Abstract.
In pregnancy-associated malaria, infected erythrocytes accumulate in the placenta. It is unclear if in polyclonal infections this results in distinct peripheral and placental parasite populations. We used long amplicon deep sequencing of Plasmodium falciparum var2csa ID1-DBL2X from 15 matched peripheral and placental samples collected at delivery from a high transmission area to determine genetic homology. Despite substantial sequence variation and detecting 23 haplotypes, the matched pairs mostly contained the same genetic variants, with 11 pairs sharing 100% of their variants, whereas others showed some heterogeneity. Thus, at delivery, peripheral and placental parasites appear to intermix and placental genotypes can be inferred through peripheral sampling.
INTRODUCTION
Malaria is one of the most common preventable causes of poor birth outcomes.1 Pregnancy-associated malaria (PAM), most commonly caused by infection with Plasmodium falciparum,2 is common in endemic regions and often leads to maternal anemia and low birth weight,3 which are important risk factors for neonatal and infant mortality.3,4 In sub-Saharan Africa, it is estimated that one in four mothers are infected with parasites at delivery and/or develop severe anemia as a result of PAM.5 In addition, one in five low birth weight deliveries can be attributed to PAM.3
The pathology of PAM is mediated by the accumulation of infected erythrocytes (IEs) in the placenta via binding of the parasite protein VAR2CSA to placental chondroitin sulfate A (CSA).6,7 Typically, only erythrocytes infected with mature asexual P. falciparum forms (schizonts and trophozoites) are observed in the placenta, whereas immature rings are most commonly found in circulating blood.8 There is some evidence to suggest that erythrocytes infected with ring stages may also be able to adhere to the placenta.9–11
VAR2CSA is a large, highly variable protein12,13 and expressed only when the infected host is pregnant.7 In areas of higher transmission, primigravid women are most susceptible to PAM and risk decreases with gravidity.14,15 Multigravid women have a higher prevalence and titer of anti-VAR2CSA antibodies16, and high levels of VAR2CSA-specific IgG are associated with a decreased risk of delivering a low-birth weight baby,7 indicating that a strong immune response targets VAR2CSA. The minimum binding region of VAR2CSA to CSA is the 54 kDa ID1-DBL2X epitope, part of the extracellular region of the protein and encoded by a 1,600 bp segment of the var2csa gene.17 ID1-DBL2X was shown to induce the cross-reactive antibodies that block adhesion of IEs to CSA18 and is being considered as a PAM vaccine candidate.19,20 The two leading PAM vaccines, PlacMalVac and PRIMALVAC target overlapping constructs of ID1-DBL2X, are based on different parasite strains (FCR3 and 3D7, respectively).19,20
There are facets of VAR2CSA function that remain poorly understood. For example, it is not known whether certain VAR2CSA variants accumulate in the placenta more than others. Furthermore, when there is a polyclonal infection, it is unclear whether the placenta acts as a separate compartment from the peripheral blood; i.e., are placental VAR2CSA variants the same or different from those in circulation. In our previous study of placental parasites in Benin, we found 57 distinct placental haplotypes in 56 pregnant women.21 Furthermore, most women were infected with more than one haplotype, with a mean number of variants per woman of 1.88 (range: 1–7).21 Given its sequence diversity, the var2csa gene could serve as a marker to distinguish parasite haplotypes in the different compartments in the context of polyclonal infections.
Studies have attempted to compare parasites in the periphery with parasites in the placenta with variable results, as the range of allele overlap between placental and peripheral compartments ranged from 10% to > 90%.22–30 These studies used methods that were insensitive to minority variants and measured variations in small genetic regions which did not capture a high degree of variation or regions which do not encode domains directly implicated in CSA binding. Comparison of circulating versus placental genotypes has not been investigated at the ID1-DBL2X locus or with deep sequencing. Here, we use long amplicon deep sequencing of the 1.6 kb region encoding the minimum binding epitope of P. falciparum VAR2CSA to human CSA18 in matched placental and peripheral samples at delivery to determine whether the same parasite haplotypes found in the placenta are also found in the circulation or whether the placenta selects for specific variants.
MATERIALS AND METHODS
Study population.
Matched peripheral and placental blood samples were collected at the time of delivery from P. falciparum-infected women, as part of a prospective cohort study (“Strategies to Prevent Pregnancy Associated Malaria”) in Comé province, Benin (2008–2010).31 At the time of collection, the study area had high malaria transmission with an estimated entomological inoculation rate of 35–60 infective bites per person per year.31 Mother and newborn demographic and clinical data were recorded31 and are shown in Supplemental Table 1. The current and parent studies have been approved by the institutional review boards of the Research Institute for Development in France, the Science and Health Faculty at the University of Abomey Calavi in Benin, and the University of North Carolina Chapel Hill in United States.
P. falciparum samples.
The placental samples used in this study have been previously sequenced by Patel et al.,21 (Genbank accession numbers MF061812-MF061963). Perfused placental blood sampling was performed as described elsewhere, with precautions to minimize contamination with peripheral maternal blood.32,33 In brief, the placenta was flushed as described by a published protocol34 with the following modifications: 50 mL of one part citrate phosphate dextrose adenine (CPDA) and nine parts phosphate buffered saline with 0.05 g/L gentamycin was drawn in a 50 mL syringe. Maternal blood was flushed out of the placenta by injecting the buffer into the placenta with a moderate flow rate changing places for every 10 mL injected. This was repeated two more times, and the blood accumulating at the edge of the dish was retained. Smears of perfused placental blood were prepared in parallel, which confirmed that at least 95% of parasitized erythrocytes collected were mature and therefore sequestered forms.32,33 The matching peripheral samples from the same patients were collected by venepucture in CPDA31 and processed for the current study, using the same DNA extraction, polymerase chain reaction (PCR) amplification, sequencing methodologies, and bioinformatics as performed on the placental samples.21
Deep sequencing.
Genomic DNA was extracted from red blood cell pellets of both peripheral and placental samples using the Thermo Scientific GeneJET DNA extraction kit (Thermo Fisher, Waltham, MA) following manufacturer’s protocols. The 1.6 kb var2csa gene region encoding ID1-DBL2X was amplified in technical duplicates for each sample using a hemi-nested approach.21 Primer details and amplification conditions are given in Supplemental Text 1. Primer barcodes specific to each PCR replicate and patient allowed pooling of amplicons before PacBio deep sequencing using the P5-C4 chemistry at the University of North Carolina High Throughput Sequencing Facility. To determine haplotypes, the raw sequences were analyzed using a novel deep sequencing clustering algorithm integrated in the SeekDeep software program (baileylab.umassmed.edu/SeekDeep) (Supplemental Text 2). The robustness of the PCR assay and clustering algorithm has been previously validated by our group,21 by mixing seven P. falciparum reference lines (3D7, FCR3, 7G8, DD2, K1, RO33, and V1/S) in varying frequencies (1–50%), which were then amplified at the var2csa-ID1-DBL2X locus, and deep sequenced on the PacBio CCS platform. The PacBio-derived sequences were clustered using the SeekDeep k-mer clustering algorithm. The results from k-mer clustering showed reliable variant calling and clustering of ID1-DBL2x variants.21
Analyses of var2csa ID1-DBL2X haplotype diversity.
The distribution of variants was visualized in Graphpad Prism (La Jolla, CA). The correlation between haplotype distribution and frequency between matched samples was visualized using a quantile-quantile plot in STATA 13 (Release 13, College Station, TX). Differences in haplotype proportions between matched samples were investigated using the Fisher’s exact test.
To estimate haplotypes richness in the placenta and peripheral groups, we computed haplotype accumulation curves (or rarefaction curves) with the package vegan in R using the rarefy function,35,36 with 1,000 subsamplings. The output was visualized in GraphPad Prism. We also evaluated estimators of haplotype richness (i.e., expected number of haplotype variants) and haplotype diversity (these indices combine the number and frequency of variants) for each of the placenta and peripheral groups of samples (Chao1 and Chao2, Fisher’s alpha, Shannon, Simpson, abundance cover estimator [ACE], and incidence cover estimator [ICE]) with 500 randomizations. Genetic relatedness among haplotypes was investigated with principal components analysis using the adegenet37 and PopGenome38 packages in R.
Associations with clinical outcomes.
Associations between haplotypic concordance between placenta and periphery parasites within-patient and clinical outcomes (gravidity, low birth weight, preterm birth, anemia, number of P. falciparum diagnoses during pregnancy, and intermittent preventive treatment during pregnancy with sulfadoxine-pyrimethamine, IPTp-SP) were evaluated with regression models.
RESULTS
Haplotype determination.
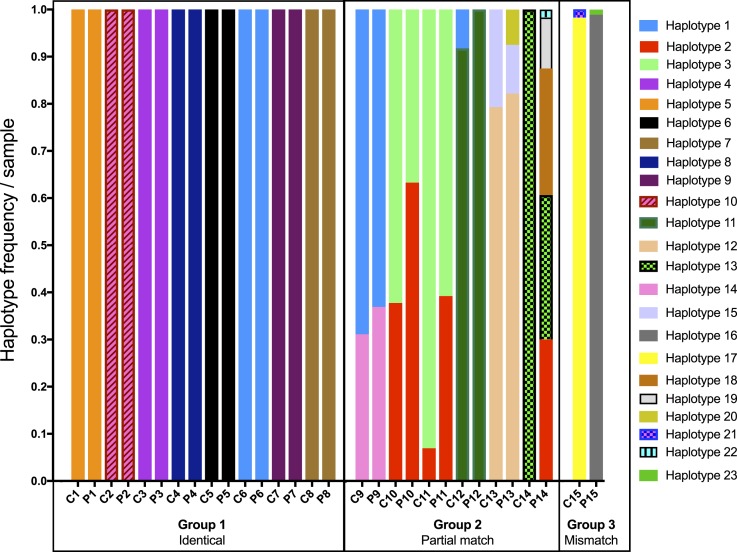
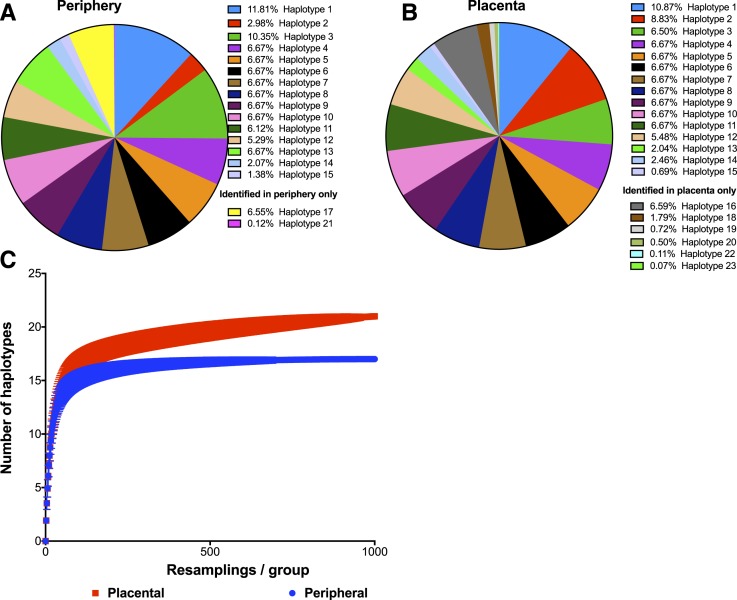
The var2csa-ID1-DBL2X locus was successfully amplified and deep sequenced using the PacBio platform in 15 peripheral samples, where matched placental sequences were available,21 resulting in 30 total samples. The raw data from previously sequenced and newly sequenced samples were combined for k-mer clustering and haplotype identification. The raw data of the newly sequenced samples were deposited at the National Center for Biotechnology Information (NCBI) Sequence Read Archive under the accession PRJNA398389. A total of number of 43,899 reads were obtained from PacBio sequencing. After quality filtering, 27,151 reads (61.8%) with a mean number of 707.3 reads per sample (range = 180–2,136 reads/sample; % reads used/sample = 84.8–100.0%) contributed to k-mer clustering and haplotype identification. Read frequencies between replicates correlated well (R2 = 0.97) (Supplemental Figure 1). Among the 30 samples, we identified 23 unique variants or haplotypes (Figure 1, Supplemental Text 3). These 23 variants represented frequencies of ≥ 1.3% in individual samples (Figure 1) and were supported by both independent PCR amplifications of each sample. Within sample populations (i.e., circulating and placental parasite groups), the frequencies of the variants ranged from 0.07% to 11.81% (Figure 2A and B). Population abundances are estimated by the sum of fractional representation across the 30 individual samples in each of the circulating and placental groups. The five phylogenetic clades, noted in placental infections from Benin in a previous analysis by our group,21 were also represented in this dataset (Supplemental Figure 2).
Figure 1.
Within-patient var2csa ID1-DBL2X haplotype composition and frequencies. Matched samples from circulation (C1–C15) and placenta (P1–P15) of 15 patients. Within-patient haplotype comparisons revealed the following situations: identical matches (Group 1, patients 1–8), partial matches with 100% shared haplotypes at different frequencies (Group 2, patients 9–11), partial matches with 20.0–66.7% shared haplotypes and additional minor haplotypes in one of the tissue compartments (Group 2, patients 12–14), and finally a mismatch where the haplotypes in circulation did not overlap with those in the placenta (Group 3, patient 15).
Figure 2.
Alpha-diversity of peripheral and placental infections. (A) var2csa ID1-DBL2X genetic diversity in peripheral samples. A total of 17 haplotypes were observed. The within-population abundance of each of the haplotype is given, as percentage. Haplotypes 17 and 21 were only observed in peripheral parasites. (B) var2csa ID1-DBL2X genetic diversity in placental samples. In addition to 15 haplotypes observed in the periphery and placenta (haplotypes 1–15), six private haplotypes (haplotypes 16, 18–20, 22–23) were observed in the placental group, bringing the total number of placental haplotypes to 21. The within-population abundance of each of the haplotype is given, as percentage. (C) Rarefaction curve analysis of peripheral and placental infections. Rarefaction with 1,000 re-samplings curves were performed. Diversity was slightly higher in placental infections. Both rarefaction curves approached the diversity asymptote, indicating our methods have uncovered nearly all the variation in these samples.
The 23 haplotypes displayed 580 variable sites across the sequence (1,738 total nucleotide sites investigated, including insertions and deletions (INDELS), Supplemental Figure 3). Percent pairwise nucleotide identity between haplotypes (calculated as number of matches in the pairX100/length of sequence, including INDELS) ranged from 69.0% to 99.8% (Supplemental Table 2). Eight were private haplotypes (i.e., observed in one sample) and 15 were shared among samples, predominantly between the peripheral and placental samples of the same patient (Figure 1). Only three haplotypes were found in more than one woman: two haplotypes appeared in three women (haplotypes 1 and 2), and one haplotype was shared by two women (haplotype 3, Figure 1). The 23 haplotypes also represented 23 unique amino acid sequences (Supplemental Table 3), with amino acid differences among sequences ranging from 1.0% to 84.0%.
Multiplicity of infection (MOI) and haplotype diversity.
More than half of all 30 samples were monoclonal (18/30, 60%), and 11 (36.7%) had a clearly predominant haplotype, typically with a MOI of 2 (MOI, defined as the number of distinct haplotypes observed in an individual sample) (Figure 1). Only one sample had an MOI > 3, harboring five haplotypes with balanced abundances (Figure 1). Among circulating parasites, we observed 17 different variants, with a mean MOI = 1.4 (range = 1–2) (Figure 2A). Of these, 15 variants were also observed in the placenta. An additional six haplotypes were detected only in the placenta (mean MOI = 1.6, range = 1–5) (Figure 2B). In polyclonal infections, minority variants with a within-sample frequency as low as 1.3% (mean number of reads/haplotype/sample = 477.7, range 12–2,136) were detected. Within-sample frequencies for dominant and minor variants ranged from 60.8% to 100% and 1.3% to 39.0%, respectively. Eight haplotypes were identified only as minority variants (Figure 1).
The within-population diversity was assessed by rarefaction (Figure 2C). The rarefaction curves approached the diversity asymptote, indicating that our methods have uncovered nearly all the variation in these samples. The analysis indicated slightly greater diversity in placental samples, with four additional haplotypes detected in this group. This finding was supported by other diversity metrics (Shannon’s index, H′, ACE, ICE, and Chao richness, Supplemental Table 4).
Haplotype comparison between peripheral and placental parasites.
The haplotype frequency and distribution correlated well among within-patient peripheral and placental pairs (Supplemental Figure 4), and 14/15 pairs (93.3%) shared 20.0–100% of haplotypes. More specifically, we identified three situations (Figure 1):
-
1.
The pair was an identical haplotype match (N = 8/15, 53.3%) (Figure 1, Group 1).
-
2.
The pair had a partial haplotype match (shared 100% of haplotypes, but at different frequencies, N = 3/15, 20.0%; or in addition to shared haplotypes, additional minor variants were detected, N = 3/15, 20.0%) (Figure 1, Group 2).
-
3.
Complete mismatch (i.e., haplotypes did not overlap between the two compartments, N = 1/15, 6.7%) (Figure 1, Group 3).
The identical matches (N = 8) belonged to women monoclonally infected (patients 1–8, Figure 1). Among partial matches of Group 2 (N = 6), three patients had pairs which shared the same haplotypes with different frequencies (patients 9–11, Figure 1). The remaining three partial matches shared 20.0–66.7% of their haplotypes (patients 12–14, Figure 1). This within-patient sharing of haplotypes was calculated as the lesser number of haplotypes observed (in the peripheral or placental sample) divided by the highest number of haplotypes seen in the other sample. For example, the placental sample of patient 13 had a total of three variants, but only two of these were observed in the peripheral sample; thus, in this patient, the parasites in the two tissues shared two of three variants, 66.7% sharing. Overall, differences in haplotype composition in the partial matches were driven by minority variants. The only fully discordant pair (patient 15) comprised two distinct haplotypes in each of the peripheral and placental samples (Figure 1). The percent sequence identity of haplotypes not shared between placental and peripheral samples of the same patient ranged from 69.2% to 97.9% (patients 12–15, Supplemental Table 2). These differences were reflected in phylogenetic analyses of these unshared haplotypes (Supplemental Figure 5).
Associations with clinical outcomes.
Associations between clinical characteristics of the woman and haplotype concordance between her placental and peripheral parasites were evaluated, but low sample size may have precluded conclusions. Results are shown in Supplemental Table 5.
DISCUSSION
Here, we compare P. falciparum genotypes found in the circulation to those found in the placenta using long-read deep-sequencing of ID1-DBL2X locus, the minimal binding epitope of var2csa. A high degree of diversity was found with that in the placenta slightly greater than that in the periphery. Because VAR2CSA is involved in parasite sequestration in the placenta, we sought to determine whether specific var2csa variants are compartmentalized in the placenta or whether the same variants circulate in both the placenta and periphery. Our findings show that within an individual patient, parasite populations in the two compartments mostly contain the same parasite genetic variants. Fourteen of 15 peripheral-placental pairs (93.3%) were either an identical match or had overlapping haplotype compositions and of 23 total haplotypes observed, 16 (69.5%) were found in both. This is generally consistent with previous studies that have noted that, although not all peripheral alleles detected during pregnancy matched those found in the placenta, peripheral and placental alleles at delivery were frequently a match.22–25,30
Despite significant sequence variation among the haplotypes and minority variants being confidently called at frequencies as low as 1.3%, most infections were monoclonal (18/30, 60.0%) or had a clearly dominant haplotype (11/30, 36.7%). Previous studies have noted that placental and peripheral P. falciparum infections at delivery have a lower MOI than antenatal and nonpregnant infections22 and that over the course of pregnancy MOI may decrease.23 As MOI in pregnancy is influenced by several factors, including transmission level, gravidity, continued acquisition of immunity during pregnancy, and clinical status of the mother, our single time-point analysis presents limitations in disentangling these relationships, and thus the use of deep-sequencing to track genotype dynamics throughout pregnancy should be explored in future studies. In addition, we note the possibility of bias that only samples with relatively higher parasitaemia yielded amplicons which were able to be deep-sequenced. Nevertheless, in our study whenever infections were monoclonal, the same clone was present in both the peripheral and placental samples, and that in all polyclonal infections but one, placental and peripheral parasites shared 20.0–66.7% of their haplotypes. This is consistent with the possibility that by the time of delivery there may be a selection of genetic variants and these variants may be those which can establish placenta infection due to placenta binding-capacity. Indeed by late pregnancy, the placenta is of considerable size (mean placenta weight at 40 week gestational age = 690 ± 135 g)39 to exert selective pressure. As a result, CSA-adherent genotypes may be at a selective advantage over non CSA-adherent genotypes, and this fitness advantage may also manifest itself in the circulation. If this hypothesis is correct, variants which express var2csa bind in the placenta and then seed the peripheral circulation, meaning circulating parasites are the young asexual forms of sequestered mature asexual parasites.
In patients 9–11 (MOI = 2, Figure 1, Group 2), we observed the same haplotypes in both compartments, but at differing frequencies. One strength of deep sequencing is the ability to detect minor clones which represent as little as 1% of the population. However, the number of reads for each haplotype may not be a precise indicator of population size; thus, the within population structure may have, in reality, been identical between the periphery and placenta but yielded different sequencing read distributions.
There were three patients in whom not all placental genotypes were also observed in the periphery (patients 13 and 14, Figure 1 Group 2 and patient 15, Figure 1, Group 3). There is limited evidence from previous studies for a “cryptic placental cycle,” i.e., whereby all stage asexual forms are noncirculating and sequestering in the placenta.9–11 Patients 13 and 14 may support this hypothesis, but this is based on minority variants only. Patient 15, a primigravida, had complete discordance between peripheral and placental haplotypes. This may be the result of two independent infections—a past infection sequestering in the placenta and a more recent one in the periphery. This is consistent with two types of observations from previous studies: 1) that primigravidae are more likely to carry chronic placenta infections40 and 2) the occurrence of mixed CSA-adherent and non-CSA-adherent (i.e., CD36-adherent) parasite populations in immunological studies, suggesting the possibility of concomitant infections foci, one in the placenta and one in the periphery.41–44
In addition, previous reports have suggested the presence of duplicated var2csa copies within one genome45 The strongest evidence has been provided for the laboratory strain HB3,45 with 89.6% sequence identity between the two copies,45 and both copies being simultaneously transcribed.46 Data for duplicated var2csa in clinical isolates remain limited.24,47 If this duplication event of var2csa is common in natural infections, our interpretation of ID1-DBL2X genetic diversity, particularly in patients where multiple variants are identified, may change in that these alternative sequences would reflect variable copies of the same parasite clone and not distinct infections.
While questions remain about how P. falciparum carrying specific var2csa types establish placental malaria infections via VAR2CSA-CSA interaction, our deep sequencing of ID1-DBL2X, the minimal binding epitope of var2csa, shows that by the time of delivery in an area of high transmission, the repertoire of peripheral genetic variants is a good indicator of placental variants. Thus, the placental population can be inferred through peripheral blood sampling.
Supplementary Material
Acknowledgments:
Our sincere gratitude goes to the participating mothers, their babies, and healthcare workers for making this study possible. We would like to thank Sujata Balasubramanian, Jonathan Parr, and Nicholas Brazeau for technical assistance and scientific discussion.
Note: Supplemental text, tables, figures and reference appear at www.ajtmh.org.
REFERENCES
- 1.Taylor SM, et al. 2011. Quantification of the burden and consequences of pregnancy-associated malaria in the Democratic Republic of the Congo. J Infect Dis 204: 1762–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller LH, Baruch DI, Marsh K, Doumbo OK, 2002. The pathogenic basis of malaria. Nature 415: 673–679. [DOI] [PubMed] [Google Scholar]
- 3.Steketee RW, Nahlen BL, Parise ME, Menendez C, 2001. The burden of malaria in pregnancy in malaria-endemic areas. Am J Trop Med Hyg 64: 28–35. [DOI] [PubMed] [Google Scholar]
- 4.van Geertruyden JP, Thomas F, Erhart A, D’Alessandro U, 2004. The contribution of malaria in pregnancy to perinatal mortality. Am J Trop Med Hyg 71: 35–40. [PubMed] [Google Scholar]
- 5.Desai M, ter Kuile FO, Nosten F, McGready R, Asamoa K, Brabin B, Newman RD, 2007. Epidemiology and burden of malaria in pregnancy. Lancet Infect Dis 7: 93–104. [DOI] [PubMed] [Google Scholar]
- 6.Ayres Pereira M, et al. 2016. Placental sequestration of Plasmodium falciparum malaria parasites is mediated by the interaction between VAR2CSA and chondroitin sulfate A on Syndecan-1. PLoS Pathog 12: e1005831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salanti A, et al. 2004. Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J Exp Med 200: 1197–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beeson JG, Amin N, Kanjala M, Rogerson SJ, 2002. Selective accumulation of mature asexual stages of Plasmodium falciparum-infected erythrocytes in the placenta. Infect Immun 70: 5412–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douki JB, Sterkers Y, Lepolard C, Traore B, Costa FT, Scherf A, Gysin J, 2003. Adhesion of normal and Plasmodium falciparum ring-infected erythrocytes to endothelial cells and the placenta involves the rhoptry-derived ring surface protein-2. Blood 101: 5025–5032. [DOI] [PubMed] [Google Scholar]
- 10.Pouvelle B, Buffet PA, Lepolard C, Scherf A, Gysin J, 2000. Cytoadhesion of Plasmodium falciparum ring-stage-infected erythrocytes. Nat Med 6: 1264–1268. [DOI] [PubMed] [Google Scholar]
- 11.Scherf A, Pouvelle B, Buffet PA, Gysin J, 2001. Molecular mechanisms of Plasmodium falciparum placental adhesion. Cell Microbiol 3: 125–131. [DOI] [PubMed] [Google Scholar]
- 12.Bordbar B, et al. 2014. Genetic diversity of VAR2CSA ID1-DBL2Xb in worldwide Plasmodium falciparum populations: impact on vaccine design for placental malaria. Infect Genet Evol 25: 81–92. [DOI] [PubMed] [Google Scholar]
- 13.Salanti A, Staalsoe T, Lavstsen T, Jensen AT, Sowa MP, Arnot DE, Hviid L, Theander TG, 2003. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol 49: 179–191. [DOI] [PubMed] [Google Scholar]
- 14.Brabin BJ, 1983. An analysis of malaria in pregnancy in Africa. Bull World Health Organ 61: 1005–1016. [PMC free article] [PubMed] [Google Scholar]
- 15.Eisele TP, Larsen DA, Anglewicz PA, Keating J, Yukich J, Bennett A, Hutchinson P, Steketee RW, 2012. Malaria prevention in pregnancy, birthweight, and neonatal mortality: a meta-analysis of 32 national cross-sectional datasets in Africa. Lancet Infect Dis 12: 942–949. [DOI] [PubMed] [Google Scholar]
- 16.Rogerson SJ, Hviid L, Duffy PE, Leke RF, Taylor DW, 2007. Malaria in pregnancy: pathogenesis and immunity. Lancet Infect Dis 7: 105–117. [DOI] [PubMed] [Google Scholar]
- 17.Clausen TM, et al. 2012. Structural and functional insight into how the Plasmodium falciparum VAR2CSA protein mediates binding to chondroitin sulfate A in placental malaria. J Biol Chem 287: 23332–23345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bordbar B, Tuikue-Ndam N, Bigey P, Doritchamou J, Scherman D, Deloron P, 2012. Identification of Id1-DBL2X of VAR2CSA as a key domain inducing highly inhibitory and cross-reactive antibodies. Vaccine 30: 1343–1348. [DOI] [PubMed] [Google Scholar]
- 19.EVI , 2017. European Vaccine Initiative - PRIMALVAC Available at: http://www.euvaccine.eu/node/155/. Accessed June 15, 2017.
- 20.EVI , 2017. European Vaccine Initiative - PlacMalVac Available at: http://www.euvaccine.eu/node/156/. Accessed June 15, 2017.
- 21.Patel JC, et al. 2017. Increased risk of low birth weight in women with placental malaria associated with P. falciparum VAR2CSA clade. Sci Rep 7: 7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arango EM, Samuel R, Agudelo OM, Carmona-Fonseca J, Maestre A, Yanow SK, 2012. Genotype comparison of Plasmodium vivax and Plasmodium falciparum clones from pregnant and non-pregnant populations in North-west Colombia. Malar J 11: 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohee LM, Kalilani-Phiri L, Mawindo P, Joshi S, Adams M, Kenefic L, Jacob CG, Taylor TE, Laufer MK, 2016. Parasite dynamics in the peripheral blood and the placenta during pregnancy-associated malaria infection. Malar J 15: 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guitard J, Andersen P, Ermont C, Gnidehou S, Fievet N, Lund O, Deloron P, Ndam NT, 2010. Plasmodium falciparum population dynamics in a cohort of pregnant women in Senegal. Malar J 9: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jafari-Guemouri S, Ndam NT, Bertin G, Renart E, Sow S, Le Hesran JY, Deloron P, 2005. Demonstration of a high level of parasite population homology by quantification of Plasmodium falciparum alleles in matched peripheral, placental, and umbilical cord blood samples. J Clin Microbiol 43: 2980–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamwendo DD, Dzinjalamala FK, Snounou G, Kanjala MC, Mhango CG, Molyneux ME, Rogerson SJ, 2002. Plasmodium falciparum: PCR detection and genotyping of isolates from peripheral, placental, and cord blood of pregnant Malawian women and their infants. Trans R Soc Trop Med Hyg 96: 145–149. [DOI] [PubMed] [Google Scholar]
- 27.Kassberger F, Birkenmaier A, Khattab A, Kremsner PG, Klinkert MQ, 2002. PCR typing of Plasmodium falciparum in matched peripheral, placental and umbilical cord blood. Parasitol Res 88: 1073–1079. [DOI] [PubMed] [Google Scholar]
- 28.Mayengue PI, Rieth H, Khattab A, Issifou S, Kremsner PG, Klinkert MQ, Ntoumi F, 2004. Submicroscopic Plasmodium falciparum infections and multiplicity of infection in matched peripheral, placental and umbilical cord blood samples from Gabonese women. Trop Med Int Health 9: 949–958. [DOI] [PubMed] [Google Scholar]
- 29.Schleiermacher D, Le Hesran JY, Ndiaye JL, Perraut R, Gaye A, Mercereau-Puijalon O, 2002. Hidden Plasmodium falciparum parasites in human infections: different genotype distribution in the peripheral circulation and in the placenta. Infect Genet Evol 2: 97–105. [DOI] [PubMed] [Google Scholar]
- 30.Serra-Casas E, et al. 2011. Persistence of Plasmodium falciparum parasites in infected pregnant Mozambican women after delivery. Infect Immun 79: 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huynh BT, Fievet N, Gbaguidi G, Dechavanne S, Borgella S, Guezo-Mevo B, Massougbodji A, Ndam NT, Deloron P, Cot M, 2011. Influence of the timing of malaria infection during pregnancy on birth weight and on maternal anemia in Benin. Am J Trop Med Hyg 85: 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doritchamou J, Bertin G, Moussiliou A, Bigey P, Viwami F, Ezinmegnon S, Fievet N, Massougbodji A, Deloron P, Tuikue Ndam N, 2012. First-trimester Plasmodium falciparum infections display a typical “placental” phenotype. J Infect Dis 206: 1911–1919. [DOI] [PubMed] [Google Scholar]
- 33.Magistrado PA, et al. 2011. High efficacy of anti DBL4varepsilon-VAR2CSA antibodies in inhibition of CSA-binding Plasmodium falciparum-infected erythrocytes from pregnant women. Vaccine 29: 437–443. [DOI] [PubMed] [Google Scholar]
- 34.Moore JM, Nahlen B, Ofulla AV, Caba J, Ayisi J, Oloo A, Misore A, Nahmias AJ, Lal AA, Udhayakumar V, 1997. A simple perfusion technique for isolation of maternal intervillous blood mononuclear cells from human placentae. J Immunol Methods 209: 93–104. [DOI] [PubMed] [Google Scholar]
- 35.Work TT, Jacobs JM, Spence JR, Volney WJ, 2010. High levels of green-tree retention are required to preserve ground beetle biodiversity in boreal mixedwood forests. Ecol Appl 20: 741–751. [DOI] [PubMed] [Google Scholar]
- 36.Oksanen J, et al. 2016. Vegan: Community Ecology Package. R package version 2.4-1. Available at: https://cran.r-project.org/web/packages/vegan/index.html. Accessed November 15, 2016.
- 37.Jombart T, Ahmed I, 2011. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27: 3070–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfeifer B, Wittelsburger U, Ramos-Onsins SE, Lercher MJ, 2014. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol Biol Evol 31: 1929–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson JM, Irgens LM, Skjaerven R, Rasmussen S, 2007. Placenta weight percentile curves for singleton deliveries. BJOG 114: 715–720. [DOI] [PubMed] [Google Scholar]
- 40.Walker PG, Griffin JT, Cairns M, Rogerson SJ, van Eijk AM, ter Kuile F, Ghani AC, 2013. A model of parity-dependent immunity to placental malaria. Nat Commun 4: 1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beeson JG, Brown GV, Molyneux ME, Mhango C, Dzinjalamala F, Rogerson SJ, 1999. Plasmodium falciparum isolates from infected pregnant women and children are associated with distinct adhesive and antigenic properties. J Infect Dis 180: 464–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fried M, Duffy PE, 1996. Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science 272: 1502–1504. [DOI] [PubMed] [Google Scholar]
- 43.Ofori MF, Staalsoe T, Bam V, Lundquist M, David KP, Browne EN, Akanmori BD, Hviid L, 2003. Expression of variant surface antigens by Plasmodium falciparum parasites in the peripheral blood of clinically immune pregnant women indicates ongoing placental infection. Infect Immun 71: 1584–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doritchamou J, Sossou-tchatcha S, Cottrell G, Moussiliou A, Hounton Houngbeme C, Massougbodji A, Deloron P, Ndam NT, 2014. Dynamics in the cytoadherence phenotypes of Plasmodium falciparum infected erythrocytes isolated during pregnancy. PLoS One 9: e98577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kraemer SM, et al. 2007. Patterns of gene recombination shape var gene repertoires in Plasmodium falciparum: comparisons of geographically diverse isolates. BMC Genomics 8: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brolin KJ, Ribacke U, Nilsson S, Ankarklev J, Moll K, Wahlgren M, Chen Q, 2009. Simultaneous transcription of duplicated var2csa gene copies in individual Plasmodium falciparum parasites. Genome Biol 10: R117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sander AF, Salanti A, Lavstsen T, Nielsen MA, Magistrado P, Lusingu J, Ndam NT, Arnot DE, 2009. Multiple var2csa-type PfEMP1 genes located at different chromosomal loci occur in many Plasmodium falciparum isolates. PLoS One 4: e6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.