Culex quinquefasciatus using maximum likelihood method (Accession no. KX611381).
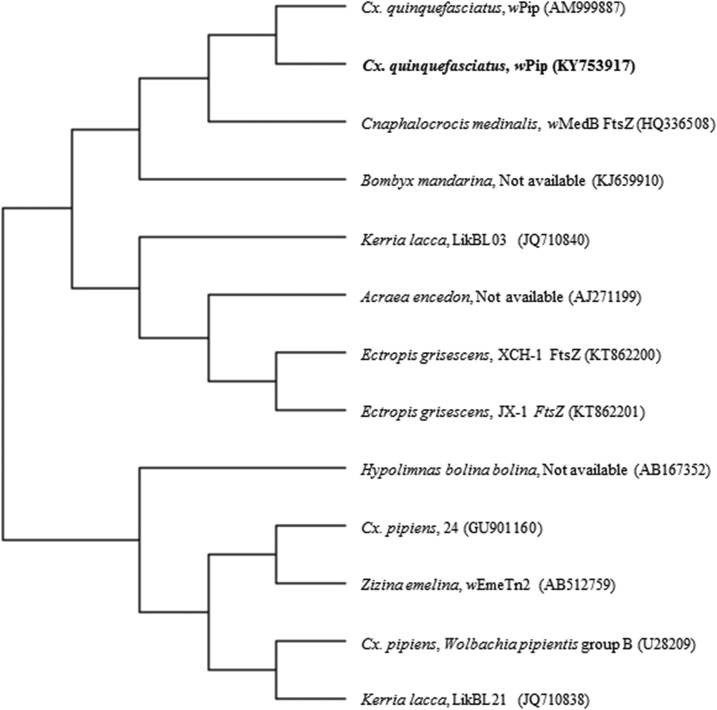
DNA sequence of the ftsZ gene was submitted to NCBI GenBank under Accession no. KY753917. NCBI BLAST results indicated 98% homology with Wolbachia of Cx. pipiens from Argentina (Accession no. GU901160) and wPip Wolbachia of Cx. quinquefasciatus (Accession no. AM99887) reported from Australia. The tree was constructed with 13 nucleotide sequences with the highest log likelihood (−6,983.9439) including 570 positions. The phylogenetic tree indicated a close relationship of the obtained ftsZ gene sequence to the supergroup B wPip isolate of Cx. quinquefasciatus (Accession no. AM99887) (Figure 5).
Figure 5.
ftsZ gene-based phylogenetic tree of Wolbachia pipientis wPip strain found in Culex quinquefasciatus using maximum likelihood method (Accession no. KY753917).
DISCUSSION
Wolbachia has received increased attention in recent years because of its high rates of distribution in several insects and its unique effects on host physiology. They are the most widely occurring intracellular bacteria in arthropods found in major insect orders, including Orthoptera, Diptera, Coleoptera, Hemiptera, Lepidoptera, and Hymenoptera.21 These obligate parasites are also found naturally in some of the medically important insects including mosquitoes such as Culex and Ae. albopictus, which are important vectors for the transmission of viruses such as West Nile Virus and Dengue viruses etc. respectively. The detection of Wolbachia strain in any given organism is done either by using staining methods3,22 or by amplification of Wolbachia-specific genes including 16S rRNA, wsp and ftsZ genes, etc.23 The molecular approach, including gene amplification and characterization, is more significant to find out the phylogenetic status of Wolbachia.7 The current study deals with the use of 16S rRNA and wsp and ftsZ genes for the detection and molecular characterization of Wolbachia isolates from Cx. quinquefasciatus collected from the different regions of Lahore, Pakistan.
In this study, 82.3% Wolbachia infection was found in Cx. quinquefasciatus female mosquitoes. Various Wolbachia infection levels have been reported in different regions of the world. In a study from Iran,24 all of the Cx. quinquefasciatus tested were found to be positive whereas other species including Culex tritaeniorynchus and Culex theileri were found to be negative for Wolbachia. It has also been reported previously that Wolbachia infection frequency in Cx. quinquefasciatus from three villages of South India was 91.2%.25 In addition, Kittayapong et al.26 conducted a survey of Wolbachia infection in mosquitoes from Thailand and found 42.1% Wolbachia infection in 29 species of the genus Culex by targeting ftsZ and wsp genes. The authors did not find any Wolbachia infection in Anopheles mosquitoes.
In another report, 14 mosquito species related to five genera (Aedes, Culex, Anopheles, Ochlerotatus, and Culiesta) from North America were tested for the presence of Wolbachia. Only species of the Cx. pipiens complex were found to be positive.27 Ricci et al.28 investigated 26 mosquito species out of which five (three Culex and two Aedes species) carried Wolbachia infection. Karami et al.29 screened Wolbachia in Cx. pipiens collected from three provinces of Iran and found 87.3% infection. In a report of Wolbachia infection in the Cx. pipiens complex of mosquitoes from Germany and the Philippines, Wolbachia infection was confirmed in all of the Culex mosquitoes tested based on the of 16S rRNA gene of Wolbachia.30
A number of valuable molecular methods have provided the basis for phylogenetic studies. General Wolbachia primers and Wolbachia supergroup A- and B-specific primers may be used for the wsp gene.
Characterization of the detected strain of Wolbachia in the current study showed that it is related to supergroup B and subgroup (strain) wPip. The current results are consistent with some previously reported studies in different regions of the world. In India, Patole and Shouche12 found Wolbachia in Cx. quinquefasciatus which was related to the wPip strain and supergroup B. In another study, it was also reported that three tested Culex species (Culex torrentium, Cx. pipiens, and Cx. modestus) fall in supergroup B.28 Recently Karami et al.29 also placed a type strain from Cx. pipiens within supergroup B. Furthermore, Kittayapong et al.26 reported Wolbachia in three Culex species (Cx. quinquefasciatus, Culex fuscocephala, and Culex sitiens) collected from the Southeast Asian region belonging to supergroup B and wPip strain. Similar findings were also reported by Yildirim et al.5 who studied Cx. pipiens.
The current study reports the detection and characterization of Wolbachia in Cx. quinquefasciatus for the first time in Pakistan. The information obtained so far about Wolbachia abundance in different arthropods found in Pakistan is inadequate. Therefore, there is dire need to determine the distribution and type of Wolbachia infections in insects. Moreover, the potential of Wolbachia strain may be evaluated as biological control agent to combat vector-borne diseases (dengue and malaria) in endemic areas of Pakistan.
Acknowledgments:
We are thankful to the Office of Research, Innovation and Commercialization (ORIC), GC University, Lahore, for funding this research. We are also thankful to Mr. Adnan Saleem for his cooperation in the fieldwork. The American Society of Tropical Medicine and Hygiene (ASTMH) assisted with publication expenses.
REFERENCES
- 1.Taylor MJ, Bandi C, Hoerauf A, 2005. Wolbachia bacterial endosymbionts of filarial nematodes. Adv Parasitol 60: 245–284. [DOI] [PubMed] [Google Scholar]
- 2.Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, Rikihisa Y, Rurangirwa FR, 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol 51: 2145–2165. [DOI] [PubMed] [Google Scholar]
- 3.Hertig M, Wolbach SB, 1924. Studies on Rickettsia-like micro-organisms in insects. J Med Res 44: 329–374. [PMC free article] [PubMed] [Google Scholar]
- 4.Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH, 2008. How many species are infected with Wolbachia?—A statistical analysis of current data. FEMS Microbiol Lett 281: 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yildirim A, Inci A, Duzlu O, Onder Z, Ciloglu A, 2013. Detection and molecular characterization of the Wolbachia endobacteria in the Culex pipiens (Diptera: Culicidae) specimens collected from Kayseri province of Turkey. Ankara Univ Vet Fak 60: 189–194. [Google Scholar]
- 6.Muturi EJ, Kim C-H, Bara J, Bach EM, Siddappaji MH, 2016. Culex pipiens and Culex restuans mosquitoes harbor distinct microbiota dominated by few bacterial taxa. Parasit Vectors 9: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasgon JL, Scott TW, 2004. Impact of population age structure on Wolbachia transgene driver efficacy: ecologically complex factors and release of genetically modified mosquitoes. Insect Biochem Mol Biol 34: 707–713. [DOI] [PubMed] [Google Scholar]
- 8.Veneti Z, Clark ME, Zabalou S, Karr TL, Savakis C, Bourtzis K, 2003. Cytoplasmic incompatibility and sperm cyst infection in different Drosophila-Wolbachia associations. Genetics 164: 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Werren JH, 1997. Wolbachia run amok. Proc Natl Acad Sci USA 94: 11154–11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann AA, Turelli M, 2013. Facilitating Wolbachia introductions into mosquito populations through insecticide-resistance selection. Proc Biol Sci 280: 20130371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glowska E, Dragun-Damian A, Dabert M, Gerth M, 2015. New Wolbachia supergroups detected in quill mites (Acari: Syringophilidae). Infect Genet Evol 30: 140–146. [DOI] [PubMed] [Google Scholar]
- 12.Patole MS, Shouche YS, 2003. Detection and phylogenetic affiliation of Wolbachia sp. from Indian mosquitoes Culex quinquefasciatus and Aedes albopictus. Curr Sci 84: 1136–1139. [Google Scholar]
- 13.Rainey SM, Shah P, Kohl A, Dietrich I, 2014. Understanding the Wolbachia-mediated inhibition of arboviruses in mosquitoes: progress and challenges. J Gen Virol 95: 517–530. [DOI] [PubMed] [Google Scholar]
- 14.Jupatanakul N, Sim S, Dimopoulos G, 2014. The insect microbiome modulates vector competence for arboviruses. Viruses 6: 4294–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordaux R, Michel-Salzat A, Frelon-Raimond M, Rigaud T, Bouchon D, 2004. Evidence for a new feminizing Wolbachia strain in the isopod Armadillidium vulgare: evolutionary implications. Heredity (Edinb) 93: 78–84. [DOI] [PubMed] [Google Scholar]
- 16.Joshi D, McFadden MJ, Bevins D, Zhang F, Xi Z, 2014. Wolbachia strain wAlbB confers both fitness costs and benefit on Anopheles stephensi. Parasit Vectors 7: 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harbach RE, 1988. The mosquitoes of the subgenus Culex in southwestern Asia and Egypt (Diptera: Culicidae). Contrib Am Entomol Inst 24: 1–240. [Google Scholar]
- 18.Reuben R, Tewari S, Hiriyan J, Akiyama J, 1994. Illustrated keys to species of Culex (Culex) associated with Japanese encephalitis in southeast Asia (Diptera: Culicidae). Mosq Syst 26: 75–96. [Google Scholar]
- 19.Zouache K, Voronin D, Tran-Van V, Mousson L, Failloux AB, Mavingui P, 2009. Persistent Wolbachia and cultivable bacteria infection in the reproductive and somatic tissues of the mosquito vector Aedes albopictus. PLoS One 4: e6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarwar MS, Jahan N, Batool F, Kalim B, 2017. Wsp gene based detection and characterization of Wolbachia in indigenous Drosophila. J Bio & Env Sci 10: 1–8. [Google Scholar]
- 21.Werren JH, Baldo L, Clark ME, 2008. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6: 741–751. [DOI] [PubMed] [Google Scholar]
- 22.Muniaraj M, Paramasivan R, Sunish IP, Arunachalam N, Mariappan T, Jerald Leo SV, Dhananjeyan KJ, 2012. Detection of Wolbachia endobacteria in Culex quinquefasciatus by Gimenez staining and confirmation by PCR. J Vector Borne Dis 49: 258–261. [PubMed] [Google Scholar]
- 23.Simoes PM, Mialdea G, Reiss D, Sagot MF, Charlat S, 2011. Wolbachia detection: an assessment of standard PCR protocols. Mol Ecol Resour 11: 567–572. [DOI] [PubMed] [Google Scholar]
- 24.Behbahani A, 2012. Wolbachia infection and mitochondrial DNA comparisons among Culex mosquitoes in south west Iran. Pak J Biol Sci 15: 54–57. [DOI] [PubMed] [Google Scholar]
- 25.Sunish IP, Rajendran R, Paramasivan R, Dhananjeyan KJ, Tyagi BK, 2011. Wolbachia endobacteria in a natural population of Culex quinquefasciatus from filariasis endemic villages of south India and its phylogenetic implication. Trop Biomed 28: 569–576. [PubMed] [Google Scholar]
- 26.Kittayapong P, Baisley KJ, Baimai V, O’Neill SL, 2000. Distribution and diversity of Wolbachia infections in southeast Asian mosquitoes (Diptera: Culicidae). J Med Entomol 37: 340–345. [DOI] [PubMed] [Google Scholar]
- 27.Rasgon JL, Scott TW, 2004. An initial survey for Wolbachia (Rickettsiales: Rickettsiaceae) infections in selected California mosquitoes (Diptera: Culicidae). J Med Entomol 41: 255–257. [DOI] [PubMed] [Google Scholar]
- 28.Ricci I, Cancrini G, Gabrielli S, D’Amelio S, Favi G, 2002. Searching for Wolbachia (Rickettsiales: Rickettsiaceae) in mosquitoes (Diptera: Culicidae): large polymerase chain reaction survey and new identifications. J Med Entomol 39: 562–567. [DOI] [PubMed] [Google Scholar]
- 29.Karami M, Moosa-Kazemi SH, Oshaghi MA, Vatandoost H, Sedaghat MM, Rajabnia R, Hosseini M, Maleki-Ravasan N, Yahyapour Y, Ferdosi-Shahandashti E, 2016. Wolbachia endobacteria in natural populations of Culex pipiens of Iran and its phylogenetic congruence. J Arthropod Borne Dis 10: 349. [PMC free article] [PubMed] [Google Scholar]
- 30.Mahilum MM, Storch V, Becker N, 2003. Molecular and electron microscopic identification of Wolbachia in Culex pipiens complex populations from the Upper Rhine Valley, Germany, and Cebu City, Philippines. J Am Mosq Control Assoc 19: 206–210. [PubMed] [Google Scholar]