Abstract
Circulating tumour cell (CTC) behaviours are distinct from those of bulk tissues. Thus, treatments to eliminate CTCs differ from the regimens followed to reduce the primary tumour and its metastases. Accordingly, comprehensively deciphering the single nucleotide variant (SNV) profiles in CTCs, which partially determine CTC behaviours, is a priority. Using viable CTCs isolated with the oHSV1-hTERT-GFP virus coupled with fluorescence-activated cell sorting (FACS), the whole genome was amplified using the multiple annealing and looping-based amplification cycle (MALBAC) method. CTC behaviours were evaluated using the SNVs found to be recurrently mutated in different cells (termed CTC-shared SNVs). Analysis of the sequencing data of 11 CTCs from 8 patients demonstrated that SNVs accumulated sporadically among CTCs and their matched primary tumours (22 co-occurring mutated genes were identified in the exomes of CTCs and their matched primary tissues and metastases), and 394 SNVs were shared by at least two CTCs. Mutated APC and LRP1B genes co-occurred in CTC-shared and bulk-tissue SNVs. Additionally, the breast-originating identity of the CTC-shared SNVs was verified, and they demonstrated the following CTC behaviours: i) intravasation competency; ii) increased migration or motility; iii) enhanced cell-cell interactions; iv) variation in energy metabolism; v) an activated platelet or coagulation system; and vi) dysfunctional mitosis. These results demonstrated that it is feasible to capture and amplify the genomes of single CTCs using the described pipeline. CTC-shared SNVs are a potential signature for identifying the origin of the primary tumour in a liquid biopsy. Furthermore, CTCs demonstrated some behaviours that are unique from those of bulk tissues. Therefore, therapies to eradicate these precursors of metastasis may differ from the existing traditional regimens.
Keywords: breast cancer, circulating tumour cells, CTC behaviours, hTERT, single cell whole genome sequencing
Introduction
In recent years, breast cancer has become the leading cause of death among Chinese females, with increasing morbidity and mortality (1). Although molecular subtype, grade and lymph node involvement all influence the prognosis of breast cancer patients, uncontrolled metastasis coupled with treatment failure, rather than the primary tumour, is the primary cause of death and the 5-year survival rate is <25% (2). The treatment of metastatic breast cancer remains a challenge. Circulating tumour cells (CTCs) are metastasis-initiating precursors and independent prognostic factors (3) that closely correlate with treatment response and tumour recurrence.
Circulating breast cancer cells possess increased mobility and epithelial-mesenchymal biphenotypic status compared with anchored tumour cells. These differences may be due to the different selection pressures placed on the local and disseminated cancer cells by the different microenvironments. Avoiding extinction through the selected clonal expansion that occurs in the tumour bulk, disseminated CTCs not only retain several genetic characteristics from primary or metastatic tumours but also exhibit a large number of unique mutations, which may be a reasonable explanation for the observed extreme genomic heterogeneity exhibited by the disseminated cancer cells before the manifestation of metastasis (4–6). Such CTC restricted mutations, which are often low-frequency ‘driver’ mutations and undetectable by bulk sequencing, are probably the primary mutations that drive CTCs to detach from their original location and survive in the entirely unfamiliar flowing blood. Thus, these driver mutations may serve as latent drug targets.
Studies on bulk breast cancer sequencing identified the following pathways altered by somatic mutations in cancer cells: DNA repair, cell cycle or proliferation and lipid metabolism defects (7). Recent studies on the molecular profiles of CTCs have focused on shared mutations via bulk tissue sequencing, rather than decoding whole genome alterations de novo (5,8), which may not be conducive to uncovering the mutation characteristics of CTCs. Hence, it is necessary to investigate the unique mutations in CTCs, which probably contribute to CTC-specific properties, such as intravasation, survival in blood, interactions with immune cells and re-attachment at the sites of metastasis. However, two major obstacles hinder the programme progression: isolating rare CTCs from millions of haematocytes and amplifying trace genomes without bias and maximum coverage of the whole genome. Therefore, CTCs have not been precisely defined until recently.
In the present study, we constructed a method for isolating viable CTCs from breast cancer using the oncolytic herpes simplex virus (oHSV1)-human telomerase reverse transcriptase (hTERT)-GFP virus coupled with fluorescence-activated cell sorting (FACS) and analysed their single nucleotide variant (SNV) profiles through single-cell genome sequencing. The CTC behaviours were then evaluated based on the SNVs found to be recurrently mutated in different cells.
Materials and methods
Study procedure
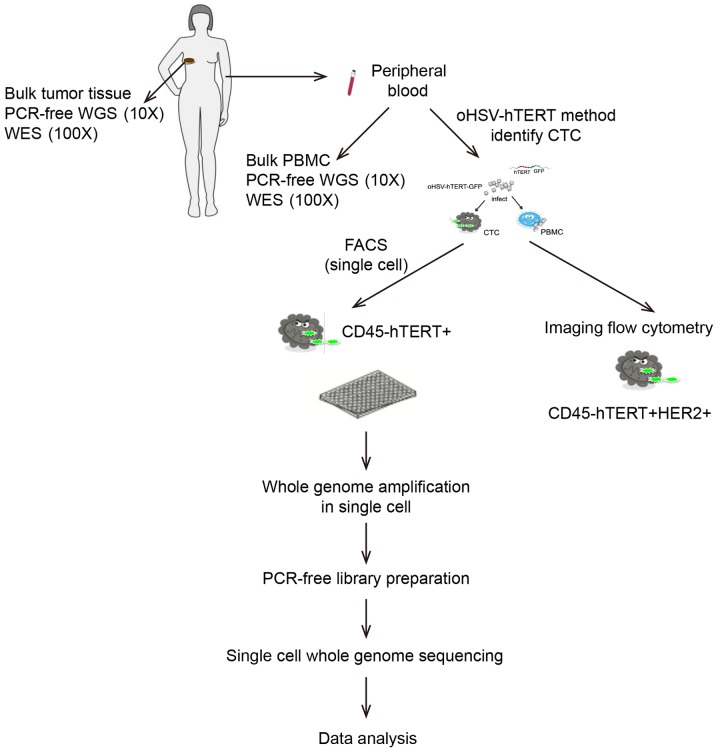
A flowchart of the study procedure is displayed in Fig. 1.
Figure 1.
A flowchart of the research procedure. PCR-free WGS (10X), PCR-free library preparation and whole genome sequencing (10X coverage); WES (100X), whole exome sequencing (100X coverage); oHSV, oncolytic herpes simplex virus; FACS, fluorescence-activated cell sorter; MALBAC, multiple annealing and looping-based amplification cycle.
Patients and specimens
The study involved 9 treatment-naïve patients who were diagnosed with invasive breast carcinoma at the Cancer Institute and Hospital of the Chinese Academy of Medical Sciences, from November 2016 to July 2017. Their clinical information is presented in Table I. A total of 8 ml of peripheral blood was collected from all patients, except patient B54, into two K2EDTA vacuum tubes (BD Biosciences, Franklin Lakes, NJ, USA; cat. no. 367844) prior to surgery, and the corresponding resected tumour tissue samples were dissected into small fragments and preserved in 3 ml of RNAlater (Invitrogen, Carlsbad, CA, USA; cat. no. AM7021) immediately after the surgical procedure. The blood and tissue samples were delivered on ice to the laboratory within 1 h. A total of 4 ml of the peripheral blood sample was used to sort CTCs. The remaining peripheral blood sample in another tube was used to isolate germline DNA. The tumour tissue fragments, which were derived from a typical region of the gross specimen, were used for DNA extraction and histological verification. In addition, only 4 ml of peripheral blood was collected from patient B54 for imaging flow cytometry (patient B54 was not included in CTC sorting). The use of human samples and the experimental procedures for this study were reviewed and approved by the Ethics Committee of the Cancer Institute and Hospital of the Chinese Academy of Medical Sciences, with the approval number 16–159/1238. The patients were informed, provided written informed consent and thoroughly understood the research.
Table I.
Patient characteristics and sample information.
| Patient ID | CTC ID | Tissue ID | Age (years) | Sex | Pathological typee | Molecular type | TNM | Countsi |
|---|---|---|---|---|---|---|---|---|
| B1 | B1CTC1 | B1Ta | 58 | Fd | BRCA, NST | Lum B/HER2(−)f | T4N1M1 | 13 |
| B1CTC3 | ||||||||
| B4 | B4CTC2 | B4T | 38 | F | BRCA, NST | Lum Ag | T1N2M0 | 27 |
| B4CTC3 | ||||||||
| B4CTC4 | ||||||||
| B5 | B5CTC3 | B5TA | 60 | F | BRCA, NST | Lum B/HER2(−) | T2N2M0 | 37 |
| B5TBb | ||||||||
| B12 | B12CTC | B12T | 34 | F | BRCA, NST | Lum B/HER2(−) | T1N1M0 | 25 |
| B15 | B15CTC | B15T | 46 | F | BRCA, NST | Lum B/HER2(−) | T2mN1M0 | 51 |
| B16 | B16CTC | B16T | 48 | F | BRCA, NST | Lum B/HER2(+) | T1N0M0 | 13 |
| B20 | B20CTC | B20T | 42 | F | BRCA, NST | Lum A | T1N2M0 | 11 |
| B37 | B37CTC | B37T | 34 | F | BRCA, NST | HER2Eh | T2N3M0 | 5 |
| B37LNc | ||||||||
| B54 | – | – | 25 | F | BRCA, NST | HER2E | T1N0M0 | – |
B1T, the primary tumour tissue of patient 1.
B5TA; B5TB, There are two tumour loci in the right breast of patient 5, named B5TA and B5TB.
B37LN, the metastatic lymph node tissue of patient 37.
F, female; M, male
BRCA, NST, breast invasive carcinoma, no special type.
Lum B/HER2(−), Luminal B/HER2-negative.
LumA, luminal A
HER2E, HER2-enriched
Counts, the counts of CTCs in 4 ml peripheral blood.
Isolating single circulating breast tumour cells using oHSV1-hTERT-GFP
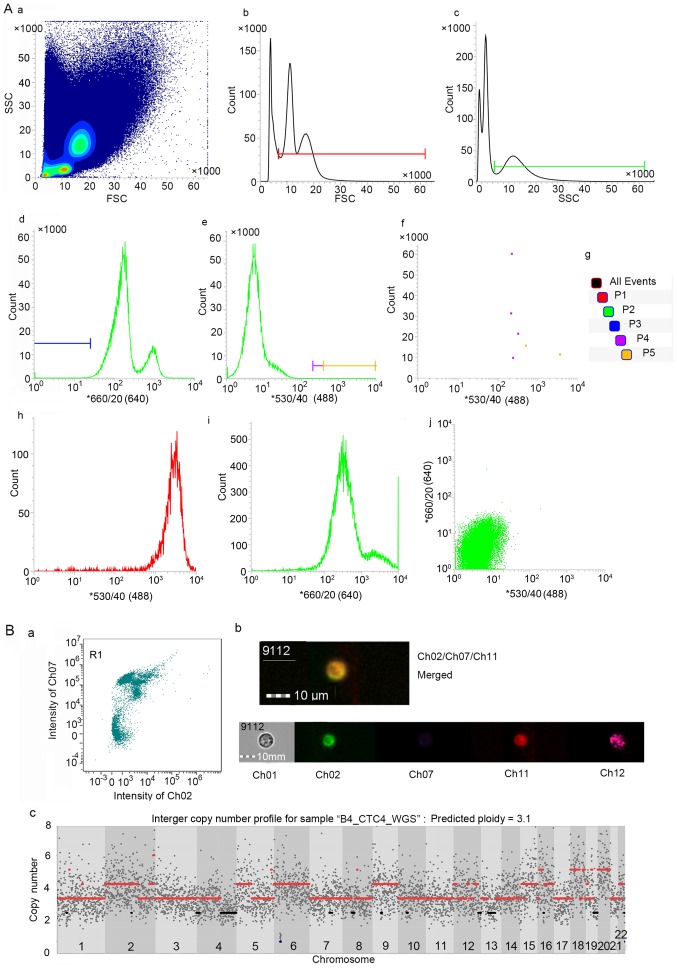
A total of 4 ml of fresh peripheral blood was centrifuged for 20 min at 120 × g, and the platelet-rich plasma phase was subsequently removed. The remaining layers, including the leucocytes and RBCs, were incubated with erythrocyte lysis buffer (Qiagen, Hilden, Germany; cat. no. 79217) for 10 min at room temperature. After centrifugation at 400 × g for 5 min, the supernatant was discarded, and the cell pellet was washed 2 times with phosphate-buffered saline (PBS). After additional centrifugation, the cells were resuspended in RPMI-1640 medium (cat. no. SH30809.01B; HyClone Laboratories, Logan, UT, USA) and infected with oHSV1-hTERT-GFP virus at an MOI=1 for 1 h at 37°C, and the culture continued for another 23 h, and then the cells were replenished in RPMI-1640 medium with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA; cat. no. 10099-141) (9). After staining the cells with 23 µl of CD45 monoclonal antibody (HI30)-APC (cat. no. 17-0459-42; eBioscience, San Diego, CA, USA), the cell suspension was evaluated and sorted using flow cytometry (BD FACSJazz; BD Biosciences) in single-cell mode. CD45−/hTERT+ cells were defined as CTCs and sorted into strips of 8 tubes (cat. no. 4316567; Life Technologies, Carlsbad, CA, USA) containing 1X ThermoPol Reaction Buffer (lysis buffer). In addition, to identify the gate of the cell population of interest, we used a single-stained or infected control (negative control, peripheral blood of a healthy person; positive control, the hTERT-expressing cancer cell line, SK-BR-3) (Fig. 2A-h-j).
Figure 2.
Feasible method to detect and isolate CTCs. (A) Gates were set to capture viable CTCs via FACS analysis. (A-b) The P1 gate was set to obtain cells with diameters larger than lymphocytes, and (A-c) the P2 gate was set to acquire granulocyte-like side scatter (SSC) cells. (A-a) P1 was combined with P2 to capture viable cells similar to tumour cells in size and SSC. (A-d and -f) The P3 and P4 gates were used to select hTERT-positive cells without leucocyte-common antigen (CD45) expression and collect highly hTERT-positive cells for further analysis. (A-e and -f) The sorted cells were detected at P5 gate (taking partial results from patient B1 for example). (A-i) The single-stained CD45-APC positive control of PBMC and (A-j) the infected control, including negative control (peripheral blood of a healthy person), (A-h) positive control (the hTERT-expressing SK-BR-3 cancer cell line). (B) Analysis of circulating breast tumour cells using imaging flow cytometry. (B-a) Cell image mapped with the point in the bottom right corner of R1 gate (high signal captured in ch02 and low signal captured in ch07, corresponding to the gate P5). Ch02 received signal emitted by GFP (hTERT), ch07 reflects the signal intensity of eFluor450 (CD45), ch11 represents the APC (ErbB2) intensity and ch12 is the SSC channel. (B-b) The merged image represents hTERT+CD45-ErbB2+ cells. (B-c) The copy number profiles of a near-triploid CTC.
Single-cell whole genomic amplification (WGA)
Single-cell WGA was performed using the MALBAC method (10), with a slight pre-amplification step modification as a time-saving measure. The prepared lysis buffer containing the sorted cells was vortexed, spun at 6,000 × g for 10 sec and immediately chilled to 4°C, instead of undergoing Qiagen Proteinase digestion. Subsequently, linear pre-amplification and amplification were performed as previously described (11). The amplified DNA product was purified using the MinElute® PCR Purification kit (Qiagen, Hilden, Germany; cat. no. 28004) according to the manufacturer's instructions. The purified PCR product was assessed using a Qubit® dsDNA HS Assay kit in a Qubit® 3.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). The molecular weight of the DNA was evaluated on 1% agarose gel. To reduce contamination, we performed the WGA-associated experiments on a clean bench and all reagents, except nucleotides and polymerases, were exposed to UV irradiation for 30 min.
Isolation of DNA from bulk specimens
DNA was extracted from breast-invasive carcinoma tissues, and germline DNA was extracted from white blood cells (WBCs) using the traditional phenol-chloroform method. The DNA was quantified using Qubit 3.0 and high-molecular-weight DNA was verified on 1% agarose gel.
PCR-free library preparation and whole genome sequencing (WGS)
A PCR-free library is preferable to minimize the base-composition bias in bulk samples and the already-existing PCR amplification bias in single-cell samples. Thus, PCR-free libraries were generated from 1.0 µg of genomic DNA from all bulk samples and 1.5 µg of amplified DNA from a single cell, based on their ladder-like distribution base sizes, using an Illumina TruSeq DNA PCR-Free LT Sample Prep kit (Illumina, San Diego, CA, USA; cat. no. FC-121-3001) according to the manufacturer's instructions. The DNA was sonicated into fragments with an average size of 350 bp, followed by end-repair, A-tailing and ligation with full-length adapter for Illumina sequencing but without further PCR amplification. The final libraries were evaluated using an Agilent 2100 Bioanalyzer and quantified by real-time PCR. Libraries were clustered on the cBot Cluster Generation System using a HiSeq X HD PE Cluster kit (Illumina) and subsequently sequenced on the Illumina HiSeq X Ten platform (paired ends, 150 bp).
Illumina target library preparation and whole exome sequencing (WES)
All bulk DNA samples, cancer-originated DNA and germline DNA were normalized to 1 µg and sheared to 150–200 bp. The fragmented DNA was captured using an Agilent SureSelect Human All Exon V6 kit (Agilent Technologies Inc., Santa Clara, CA, USA) according to the manufacturer's instructions and the ends of the enriched DNA fragments were repaired and ligated with adapters using a Paired-End DNA Sample Prep kit (Illumina). The qualified multi-regional target libraries were sequenced using an Illumina HiSeq 4000 (paired ends, 150 bp).
Sequencing of data analysis
Adapter sequences and low-quality reads were removed from the raw sequence data using Cutadapt (12) and sickle (http://github.com/najoshi/sickle/) software. After the clean data were aligned to hg19 using the Burrows-Wheeler Aligner (BWA) (13), SAM files were produced. Picard tools (http://broadinstitute.github.io/picard/) were used to transform the SAM files to a bam format and to sort and remove PCR duplications. In addition with the help of the Genome Analysis Toolkit (GATK) pipeline (14), the original bam files were processed for local Indel realignment, base quality estimation and recalibration for further analysis.
Somatic mutation calling was performed. For whole exome data, VarScan2 (15) combined with the JointSNVMix2 (16) algorithm were used to identify somatic mutations and the candidates were merged, while whole genomic data somatic mutation calling used only the VarScan2 algorithm. To reduce false-positive calls, the candidate calls were filtered using the following criteria: i) zero mutant reads detected in the germline sample; ii) at least 10X accumulated coverage for both the forward and reverse reads of the tumour sample in the mutant alleles; and iii) the nearest adjacent mutant base was at least 100 bp away in the same sample. The filtered SNVs were annotated using Oncotator (17), and the variant allele fraction (VAF) was calculated as follows: VAF = read counts that covered the mutant position/all read counts that covered the position for data from a single cell.
To determine the copy number, we used Ginkgo (http://qb.cshl.edu/ginkgo), an open-source web platform that specifically analyses single-cell copy number variants (CNVs), and two R packages (HMMcopy and DNAcopy), with hg19 as the reference genome.
Validating circulating breast tumour cells using imaging flow cytometry
Slightly different from the oHSV1-hTERT-GFP-coupled FACS method, the infected cells were stained with CD45 monoclonal antibody (HI30)-eFluor 450 (cat. no. 48-0459-42; eBioscience, San Diego, CA, USA) and human ErbB2/Her2 APC-conjugated antibody (R&D Systems, Minneapolis, MN, USA; cat. no. FAB1129A) and the cell suspension was subsequently imaged using an ImageStream®X Mark II Imaging Flow cytometer (Merck Millipore, Darmstadt, Germany).
Results
oHSV1-hTERT-GFP coupled with FACS is a feasible method to detect and isolate CTCs
The efficacy and accuracy of the oHSV1-hTERT-GFP method for detecting and isolating CTCs were validated using spike and recovery experiments, as previously described, with a recovery range between 75.5 and 87.19% (correlation r2=0.9909) when 0, 10, 20, 40, 80, 160 and 320 BGC823 cells were spiked (9). The gates set to capture viable CD45−/hTERT+ CTCs are displayed in Fig. 2A. To further validate the capacity for detecting circulating breast tumour cells, we conducted imaging flow cytometry, and the observed CD45−/hTERT+/ErbB2(Her2)+ cells were defined as CTCs from breast cancer in patient B45 (Fig. 2B-a-b).
Copy number variations of CTCs possibly confirm the capture and amplification method
A quality control was applied to the single-cell WGS data for 11 CTCs from 8 patients, resulting in a mean coverage of 5.14±1.73 and an average global mapping rate of 84.59±6.5%. Both were slightly lower than those for bulk sequencing (9.62±1.77 for mean coverage and 93.29±3.62% for the mapping rate).
Genome instability is a hallmark of human cancers (18). Amplifications of chr1q and chr16p, commonly identified in breast cancer (19,20) were discovered in 6 and 5 out of 11 CTCs, respectively. Downregulation of human leukocyte antigen (HLA) class I expression has been reported in a significant proportion of breast cancers (21) and was found to be significantly associated with a shorter disease-free interval (22). The HLA super-locus in chr6p21 was found to be lost in all 11 CTCs, indicating an escape mechanism from cytotoxic T lymphocytes in the blood. In addition to local gains and losses, whole-chromosome aneuploidy was also found in B1CTC3 (with trisomies 10 and 18), B12CTC (with trisomies 1, 3 and 22), B4CTC2 (with trisomy 22), B4CTC3 (with trisomies 14 and 21) and B4CTC4 (with triploidy of the whole set of chromosomes). The captured circulating aneuploid cells without leucocyte-common antigen (CD45) indicated that our capture and amplification procedure was satifactory.
SNVs accumulated sporadically between CTCs and their matched primary tumours
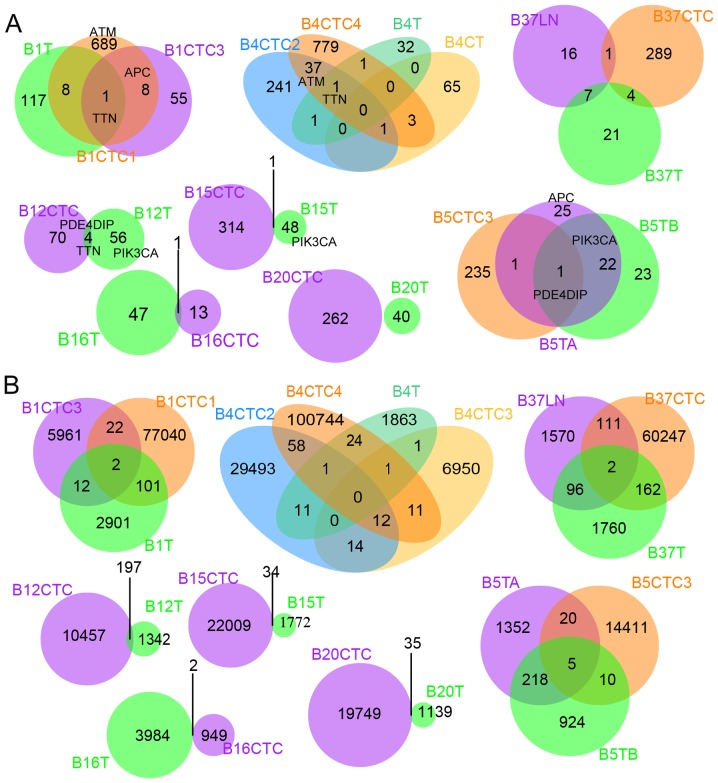
As to the 11 CTCs analysed by single-cell sequencing, on average, 299 high-confidence SNVs within exons in each cell were found passing the filters mentioned in Materials and methods. Compared with the single-cell sequencing data, the abundance of SNVs (in exons) in bulk tissues was low (52 point mutations on average for each primary or metastatic tissue), even though we merged the mutations called from both WGS and WES data (Table II). This result partially reflected differences in low-abundance mutation mining competence between single cells and bulk samples. Only 22 commonly mutated genes were identified in CTCs and their matched primary or metastatic tissues in the exome (Fig. 3A and Table III). However, the shared mutation loci were much more prevalent genome wide (Fig. 3B), indicating the importance of genomic dark matter during the evolutionary process of cancer cells. The heterogeneous mutation spectra of CTCs and matched bulk cancers observed here is consistent with previous studies (23–26).
Table II.
The point mutation counts of the exon region and whole region of all samples.
| Sample ID | Non-syn SNVc | Stop-gain | Stop-loss | Syn SNVd | Total mutation |
|---|---|---|---|---|---|
| B1CTC1_WGSa | 493 | 26 | 0 | 222 | 77,165 |
| B1CTC3_WGS | 44 | 1 | 0 | 20 | 5,997 |
| B1T_WGS | 9 | 0 | 0 | 2 | 2,424 |
| B1T_WESb | 80 | 6 | 0 | 31 | 590 |
| B4CTC2_WGS | 201 | 15 | 0 | 74 | 29,589 |
| B4CTC3_WGS | 54 | 3 | 0 | 16 | 6,989 |
| B4CTC4_WGS | 546 | 17 | 1 | 318 | 100,851 |
| B4T_WGS | 6 | 0 | 0 | 10 | 1,693 |
| B4T_WES | 15 | 0 | 0 | 5 | 192 |
| B5CTC3_WGS | 152 | 4 | 1 | 83 | 14,446 |
| B5TA_WGS | 10 | 0 | 0 | 4 | 1,303 |
| B5TA_WES | 21 | 2 | 0 | 13 | 281 |
| B5TB_WGS | 5 | 0 | 0 | 6 | 899 |
| B5TB_WES | 21 | 1 | 0 | 16 | 251 |
| B12CTC_WGS | 49 | 0 | 0 | 29 | 10,654 |
| B12T_WGS | 8 | 0 | 0 | 6 | 1,026 |
| B12T_WES | 16 | 0 | 0 | 30 | 499 |
| B15CTC_WGS | 229 | 5 | 0 | 89 | 22,043 |
| B15T_WGS | 10 | 1 | 0 | 4 | 1,383 |
| B15T_WES | 24 | 3 | 0 | 9 | 404 |
| B16CTC_WGS | 8 | 0 | 0 | 6 | 951 |
| B16T_WGS | 18 | 0 | 0 | 22 | 3,782 |
| B16T_WES | 8 | 0 | 0 | 4 | 195 |
| B20CTC_WGS | 173 | 16 | 1 | 83 | 19,784 |
| B20T_WGS | 5 | 0 | 0 | 2 | 911 |
| B20T_WES | 16 | 5 | 0 | 12 | 254 |
| B37CTC_WGS | 168 | 2 | 0 | 137 | 60,522 |
| B37T_WGS | 10 | 0 | 0 | 5 | 1,765 |
| B37T_WES | 9 | 2 | 0 | 6 | 251 |
| B37LN_WGS | 7 | 0 | 0 | 4 | 1,607 |
| B37LN_WES | 5 | 0 | 0 | 8 | 163 |
WGS, whole genomic sequencing
WES, whole exon sequencing
Non-syn SNV, non-synonymous single nucleotide variant
Syn SNV, synonymous single nucleotide variant.
Figure 3.
Mutations are shared between CTCs and their matched tissues. (A) Mutant genes are shared between CTCs and their matched tissues based on whole exome data. (B) Mutation positions are shared between CTCs and their matched tissues based on whole genome data.
Table III.
Mutation genes appeared in both CTCs and their matched primary or metastatic tissues and the concurring mutant between CTC-shared SNVs and bulk SNVs.
| Category | Gene |
|---|---|
| B1 | XIRP2, TTN, PKHD1, MUC17, C12orf29, UTP20, AHNAK2, MYO1E, TLN2, XIRP2 |
| B4 | TTN, CDCA2, TGA9 |
| B5 | PDE4DIP, RFPL4A |
| B12 | TTN, PDE4DIP, CUBN, PRH1 |
| B15 | FAM184A |
| B16 | DSPP |
| B37 | OXCT2, OR8D1, LILRA6, POTEH, FAM46A |
| Co-mutant between CTC-shared | TTN, APC, MDN1, MUC17, UTP20, AHNAK2, MYO1E, FBN1, DNAH17, KMT2C, |
| SNVs and bulk SNVs | NBPF8, LRP1B, XIRP2, KLHL41, CUBN, PZP, NAV3, CIT, PDE4DIP, ARHGEF5, FAM186A, NBPF10, COL6A6, NUMA1, DDRGK1, COL24A1, MANSC1, MUC12, TOP2B, ZFHX4 |
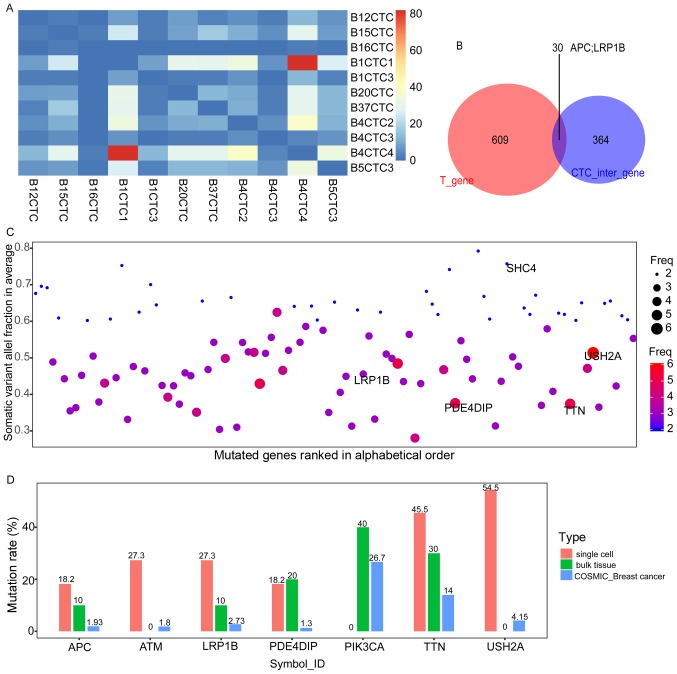
Characteristics of CTC-shared SNVs in the exome
To prevent bias and errors from the WGA, 394 concurring mutation genes (hereafter referred to as CTC-shared SNVs), which were identified in at least two CTCs, were used for further analysis (Fig. 4A). Intersecting the CTC-shared SNVs with the SNVs in the exome (hereafter, exome SNVs) of the bulk tissues, 30 CTC-bulk shared mutation genes were identified, including the well-established tumour suppressor gene APC and the putative tumour suppressor gene LRP1B (Fig. 4B and Table III). The CTC-shared SNVs comprised some prevalent mutated genes that have been reported in the Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov/) database, including USH2A, TTN, MUC16 (CA125) and other members of the mucin family (i.e., MUC17 and MUC3A). In particular, mutations with high frequencies in metastatic samples reported in TCGA, such as mutations in LRP1B, were frequently observed among the CTCs (Fig. 4C) (27). We further compared the allele frequencies of seven relatively prominent genes between our samples and another published database, the Catalogue of Somatic Mutations in Cancer (COSMIC) (28). The frequencies of PIK3CA, a mutation driver in breast cancer (29), were higher in the bulk sample than the COSMIC record, and the mutation rate of TTN exceeded 10% in all three datasets (Fig. 4D).
Figure 4.
The CTCs-shared SNVs. (A) The heat map of the number of CTCs-shared SNVs contributed by each cell. The majority of CTCs-shared SNVs are offered by B1CTC1 and B4CTC4, and a few overlapping SNVs in B16CTC and other single cells. The diagonal of this heat map represents the overlap of the same cells, which were set to zero. (B) Intersecting the CTC-shared SNVs with the exome SNVs of the bulk tissues, 30 CTC-bulk shared mutation genes were identified, including the tumour suppressor gene APC and the putative tumour suppressor gene LRP1B. (C) The somatic variant allele fraction (VAF) distribution and mutation frequency in the CTCs-shared SNVs. The CTCs-shared SNVs demonstrated here are covered with more than ten reads without any mutations in the normal tissue, with VAF ≥0.6 or frequency ≥3. The vertical axis shows the average somatic VAF within a gene. The horizontal axis depicts the mutated genes (official_symbol_ID) in alphabetical order. The diameter or colour of the circle represents the observed mutation frequency in the 11 cells. (D) The frequency of several prominent mutations in our data and COSMIC data. The vertical axis presents the prevalence of the mutations in each group, including the 11 cells, 10 bulk tissues, and the COSMIC breast cancer samples. The horizontal axis depicts APC, ATM (DNA-damage response gene that regulates the tumour suppressor proteins p53 and BRCA1 and some checkpoint or DNA repair proteins), LRP1B (LDL receptor-related protein 1 B, which is ranked in the top 3 mutated genes in at least one cancer type, particularly in the metastatic samples of TCGA), PDE4DIP, PIK3CA, TTN and USH2A (with mutants occurring in 6 cells, GO annotation relates to collagen binding and myosin binding).
Verification of the breast-originating identity of CTC-shared SNVs
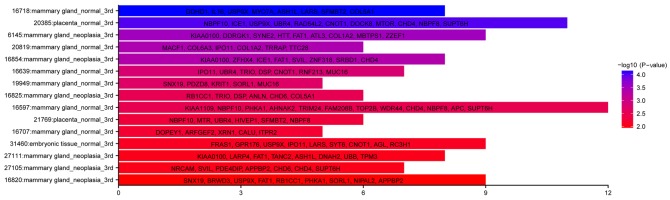
Although the relationship between SNVs and the cDNA library is obscure, it is believed that SNVs can alter the expression level of mutated genes in several ways. For example, some SNVs located in the promoter or 3′UTR of genes, can influence expression levels by altering the regulatory influence of the transcription factors on the promoter region or some regulatory non-coding RNAs on their mRNA targets. To explore the general classification of CTC-shared SNVs, the SNV-bearing genes were identified in the Cancer Genome Anatomy Project library (CGAP, https://cgap.nci.nih.gov/), particularly the EST database (dbEST) with the tissue tool. In total, 285 annotated genes were included in the dbEST, and these could be divided into three major categories including mammary gland_normal, mammary gland_neoplasia and placenta_normal (Fisher's exact test, P<0.05; Fig. 5). The former two categories uncovered the origin of these CTCs. As to the category referring to normal placenta, it may represent a cluster of genes with hormone-associated, invasive or low-immunogenicity functions, which are known as features of the trophoblast model of cancer (30).
Figure 5.
CTCs-shared SNVs indicate the breast-associated derivation of the primary cancer. The vertical axis displays the tissue-originated terms from the CGAP database. The horizontal axis of the bar diagrams shows the gene counts enriched for each term. The colour indicates the -log10 (P-value).
CTC-shared SNVs and their CTC-associated functions
To further understand the potential biological functions of CTC-shared SNVs, KEGG pathway (http://www.kegg.jp/) and GO (http://www.geneontology.org/) biological process analyses were used to perform enrichment analysis on the genes bearing the 394 CTC-shared SNVs. As displayed in Tables IV and V, the data indicated that these genes alter the following functions of the CTCs: i) intravasation competency through ECM remodelling and collagen catabolism; ii) increased migration or motility dependent on cilia motility, actin cytoskeleton organization and other myocardium-like movement, indicating that captured viable CTCs undergo EMT; iii) initiation of cell-cell interactions, such as integrin-mediated cell adhesion; iv) alterations to energy metabolism, including lipoprotein transport, glycan processing and ABC transporters; v) platelet or coagulation system activation, affecting wound healing; and vi) dysfunctional mitosis, exhibited through DNA repair deficiency (Fanconi anaemia pathway enriched in the KEGG pathway analysis) (31) and dysfunction in DNA double-strand break repair by non-homologous end-joining (mutant genes enriched in GO: histone H3-K36 dimethylation, H3K36me2) (32).
Table IV.
CTCs-shared SNVs involved in KEGG pathway.
| Category | Term | Count | P-value |
|---|---|---|---|
| KEGG_PATHWAY | hsa04512: ECM-receptor interaction | 14 | 9.21E-09 |
| KEGG_PATHWAY | hsa04510: Focal adhesion | 17 | 1.98E-06 |
| KEGG_PATHWAY | hsa04974: Protein digestion and absorption | 11 | 6.71E-06 |
| KEGG_PATHWAY | hsa04151: PI3K-Akt signaling pathway | 16 | 0.002665514 |
| KEGG_PATHWAY | hsa05410: Hypertrophic cardiomyopathy (HCM) | 6 | 0.017366873 |
| KEGG_PATHWAY | hsa05414: Dilated cardiomyopathy | 6 | 0.02318041 |
| KEGG_PATHWAY | hsa02010: ABC transporters | 4 | 0.053062934 |
| KEGG_PATHWAY | hsa04260: Cardiac muscle contraction | 5 | 0.05722822 |
| KEGG_PATHWAY | hsa03460: Fanconi anemia pathway | 4 | 0.082951282 |
Table V.
CTCs-shared SNVs confer biology process on free-floating tumour cells (GO biology process).
| Term_ID | Term | P-value |
|---|---|---|
| GO:0060285a | Cilium-dependent cell motility | 1.30E-06 |
| GO:0060048a | Cardiac muscle contraction | 2.59E-04 |
| GO:0002027a | Regulation of heart rate | 4.66E-04 |
| GO:0030198b | Extracellular matrix organization | 6.02E-04 |
| GO:0030049a | Muscle filament sliding | 9.09E-04 |
| GO:0030574b | Collagen catabolic process | 0.001734612 |
| GO:0006941a | Striated muscle contraction | 0.001956422 |
| GO:0007155c | Cell adhesion | 0.002348521 |
| GO:0007507 | Heart development | 0.003843656 |
| GO:0007010a | Cytoskeleton organization | 0.005175679 |
| GO:0010880a | Regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 0.005183546 |
| GO:0031119 | tRNA pseudouridine synthesis | 0.005677556 |
| GO:0045214a | Sarcomere organization | 0.019748792 |
| GO:0045666 | Positive regulation of neuron differentiation | 0.020028672 |
| GO:0006936a | Muscle contraction | 0.020559102 |
| GO:0042060d | Wound healing | 0.022090779 |
| GO:0031581 | Hemidesmosome assembly | 0.023078156 |
| GO:0051480 | Regulation of cytosolic calcium ion concentration | 0.025660253 |
| GO:0030239a | Myofibril assembly | 0.026918471 |
| GO:0007512 | Adult heart development | 0.026918471 |
| GO:0060402 | Calcium ion transport into cytosol | 0.030995997 |
| GO:0071044 | Histone mRNA catabolic process | 0.030995997 |
| GO:0016266e | O-glycan processing | 0.031968827 |
| GO:0030335a | Positive regulation of cell migration | 0.032042051 |
| GO:0010951 | Negative regulation of endopeptidase activity | 0.034801689 |
| GO:0033627c | Cell adhesion mediated by integrin | 0.035299764 |
| GO:0042953e | Lipoprotein transport | 0.035299764 |
| GO:0034728 | Nucleosome organization | 0.039619843 |
| GO:0030036a | Actin cytoskeleton organization | 0.046670842 |
| GO:0002026a | Regulation of the force of heart contraction | 0.054569593 |
| GO:0010881a | Regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion | 0.054569593 |
| GO:0097676f | Histone H3-K36 dimethylation | 0.058838886 |
| GO:0002331 | Pre-B cell allelic exclusion | 0.058838886 |
| GO:0032439 | Endosome localization | 0.058838886 |
| GO:0000472 | Endonucleolytic cleavage to generate mature 5′-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 0.058838886 |
| GO:0007030 | Golgi organization | 0.060936288 |
| GO:0007067f | Mitotic nuclear division | 0.06220872 |
| GO:0022617b | Extracellular matrix disassembly | 0.065918416 |
| GO:0006816 | Calcium ion transport | 0.065918416 |
| GO:0090287 | Regulation of cellular response to growth factor stimulus | 0.077674441 |
| GO:0007018a | Microtubule-based movement | 0.079266502 |
| GO:0035023 | Regulation of Rho protein signal transduction | 0.079266502 |
| GO:0001701 | In utero embryonic development | 0.082288621 |
| GO:0060070 | Canonical Wnt signaling pathway | 0.084954447 |
| GO:0006887 | Exocytosis | 0.084954447 |
| GO:0022604 | Regulation of cell morphogenesis | 0.088600004 |
| GO:0008544 | Epidermis development | 0.09083611 |
| GO:0003356a | Regulation of cilium beat frequency | 0.096134138 |
| GO:0002634 | Regulation of germinal center formation | 0.096134138 |
| GO:0042415 | Norepinephrine metabolic process | 0.096134138 |
The GO biology process terms are associated with
cell mobility
extracellular matrix degradation
cell adhesion
coagulation processes
material metabolism
mitosis processes.
Discussion
More than a decade has passed since the first trial enumerated CTCs through EpCAM-based detection to estimate the prognosis of patients with breast cancer (3). In addition, following the validation of their prognostic power in breast cancer, CTC counts have been correlated to poor prognosis in other high-morbidity cancers, including lung (33), prostate (34) and colorectal cancer (35). In addition, single-nucleotide mutation characteristics have been reported by whole genome sequencing in kidney tumours (23), prostate (36) and lung cancer (24). Although invasive breast carcinoma (no special type) is known to derive from the ductal epithelium and prognosis prediction through EpCAM-phenotype CTCs has been long validated (37), the molecular characterization of these EpCAM-based free-floating tumour cells had, until recently, only been performed using whole genomes (20). However, the present study is the first to profile the mutant characteristics of hTERT-based CTCs in the whole genome. In addition, the status and phenotypes of circulating breast cancer cells are diverse and include stem-like cells (38), dormant states (39,40) and dynamic EMT changes (6,41). Thus, it is imperative to determine the molecular characteristics of specific cell populations, as they exhibit a great deal of genomic disparity among them that can lead to the initiation of metastasis or can act as a decisive factor in treatment failure. Unfortunately, molecular profiling has been hindered by the rarity and heterogeneity of CTCs, coupled with unsophisticated technology for their enrichment, isolation and genomic amplification. Compared with several studies that have analysed some typical mutations (TP53, PIK3CA, KRAS, RB1 and ErbB2) in CTCs and bulk tissues (8,42,43), which simply represent the tip of the iceberg, this study is the first to examine the whole genome of hTERT-positive CTCs. In addition, this is the first study to profile SNVs in viable hTERT-positive CTCs, rather than EpCAM-CTCs, to define CTC behaviours.
In contrast with the reported bulk sequencing data, one mutant gene (SHC4/RaLP/SHCD) was highlighted in our single-CTC data. Recent studies concerning SHC4 have focused on metastatic melanoma and reproductive organ cancer (particularly prostate and ovarian cancers involving the BRCA1 pathway) and have identified several plausible roles for SHC4, such as regulating apoptosis (44), stem cell differentiation (45), cell invasion and migration (44). It appears, therefore, that although considered a ‘low-probability event’, when the specific cell population acquired the essential mutation during the random accumulation process, they began to detach and survive in the bloodstream.
We extracted 394 CTC-shared SNVs from all the identified CTC mutations, and these may confer primary tumour cells with the following alterations. During the metastasis initiation phase, the selected population interacts with ECM receptors (hsa04512), followed by extracellular matrix organization (GO:0030198) via collagen catabolism (GO:0030574). Similarly, the results from single-cell RNA sequencing data in a pancreatic cancer experimental animal model revealed that the aberrant expression of stromal ECM genes by EM-CTCs are liable to aid spread into the bloodstream (46). Indeed, the five-step model for proteolytic cell migration has been suggested for many years. This model proposes that the elongated cell morphology and dynamic membrane protrusions that are tailored for ECM degradation promote cell detachment from one spot (47,48). Specifically, von Nandelstadh et al (48) confirmed that the actin-associated protein paladin functions in actin-based pseudopods (cell cytoskeleton alteration) to drive invasion through ECM degradation. Thus, the ever-increasing motility (GO:0060285, cilium-dependent cell motility; GO:0030036, actin cytoskeleton organization) and ECM degradation of CTCs may co-occur in CTC behaviours, similar to our data. The motility-enhanced EM-CTCs not only tend to penetrate into vessels, but also are prone to escape immune attack through autophagy when encountering T cells (49). Therefore, the increased mobility of CTCs ensures both detachment and survival.
To ensure survival in the circulation, the detached tumour cells are equipped not only with increased motility but also enhanced focal adhesion (KEGG, hsa04510), cell adhesion (GO:0007155), and cell adhesion mediated through integrins (GO:0033627). Cell adhesion can be classified as follows: CTC-CTC interactions; CTC-non-CTC interactions, which include all cell types except CTCs and various molecules, such as hormones, cytokines and growth factors in the bloodstream; and CTC-CTC interactions bridged by non-CTCs in the circulatory system. In contrast with isolated, floating CTCs, CTCs and their binding partners favour resistance to anoikis, and the counterparts of cell adhesion, wound healing (GO:0042060) and its child term, blood coagulation (GO:0007596), play significant roles in survival. Collectively, the terms associated with cell adhesion and wound healing highlight a novel theory. A secure shelter of CTCs is built when neutrophil extracellular traps (NETs) sequester CTCs via β1-integrin-mediated interactions (50) and act as a scaffold to recruit platelets and coagulation factors (51). Activating platelets and the coagulation system predisposes the tumour to dissemination (52).
Additional distinguishing behaviours of CTCs are the dysfunction of energy metabolism, ABC transporters (KEGG, hsa02010) and lipoprotein transport (GO:0042953). High-fat diets increase the frequency and reduce the time to occurrence of breast cancer (53). LRP1B, which encodes a member of the low-density lipoprotein (LDL) receptor family, affects LDL receptor activity (54) and acquired chemotherapeutic resistance (55), with mutations detected in three CTCs.
The enumerated CTC behaviours facilitate metastasis initiation, and survival is rooted in genomic alterations, including DNA repair dysfunction (KEGG, hsa03460; GO:0097676) and mitotic nuclear division resulting in aneuploidy (GO:0007067). Genomic (DNA repair-associated genes) and epigenomic (histone methylation, particularly H3K36me2) alterations regulate DNA repair to destabilize the genome and produce a series of CTC-shared SNVs.
Although the CTC behaviours deduced from the 394 CTC-shared SNVs are supported by previous studies, there are no sufficiently amplified DNA samples to validate the SNVs detected through next-generation sequencing via Sanger sequencing since more DNA is needed to prepare PCR-free libraries than traditional sequencing libraries. However, the PCR-free library reduced the bias from PCR. Reflecting the limitations of the unsophisticated isolation and amplification technologies, the same sorting and amplification work flow deficiencies were observed in the present study, and only a few CTCs were successfully amplified and sequenced from each patient, which is far from covering all the heterogeneous CTCs. There were two reasons for only 1–3 CTCs being sorted for one patient in the present study. Firstly, to balance the sorted speed and efficiency in our flow cytometry (BD FACSJazz; BD Biosciences), we sorted the peripheral blood until 5 to 10 CTCs collected, and counted the CTC number in the rest. However, in most cases, less than 5 CTCs could be captured successfully into a PCR tube for one patient, even there were dozens of CTCs counted by FACS. The limitations of the sorting accuracy of the flow cytometry are the main reason. Secondly, the amount of DNA needed to prepare the PCR-free library are much more than that for the traditional library. Not all single cell DNA amplificated by the MALBAC method could pass the quality control for PCR-free library construction. Recently, one protocol combining immunomagnetic enrichment and fluorescence-activated cell sorting (IE/FACS) increased the yield of CTCs (56). As for the isotype control in flow cytometry, more and more researchers abandoned the isotype control as it indicated that such controls may be unreliable to be used as gating controls or to determine background signal (57). The application of Fc-blocking reagent is recommended instead. In addition, during our CTC isolated period, 10% FBS was added into the medium as the Fc-blocking reagent. Thus, considering the convenience and the blood consumption, we did not use the isotype control to detect the specificity of the CD45 antibody in the group of these breast cancer patients.
The genomic data in the present study provided clues to the behaviour of the hTERT-positive metastasis initiation cells in peripheral blood. There are many interesting events to be investigated in the future, for example, the function of the high-frequency mutant SHC4/RaLP/SHCD in CTCs, the relationship of the mutant of LRP1B and their distinguished metabolic way. Uncovering the specific properties of CTCs as the major drivers of metastasis initiation and the pool of treatment resistance and tumour recurrence is urgently needed.
Acknowledgements
We thank the National Center for Protein Sciences Beijing (Peking University) and Ms Fei Wang for her technical assistance with the imaging flow cytometry. We thank the National Cancer Center/Cancer Hospital (CAMS and PUMC) and Professor Shujun Cheng.
Glossary
Abbreviations
- APC
allophycocyanin
- CTC
circulating tumour cell
- CNV
copy number variant
- ECM
extracellular matrix
- EM-CTC
epithelial and mesenchymal biphenotypic CTC
- ErbB2
v-erb-b2 avian erythroblastic leukaemia viral
- FACS
fluorescence-activated cell sorting
- hTERT
human telomerase reverse transcriptase
- Her2
oncogene homologue 2
- MALBAC
multiple annealing and looping-based amplification cycles
- oHSV
oncolytic herpes simplex virus
- SNV
single nucleotide variant
- WGA
whole genome amplification
- WGS
whole genome sequencing
Funding
The present study was funded by the National Key Research and Development Program of the Ministry of Science and Technology of China (2016YFC0905301), the CAMS Innovation Fund for Medical Sciences (CIFMS, 2016-I2M-3-005) and the National Key Basic Research Program (NKBRP, 2015CB553901).
Availability of data and materials
The datasets used during the present study are available from the corresponding authors upon reasonable request.
Authors' contributions
KZ, GC, LF and WZ designed and supervised the project. YW and GC provided the samples and clinical information. LG performed the experiments. TX and XD provided instruction of the experiments. LG and YW analyzed the data and wrote the manuscript. LF and KZ edited the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Human samples and the experimental procedures involved in this study were reviewed and approved by the Ethics Committee of the Cancer Institute and Hospital of the Chinese Academy of Medical Sciences, with the approval number 16–159/1238. The patients were informed, provided written informed consent and thoroughly understood the research.
Consent for publication
All authors are in agreement with the content of the proof, and consent for publication.
Competing interests
The authors have no conflict of interest.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Giuliano M, Giordano A, Jackson S, De Giorgi U, Mego M, Cohen EN, Gao H, Anfossi S, Handy BC, Ueno NT, et al. Circulating tumor cells as early predictors of metastatic spread in breast cancer patients with limited metastatic dissemination. Breast Cancer Res. 2014;16:440. doi: 10.1186/s13058-014-0440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Magbanua MJ, Carey LA, DeLuca A, Hwang J, Scott JH, Rimawi MF, Mayer EL, Marcom PK, Liu MC, Esteva FJ, et al. Circulating tumor cell analysis in metastatic triple-negative breast cancers. Clin Cancer Res. 2015;21:1098–1105. doi: 10.1158/1078-0432.CCR-14-1948. [DOI] [PubMed] [Google Scholar]
- 4.Klein CA. Selection and adaptation during metastatic cancer progression. Nature. 2013;501:365–372. doi: 10.1038/nature12628. [DOI] [PubMed] [Google Scholar]
- 5.Deng G, Krishnakumar S, Powell AA, Zhang H, Mindrinos MN, Telli ML, Davis RW, Jeffrey SS. Single cell mutational analysis of PIK3CA in circulating tumor cells and metastases in breast cancer reveals heterogeneity, discordance, and mutation persistence in cultured disseminated tumor cells from bone marrow. Bmc Cancer. 2014;14:456. doi: 10.1186/1471-2407-14-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng T, Wang Y, Seiler M, Hu Z. Functional characterization of breast cancer using pathway profiles. Bmc Med Genomics. 2014;7:45. doi: 10.1186/1755-8794-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polzer B, Medoro G, Pasch S, Fontana F, Zorzino L, Pestka A, Andergassen U, Meier-Stiegen F, Czyz ZT, Alberter B, et al. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO Mol Med. 2014;6:1371–1386. doi: 10.15252/emmm.201404033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W, Bao L, Yang S, Qian Z, Dong M, Yin L, Zhao Q, Ge K, Deng Z, Zhang J, et al. Tumor-selective replication herpes simplex virus-based technology significantly improves clinical detection and prognostication of viable circulating tumor cells. Oncotarget. 2016;7:39768–39783. doi: 10.18632/oncotarget.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Bourcy CF, De Vlaminck I, Kanbar JN, Wang J, Gawad C, Quake SR. A quantitative comparison of single-cell whole genome amplification methods. PLoS One. 2014;9:e105585. doi: 10.1371/journal.pone.0105585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 13.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The genome analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roth A, Ding J, Morin R, Crisan A, Ha G, Giuliany R, Bashashati A, Hirst M, Turashvili G, Oloumi A, et al. JointSNVMix: A probabilistic model for accurate detection of somatic mutations in normal/tumour paired next-generation sequencing data. Bioinformatics. 2012;28:907–913. doi: 10.1093/bioinformatics/bts053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G, Meyerson M, Getz G. Oncotator: Cancer variant annotation tool. Hum Mutat. 2015;36:E2423–E2429. doi: 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Silva GO, He X, Parker JS, Gatza ML, Carey LA, Hou JP, Moulder SL, Marcom PK, Ma J, Rosen JM, Perou CM. Cross-species DNA copy number analyses identifies multiple 1q21-q23 subtype-specific driver genes for breast cancer. Breast Cancer Res Treat. 2015;152:347–356. doi: 10.1007/s10549-015-3476-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gulbahce N, Magbanua MJM, Chin R, Agarwal MR, Luo X, Liu J, Hayden DM, Mao Q, Ciotlos S, Li Z, et al. Quantitative whole genome sequencing of circulating tumor cells enables personalized combination therapy of metastatic cancer. Cancer Res. 2017;77:4530–4541. doi: 10.1158/0008-5472.CAN-17-0688. [DOI] [PubMed] [Google Scholar]
- 21.Palmisano GL, Pistillo MP, Capanni P, Pera C, Nicolò G, Salvi S, Perdelli L, Pasciucco G, Ferrara GB. Investigation of HLA class I downregulation in breast cancer by RT-PCR. Hum Immunol. 2001;62:133–139. doi: 10.1016/S0198-8859(00)00241-X. [DOI] [PubMed] [Google Scholar]
- 22.Kaneko K, Ishigami S, Kijima Y, Funasako Y, Hirata M, Okumura H, Shinchi H, Koriyama C, Ueno S, Yoshinaka H, Natsugoe S. Clinical implication of HLA class I expression in breast cancer. BMC Cancer. 2011;11:454. doi: 10.1186/1471-2407-11-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, Li F, Tsang S, Wu K, Wu H, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ni X, Zhuo M, Su Z, Duan J, Gao Y, Wang Z, Zong C, Bai H, Chapman AR, Zhao J, et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc Natl Acad Sci USA. 2013;110:21083–21088. doi: 10.1073/pnas.1320659110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, Chen K, Scheet P, Vattathil S, Liang H, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Y, Ni X, Guo H, Su Z, Ba Y, Tong Z, Guo Z, Yao X, Chen X, Yin J, et al. Single-cell sequencing deciphers a convergent evolution of copy number alterations from primary to circulating tumour cells. Genome Res. 2017;27:1312–1322. doi: 10.1101/gr.216788.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim N, Hong Y, Kwon D, Yoon S. Somatic mutaome profile in human cancer tissues. Genomics Inform. 2013;11:239–244. doi: 10.5808/GI.2013.11.4.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. COSMIC: Mining complete cancer genomes in the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Network: Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross CA. The Trophoblast Model of Cancer. Nutr Cancer. 2015;67:61–67. doi: 10.1080/01635581.2014.956257. [DOI] [PubMed] [Google Scholar]
- 31.Alix-Panabieres C, Cayrefourcq L, Mazard T, Maudelonde T, Assenat E, Assou S. Molecular portrait of metastasis-competent circulating tumor cells in colon cancer reveals the crucial role of genes regulating energy metabolism and DNA repair. Clin Chem. 2017;63:700–713. doi: 10.1373/clinchem.2016.263582. [DOI] [PubMed] [Google Scholar]
- 32.Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, Radhakrishnan K, Lee SH, Nickoloff JA, Hromas R. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci USA. 2011;108:540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Wang HT, Li BG. Prognostic significance of circulating tumor cells in small-cell lung cancer patients: A meta-analysis. Asian Pac J Cancer Prev. 2014;15:8429–8433. doi: 10.7314/APJCP.2014.15.19.8429. [DOI] [PubMed] [Google Scholar]
- 34.Goldkorn A, Ely B, Quinn DI, Tangen CM, Fink LM, Xu T, Twardowski P, Van Veldhuizen PJ, Agarwal N, Carducci MA, et al. Circulating tumor cell counts are prognostic of overall survival in SWOG S0421: A phase III trial of docetaxel with or without atrasentan for metastatic castration-resistant prostate cancer. J Clin Oncol. 2014;32:1136–1142. doi: 10.1200/JCO.2013.51.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romiti A, Raffa S, Di Rocco R, Roberto M, Milano A, Zullo A, Leone L, Ranieri D, Mazzetta F, Medda E, et al. Circulating tumor cells count predicts survival in colorectal cancer patients. J Gastrointestin Liver Dis. 2014;23:279–284. doi: 10.15403/jgld.2014.1121.233.arom1. [DOI] [PubMed] [Google Scholar]
- 36.Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruz-Gordillo P, Francis JM, Zhang CZ, Shalek AK, Satija R, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol. 2014;32:479–484. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weissenstein U, Schumann A, Reif M, Link S, Toffol-Schmidt UD, Heusser P. Detection of circulating tumor cells in blood of metastatic breast cancer patients using a combination of cytokeratin and EpCAM antibodies. BMC Cancer. 2012;12:206. doi: 10.1186/1471-2407-12-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papadaki MA, Kallergi G, Zafeiriou Z, Manouras L, Theodoropoulos PA, Mavroudis D, Georgoulias V, Agelaki S. Co-expression of putative stemness and epithelial-to-mesenchymal transition markers on single circulating tumour cells from patients with early and metastatic breast cancer. BMC Cancer. 2014;14:651. doi: 10.1186/1471-2407-14-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanwar N, Hu P, Bedard P, Clemons M, McCready D, Done SJ. Identification of genomic signatures in circulating tumor cells from breast cancer. Int J Cancer. 2015;137:332–344. doi: 10.1002/ijc.29399. [DOI] [PubMed] [Google Scholar]
- 40.Meng S, Tripathy D, Frenkel EP, Shete S, Naftalis EZ, Huth JF, Beitsch PD, Leitch M, Hoover S, Euhus D, et al. Circulating tumor cells in patients with breast cancer dormancy. Clin Cancer Res. 2004;10:8152–8162. doi: 10.1158/1078-0432.CCR-04-1110. [DOI] [PubMed] [Google Scholar]
- 41.Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009;11:R46. doi: 10.1186/bcr2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bingham C, Fernandez SV, Fittipaldi P, Dempsey PW, Ruth KJ, Cristofanilli M, Alpaugh Katherine R. Mutational studies on single circulating tumor cells isolated from the blood of inflammatory breast cancer patients. Breast Cancer Res Treat. 2017;163:219–230. doi: 10.1007/s10549-017-4176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mu Z, Benali-Furet N, Uzan G, Znaty A, Ye Z, Paolillo C, Wang C, Austin L, Rossi G, Fortina P, et al. Detection and characterization of circulating tumor associated cells in metastatic breast cancer. Int J Mol Sci. 2016;17:E1665. doi: 10.3390/ijms17101665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fagiani E, Giardina G, Luzi L, Cesaroni M, Quarto M, Capra M, Germano G, Bono M, Capillo M, Pelicci P, Lanfrancone L. RaLP, a new member of the Src homology and collagen family, regulates cell migration and tumor growth of metastatic melanomas. Cancer Res. 2007;67:3064–3073. doi: 10.1158/0008-5472.CAN-06-2301. [DOI] [PubMed] [Google Scholar]
- 45.Turco MY, Furia L, Dietze A, Diaz Fernandez L, Ronzoni S, Sciullo A, Simeone A, Constam D, Faretta M, Lanfrancone L. Cellular heterogeneity during embryonic stem cell differentiation to epiblast stem cells is revealed by the ShcD/RaLP adaptor protein. Stem Cells. 2012;30:2423–2436. doi: 10.1002/stem.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ting DT, Wittner BS, Ligorio M, Jordan Vincent N, Shah AM, Miyamoto DT, Aceto N, Bersani F, Brannigan BW, Xega K, et al. Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep. 2014;8:1905–1918. doi: 10.1016/j.celrep.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedl P, Wolf K. Proteolytic interstitial cell migration: A five-step process. Cancer Metastasis Rev. 2009;28:129–135. doi: 10.1007/s10555-008-9174-3. [DOI] [PubMed] [Google Scholar]
- 48.von Nandelstadh P, Gucciardo E, Lohi J, Li R, Sugiyama N, Carpen O, Lehti K. Actin-associated protein palladin promotes tumor cell invasion by linking extracellular matrix degradation to cell cytoskeleton. Mol Biol Cell. 2014;25:2556–2570. doi: 10.1091/mbc.E13-11-0667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akalay I, Janji B, Hasmim M, Noman MZ, André F, De Cremoux P, Bertheau P, Badoual C, Vielh P, Larsen AK, et al. Epithelial-to-mesenchymal transition and autophagy induction in breast carcinoma promote escape from T-cell-mediated lysis. Cancer Res. 2013;73:2418–2427. doi: 10.1158/0008-5472.CAN-12-2432. [DOI] [PubMed] [Google Scholar]
- 50.Najmeh S, Cools-Lartigue J, Rayes RF, Gowing S, Vourtzoumis P, Bourdeau F, Giannias B, Berube J, Rousseau S, Ferri LE, Spicer JD. Neutrophil extracellular traps sequester circulating tumor cells via β1-integrin mediated interactions. Int J Cancer. 2017;140:2321–2330. doi: 10.1002/ijc.30635. [DOI] [PubMed] [Google Scholar]
- 51.Demers M, Krause DS, Schatzberg D, Martinod K, Voorhees JR, Fuchs TA, Scadden DT, Wagner DD. Cancer predispose neutrophils to release extracellular DNA traps that contribute to cancer-associated thrombosis. Proc Natl Acad Sci USA. 2012;109:13076–13081. doi: 10.1073/pnas.1200419109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goubran HA, Burnouf T, Radosevic M, El-Ekiaby M. The platelet-cancer loop. Eur J Intern Med. 2013;24:393–400. doi: 10.1016/j.ejim.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 53.Hulka BS, Stark AT. Breast cancer: Cause and prevention. Lancet. 1995;346:883–887. doi: 10.1016/S0140-6736(95)92713-1. [DOI] [PubMed] [Google Scholar]
- 54.May P, Woldt E, Matz RL, Boucher P. The LDL receptor-related protein (LRP) family: An old family of proteins with new physiological functions. Ann Med. 2007;39:219–228. doi: 10.1080/07853890701214881. [DOI] [PubMed] [Google Scholar]
- 55.Cowin PA, George J, Fereday S, Loehrer E, Van Loo P, Cullinane C, Etemadmoghadam D, Ftouni S, Galletta L, Anglesio MS, et al. LRP1B deletion in high-grade serous ovarian cancers is associated with acquired chemotherapy resistance to liposomal doxorubicin. Cancer Res. 2012;72:4060–4073. doi: 10.1158/0008-5472.CAN-12-0203. [DOI] [PubMed] [Google Scholar]
- 56.Magbanua MJ, Park JW. Isolation of circulating tumor cells by immunomagnetic enrichment and fluorescence-activated cell sorting (IE/FACS) for molecular profiling. Methods. 2013;64:114–118. doi: 10.1016/j.ymeth.2013.07.029. [DOI] [PubMed] [Google Scholar]
- 57.Andersen MN, Al-Karradi SN, Kragstrup TW, Hokland M. Elimination of erroneous results in flow cytometry caused by antibody binding to Fc receptors on human monocytes and macrophages. Cytometry A. 2016;89:1001–1009. doi: 10.1002/cyto.a.22995. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding authors upon reasonable request.