Abstract
Bladder cancer is the most common tumor of the urinary tract. Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ), a gene encoding the 14-3-3ζ protein, has been observed to be frequently amplified in bladder cancer. However, the role of 14-3-3ζ in various types of cancer is controversial. With reproduction of The Cancer Genome Atlas database, we searched the correlation of YWHAZ expression with survival outcomes of multiple cancers in silico. Our results revealed that only in bladder cancer was there a positive survival trend with YWHAZ overexpression. To study its role in bladder cancer, YWHAZ was successfully downregulated by lentivirus in 5637 and T24 cells. MTT and colony-formation assays showed that YWHAZ downregulation increased cell proliferation. Wound healing and Transwell assays showed that YWHAZ downregulation enhanced cell migration and invasiveness. FACS analysis showed that YWHAZ induced cell cycle arrest, but not apoptosis. A xenograft tumor model revealed that YWHAZ knockdown enhanced tumor growth. Gene set enrichment analysis confirmed that the cell cycle pathway plays a vital role in the function of the YWHAZ gene. In conclusion, knockdown of YWHAZ promoted both in vitro and in vivo tumorigenesis in bladder cancer and may be a novel biomarker for bladder cancer deserving further study.
Keywords: YWHAZ, 14-3-3ζ, bladder cancer, cell cycle
Introduction
Bladder cancer remains one of the most commonly diagnosed urological malignancies worldwide (1). Approximately one-fourth of patients present with muscle-invasive bladder cancer (MIBC) at diagnosis. The outcomes for MIBC are poor and progress in regards to the treatment of bladder cancer has been stagnant for decades (2). Recently, The Cancer Genome Atlas (TCGA) project revealed the whole genomic characteristics of bladder cancer based on next generation sequencing (3). This provided new insight into the understanding of the molecular features of bladder cancer.
YWHAZ, a gene that encodes 14-3-3ζ, has been demonstrated to be overexpressed in some MIBC patients (4). 14-3-3 proteins are a family of conserved regulatory molecules that bind to phosphorylate serine or threonine residue on their target molecules. Through 14-3-3 protein's binding ability, it regulates critical proteins in a variety of cell processes. To date, seven different isoforms (β, γ, ε, σ, η, τ and ζ) have been identified (5). The role of 14-3-3ζ in cancer remains controversial. The overexpression of 14-3-3ζ is generally associated with malignant biological behavior and worse survival outcomes in many other tumor types, such as prostate, lung and breast cancer (6–8). However, in other cancer types such as squamous cell carcinoma of the tongue, 14-3-3ζ silencing also retards tumor growth (9).
In our previous study, we revealed that the amplification of YWHAZ, along with TP53 or CDKN2A loss predicted better survival (10). This finding demonstrated that YWHAZ may play an antitumor effect in bladder cancer development. Therefore, in the present study, we aimed to investigate the function of YWHAZ by downregulation of its expression both in vitro and in vivo, and its related pathway was also explored in silico through gene set enrichment analysis (GSEA).
Materials and methods
Data mining and analysis in the TCGA database
An in silico reproduction using the TCGA database was performed in the present study, as previously described (11–13). The TCGA bladder cancer (provisional), pan lung cancer (provisional), invasive breast cancer (provisional) and prostate adenocarcinoma (provisional) was respectively chosen on the cBioPortal online platform (www.cbioportal.org). Cases with YWHAZ alteration were queried. The ‘Survival’ function was used to plot Kaplan-Meier curves for overall survival and disease-free survival.
Cell culture
Two MIBC cell lines, T24 and 5637, were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). The cells were cultured in RPMI-1640 medium (Life Technologies; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS), 100 U/ml penicillin and 0.1 mg/ml streptomycin. The cells were maintained in an atmosphere of 5% CO2 at 37°C.
RNA extraction and real-time quantitative PCR (qPCR)
Total cellular RNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Reverse transcription of RNA was carried out using a M-MLV Reverse Transcription kit (Promega, Madison, WI, USA) according to the manufacturer's instructions. The relative expression level of YWHAZ was determined by the real-time quantitative PCR using SYBR Premix Ex Taq II (Takara, Tokyo, Japan). The primer was synthesized by Shanghai Genechem Co., Ltd. (Shanghai, China) for YWHAZ (forward primer, AGCCATTGCTGAACTTGATACA and reverse primer, AATTTTCCCCTCCTTCTCCTG); and reference gene (GAPDH) (forward primer, TGACTTCAACAGCGACACCCA and reverse primer, CACCCTGTTGCTGTAGCCAAA). The 2−ΔΔCt method was used to calculate the relative expression ratio of YWHAZ.
Lentiviral RNA interference
Lentivirus encoding YWHAZ shRNA was generated by Shanghai Genechem Co., Ltd. Lentivirus expressing scramble shRNA were used as negative control (NC). Cells were plated in 12-well plates (1×105 cells/well), transduced with 5 MOI lentiviral particles [using 8 µg/ml hexadimethrine bromide (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)] and incubated at 37°C with 5% CO2. Suppression of YWHAZ in stable cells was confirmed by qPCR.
Cell proliferation assay
Cell viability in each group was measured by using MTT assay. The cells were seeded into 96-well plates at a density of 2×104 cells/well, cultured overnight and treated as mentioned above. Next, 10 µl of 5 mg/ml MTT was added to each well. After 4 h of further incubation, dimethyl sulfoxide (DMSO) was added to solve the formazan, then the optical density at 490 nm of each well was measured with a microplate reader. Cell proliferation ability was analyzed daily for 5 consecutive days.
Colony-formation assay
A total of 10 ml complete medium containing 200 cells were added to each well of a 6-well plate. The plates were incubated at 37°C in 5% CO2 for 2 weeks. Next, the cells were washed with phosphate-buffered saline (PBS) solution and fixed by 4% paraformaldehyde, and then were stained with Giemsa for 10 min. Colonies containing at least 50 cells were counted under a microscope (XDS-100; Carl Zeiss, Göttingen, Germany).
Cell migration and invasion assays
Migration and invasive abilities were assessed using a cell invasion assay kit (Chemicon International, Inc., Temecula, CA, USA). Cells (5×104 cells/well) were cultured in the upper chamber of the 8.0-µm pore-size cell culture inserts that were either coated or uncoated with Matrigel for migration and invasive assays, respectively. The insets were then placed in a 24-well plate filled with medium with 5% FBS. The cells that penetrated to the underside of the surfaces of the inserts were fixed and stained with the Diff-Quick method (Thermo Fisher Scientific, Inc.) and were counted under the microscope (XDS-100; Carl Zeiss). The mean cell number of three high power fields at ×100 magnification for each condition was calculated. Assays were repeated at least three times.
Wound healing assay
When the cells reached 95% confluence in 6-well plates after transfection, a 200-µl pipette tip was used to draw a wound with the same width. After washing and removing the cell debris, the cells were cultured using a serum-free medium under standard conditions. The same area of the gap was taken at ×100 magnifications using a microscope equipped with a digital camera (Olympus Corp., Centre Valley, PA, USA) at 0, 4, 8 and 24 h after scratching, and the scratch width was quantified using ImageJ software (http://rsbweb.nih.gov/ij/). The migration rate was calculated as the difference between the initial width and the width at the different time-points.
Cell cycle assay
Cells were plated in 25-cm2 flasks and incubated overnight. The cells were then collected and fixed in pre-cold 70% ethanol for 1.5 h at 4°C. After fixation, the cells were washed in PBS again and centrifuged for 5 min at 1,000 rpm. The PBS was discarded and propidium iodide (PI) was added to a final concentration of 50 µg/ml in dark at 4°C for 30 min. Flow cytometric analysis was performed on the FACSCalibur flow cytometer (Becton-Dickinson; BD Biosciences, San Jose, CA, USA). The cell cycle was analyzed using CellQuest software (BD Biosciences).
Cell apoptosis assay
The cells were plated in 25-cm2 flasks and were incubated overnight. The cells were then collected and adjusted with staining buffer at a density of 106 cells/ml. After incubation with 5 µl of Annexin V-FITC and 10 µl of PI in dark for 15 min, the cells were analyzed by flow cytometry (Becton-Dickinson). Apoptotic cells were recognized as having a high Annexin V fluorescence signal with a low PI signal. The percentages of apoptotic cells were calculated by data from FACS analysis.
Bladder cancer xenograft model
Twelve male BALB/c nude mice at 6 weeks of age, weighing 18–22 g, were obtain from SLAC Laboratory Animal Co., Ltd. (Shanghai, China) and were bred in a special pathogen-free (SPF) grade laboratory in Fudan University. The mice were housed 5–10/cage, in a 12:12-h light:dark cycle environment, with ad libitum access to food and water. Mice were randomly divided into 2 groups (T24; treatment vs. control). A total of 107 cancer cells resuspended in 100 ml of PBS were injected subcutaneously at the left axilla of each mouse. Tumors became perceptible at ~5 mm in diameter on day 7. Thereafter, intratumoral injection of 50 µl of the lentivirus at 108 U/ml or control (PBS) with the same volume was administered. All mice were sacrificed on day 35 and the tumors were extracted. The tumor size was calculated with the following formula: Volume = length × width2/2. All experimental protocols were approved by the Institutional Review Board of the Department of Laboratory Animal Science of Fudan University (Shanghai, China).
Immunohistological analysis and assessment of Ki-67
Firstly, 2-µm unstained xenograft tumor sections from formalin-fixed paraffin embedded (FFPE) tissue were deparaffinized in xylene and rehydrated through graded alcohols. After antigen retrieval, the sections were stained with primary antibody, Ki-67 (dilution 1:200; 28-9; Abcam), followed by antibody localization using the Dako EnVision+ System-HRP labelled polymer (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA). Staining was visualized by a 5-min incubation with diaminobenzidine.
The staining patterns were assessed by two independent pathologists using standard light microscopy (XDS-100; Carl Zeiss). Positive staining was considered when >10% of tumor cells showed reactivity. Stain intensity and the percentage of tumor cells stained were categorized as <25%, 25–50%, >50–75% and >75% according to the number of positive tumor cells stained.
GSEA of TCGA data
To investigate the YWHAZ overexpression and the cancer-related pathway, GSEA was performed using TCGA bladder cancer (provisional) level 3 RNASeq V2 datasets. This analytical technique is designed to test a priori defined gene sets for association with phenotypes (14). KEGG gene sets (v6.0) and phenotype label files were created and loaded into GSEA software (v2.0.13; Broad Institute, Cambridge, MA, USA). Cases with YWHAZ mRNA expression were queried on the cBioPortal online platform. The phenotype label was YWHAZ overexpression vs. YWHAZ normal expression based on the cBioPortal platform. The number of permutations was set to 1,000. A ranked-list metric was generated by calculating the signal-to-noise ratio, which is based on the difference of means scaled according to the standard deviation.
Statistical analysis
Statistical analysis was performed using IBM SPSS version 20.0 (IBM SPSS, Armonk, NY, USA). Data are shown as means ± SD. The difference between two groups was analyzed using the Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
YWHAZ amplification indicates a better survival trend in bladder cancer
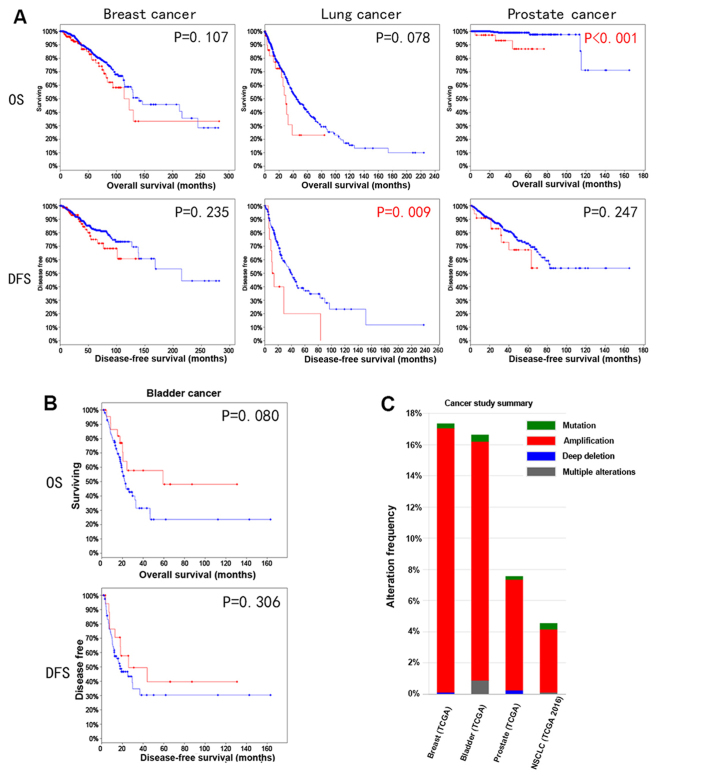
Since YWHAZ amplification is commonly observed in many types of cancer, such as breast, lung and prostate cancer, we therefore investigated its correlation with survival outcomes among these types of cancer. Amplification was determined using algorithm GISTIC value as ≥2 according to the TCGA definition. Amplification was found in 35/442 (7%) patients with prostate cancer, 47/1,144 (4%) patients with lung cancer, 180/1,080 (17%) patients with breast cancer and 69/408 (17%) patients with bladder cancer (Fig. 1C). A worse survival trend among breast, lung and prostate cancer both in overall survival and disease-free survival was noted (Fig. 1A). Of note, lung cancer patients with YWHAZ amplification had significantly worse disease-free survival compared with those without YWHAZ amplification (P=0.009). Similarly, the overall survival defects in prostate cancer patients with YWHAZ amplification also reached statistical significance (P<0.001). In bladder cancer, there was a better survival trend for patients with YWHAZ amplification. Specifically, the significance for overall survival was marginal (P=0.08) (Fig. 1B).
Figure 1.
Reproduction of TCGA database using cBioPortal shows that the association with YWHAZ amplification had (A) relatively worse survival outcomes among breast, lung and prostate cancer, but (B) a better survival trend in bladder cancers. (C) YWHAZ amplification was the dominant gene alteration found among breast, bladder, prostate and lung cancer. TCGA, The Cancer Genome Atlas; OS, overall survival; DFS, disease-free survival.
Downregulation of YWHAZ enhances bladder cancer cell proliferation and colony number
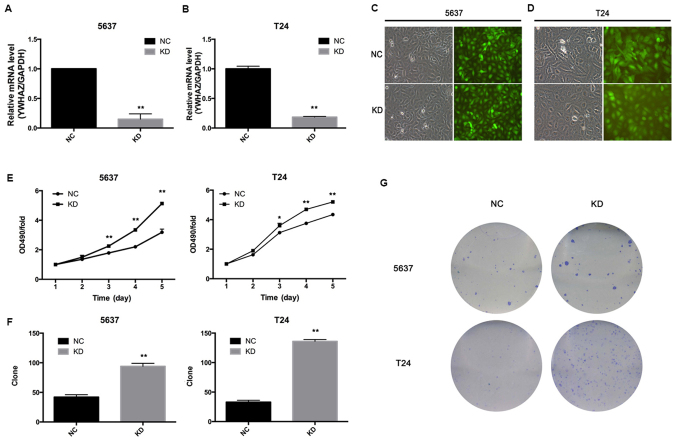
Next, we investigated the effect of YWHAZ on bladder cancer cell growth, and we infected T24 and 5637 cells with Lenti-YWHAZ (knockdown, KD group) or Lenti-NC (negative control, NC group). Transfection of 5637 and T24 cells with Lenti-YWHAZ significantly inhibited YWHAZ gene expression (P<0.05) (Fig. 2A and B). Fluorescence was observed using fluorescence microscope 96 h after infection with the indicated lentivirus (Fig. 2C and D). MTT assay demonstrated that the proliferation ability of the T24 and 5637 cells infected with Lenti-YWHAZ was markedly enhanced compared with the the Lenti-NC-infected group 3 days after infection (Fig. 2E). Colony-formation assay showed that the colony numbers of the T24 (136±3 vs. 33±3; P<0.01) and 5637 (94±5 vs. 42±4; P<0.01) cells infected with Lenti-YWHAZ were significantly higher compared with these values in the Lenti-NC-infected group (Fig. 2F and G). Therefore, it is suggested that knockdown of YWHAZ enhances bladder cancer cell proliferation.
Figure 2.
YWHAZ downregulation increased cell proliferation and colony forming ability. (A and B) Transfection of Leni-YWHAZ (KD group) successfully downregulated YWHAZ expression in the 5637 and T24 bladder cancer cell lines. (C and D) Fluorescence showed that cells were successfully transfected with the lentivirus. (E) MTT assay showed that cell viability with OD490 value was significantly elevated in the Leni-YWHAZ (KD) groups after 3 days. Clone formation assay showed that a number of colonies were increased in the KD groups compared with the control groups after 14 days in the 5637 and T24 cells. (F) Quantitation of the colony numbers and (G) representative images of the colony formation assay in the KD and NC groups. KD, knockdown; NC, negative control. *P<0.05, **P<0.01.
Downregulation of YWHAZ enhances the migration and invasiveness of bladder cancer cells
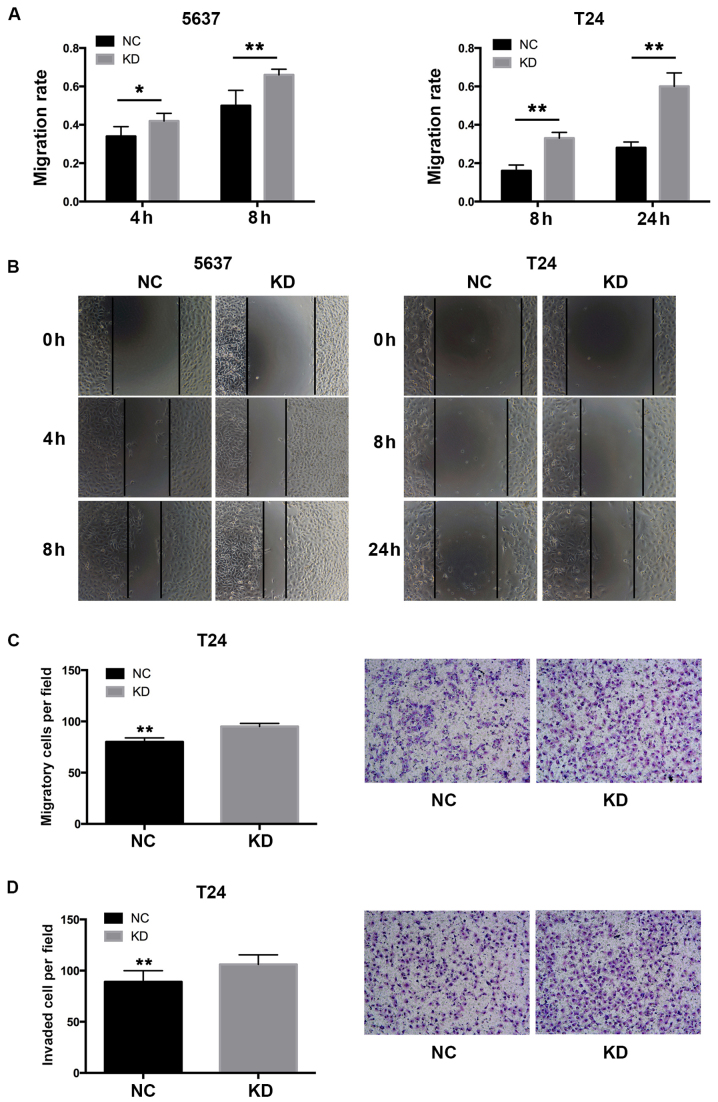
To investigate whether the overexpression of YWHAZ has any influence on the migration ability of bladder cancer cells, wound healing assays were performed. Knockdown of YWHAZ significantly enhanced the migration rate of the bladder cancer cells in both the T24 (0.33±0.03 vs. 0.16±0.03 at 8 h, P<0.01; 0.6±0.07 vs. 0.28±0.03 at 24 h, P<0.01) and 5637 (0.42±0.04 vs. 0.34±0.05 at 4 h, P=0.03; 0.66±0.03 vs. 0.5±0.08 at 8 h, P<0.01) cells (Fig. 3). Of note, T24 cells with downregulated YWHAZ demonstrated almost doubled migration capacity compared to the respective controls. We further performed Transwell assays to investigate the migration and invasive ability of the T24 bladder cancer cells. The result showed that the Lenti-YWHAZ group had significantly elevated migrated (95±3.02 vs. 80±3.94; P<0.01) and invasive cells (88±7.02 vs. 106±5.76; P<0.01) compared with the Lenti-NC infected group (Fig. 3). These results demonstrated that YWHAZ downregulation significantly contributed to the migration and invasiveness of bladder cancer cells.
Figure 3.
YWHAZ downregulation enhances the migration and invasiveness of bladder cancer cells. (A) Wound healing assays showed that the migration rate was significantly elevated in the 5637 and T24 KD cells when compared to the NC groups. (B) Representative images of the wound healing assay in the 5637 and T24 cells. (C) Transwell assay showed that the number of migrated cells was significantly increased in the T24 KD cells compared with the NC group and representative images of T24 cells are shown. (D) Transwell assay showed that the number of invaded cells was significantly increased in the T24 KD cells compared with the NC group and representative images of T24 cells are shown. KD, knockdown; NC, negative control. *P<0.05, **P<0.01.
YWHAZ knockdown promotes cell cycle progression
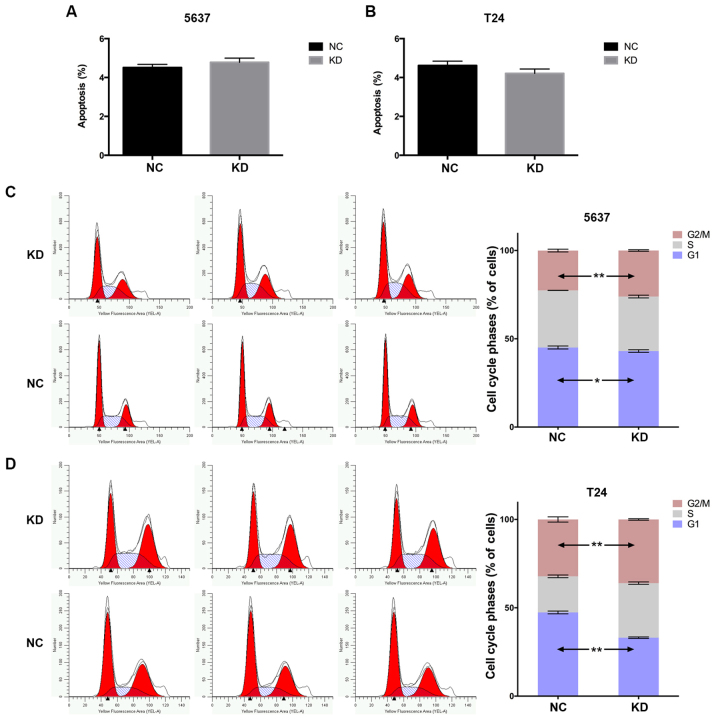
To address the mechanisms underlying YWHAZ downregulation of proliferation and migration of bladder cancer cells, we further investigated the influence of YWHAZ knockdown on apoptosis. The percentage of apoptotic cells were comparable with those of the Lenti-NC infected group (P>0.05) (Fig. 4A and B). Furthermore, the cell cycle profile was analyzed through flow cytometry. The distribution of cell cycle phase was significantly different between two groups in both the T24 and 5637 cell lines. The percentage of cells in G1 phase infected with Lenti-YWHAZ were significantly lower than those of cells infected with Lenti-NC. An obvious decrease in the G1 phase (45.08±0.84 vs. 43.06±0.73%; P=0.03) and a significant increase in G2/M (26.07±0.41 vs. 22.58±0.78%; P<0.01) cell populations were observed compared with the control group in the 5637 cell line. Similarly, a significant decrease in the G1 phase (45.08±0.84 vs. 43.06±0.73%; P<0.01), while a significant increase in S phase (30.85±0.68 vs. 20.45±0.75%; P<0.001) and G2/M (36.04±0.41 vs. 32.19±1.48%; P=0.001) cell populations were observed compared with the control group in the T24 cell line (Fig. 4C and D). These results demonstrated that YWHAZ knockdown significantly promoted G1/S transition.
Figure 4.
Effects of Lenti-YWHAZ on cell apoptosis and cell cycle distribution as determined by flow cytometric analysis. Flow cytometric analysis showed that Lenti-YWHAZ did not affect the apoptosis of the (A) 5637 and (B) T24 cells compared with the NC groups. Flow cytometric analysis showed the distribution of cell cycle phases in the (C) 5637 and (D) T24 cells. KD, knockdown; NC, negative control. *P<0.05, **P<0.01.
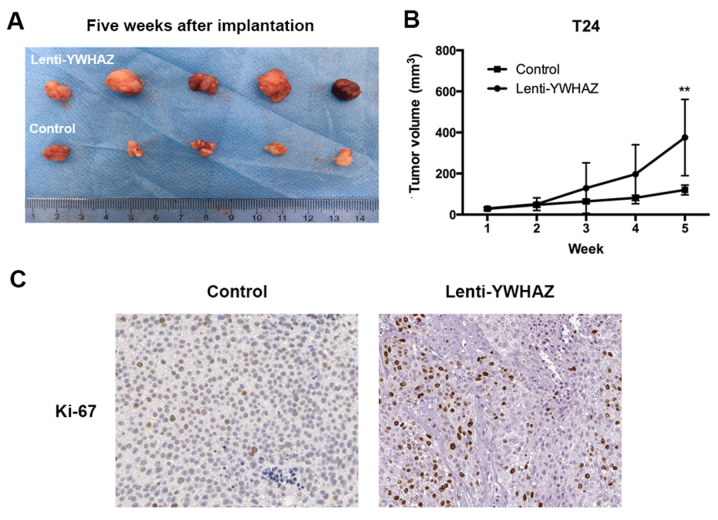
YWHAZ downregulation promotes tumor growth in vivo
To investigate the effect of YWHAZ downregulation on antitumor activities in vivo, we established a T24 xenograft tumor model in nude mice. Assessment of tumor volume indicated that the YWHAZ downregulation group had enhanced tumor growth compared to the control group and the group treated with PBS (Fig. 5A and B). The xenografts were further evaluated by immunohistology in relation to the proliferation marker Ki-67. The results demonstrated that the percentage of Ki-67-positive cells was higher in the YWHAZ downregulation group than that noted in the control group (Fig. 5C and Table I).
Figure 5.
YWHAZ knockdown promotes bladder cancer tumor growth in the xenograft tumor model. (A) Representative images of tumors after 5 weeks of implantation. (B) Tumor volume was significance increased after 5 weeks of implantation in the T24 Lenti-YWHAZ group. (C) Representative images of Ki-67 staining of xenograft tumors after 5 weeks of implantation.
Table I.
Comparison of immunoexpression of tumor marker Ki-67 in the YWHAZ-knockdown and control group in the xenograft model.
| Ki-67 expression levels (%) | Lenti-YWHAZ group (no. of tumors) | Control group (no. of tumors) | P-value |
|---|---|---|---|
| <25 | 0 | 0 | 0.02 |
| 25–50 | 1 | 5 | |
| >50–75 | 3 | 1 | |
| >75 | 2 | 0 |
YWHAZ may exert its function through the cell cycle pathway
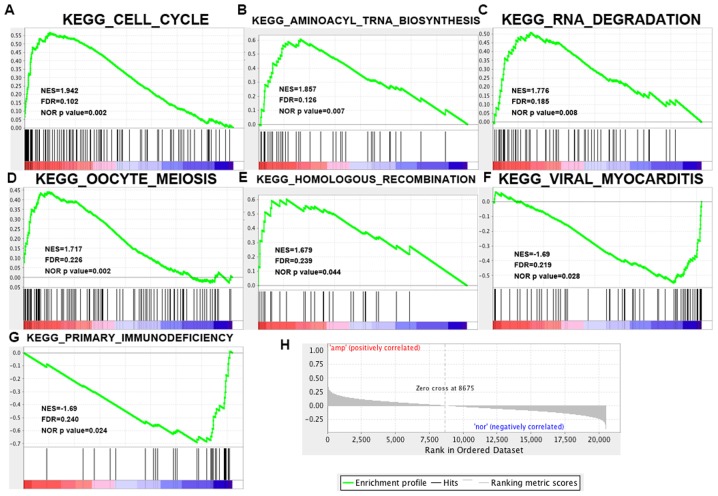
GSEA was performed to determine the associations between YWHAZ expression and cancer-related pathways based on KEGG gene sets. As a result, 60/178 gene sets were found to be upregulated in the YWHAZ overexpression phenotype with 5 gene sets significant at FDR <25% and 4 gene sets significantly enriched at nominal P<0.01. Of the YWHAZ normal-expression phenotype, 118/178 gene sets were upregulated with 2 gene sets significantly enriched at FDR <25% and 2 gene sets significantly enriched at nominal P<0.01. The enrichment plot of 7 upregulated gene sets is shown in Fig. 6. Of these, the cell cycle pathway shows the most relative correlation with the smallest normal P-value 0.002 and FDR 0.102.
Figure 6.
Gene expression profiling identifies pathways upregulated from the TCGA bladder cancer database (provisional). (A-G) The 7 significantly enriched gene sets in bladder cancer. The normalized enrichment score, the false discovery rates (FDR) and the nominal P-value score (NES) are indicated for each gene set. Each bar at the bottom of each panel represents a member gene of the respective pathway from plot A-G and (H) shows its relative location in the ranked list of genes. TCGA, The Cancer Genome Atlas.
Discussion
In the present study, we found that YWHAZ downregulation dramatically promoted tumorigenesis both in vitro and in vivo. This finding suggests that YWHAZ functions as a tumor-suppressor gene and it could be a potential biomarker for bladder cancer. To the best of our knowledge, scarce studies have focused on YWHAZ and its coding protein, 14-3-3ζ on bladder cancer. Thus, this study was an innovative attempt and these findings were also in accordance with the results of our previous study that YWHAZ amplification predicts better survival, under the background of some anti-oncogene loss such as TP53 and CDKN2A (10).
However, previous studies have reported that in some other types of cancer, 14-3-3ζ may serve as a pro-tumor molecule (6,7,15,16). Thus, in our first step, we verified the role of YWHAZ in three typical cancers that have been commonly reported before, breast, lung and prostate cancer. Our results in these three types of cancer showed that although some differences did not reach statistically significance, there was always a trend toward worse survival outcomes when YWHAZ was overexpressed. These demonstrated that 14-3-3ζ may function differentially in bladder cancer than in other types of cancer.
A previous study demonstrated that 14-3-3 protein exerts its function through binding ability. The structure of 14-3-3 contains two subunits that are strongly associated with each other to form C-shaped dimers. In the dimers, the N-terminal helices of two subunits contact one another and form the floor of a central groove. Phosphorylated sites on target peptides and proteins dock directly into the groove (17). The binding partner decides the functions that 14-3-3 may play in multiple cellular process. The majority of studies have demonstrated that 14-3-3 could bind to multiple pro-apoptosis proteins, such as Bax, FOXO3 and AP-1, and thus may affect cell apoptosis (18,19). However, in the present study, we found that YWHAZ downregulation had no effects on cell apoptosis. This suggests that in bladder cancer 14-3-3 may not mainly function in cell apoptosis.
Our results indicated that both in 5637 and T24 cells, YWHAZ knockdown promoted cell cycle transition from G1 to S phase. This suggest that 14-3-3ζ may exert its antitumor function through blocking the cell cycle. Further in silico GSEA analysis also confirmed the hypothesis. Studies have also shown that 14-3-3 family protein could bind to CDC25 family protein, which plays an important role in transitions between cell cycle phases by dephosphorylating and activating CDKs (20). CDC25B and CDC25C play a major role in G2/M progression, whereas CDC25A assists in G1/S transition (21,22). Combining the established facts and our findings, we speculated that 14-3-3 could arrest cell cycle by sequestering the critical stage, thus exerting a tumor-suppressor function. One study has shown that high expression of CDC25B and low expression of 14-3-3σ is associated with the development and poor prognosis in urothelial carcinoma of bladder (23). Since CDC25 have varieties of isoforms, the affinity of 14-3-3ζ to different CDC25 isoforms remains to be explored. Moreover, this could explain our findings in a previous study that YWHAZ amplification significantly benefited survival outcomes under the background of CDKN2A or TP53 loss, both critical factors in cell cycle regulation (10). The malfunction of CDKN2A or TP53 causes disruption of cell cycle control and enhancement of proliferation. Thus, YWHAZ amplification could offset the cell cycle acceleration effect by the reason of critical cell cycle regulator disorder.
Since 14-3-3 proteins have seven isoforms, the affinity of their binding ability to CDC25 protein should be distinct. A study has shown that CDC25B has a strong interaction with 14-3-3ζ and 14-3-3η, but poor with 14-3-3ε (24). The different binding ability may contribute to the contrasting role of different isoforms in a certain cancer type. As an instance, 14-3-3ζ promoted tumor invasion, while 14-3-3σ serves as a tumor-suppressor gene in non-small cell lung cancer (16,25). In addition, the expression level of 14-3-3 isoforms in tissue may be different (26). This may explain why 14-3-3ζ plays a pro-tumor role in various types of cancer, but exerts antitumor function in bladder cancer.
In conclusion, our results indicated that YWHAZ knockdown promoted the tumorigenesis of bladder cancer both in vitro and in vivo, and revealed its correlation with the cell cycle pathway. Importantly, our study highlighted for the first time that YWHAZ may be a novel beneficial prognostic factors in bladder cancer warranting further study.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
SL and CF conceived and designed the study. SL, HJ, QD and HW performed the experiments. SL and HJ wrote the paper. SL, HJ, QD and CF reviewed and edited the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
All experimental protocols were approved by the Institutional Review Board of the Department of Laboratory Animal Science of Fudan University (Shanghai, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Abdollah F, Gandaglia G, Thuret R, Schmitges J, Tian Z, Jeldres C, Passoni NM, Briganti A, Shariat SF, Perrotte P, et al. Incidence, survival and mortality rates of stage-specific bladder cancer in United States: A trend analysis. Cancer Epidemiol. 2013;37:219–225. doi: 10.1016/j.canep.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network: Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matta A, Siu KW, Ralhan R. 14-3-3 zeta as novel molecular target for cancer therapy. Expert Opin Ther Targets. 2012;16:515–523. doi: 10.1517/14728222.2012.668185. [DOI] [PubMed] [Google Scholar]
- 5.Aghazadeh Y, Papadopoulos V. The role of the 14-3-3 protein family in health, disease, and drug development. Drug Discov Today. 2016;21:278–287. doi: 10.1016/j.drudis.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Neal CL, Yao J, Yang W, Zhou X, Nguyen NT, Lu J, Danes CG, Guo H, Lan KH, Ensor J, et al. 14-3-3zeta overexpression defines high risk for breast cancer recurrence and promotes cancer cell survival. Cancer Res. 2009;69:3425–3432. doi: 10.1158/0008-5472.CAN-08-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murata T, Takayama K, Urano T, Fujimura T, Ashikari D, Obinata D, Horie-Inoue K, Takahashi S, Ouchi Y, Homma Y, et al. 14-3-3ζ, a novel androgen-responsive gene, is upregulated in prostate cancer and promotes prostate cancer cell proliferation and survival. Clin Cancer Res. 2012;18:5617–5627. doi: 10.1158/1078-0432.CCR-12-0281. [DOI] [PubMed] [Google Scholar]
- 8.Zhao GY, Ding JY, Lu CL, Lin ZW, Guo J. The overexpression of 14-3-3ζ and Hsp27 promotes non-small cell lung cancer progression. Cancer. 2014;120:652–663. doi: 10.1002/cncr.28452. [DOI] [PubMed] [Google Scholar]
- 9.Jin LM, Han XH, Jie YQ, Meng SS. 14-3-3ζ silencing retards tongue squamous cell carcinoma progression by inhibiting cell survival and migration. Cancer Gene Ther. 2016;23:206–213. doi: 10.1038/cgt.2016.15. [DOI] [PubMed] [Google Scholar]
- 10.Liu S, Wu Y, Yang T, Feng C, Jiang H. Coexistence of YWHAZ amplification predicts better prognosis in muscle-invasive bladder cancer with CDKN2A or TP53 loss. Oncotarget. 2016;7:34752–34758. doi: 10.18632/oncotarget.9158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng C, Xiong Z, Jiang H, Ding Q, Fang Z, Hui W. Genetic alteration in notch pathway is associated with better prognosis in renal cell carcinoma. Biofactors. 2016;42:41–48. doi: 10.1002/biof.1250. [DOI] [PubMed] [Google Scholar]
- 12.Feng C, Ding G, Jiang H, Ding Q, Wen H. Loss of MLH1 confers resistance to PI3Kβ inhibitors in renal clear cell carcinoma with SETD2 mutation. Tumour Biol. 2015;36:3457–3464. doi: 10.1007/s13277-014-2981-y. [DOI] [PubMed] [Google Scholar]
- 13.Feng C, Sun Y, Ding G, Wu Z, Jiang H, Wang L, Ding Q, Wen H. PI3Kβ inhibitor TGX221 selectively inhibits renal cell carcinoma cells with both VHL and SETD2 mutations and links multiple pathways. Sci Rep. 2015;5:9465. doi: 10.1038/srep09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: A desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Li Y, Lin C, Ding J, Liao G, Tang B. Aberrant upregulation of 14-3-3σ and EZH2 expression serves as an inferior prognostic biomarker for hepatocellular carcinoma. PLoS One. 2014;9:e107251. doi: 10.1371/journal.pone.0107251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun N, Wu Y, Huang B, Liu Q, Dong Y, Ding J, Liu Y. Decreased expression of 14-3-3σ, an early event of malignant transformation of respiratory epithelium, also facilitates progression of squamous cell lung cancer. Thorac Cancer. 2015;6:715–721. doi: 10.1111/1759-7714.12246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem J. 2004;381:329–342. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sunayama J, Tsuruta F, Masuyama N, Gotoh Y. JNK antagonizes Akt-mediated survival signals by phosphorylating 14-3-3. J Cell Biol. 2005;170:295–304. doi: 10.1083/jcb.200409117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida K, Yamaguchi T, Natsume T, Kufe D, Miki Y. JNK phosphorylation of 14-3-3 proteins regulates nuclear targeting of c-Abl in the apoptotic response to DNA damage. Nat Cell Biol. 2005;7:278–285. doi: 10.1038/ncb1228. [DOI] [PubMed] [Google Scholar]
- 20.Chen MS, Ryan CE, Piwnica-Worms H. Chk1 kinase negatively regulates mitotic function of Cdc25A phosphatase through 14-3-3 binding. Mol Cell Biol. 2003;23:7488–7497. doi: 10.1128/MCB.23.21.7488-7497.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavecchia A, Di Giovanni C, Novellino E. Inhibitors of Cdc25 phosphatases as anticancer agents: A patent review. Expert Opin Ther Pat. 2010;20:405–425. doi: 10.1517/13543771003623232. [DOI] [PubMed] [Google Scholar]
- 22.Sur S, Agrawal DK. Phosphatases and kinases regulating CDC25 activity in the cell cycle: Clinical implications of CDC25 overexpression and potential treatment strategies. Mol Cell Biochem. 2016;416:33–46. doi: 10.1007/s11010-016-2693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z, Zhang G, Kong C. High expression of Cdc25B and low expression of 14-3-3σ is associated with the development and poor prognosis in urothelial carcinoma of bladder. Tumour Biol. 2014;35:2503–2512. doi: 10.1007/s13277-013-1331-9. [DOI] [PubMed] [Google Scholar]
- 24.Mils V, Baldin V, Goubin F, Pinta I, Papin C, Waye M, Eychene A, Ducommun B. Specific interaction between 14-3-3 isoforms and the human CDC25B phosphatase. Oncogene. 2000;19:1257–1265. doi: 10.1038/sj.onc.1203419. [DOI] [PubMed] [Google Scholar]
- 25.Tong S, Xia T, Fan K, Jiang K, Zhai W, Li JS, Wang SH, Wang JJ. 14-3-3ζ promotes lung cancer cell invasion by increasing the Snail protein expression through atypical protein kinase C (aPKC)/NF-κB signaling. Exp Cell Res. 2016;348:1–9. doi: 10.1016/j.yexcr.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 26.MacKay RK, Colson NJ, Dodd PR, Lewohl JM. Differential expression of 14-3-3 isoforms in human alcoholic brain. Alcohol Clin Exp Res. 2011;35:1041–1049. doi: 10.1111/j.1530-0277.2011.01436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.