Abstract
Competing endogenous RNAs (ceRNAs) render the functions of long non-coding RNAs (lncRNAs) more complicated during cancer processes. Potential lncRNA biomarkers and their roles as ceRNAs have not been clearly described for rectal adenocarcinoma (READ). In the present study, we extracted data from The Cancer Genome Atlas (TCGA) including data from 167 tumor samples and 10 adjacent non-tumor samples. A total of 202 lncRNAs, 190 microRNAs (miRNAs) and 1,530 mRNAs were identified as READ-specific RNAs [log2(fold-change)>2, FDR<0.01]. The Gene Ontology (GO) biological processes and the Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways were analysed for 1,530 specific mRNAs. Among 202 READ-specific lncRNAs, 7 lncRNAs were identified as being associated with overall survival of READ patients. Then, a ceRNA network was constructed with 34 key lncRNAs, 25 miRNAs and 65 mRNAs. A total of 7 lncRNAs from the network were revealed to be linked to clinical features. The results of qRT-PCR ascertained that our analysis was credible. Overall, this research provides a novel perspective from which to study the lncRNA-related ceRNA network in READ and assists in the identification of new potential biomarkers to be used for diagnostic and prognostic purposes.
Keywords: rectal adenocarcinoma, long non-coding RNA, competing endogenous RNA, biomarkers, The Cancer Genome Atlas, bioinformatics analysis
Introduction
Colorectal cancer (CRC) is a cancer type that has high incidence and mortality worldwide. Rectal adenocarcinoma (READ) is a type of CRC (1). Many studies have focused on READ. However, its mortality remains high due to a lack of efficient biomarkers. Previous studies have demonstrated that various types of RNAs play important roles in cancer development and progression by acting in multiple ways (2,3). To further investigate the relationship among various types of RNAs and to obtain more efficient biomarkers, we identified cancer-specific RNAs and developed a competing endogenous RNA (ceRNA) network based on three types of RNAs, including long non-coding RNAs (lncRNAs), microRNAs (miRNAs) and mRNAs, that are differentially expressed in READ.
Non-coding RNA (ncRNA) is a type of RNA molecule that exists ubiquitously in organisms but lacks protein-coding ability (4). lncRNAs, once viewed as transcriptional ‘noise’, are one subtype of ncRNA and are identified as ncRNAs with >200 bp. Previous studies have revealed that lncRNAs play key roles in cancer processes including proliferation, invasion and metastasis (5,6). For instance, homeobox transcript antisense intergenic RNA (HOTAIR) has been identified as having higher expression levels in the plasma of CRC patients than in the plasma of healthy controls, and its high expression level predicts a poor prognosis (7).
lncRNAs function in multiple ways including interacting with mRNAs and miRNAs. In 2011, Salmena et al (8) reported the ceRNA hypothesis and demonstrated that RNA transcripts communicate with each other by miRNA response elements (MREs). mRNAs and lncRNAs use MREs to compete for miRNA-binding sites, further affecting the expression of miRNAs and the competition between mRNAs, lncRNAs and pseudogene transcripts and playing a crucial role in tumor processes (9,10).
Some studies regarding lncRNA profiles in CRC have been performed, and the functions of some lncRNAs in CRC have already been demonstrated (11–14), however studies with large sample sizes and high throughput detection methods on specific READ lncRNAs are still lacking. In addition, few studies have focused on dysregulated lncRNAs that are associated with sex, TNM, survival or other clinical features, and even fewer studies have been designed to identify the potential ceRNA network in READ. To provide answers to the questions mentioned, we used bioinformatic tools and analysis data from The Cancer Genome Atlas (TCGA), which is a public database with expression data of lncRNAs, miRNAs and mRNAs of READ. TCGA contains RNA sequencing data of a total of 167 READ tumor tissues and 10 adjacent non-tumor tissue samples. To the best of our knowledge, the present study is the first to identify lncRNAs that can be potential biomarkers and to further identify the ceRNA network in READ. To ascertain the credibility of our results, we randomly selected several lncRNAs from the ceRNA network and confirmed their profiles by qRT-PCR. The relationship between the expression pattern and the clinical features of seven lncRNAs were also confirmed by qRT-PCR. This approach aided in revealing the functions of lncRNAs and constructed a ceRNA network for READ.
Materials and methods
Data of patients and samples
RNA expression data with clinical data such as pathological stage, sex, and TNM information were all downloaded from TCGA database. We set exclusion criteria as follows: i) histological diagnosis revealing that the tissue was not READ; ⅱ) suffering malignancies other than READ; ⅲ) samples without enough data for analysis; and ⅳ) patients who underwent preoperative radiotherapy and chemotherapy. Ultimately, 167 tumor tissues and 10 adjacent non-tumor tissues were used in the study. The number of tumor tissues in tumor stages I, II, III, and IV were 30, 51, 51, and 24, respectively, based on the pathological stage. The study followed the guidelines of TCGA, thus, the approval of an ethics committee was not required.
The READ specimens and their paired adjacent non-tumor tissues of 90 patients were selected from the First Affiliated Hospital of Nanjing Medical University (Jiangsu, China) for qRT-PCR analysis. Their ages ranged from 45–80 years, and they were diagnosed as having rectal cancer based on histopathology and clinical history. The tissues were stored in RNAlater (Ambion; Thermo Fisher Scientific, Inc., Austin, TX, USA) at −80°C until RNA extraction and further analyses were performed. The clinical features of 60 patients were also obtained.
RNA sequence data and further analysis
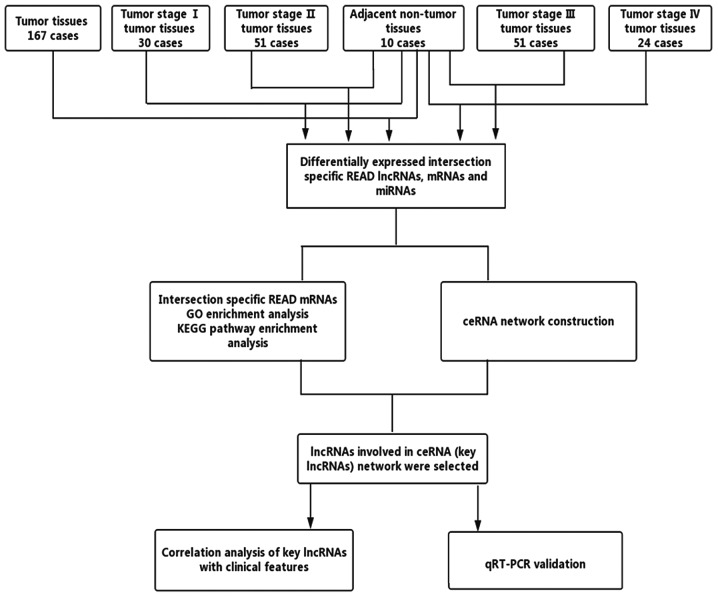
RNA expression pattern data (level 3) from patients with READ was obtained from TCGA database (September 2017), which provides normalized data from RNA sequencing by the RNASeqV2 system, including lncRNAs and mRNA expression profiles. We used an Illumina HiSeq 2000 miRNA sequencing (miRNAseq) platform (Illumina, Inc., Hayward, CA, USA) to obtain STAD level 3 miRNAseq data from TCGA. The downloaded data included a number of individual data files, with each file representing one tissue sample. We then divided the tumor samples into four groups (tumor stages I, II, III, IV) and analyzed the differences in the expression levels between each tumor stage (tumor stage I, II, III, IV) and adjacent non-tumor tissues, and between all tumor tissues and all adjacent non-tumor tissues using the Empirical Analysis of Digital Gene Expression Data package in R (edgeR, R version 3.4.1) [absolute log2(fold-change)>2.0, FDR<0.01]. Then, we chose an intersection of differentially expressed READ lncRNAs, mRNAs and miRNAs for further analysis. The process is shown in Fig. 1.
Figure 1.
Flowchart of bioinformatics analysis. READ, rectal adenocarcinoma; lncRNAs, long non-coding RNAs; miRNAs, microRNAs; GO, Gene Ontology; KEGG, Kyoto Encyclopaedia of Genes and Genomes; ceRNAs, competing endogenous RNAs.
Functional enrichment analysis
Database for Annotation, Visualization, and Integrated Discovery (DAVID) bioinformatics resources (https://david.ncifcrf.gov/) was used for the functional enrichment analysis, and we only researched the Gene Ontology (GO) biological processes and the Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways. The criteria were set as P<0.05 and an enrichment score >1.5.
Construction of a ceRNA network
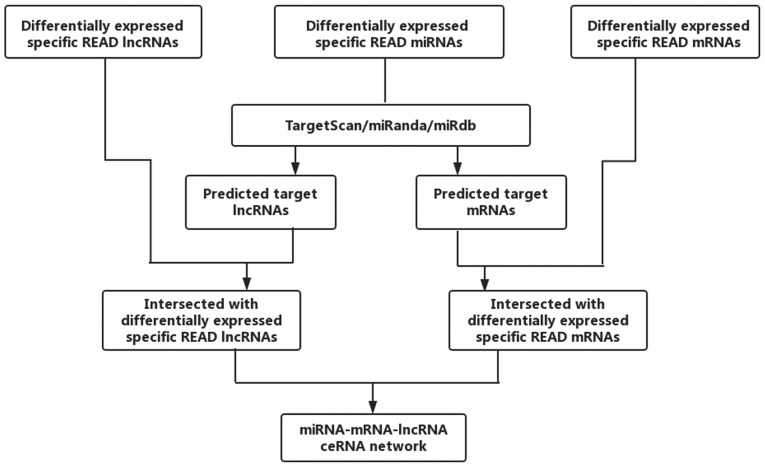
According to the theory that lncRNAs can affect miRNAs and can act as miRNA sponges to further regulate mRNAs, we constructed a ceRNA network. The miRcode (http://www.mircode.org/) was used to predict the lncRNA/miRNA interactions based on specific READ miRNAs. We predicted miRNA-targeted mRNA by using TargetScan (http://www.targetscan.org/), miRDB (http://www.mirdb.org/) and miRanda (http://www.microrna.org/microrna/home.do). We retained the intersection with the differentially expressed lncRNAs and mRNAs. Cytoscape v3.0 was used to construct the lncRNA/miRNA/mRNA ceRNA network. The flowchart of the ceRNA network is presented in Fig. 2.
Figure 2.
Flowchart of the construction of a ceRNA network. READ, rectal adenocarcinoma; lncRNAs, long non-coding RNAs; miRNAs, microRNAs; ceRNAs, competing endogenous RNA.
Clinical feature analysis of key lncRNAs
We extracted clinical information from TCGA database. Based on the established ceRNA network in this study, we selected lncRNAs from the network for further clinical analysis, and clinical features such as sex, age, tumor staging, TNM staging and lymphatic metastasis were chosen to analyze the correlation between these features and key lncRNAs. We also investigated the association between specific READ lncRNAs and the overall survival time of patients.
RNA extraction and qRT-PCR validation
We extracted RNA from tissue samples using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Complementary DNA (cDNA) was synthesized using the PrimeScript RT kit (Takara, Dalian, China). qRT-qPCR was performed with a SYBR-Green PCR kit (Roche Diagnostics, Indianapolis, IN, USA) via a StepOnePlus Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). The sequences of the primers used for the PCR are presented in Table I. The PCR cycling conditions were as follows: 95°C for 30 sec, 40 cycles of 95°C for 5 sec, 60°C for 30 sec, dissociation at 95°C for 15 sec, 60°C for 1 min and 95°C for 15 sec. The results were analysed using the 2−ΔΔCt method. The qRT-PCR reactions were all repeated three times.
Table I.
Primer sequences used for qRT-PCR.
| Primer | Sequence |
|---|---|
| HULC | F 5′-ATCTGCAAGCCAGGAAGAGTC-3′ |
| R 5′-CTTGCTTGATGCTTTGGTCTGT-3′ | |
| CRNDE | F 5′-TGGATGCTGTCAGCTAAGTTCAC-3′ |
| R 5′-TTCCAGTGGCATCCTCCTTATC-3′ | |
| PVT1 | F 5′-TGAGAACTGTCCTTACGTGACC-3′ |
| R 5′-AGAGCACCAAGACTGGCTCT-3′ | |
| ADAMTS9-AS2 | F 5′-TAAGACCCACGAACGACAGC-3′ |
| R 5′-CGTCATGCTTCGGCTTTCAG-3′ | |
| GAPDH | F 5′-ACAGTCAGCCGCATCTTCTT-3′ |
| R 5′-GACAAGCTTCCCGTTCTCAG-3′ |
Statistical analysis
Statistical analysis was performed using R Studio (R version 3.4.1), Statistical Programme for Social Sciences 20.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA). The lncRNA data set and the overall survival information were profiled using the univariate Cox proportional hazards regression model. The results were presented as Kaplan-Meier survival curves and multivariate Cox regression analysis was applied for further study. Paired t-tests were used to compare the differences in the qRT-PCR results. P<0.05 was considered to indicate a statistically significant difference.
Results
Specific READ lncRNAs in patients
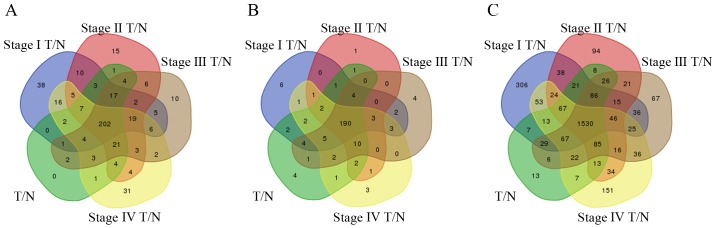
In total, 272 lncRNAs were identified that were differentially expressed between READ tissues and adjacent non-tumor tissues from TCGA database [absolute log2(fold-change)>2, FDR<0.01], of which 143 lncRNAs were upregulated, and 129 lncRNAs were downregulated. Further analysis of the differences between tumor tissues and adjacent non-tumor tissues in patients with stage I, II, III and IV cancer was performed. Then, 337, 323, 307 and 330 differentially expressed lncRNAs were identified between adjacent non-tumor tissues and READ stage I, II, III and IV tumor tissues, respectively. To enhance the credibility of the data, we used 202 differentially expressed lncRNAs from the intersection of the aforementioned five groups for further analysis (Fig. 3A). These 202 lncRNAs were named READ-specific lncRNAs. Finally, 34 lncRNAs were used to construct the lncRNA/miRNA/mRNA ceRNA network (Table II).
Figure 3.
Venn diagram analysis of differentially expressed (A) lncRNAs, (B) miRNAs and (C) mRNAs between T/N stages I, II, III and IV. lncRNAs, long non-coding RNAs; miRNAs, microRNAs; T, tumor tissues; N, adjacent non-tumor tissues.
Table II.
Key lncRNAs involved in the ceRNA network.
| lncRNAs | Log2(fold-change) | -Log(FDR) | lncRNAs | Log2(fold-change) | -Log(FDR) |
|---|---|---|---|---|---|
| HULC | 8.48 | 2.92 | LINC00092 | −2.59 | 17.31 |
| ERVMER61-1 | 6.41 | 2.17 | LINC00402 | −2.85 | 9.49 |
| LINC00460 | 6.02 | 7.76 | LIFR-AS1 | −2.88 | 16.09 |
| CLDN10-AS1 | 5.37 | 4.35 | LINC00163 | −2.93 | 10.21 |
| POU6F2-AS1 | 5.20 | 2.65 | GDNF-AS1 | −2.96 | 16.35 |
| UCA1 | 4.84 | 5.31 | SFTA1P | −3.09 | 13.19 |
| CRNDE | 3.91 | 10.16 | HCG23 | −3.14 | 17.60 |
| DLX6-AS1 | 3.83 | 3.26 | CHL1-AS2 | −3.24 | 16.69 |
| GAS6-AS1 | 3.33 | 5.68 | RBMS3-AS3 | −3.34 | 25.42 |
| MIR17HG | 2.85 | 8.11 | LINC00473 | −3.48 | 17.51 |
| PVT1 | 2.76 | 17.27 | JAZF1-AS1 | −3.60 | 25.95 |
| PRSS30P | 2.70 | 3.94 | ADAMTS9-AS2 | −3.80 | 35.18 |
| PCAT1 | 2.13 | 4.21 | LINC00461 | −3.88 | 25.86 |
| SOX2-OT | −2.16 | 9.37 | C20orf166-AS1 | −4.30 | 35.28 |
| GRIK1-AS1 | −2.30 | 10.22 | LINC00507 | −4.33 | 17.01 |
| LINC00484 | −2.43 | 17.18 | FRMD6-AS2 | −4.61 | 23.38 |
| LINC00472 | −2.47 | 14.45 | ADAMTS9-AS1 | −5.09 | 40.40 |
FDR using Benjamini and Hochberg (1995) method. lncRNA, long non-coding RNA; ceRNA, competing endogenous RNA; FDR, false discovery rate.
GO enrichment and KEGG pathway analyses of differentially expressed genes
For further study of the functions of the differentially expressed genes, we selected intersecting mRNAs across all READ stages from the differentially expressed mRNAs for analysis. We first identified 2,000 differentially expressed mRNAs between the tumor tissues of READ patients and the adjacent non-tumor tissues [absolute log2(fold-change)>2, FDR<0.01]. We also identified 2,363; 2,124; 2,113; and 2,189 differentially expressed mRNAs between adjacent non-tumor tissues and READ tumor tissues (stage I, II, III and IV, respectively). Then, we chose the mRNAs that were differentially expressed in all five comparison groups, finally obtaining 1,530 differentially expressed mRNAs that were used for further analysis (Fig. 3C). These mRNAs were identified as READ-specific mRNAs.
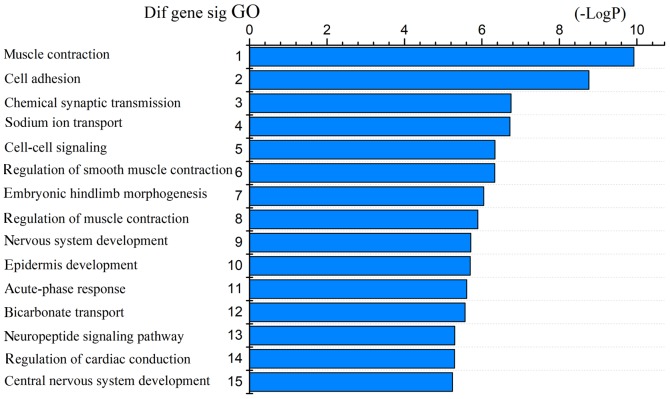
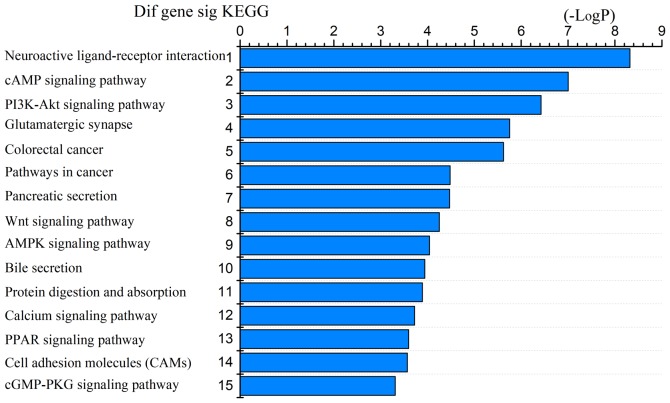
The 1,530 differentially expressed mRNAs were analyzed with DAVID bioinformatics resources. We chose to show the top 15 GO biological processes and 15 KEGG pathways of the differentially expressed genes based on the P-values (Figs. 4 and 5). Among these pathways, the PI3K-Akt, Wnt, AMPK, and cGMP-PKG signaling pathways and the cell adhesion molecules (CAMs) were confirmed as CRC-associated pathways.
Figure 4.
Top 15 GO terms for intersection mRNAs [-LogP represents -Log(p-value)]. GO, Gene Ontology.
Figure 5.
Top 15 KEGG terms for intersection mRNAs [-LogP represents -Log(p-value)]. KEGG, Kyoto Encyclopaedia of Genes and Genomes.
Prediction of miRNA targets and construction of a ceRNA network
The 31 miRNAs that were differentially expressed between READ tumor tissues and adjacent non-tumor tissues were identified. For further investigation, we then divided the tumor tissues into four groups (stages I, II, III and IV) according to the pathological stage, and then compared each group with adjacent non-tumor tissues in turn. Subsequently, we obtained 190 specific miRNAs by selecting the intersection of miRNAs differentially expressed across the five comparison groups (Fig. 3B). These miRNAs were identified as READ-specific miRNAs. In the next step, we investigated the interactions between these intersecting miRNAs and the READ-specific lncRNAs based on miRcode (http://www.mircode.org/), and 25 key miRNAs were predicted to target 34 key lncRNAs (Table III).
Table III.
miRNAs that may target READ lncRNAs.
| miRNAs | lncRNAs |
|---|---|
| hsa-mir-107 | C20orf166-AS1, UCA1, DLX6-AS1, LINC00472, LINC00460, LINC00163, LINC00402, LINC00484, ADAMTS9-AS2, LINC00461, LINC00507, FRMD6-AS2 |
| hsa-mir-141 | DLX6-AS1, LINC00472, LINC00402, LINC00484, ADAMTS9-AS2, SOX2-OT, LINC00461 |
| hsa-mir-143 | PRSS30P, UCA1, CLDN10-AS1, SFTA1P, LINC00472, LINC00460, JAZF1-AS1, LINC00163, LINC00402, LINC00484, ADAMTS9-AS2, SOX2-OT, LINC00461, CRNDE, GDNF-AS1, PVT1, FRMD6-AS2 |
| hsa-mir-144 | POU6F2-AS1, DLX6-AS1, ADAMTS9-AS1, ADAMTS9-AS2, LIFR-AS1, LINC00461, CRNDE |
| hsa-mir-150 | PRSS30P, C20orf166-AS1, CLDN10-AS1, LINC00473, LINC00092, DLX6-AS1, LINC00460, JAZF1-AS1, LINC00402, ADAMTS9-AS1, ADAMTS9-AS2, LIFR-AS1, LINC00461, PVT1, HULC |
| hsa-mir-152 | DLX6-AS1, LINC00484, ADAMTS9-AS2, PVT1 |
| hsa-mir-155 | DLX6-AS1, LINC00472, LINC00402, ADAMTS9-AS1, ADAMTS9-AS2, LIFR-AS1, CRNDE, HULC |
| hsa-mir-17 | C20orf166-AS1, HCG23, DLX6-AS1, JAZF1-AS1, LINC00402, PVT1, PCAT1 |
| hsa-mir-182 | UCA1, SFTA1P, ERVMER61-1, GAS6-AS1, LINC00402, RBMS3-AS3, ADAMTS9-AS1, ADAMTS9-AS2, SOX2-OT, LIFR-AS1, PCAT1, FRMD6-AS2 |
| hsa-mir-183 | C20orf166-AS1, CHL1-AS2, LINC00163, ADAMTS9-AS2, CRNDE, PVT1, LINC00507 |
| hsa-mir-192 | POU6F2-AS1, HCG23, DLX6-AS1, SOX2-OT, LIFR-AS1, LINC00461, PCAT1 |
| hsa-mir-200a | DLX6-AS1, LINC00472, LINC00402, LINC00484, ADAMTS9-AS2, SOX2-OT, LINC00461 |
| hsa-mir-21 | PRSS30P, ERVMER61-1, JAZF1-AS1, ADAMTS9-AS1, SOX2-OT, PVT1 |
| hsa-mir-215 | POU6F2-AS1, HCG23, DLX6-AS1, SOX2-OT, LIFR-AS1, LINC00461, PCAT1 |
| hsa-mir-217 | LINC00402, LINC00484, CRNDE, PVT1 |
| hsa-mir-22 | C20orf166-AS1, DLX6-AS1, LINC00472, JAZF1-AS1, LINC00402, LINC00484, ADAMTS9-AS2, LIFR-AS1, LINC00461, CRNDE, PCAT1, FRMD6-AS2 |
| hsa-mir-223 | DLX6-AS1, GAS6-AS1, LINC00484, ADAMTS9-AS2, CRNDE |
| hsa-mir-32 | CLDN10-AS1, POU6F2-AS1, GAS6-AS1, JAZF1-AS1, LINC00484, ADAMTS9-AS2, LIFR-AS1, LINC00461, CRNDE, PCAT1 |
| hsa-mir-375 | C20orf166-AS1, GRIK1-AS1, ADAMTS9-AS2, SOX2-OT, LIFR-AS1, PCAT1, LINC00507, FRMD6-AS2 |
| hsa-mir-424 | PRSS30P, LINC00473, LINC00092, SFTA1P, DLX6-AS1, LINC00472, LINC00484, LINC00461, GDNF-AS1, PVT1, PCAT1 |
| hsa-mir-425 | C20orf166-AS1, MIR17HG, LINC00472, LINC00460, LINC00461 |
| hsa-mir-429 | C20orf166-AS1, DLX6-AS1, LINC00460, LINC00402, SOX2-OT |
| hsa-mir-454 | C20orf166-AS1, ADAMTS9-AS1, ADAMTS9-AS2, SOX2-OT |
| hsa-mir-96 | UCA1, ERVMER61-1, GAS6-AS1, RBMS3-AS3, ADAMTS9-AS1, ADAMTS9-AS2, SOX2-OT, LIFR-AS1, LINC00461, FRMD6-AS2 |
| hsa-mir-98 | JAZF1-AS1, LINC00484, ADAMTS9-AS2 |
miRNAs, microRNAs; READ, rectal adenocarcinoma; lncRNAs, long non-coding RNAs.
The 25 key miRNAs mentioned in Table III were then used to predict key mRNAs using Targetscan (http://www.targetscan.org/), miRDB (http://www.mirdb.org/) and miRanda (http://www.microrna.org/microrna/home.do). We then compared the predicted mRNAs with the 1,530 READ-specific mRNAs and chose mRNAs that were found in both groups. Finally, 65 mRNAs were found to interact with 25 miRNAs (Table IV). Among these mRNAs, some have been verified to be transcribed from cancer-related genes.
Table IV.
miRNAs that may target READ mRNAs.
| miRNAs | mRNAs |
|---|---|
| hsa-mir-107 | SALL4, AXIN2, FGF2, FGFRL1 |
| hsa-mir-141 | MACC1, ZEB1, EPHA7, KIAA1549, ELAVL4 |
| hsa-mir-143 | COL1A1 |
| hsa-mir-144 | FGF2, GRIK3, NR3C1, KCNQ5 |
| hsa-mir-150 | HILPDA, ZEB1 |
| hsa-mir-152 | BMP3, NPTX1 |
| hsa-mir-155 | MEIS1, CD36, GPM6B, NOVA1, PCDH9 |
| hsa-mir-17 | FAM46C, CLIP4, CFL2, FAXC, NPAS3, FOXQ1, SLC16A9, FJX1, FAM129A, CADM2 |
| hsa-mir-182 | NR3C1, NPTX1, ULBP2, CHL1, TCEAL7 |
| hsa-mir-183 | ZEB1, NR3C1, AKAP12 |
| hsa-mir-192 | GRHL1, TCF7 |
| hsa-mir-200a | EPHA7, ZEB1, MACC1, KIAA1549 |
| hsa-mir-21 | ATP2B4, PRICKLE2, CALD1, EDIL3, EPM2A, OSR1, TGFBI |
| hsa-mir-215 | TCF7 |
| hsa-mir-217 | DACH1 |
| hsa-mir-22 | NR3C1, RGS2 |
| hsa-mir-223 | EPB41L3 |
| hsa-mir-32 | ATP2B4, UGP2, |
| hsa-mir-375 | ELAVL4 |
| hsa-mir-424 | AMOTL1, TPM2, CBX2, TMEM100, AXIN2, PSAT1, FGF2 |
| hsa-mir-425 | THRB |
| hsa-mir-429 | ZEB1 |
| hsa-mir-454 | SPG20, CFL2, RBM20 |
| hsa-mir-96 | JAZF1, TRIB3, ZEB1 |
| hsa-mir-98 | PRSS22, IGF2BP1, HAND1, SLC5A6, IGF2BP3, TRIM71, CPA4 |
miRNAs, microRNAs; READ, rectal adenocarcinoma.
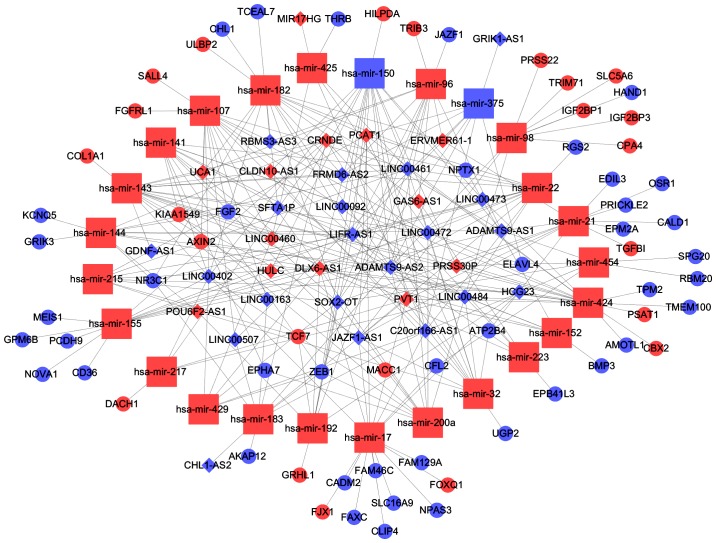
According to information provided in Tables III and IV, we constructed a miRNA/lncRNA/mRNA ceRNA network using Cytoscape 3.0. In conclusion, 25 miRNAs, 65 mRNAs and 34 lncRNAs were involved in the network (Fig. 6). We called lncRNAs involved in the ceRNA network key lncRNAs.
Figure 6.
The lncRNA/miRNA/mRNA ceRNA network. Red balls, upregulated mRNAs; blue balls, downregulated mRNAs; red squares, upregulated miRNAs; blue squares, downregulated miRNAs; red diamonds, upregulated lncRNAs; blue diamonds, downregulated lncRNAs. lncRNAs, long non-coding RNAs; miRNAs, microRNAs; ceRNA, competing endogenous RNA.
Clinical feature analysis of key READ-specific lncRNAs
To further study the lncRNAs, the correlation between the lncRNAs involved in the ceRNA network and clinical features including sex, age, tumor stage, TNM stage and lymphatic metastasis status in TCGA database were analysed. We identified 7 lncRNAs associated with clinical features (P<0.05). The results revealed that UCA1 and HULC were age-related, CHL1-AS2, LINC00484 and HULC were sex-related, LINC00484 and ADAMTS9-AS1 were associated with TNM stage, CLDN10-AS1 was associated with tumor stage, and UCA1 was associated with lymphatic metastasis (Table V).
Table V.
The correlation between COAD key lncRNAs involved in the ceRNA network and their clinical features.
| Comparisons | Upregulated | Downregulated |
|---|---|---|
| Age (<50 vs. >50 years) | UCA1, HULC | |
| Sex (female vs. male) | CHL1-AS2, LINC00484 | HULC |
| Lymphatic metastasis (no vs. yes) | UCA1 | |
| Tumor stage (stage I, II vs. stage III, IV) | CLDN10-AS1 | DLX6-AS1 |
| TNM staging system (T1+T2 vs. T3+T4) | LINC00484, ADAMTS9-AS1 |
COAD, colon adenocarcinoma; lncRNAs, long non-coding RNAs; ceRNA, competing endogenous RNA.
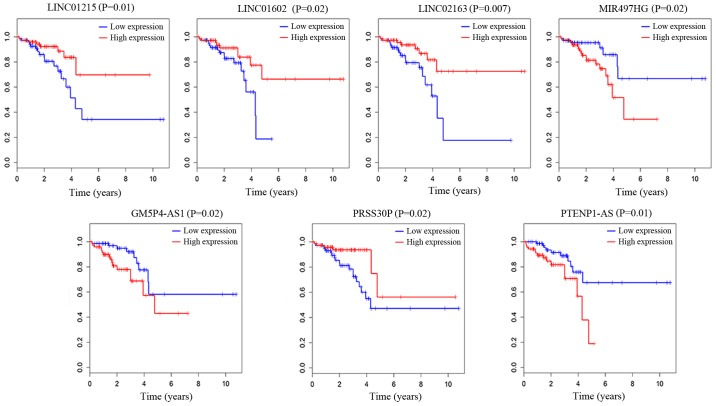
We also analyzed the association of the overall survival of patients with the 202 specific lncRNAs based on the clinical data of 177 samples from TCGA database. To carry out this research, we used a univariate Cox proportional hazards regression model and finally found 7 lncRNAs that were significantly associated with the overall survival of READ patients (log-rank P<0.05). As described in Fig. 7, LINC01215, LINC01602, LINC02163 and PRSS30P were positively correlated with overall survival (P<0.05). MIR497HG, PGM5P4-AS1 and PTENP1-AS were negatively correlated with overall survival (P<0.05). Based on the results of the univariate Cox regression analysis, 7 lncRNAs were associated with overall survival. We performed a multivariate Cox regression analysis with these 7 lncRNAs. LINC01602, LINC02163 and MIR497HG were found to be independent influencing factors of survival time, LINC01602 and MIR497HG were negatively related to overall survival, and LINC02163 was positively related to overall survival. We present these results in Table VI.
Figure 7.
Kaplan-Meier survival curves for 7 lncRNAs associated with overall survival. Horizontal axis, overall survival time (years); vertical axis, survival function. lncRNAs, long non-coding RNAs.
Table VI.
Results of multivariate cox regression analysis.
| lncRNAs | β | OR (95CI) | P-value |
|---|---|---|---|
| LINC01215 | −0.016 | 0.984 (0.951–1.019) | 0.368 |
| LINC01602 | 0.004 | 1.004 (1.001–1.007) | 0.011 |
| LINC02163 | −0.042 | 0.959 (0.929–0.991) | 0.011 |
| PRSS30P | −0.005 | 0.995 (0.989–1.002) | 0.144 |
| MIR497HG | 0.046 | 1.048 (1.006–1.090) | 0.023 |
| PGM5P4-AS1 | −0.036 | 0.965 (0.837–1.112) | 0.621 |
| PTENP1-AS | 0.320 | 1.377 (0.968–1.960) | 0.075 |
lncRNAs, long non-coding RNAs.
qRT-PCR validation
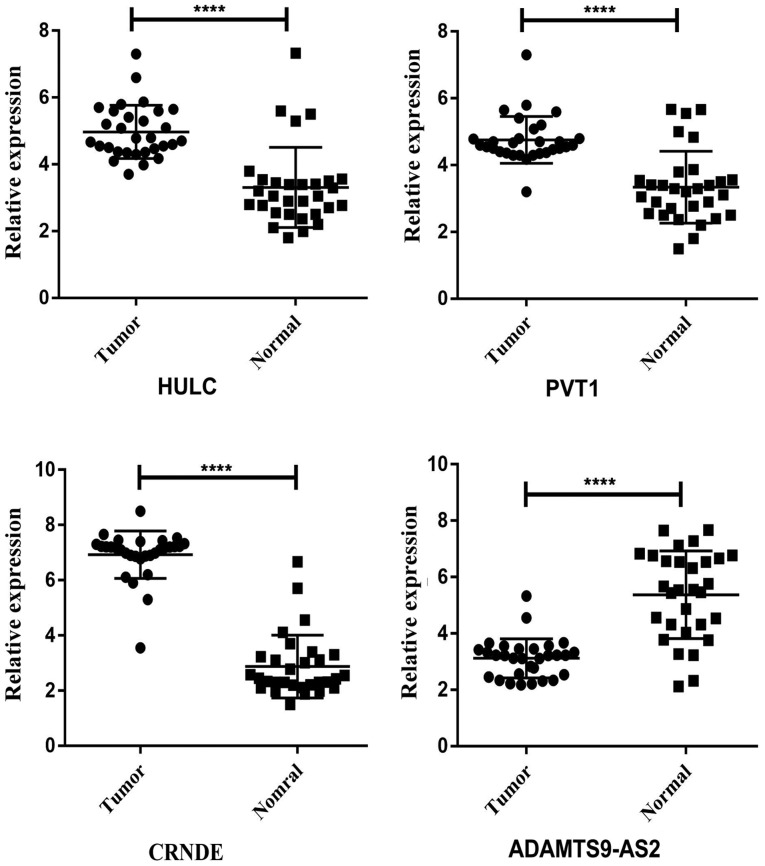
To check the credibility of the bioinformatics results, we randomly selected 4 key lncRNAs (HULC, CRNDE, PVT1 and ADAMTS9-AS2) from the network. Paired t-tests were applied to analyse the differences in expression of the selected lncRNAs between READ tumor tissues and the adjacent non-tumor tissues. HULC, CRNDE and PVT1 expression levels were increased, and the expression level of ADAMTS9-AS2 was decreased in READ tumor tissues compared to adjacent non-tumor tissues. The qRT-PCR results from 30 READ patients were consistent with the bioinformatics results (Fig. 8). We also confirmed the correlation between the expression pattern and clinical features of 7 lncRNAs. Sixty patients were divided into two groups according to the expression level of the lncRNA concerned. Patients with higher and lower than the median expression of the lncRNA concerned were allocated into high and low expression groups, respectively. The results are displayed in Table VII. Thus, our bioinformatics analysis was reliable.
Figure 8.
lncRNA levels in READ and normal tissues (n=30) were assessed by real-time polymerase chain reaction (****P<0.0001 compared with the control adjacent non-tumorous tissues). lncRNA, long non-coding RNA; READ, rectal adenocarcinoma.
Table VII.
Expression of lncRNAs related to clinical features according to the clinicopathological characteristics of patients.
| UCA1 expression | HULC expression | ||||||
|---|---|---|---|---|---|---|---|
| Characteristics | No. | High group | Low group | P-value | High group | Low group | P-value |
| Age (years) | |||||||
| <50 | 18 | 5 | 13 | 0.024 | 5 | 13 | 0.024 |
| >50 | 42 | 25 | 17 | 25 | 17 | ||
| Sex | |||||||
| Female | 32 | 17 | 15 | 0.605 | 12 | 20 | 0.038 |
| Male | 28 | 13 | 15 | 18 | 10 | ||
| Lymphatic metastasis | |||||||
| No | 36 | 14 | 22 | 0.035 | 15 | 21 | 0.113 |
| Yes | 24 | 16 | 8 | 15 | 9 | ||
| Tumor stage | |||||||
| Stage I, II | 33 | 12 | 21 | 0.020 | 15 | 18 | 0.436 |
| Stage III, IV | 27 | 18 | 9 | 15 | 12 | ||
| TNM staging system | |||||||
| T1+T2 | 32 | 17 | 15 | 0.605 | 14 | 18 | 0.301 |
| T3+T4 | 28 | 13 | 15 | 16 | 12 | ||
| CHL1-AS2 expression | LINC00484 expression | ||||||
| Characteristics | No. | High group | Low group | P-value | High group | Low group | P-value |
| Age (years) | |||||||
| <50 | 18 | 8 | 10 | 0.573 | 9 | 9 | 1.000 |
| >50 | 42 | 22 | 20 | 21 | 21 | ||
| Sex | |||||||
| Female | 32 | 20 | 12 | 0.038 | 21 | 11 | 0.009 |
| Male | 28 | 10 | 18 | 9 | 19 | ||
| Lymphatic metastasis | |||||||
| No | 36 | 17 | 19 | 0.598 | 19 | 17 | 0.598 |
| Yes | 24 | 13 | 11 | 11 | 13 | ||
| Tumor stage | |||||||
| Stage I, II | 33 | 16 | 17 | 0.796 | 18 | 15 | 0.436 |
| Stage III, IV | 27 | 14 | 13 | 12 | 15 | ||
| TNM staging system | |||||||
| T1+T2 | 32 | 13 | 19 | 0.120 | 20 | 12 | 0.038 |
| T3+T4 | 28 | 17 | 11 | 10 | 18 | ||
| CLDN10-AS1 expression | DLX6-AS1 expression | ||||||
| Characteristics | No. | High group | Low group | P-value | High group | Low group | P-value |
| Age (years) | |||||||
| <50 | 18 | 8 | 10 | 0.573 | 7 | 11 | 0.260 |
| >50 | 42 | 22 | 20 | 23 | 19 | ||
| Sex | |||||||
| Female | 32 | 16 | 16 | 1.000 | 15 | 17 | 0.605 |
| Male | 28 | 14 | 14 | 15 | 13 | ||
| Lymphatic metastasis | |||||||
| No | 36 | 21 | 15 | 0.113 | 20 | 16 | 0.292 |
| Yes | 24 | 9 | 15 | 10 | 14 | ||
| Tumor stage | |||||||
| Stage I, II | 33 | 21 | 12 | 0.020 | 11 | 22 | 0.004 |
| Stage III, IV | 27 | 9 | 18 | 19 | 8 | ||
| TNM staging system | |||||||
| T1+T2 | 32 | 18 | 14 | 0.301 | 15 | 17 | 0.605 |
| T3+T4 | 28 | 12 | 16 | 15 | 13 | ||
| ADAMTS9-AS1 expression | |||||||
| Characteristics | No. | High group | Low group | P-value | |||
| Age (years) | |||||||
| <50 | 18 | 6 | 12 | 0.091 | |||
| >50 | 42 | 24 | 18 | ||||
| Sex | |||||||
| Female | 32 | 17 | 15 | 0.605 | |||
| Male | 28 | 13 | 15 | ||||
| Lymphatic metastasis | |||||||
| No | 36 | 20 | 16 | 0.291 | |||
| Yes | 24 | 10 | 14 | ||||
| Tumor stage | |||||||
| Stage I, II | 33 | 18 | 15 | 0.436 | |||
| Stage III, IV | 27 | 12 | 15 | ||||
| TNM staging system | |||||||
| T1+T2 | 32 | 21 | 11 | 0.010 | |||
| T3+T4 | 28 | 9 | 19 | ||||
lncRNAs, long non-coding RNAs.
Discussion
CRC is one of the most common cancers around the world, and its incidence and mortality remain high. READ is a type of CRC and has a higher incidence than the other types of colon adenocarcinoma (COAD) (15). Although great progress in the treatment, prognosis and diagnosis of READ has been achieved through studies, its mortality still remains high, which may be due to the lack of efficient biomarkers and the unclear mechanisms underlying READ.
Recent studies have demonstrated that lncRNAs play vital roles during tumor progression, and some of them may be biomarkers for better prognosis and diagnosis (16–18). lncRNAs function in many ways. The ceRNA hypothesis postulates that lncRNAs may compete with mRNAs for the binding sites of miRNAs and may further affect mRNA expression through MREs (8). The hypothesis makes the relationship of miRNAs, mRNAs and lncRNAs more complicated and better explains the interaction among a variety of types of RNAs at the genetic level.
There are many studies on lncRNAs in CRC (19,20), but few of them have focused on READ. Additionally, the sample sizes of previous studies were not large enough, and almost none of the studies focused on the potential ceRNA network. In the present study, we aimed to research the lncRNAs that may have the ability to be better biomarkers and further explored the interaction among lncRNAs, miRNAs and mRNAs by constructing a ceRNA network in READ.
In this study, we identified READ-specific lncRNAs, mRNAs and miRNAs based on the intersecting differential expression between tumor tissues from all four stages and adjacent non-tumor tissues. We further analyzed the functions and pathways involving the differentially expressed genes by GO and KEGG. Then, with the use of bioinformatics tools, we constructed a ceRNA network with READ-specific mRNAs, miRNAs and lncRNAs. We further analyzed the lncRNAs involved in the ceRNA network, examining their correlations with clinical features, and we identified specific lncRNAs that were correlated with overall survival. We finally confirmed our findings in the tissues of 30 READ patients using qRT-PCR.
Some lncRNAs from the READ-specific lncRNAs have been reported as playing vital roles in cancers. CCAT1 has been reported to promote gallbladder cancer development (21), and CCAT2 has been identified to play a significant role in the progression of colon cancer (22). This also supported the reliability of our analysis. Through univariate and multivariate Cox regression analyses, 3 lncRNAs (LINC01602, LINC02163 and MIR497HG) from the READ-specific lncRNAs were identified as being associated with overall survival. LINC01602 and MIR497HG were negatively related to overall survival, and LINC02163 was positively related to overall survival. These 3 lncRNAs were not involved in the ceRNA network but may still play vital roles in READ and be potential indicators of the prognosis of READ since they were related to overall survival.
Using GO and KEGG, we analysed the functions and pathways of READ-specific mRNAs. The GO results revealed that the functions enriched, involved aspects of immune function, metabolism and cellular functions. Among the results of the KEGG pathway analysis, several were confirmed to be cancer-associated. PI3K/AKT signaling plays crucial roles in reducing apoptosis, stimulating cell growth and increasing proliferation, as reported in previous studies (23). It has also been reported that many lncRNAs including PlncRNA-1 and AB073614 affect CRC through the PI3K/AKT signaling pathway (24,25). Studies have demonstrated that AMPK can also be activated by some lncRNAs. Li et al revealed that liver kinase B1 (LKB1) phosphorylates and promotes AMPK and then reduces cancer cell proliferation and metabolism (26). Available data indicate that Wnt signaling substantially impacts non-small cell lung cancer (NSCLC) tumorigenesis, prognosis, and resistance to therapy (27). Previous studies have reported that cell adhesion molecules (CAMs) play a significant role in the progression of metastasis (28). Li et al found that by activating the cGMP/PKG pathway, Wnt/β-catenin signaling can be suppressed (29). Some other cancer-associated pathways such as those known to be involved in CRC were also revealed by our results, further demonstrating the reliability of the results. These pathways and functions may also be related to READ-specific lncRNAs due to different interactions between lncRNAs and mRNAs in READ.
With the ceRNA network in READ, we can further research the underlying mechanism of the intersections among lncRNAs, mRNAs and miRNAs in READ. Several interactions between RNAs in our network have been previously confirmed. UCA1 interacts with miR-182 to modulate glioma proliferation (30), and UCA1 regulates miR-143 to promote the invasion and EMT of bladder cancer (31). These previous studies strongly demonstrate that our analysis was reliable. In addition, the network can aid in understanding the interactions of miRNAs, lncRNAs and mRNAs in READ from various perspectives. Some lncRNAs that exist in the ceRNA network were previously reported to be READ-related lncRNAs. For example, Yang et al discovered that HULC promotes colorectal carcinoma progression by epigenetically repressing NKD2 expression (32). Han et al demonstrated that the lncRNA CRNDE can regulate the progression and chemoresistance of CRC via modulation of the expression levels of miR-181a-5p and Wnt/β-catenin signaling activity (33). High expression levels of PCAT-1 were involved in CRC progression and it can be a novel biomarker of poor prognosis in patients with CRC (34). PVT1 may be a new oncogene co-amplified with c-Myc in CRC tissues and extracellular vesicles and functionally correlated with the proliferation and apoptosis of CRC cells (35). Most lncRNAs that interacted with other RNAs may play significant roles in READ processes Through further investigation of the lncRNAs involved in the network, 7 lncRNAs were identified as being associated with clinical features. Among these 7 lncRNAs, UCA1, HULC and LINC00484 appeared often, and these 3 lncRNAs may be the most clinically relevant lncRNAs, potentially acting as biomarkers. Further study still needs to be performed to understand their correlation with clinical features and their potential as efficient biomarkers.
To verify our bioinformatic analysis, we randomly selected 4 lncRNAs from the network to assess their expression levels in 30 paired READ tumor tissues and adjacent non-tumor tissues. We also confirmed the correlation between the expression pattern and the clinical features of 7 lncRNAs using qRT-PCR. The results revealed that our analysis was credible.
Our analysis has great meaning in the study of a ceRNA network in READ and some results were confirmed by qRT-PCR. However, there were still several limitations in our study. Firstly, the sample number of normal tissues was not very large although previous studies with a small sample size also exist (36). Secondly the follow-up information of our patients was not enough to study the overall survival, most follow-up information after surgery was ~2–3 years. In the future, we will validate our analysis results as soon as we obtain enough follow-up information. Thirdly, further research is also warranted on the functions of key lncRNAs in vivo and in vitro.
In conclusion, our study revealed READ-specific lncRNAs using bioinformatics analysis and studied their associations with clinical features based on data from TCGA database. To the best of our knowledge, studies about lncRNA profiles with such a large sample size are rare. Some key lncRNAs may become efficient biomarkers for READ diagnosis and prognosis. In addition, we constructed a ceRNA network with READ-specific lncRNAs, miRNAs and mRNAs, and it revealed the relationship among these three types of RNAs and aided in elucidating the mechanisms underlying READ on the genetic level.
Acknowledgements
Not applicable.
Funding
The study was funded by Jiangsu Key Medical Discipline (General Surgery) (no. ZDXKA2016005).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
ZZ conceived, designed and further performed the study. SeW, DJ, WQ, QW, JL, JG, WP, TH, BJ, YZ, ShW and YS were also involved in the conception of the study and gave their advice in the process of the research. SW and DJ helped ZZ did the data analysis. ZZ wrote the paper. ZZ and DJ reviewed and edited the manuscript. All authors have read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
All experimental protocols were approved by the Nanjing Medical University (Nanjing, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Lee YJ, Kang YR, Lee SY, Jin Y, Han DC, Kwon BM. Ginkgetin induces G2-phase arrest in HCT116 colon cancer cells through the modulation of b-Myb and miRNA34a expression. Int J Oncol. 2017;51:1331–1342. doi: 10.3892/ijo.2017.4116. [DOI] [PubMed] [Google Scholar]
- 3.Wu S, Wu F, Jiang Z. Identification of hub genes, key miRNAs and potential molecular mechanisms of colorectal cancer. Oncol Rep. 2017;38:2043–2050. doi: 10.3892/or.2017.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamura S, Imai-Sumida M, Tanaka Y, Dahiya R. Interaction and cross-talk between non-coding RNAs. Cell Mol Life Sci. 2018;75:467–484. doi: 10.1007/s00018-017-2626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sana J, Faltejskova P, Svoboda M, Slaby O. Novel classes of non-coding RNAs and cancer. J Transl Med. 2012;10:103. doi: 10.1186/1479-5876-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi K, Yan I, Haga H, Patel T. Long noncoding RNA in liver diseases. Hepatology. 2014;60:744–753. doi: 10.1002/hep.27043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svoboda M, Slyskova J, Schneiderova M, Makovicky P, Bielik L, Levy M, Lipska L, Hemmelova B, Kala Z, Protivankova M, et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis. 2014;35:1510–1515. doi: 10.1093/carcin/bgu055. [DOI] [PubMed] [Google Scholar]
- 8.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bassett AR, Azzam G, Wheatley L, Tibbit C, Rajakumar T, McGowan S, Stanger N, Ewels PA, Taylor S, Ponting CP, et al. Understanding functional miRNA-target interactions in vivo by site-specific genome engineering. Nat Commun. 2014;5:4640. doi: 10.1038/ncomms5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L, Qiu M, Xu Y, Wang J, Zheng Y, Li M, Xu L, Yin R. Upregulation of long non-coding RNA PRNCR1 in colorectal cancer promotes cell proliferation and cell cycle progression. Oncol Rep. 2016;35:318–324. doi: 10.3892/or.2015.4364. [DOI] [PubMed] [Google Scholar]
- 12.Cao Y, Lin M, Bu Y, Ling H, He Y, Huang C, Shen Y, Song B, Cao D. p53-inducible long non-coding RNA PICART1 mediates cancer cell proliferation and migration. Int J Oncol. 2017;50:1671–1682. doi: 10.3892/ijo.2017.3918. [DOI] [PubMed] [Google Scholar]
- 13.Chen DL, Chen LZ, Lu YX, Zhang DS, Zeng ZL, Pan ZZ, Huang P, Wang FH, Li YH, Ju HQ, et al. Long noncoding RNA XIST expedites metastasis and modulates epithelial-mesenchymal transition in colorectal cancer. Cell Death Dis. 2017;8:e3011. doi: 10.1038/cddis.2017.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding J, Li J, Wang H, Tian Y, Xie M, He X, Ji H, Ma Z, Hui B, Wang K, et al. Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis. 2017;8:e2997. doi: 10.1038/cddis.2017.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turati F, Bravi F, Di Maso M, et al. Adherence to the World Cancer Research Fund/American Institute for Cancer Research recommendations and colorectal cancer risk. Eur J Cancer. 2017;85:86–94. doi: 10.1016/j.ejca.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Liang X, Li Y. Long non-coding RNA MEG3 inhibits cell growth of gliomas by targeting miR-93 and inactivating PI3K/AKT pathway. Oncol Rep. 2017;38:2408–2416. doi: 10.3892/or.2017.5871. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Huang S, Li Y, Zhang W, He K, Zhao M, Lin H, Li D, Zhang H, Zheng Z, et al. Decreased expression of LncRNA SLC25A25-AS1 promotes proliferation, chemoresistance, and EMT in colorectal cancer cells. Tumor Biol. 2016;37:14205–14215. doi: 10.1007/s13277-016-5254-0. [DOI] [PubMed] [Google Scholar]
- 18.Qiu JJ, Zhang XD, Tang XY, Zheng TT, Zhang Y, Hua KQ. ElncRNA1, a long non-coding RNA that is transcriptionally induced by oestrogen, promotes epithelial ovarian cancer cell proliferation. Int J Oncol. 2017;51:507–514. doi: 10.3892/ijo.2017.4030. [DOI] [PubMed] [Google Scholar]
- 19.Ozawa T, Matsuyama T, Toiyama Y, Takahashi N, Ishikawa T, Uetake H, Yamada Y, Kusunoki M, Calin G, Goel A. CCAT1 and CCAT2 long noncoding RNAs, located within the 8q.24.21 ‘gene desert’, serve as important prognostic biomarkers in colorectal cancer. Ann Oncol. 2017;28:1882–1888. doi: 10.1093/annonc/mdx248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang L, Xu L, Wang Q, Wang M, An G. Dysregulation of long non-coding RNA profiles in human colorectal cancer and its association with overall survival. Oncol Lett. 2016;12:4068–4074. doi: 10.3892/ol.2016.5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY, Gong W, Quan ZW. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e1583. doi: 10.1038/cddis.2014.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafà R, Song J, Guo Z, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–1461. doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Danielsen SA, Eide PW, Nesbakken A, Guren T, Leithe E, Lothe RA. Portrait of the PI3K/AKT pathway in colorectal cancer. Biochim Biophys Acta. 2015;1855:104–121. doi: 10.1016/j.bbcan.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Song W, Mei JZ, Zhang M. LncRNA PlncRNA-1 promotes colorectal cancer cell progression by regulating PI3K/Akt signaling pathway. Oncol Res. 2017 Aug 23; doi: 10.3727/096504017X15031557924132. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Kuang H, Xue J, Liao L, Yin F, Zhou X. LncRNA AB073614 regulates proliferation and metastasis of colorectal cancer cells via the PI3K/AKT signaling pathway. Biomed Pharmacother. 2017;93:1230–1237. doi: 10.1016/j.biopha.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 26.Li N, Huang D, Lu N, Luo L. Role of the LKB1/AMPK pathway in tumor invasion and metastasis of cancer cells (Review) Oncol Rep. 2015;34:2821–2826. doi: 10.3892/or.2015.4288. [DOI] [PubMed] [Google Scholar]
- 27.Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst. 2014;106:djt356. doi: 10.1093/jnci/djt356. [DOI] [PubMed] [Google Scholar]
- 28.Saadatmand S, de Kruijf EM, Sajet A, Dekker-Ensink NG, van Nes JG, Putter H, Smit VT, van de Velde CJ, Liefers GJ, Kuppen PJ. Expression of cell adhesion molecules and prognosis in breast cancer. Br J Surg. 2013;100:252–260. doi: 10.1002/bjs.8980. [DOI] [PubMed] [Google Scholar]
- 29.Li N, Xi Y, Tinsley HN, Gurpinar E, Gary BD, Zhu B, Li Y, Chen X, Keeton AB, Abadi AH, et al. Sulindac selectively inhibits colon tumor cell growth by activating the cGMP/PKG pathway to suppress Wnt/β-catenin signaling. Mol Cancer Ther. 2013;12:1848–1859. doi: 10.1158/1535-7163.MCT-13-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Z, Wang Y, Huang G, Wang Q, Zhao D, Chen L. The lncRNA UCA1 interacts with miR-182 to modulate glioma proliferation and migration by targeting iASPP. Arch Biochem Biophys. 2017;623–624:1–8. doi: 10.1016/j.abb.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Luo J, Chen J, Li H, Yang Y, Yun H, Yang S, Mao X. LncRNA UCA1 promotes the invasion and EMT of bladder cancer cells by regulating the miR-143/HMGB1 pathway. Oncol Lett. 2017;14:5556–5562. doi: 10.3892/ol.2017.6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang XJ, Huang CQ, Peng CW, Hou JX, Liu JY. Long noncoding RNA HULC promotes colorectal carcinoma progression through epigenetically repressing NKD2 expression. Gene. 2016;592:172–178. doi: 10.1016/j.gene.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY, Yu ZW, Jia YH, Bai XF, Li L, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol Cancer. 2017;16:9. doi: 10.1186/s12943-017-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ge X, Chen Y, Liao X, Liu D, Li F, Ruan H, Jia W. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Med Oncol. 2013;30:588. doi: 10.1007/s12032-013-0588-6. [DOI] [PubMed] [Google Scholar]
- 35.Guo K, Yao J, Yu Q, Li Z, Huang H, Cheng J, Wang Z, Zhu Y. The expression pattern of long non-coding RNA PVT1 in tumor tissues and in extracellular vesicles of colorectal cancer correlates with cancer progression. Tumor Biol. 2017;39:1010428317699122. doi: 10.1177/1010428317699122. [DOI] [PubMed] [Google Scholar]
- 36.Wang S, Zhang C, Zhang Z, Qian W, Sun Y, Ji B, Zhang Y, Zhu C, Ji D, Wang Q, et al. Transcriptome analysis in primary colorectal cancer tissues from patients with and without liver metastases using next-generation sequencing. Cancer Med. 2017;6:1976–1987. doi: 10.1002/cam4.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.