Abstract
Purpose
NSD3 has been implicated as a candidate driver oncogene from the 8p11-p12 locus, and we have previously published evidence for its amplification and overexpression in human breast cancer. This aim of this study was to further characterize the transforming function of NSD3 in vivo.
Methods
We generated a transgenic mouse model in which NSD3 gene expression was driven by the MMTV promoter and expressed in mammary epithelium of FVB mice. Mammary glands were fixed and whole mounts were stained with carmine to visualize gland structure. Mammary tumors were formalin-fixed, and paraffin embedded (FFPE) tumors were stained with hematoxylin and eosin.
Results
Pups born to transgenic females were significantly underdeveloped compared to pups born to WT females due to a lactation defect in transgenic female mice. Whole mount analysis of the mammary glands of transgenic female mice revealed a profound defect in functional differentiation of mammary gland alveoli that resulted in the lactation defect. We followed parous and virgin NSD3 transgenic and control mice to 50 weeks of age and observed that several NSD3 parous females developed mammary tumors. Whole mount analysis of the mammary glands of tumor-bearing mice revealed numerous areas of mammary hyperplasia and ductal dysplasia. Histological analysis showed that mammary tumors were high-grade ductal carcinomas, and lesions present in other mammary glands exhibited features of alveolar hyperplasia, ductal dysplasia, and carcinoma in situ.
Conclusions
Our results are consistent with our previous studies and demonstrate that NSD3 is a transforming breast cancer oncogene.
Keywords: Breast cancer, Transgenic mouse model, Amplicon, Oncogene, WHSC1L1, NSD3, ER positive
Introduction
The genomic region at the 8p11-p12 locus is commonly amplified in human breast cancer, human squamous cell lung cancer, and several other cancer types [1–3]. This amplicon is generally focal in nature, but has several discreet regions that can be amplified independently of each other [4]. Accordingly, it is likely that this genomic region harbors several driving oncogenes, with at least one in each sub-region. Indeed, there are numerous reports in the literature documenting the transforming potential of at least six different genes from this amplicon [5–14]. For several years, our laboratory has been interested in the role played by one of these candidate oncogenes, the histone methyl transferase, NSD3 (formerly known as WHSC1L1), and we have published evidence for its amplification and overexpression in the SUM-44 human breast cancer cell line, primary human breast cancers, and its transforming function when overexpressed in MCF-10A cells. More recently, we demonstrated that NSD3 can drive high level expression of ERa and induce estrogen-independent, but estrogen receptor-dependent proliferation and survival of SUM-44 breast cancer cells [2, 4, 15, 16]. These findings are consistent with data obtained by other labs that have identified NSD3 as one of several genes in the amplicon that are associated with a poor outcome in ERa-positive breast cancer [6, 17–22]. NSD3 has also been shown to have transforming ability in other model systems, and has been implicated as a driving oncogene by GISTIC analysis of large datasets derived from human breast cancer specimens [23]. Thus, there is a significant body of literature supporting the notion that NSD3 is an important oncogene in breast and other cancers in which the 8p11 region is focally amplified. However, because of the complexity of this genomic region, and because FGFR1 is a neighboring gene on the amplicon that is almost always co-amplified and overexpressed with NSD3, the oncogenic role of NSD3 in human breast cancer has not yet been widely accepted [24]. To investigate further the potential of NSD3 to act as a breast cancer oncogene, we developed a transgenic mouse model with targeted expression of NSD3 to the mammary epithelium. In this report, we demonstrate that NSD3 transgenic FVB mice exhibit altered mammary gland morphology during development, differentiation, and involution, resulting in a profound lactation defect caused by the failure of the alveoli to undergo full functional differentiation. By 40 weeks of age, many of these mice developed mammary gland hyperplasias, dysplasias, carcinoma in situ, and mammary carcinomas. Thus, these results extend our in vitro results and show that targeted expression of NSD3 to the mammary gland of FVB mice is oncogenic, consistent with the hypothesis that NSD3 is an important driver oncogene in human breast cancer.
Materials and methods
Chemicals and antibodies
All reagents used for carmine staining of whole mounts, including ethanol, xylene, and glacial acetic acid, were purchased from Fisher Scientific (Hampton, NH) unless stated otherwise. The NSD3 antibody used for western blotting and immunostaining was purchased from ProteinTech Group Inc. (Cat. No. 11345-1-AP) and the ERa antibody was purchased from Santa Cruz (Cat. No. sc-7207). Primers used for genotyping were purchased from IDT.
Transgenic mice
The Institutional Animal Care and Use Committee (IACUC) at the Medical University of SC (MUSC) approved all animal experiments (AR#3397). For the purposes of this study, FVB/N mice were used to generate NSD3 and KAT6A transgenic mouse colonies. The inbred FVB/N strain is commonly used for the production of transgenic mice, as they produce large litters and their large pronuclei can withstand microinjection of the transgene. Male and female WT FVB/N mice were purchased from Jackson Laboratories (Bar Harbor, ME) and bred under typical conditions with unlimited access to food and water. Targeted expression of NSD3 and KAT6A to the mammary gland was achieved by cloning each transgene into the MKbpAII vector with an MMTV promoter. Cloning and preparation of NSD3 and KAT6A constructs as well as generation of transgenic mice was performed by the Gene Function Core at MUSC following the standard protocol for production of transgenic mice.
NSD3 and KAT6A genotyping
To identify positive transgenic pups, tail snips were obtained from 3 week-old pups before weaning. Tails were incubated overnight at 55 °C in digestion buffer and DNA was purified using the ZR Genomic DNA™ Mini Prep Kit (Zymo Research, Irvine, CA). Genotyping by PCR was performed with the following primers: NSD3 For, 5′-ATACTCTGAGTCCAAACCGGG-3′ and NSD3 Rev, 5′-GTGTGAGCGCTCTTCAG-3′; KAT6A For; 5′-GGGGCAAAGTTTGGGTG-3′ and KAT6A Rev, 5′-CAGGATGGATGGCCACTGTT-3′. PCR amplification was performed with Taq polymerase from Qiagen under the following conditions: 30 cycles of 94 °C 3 min, 94 °C 30 s, 62 °C 30 s, 72 °C 1 min 30 s, 72 °C 10 min. PCR products were visualized on a 1% agarose gel.
Whole mount preparation
Mammary glands were harvested from female mice at various time points following euthanasia of the animals per an IACUC approved protocol. Virgin mammary glands were harvested at 12 weeks of age and mammary glands of females at mid-pregnancy were harvested between day 12 and 14 post-mating. Lactating mammary glands were taken 7–10 days following the birth of litters, and at least 2 weeks were allowed for involution to occur before harvesting mammary glands post-weaning. The glands were spread on microscope slides (Polysine pre-cleaned microscope slides, Thermo Scientific, MA) and prepared for carmine staining. Briefly, once the glands adhered to the slide (approximately 1–2 min), they were fixed in 100% ethanol/30% acetic acid (3:1) at room temperature overnight. The following day, slides were washed in 70% ethanol for 15 min, then in distilled water for 5 min. The glands were then stained overnight with carmine alum [1 g Carmine (Sigma-Aldrich C-6125), 2.5 g Aluminum potassium sulfate (Sigma-Aldrich C-237,086) in 500 ml water]. The tissues were then washed in 75, 95, and 100% ethanol for 15 min each and cleared in xylene overnight. For long-term storage, whole mounts were stored in methyl salicylate (Sigma-Aldrich, St. Louis, MO). Tumors harvested from NSD3 transgenic females were cleanly excised and fixed in formalin overnight.
Tissue processing and staining
Whole mounts and tumors were processed by the Biorepository and Tissue Analysis Shared Resource at the Medical University of South Carolina. Flash frozen paraffin embedded (FFPE) Sects. (5 lm) were deparaffinized in xylene, rehydrated in alcohol, and processed as follows. The sections were incubated with target retrieval solution (Dako Glostrup, Denmark) in a steamer (Oster CKSTSTMD5-W) for 30 min and then 3% hydrogen peroxide solution for 10 min followed by protein block (Dako ×0909) for 20 min at room temperature. The sections were then incubated with NSD3 and ERa primary antibodies overnight in a humid chamber at 4 °C followed by incubation with biotinylated secondary antibody (Vector, CA, USA) for 30 min and ABC reagent (Vector, CA USA) for 30 min. Immunocomplexes were visualized by DAB (Dako, Glostrup, Denmark) reaction and sections were counterstained with hematoxylin before mounting.
Results
Delayed development in pups from NSD3 transgenic female mice
To further our studies on the oncogenic role of the breast cancer candidate oncogene NSD3, we generated a transgenic mouse line in which NSD3 expression was targeted to the mammary gland. To identify potentially positive transgenic pups, tail biopsies were taken from 2 week-old mice and PCR was performed to detect the transgene, thereby identifying founder (F0) mice. Once founder mice were of breeding age, both male and female F0 mice were mated with wild-type (WT) FVB/N mice to establish F1 offspring.
The first goal of these experiments was to determine if targeted expression of NSD3 in the mammary gland would have observable effects on mammary gland biology through the normal growth, differentiation, and involution cycle of the mouse mammary gland. Early on in our observations, we noticed that pups born to NSD3 transgenic female mice were underdeveloped. By contrast, pups born to WT females mated with NSD3 transgenic males did not display any developmental defects (Fig. 1). This was readily apparent at day 9, at which point pups nursed by NSD3 transgenic females were not only significantly smaller than pups born to wild-type mothers, but also showed delayed hair growth and eye opening compared to pups nursed by WT females. However, pups born to WT females mated with NSD3 transgenic males and nursed by WT females did not display this developmental lag. The observed developmental lag displayed in pups nursed by NSD3 transgenic females was evident in both WT and NSD3 transgenic pups, giving the first indication that this developmental delay was due to a lactation defect present in the NSD3 transgenic females and not a result of NSD3 expression in the pups.
Fig. 1.
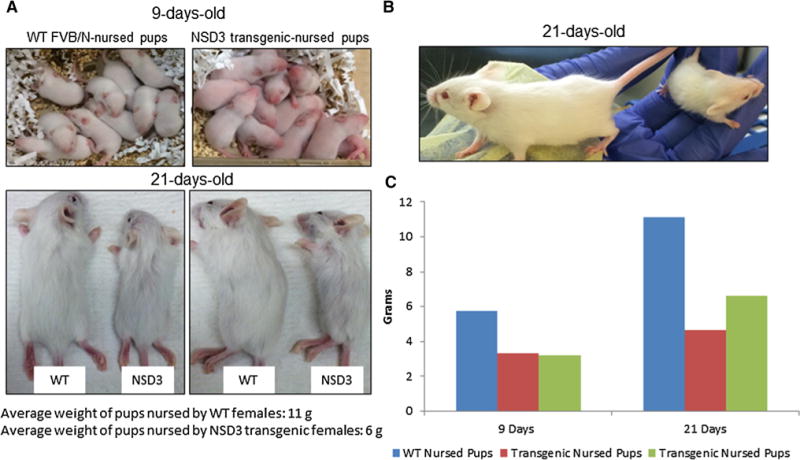
Underdevelopment in pups from NSD3 transgenic females. a WT FVB/N and NSD3 transgenic females were mated with WT FVB/N males. All females produced normal size litters of 8–10 pups. At 9 days of age, pups nursed by NSD3 transgenic females (right) were already lagging behind developmentally compared to pups nursed by WT FVB/N females (left). b Side by side comparison photos show the underdevelopment of pups nursed by NSD3 transgenic females at the time of weaning or at 21 days of age. As seen on the left in all photos, these pups remained developmentally behind pups nursed by WT FVB/N females. c The bar graph shows the average weights of two litters of pups nursed by NSD3 transgenic females at days 9 and 21 compared to one litter of WT-nursed pups. NSD3-nursed pups were approximately half the weight of healthy pups at both time points compared to WT-nursed pups
Lactation deficiency in NSD3 transgenic females
To further investigate the influence of mammary gland-targeted expression of NSD3 on mammary gland biology, mammary glands from virgin, mid-pregnant, lactating, and post-lactating mice were harvested for whole mount and histologic analysis. As seen in Fig. 2a, mammary glands from 12 week-old virgin transgenic females displayed increased ductal branching and terminal bud formation compared to WT female virgins. Additionally, mammary glands taken from NSD3 transgenic females mid-pregnancy (12–14 days; Fig. 2b) showed more numerous and densely packed alveolar buds compared to WT females during pregnancy. Although these phenotypes were subtle, blind scoring of these mammary gland whole mounts identified mammary glands from transgenic mice 100% of the time. The phenotypic differences between mammary glands from lactating transgenic females versus wild-type mice were not subtle and were consistent with the lactation defect hypothesized based on the growth of the pups as described above. Mammary glands from lactating transgenic mice showed evidence of gross defects in the full functional differentiation of many of the alveoli in these mammary glands (Fig. 2c). This failure to achieve full functional differentiation was observed in every lactation-state mammary gland we observed in transgenic females, and is consistent with the growth retardation observed in the pups of these mice. In addition, examination of post-lactation mammary glands from wild-type and transgenic mice showed that while post-lactation mammary glands from WT female mice had undergone involution, glands from transgenic females failed to completely involute and showed evidence of hyperplasia (Fig. 2d).
Fig. 2.
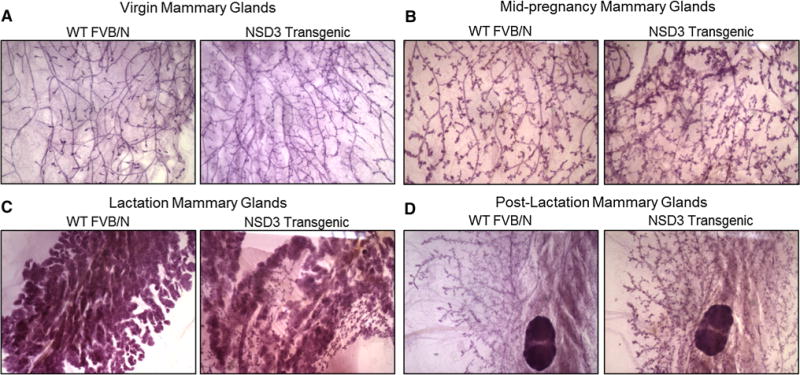
Lactation deficiency in NSD3 transgenic female mice. a After euthanization, mammary glands were harvested from female mice at various time points. Virgin mammary glands were harvested from WT and NSD3 transgenic females at 12 weeks of age. NSD3 mammary glands showed increased ductal branching and terminal bud formation. b Mammary glands were harvested from females at mid-pregnancy between day 12 and 14 post-mating. Mammary glands from pregnant NSD3 mice showed more numerous and densely packed alveolar buds compared to mammary glands harvested from pregnant WT females. c Lactating mammary glands were harvested from WT and NSD3 transgenic females 7–10 days following the birth of litters. NSD3 mammary glands showed failure of the glands to functionally differentiate compared to WT mammary glands. d Two weeks were allowed post-weaning for involution to occur before harvesting mammary glands from WT and NSD3 females. Staining shows that mammary glands from NSD3 transgenic females failed to involute, while the WT mammary glands had undergone involution
Mammary hyperplasia, dysplasia, and tumorigenesis in NSD3 transgenic females
To examine the long-term effects of mammary gland-targeted expression of NSD3, a group of 25 transgenic and wild-type females were maintained until they reached one year of age and monitored weekly for tumor development. We first palpated mammary tumors in two transgenic mice at 40 weeks of age, and thereafter, several additional transgenic parous females developed mammary tumors at ages between 40 and 50 weeks. Overall, five of the 25 transgenic mice, all of which were parous females, developed mammary tumors. By contrast, none of the WT mice developed tumors or any other morphologic abnormalities. Once tumors reached a size of 1 cm3, the mice were euthanized and the tumors were excised and fixed in formalin for histological analysis. At the same time, the remaining mammary glands were harvested from tumor-bearing females and whole mounts were prepared. Interestingly, mammary glands from tumor-bearing mice exhibited numerous ductal and alveolar hyperplasias and ductal dysplasias (Fig. 3). Histological analysis of these mammary lesions revealed a wide variety of abnormalities including areas that would be identified as proliferative and non-proliferative fibrocystic disease in humans, such as alveolar and ductal hyperplasia, adenosis, microcyst formation, and columnar cell change. Papillary lesions were also identified. In addition, areas of epithelial atypia, dysplasia, and ductal carcinoma in situ (DCIS) were commonly observed. Patterns of DCIS were similar to those seen in humans with identifiable micropapillary, cribriform, and clinging-type carcinoma. Patterns of invasive carcinoma markedly resembled that seen in high-grade invasive ductal carcinoma in humans with often solid or trabecular type growth patterns that exhibit a high mitotic rate and increasing degrees of necrosis as the tumors increased in size. No lesions or tumors were observed in mammary glands of age-matched wild-type female mice, consistent with published reports on the background incidence of mammary tumors in FVB mice. A total of five of the 25 transgenic mice followed to 50 weeks of age developed frank mammary tumors. All of these mice also exhibited areas of mammary hyperplasia, dysplasia, and carcinoma in situ in other mammary glands. In addition, three mice that did not develop palpable tumors during this time exhibited mammary hyperplasias and dysplasias. In all, 95 mammary glands from the 25 mice were examined, with structural abnormalities identified in 18 of the 95 whole mounts (Fig. 4).
Fig. 3.
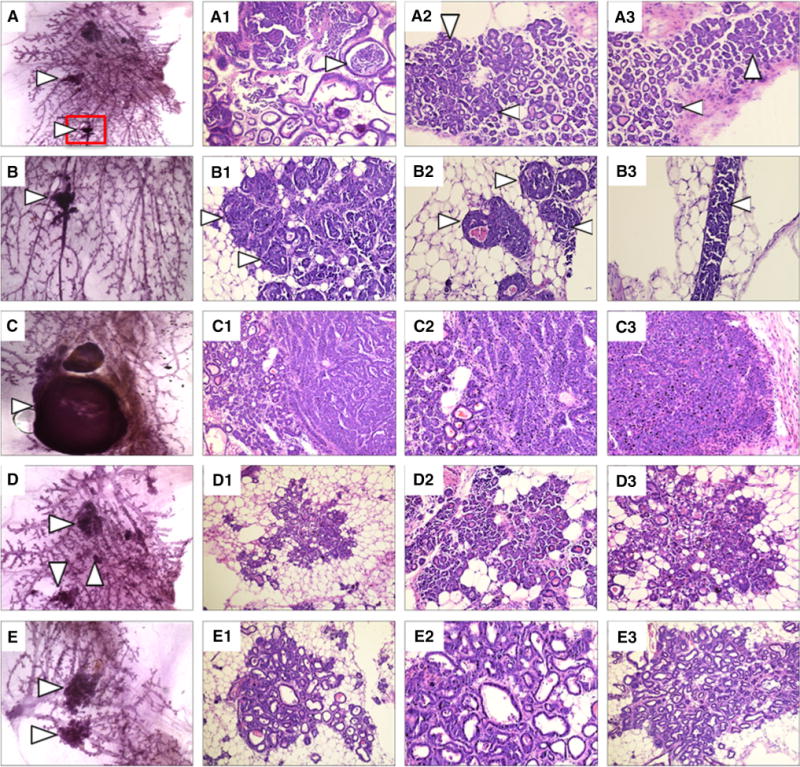
Mammary gland hyperplasia and dysplasia in NSD3 transgenic females. a Carmine staining of a whole mount of a right thoracic mammary gland harvested from an NSD3 female showing numerous areas of hyperplasia and dysplasia. A1–A3. H&E staining of sections cut from the whole mount shown in panel A detailing cystic lesions and ductal carcinoma in situ (DCIS). b Higher magnification of the area boxed in red from panel A. B1-3. Higher magnification images of H&E staining of the area shown in panel B. c Carmine-stained whole mount of the right inguinal mammary gland taken from an NSD3 female displaying a small mammary carcinoma. C1-3. H&E staining of the whole mount shown in panel C. d Carmine-stained whole mount of a right thoracic mammary gland harvested from an NSD3 female displaying numerous hyperplastic lesions. D1-3. H&E staining of the whole mount shown in panel D. e Carmine-stained whole mount of a left thoracic mammary gland harvested from an NSD3 transgenic female. E1-3. H&E staining of the whole mount shown in panel E
Fig. 4.
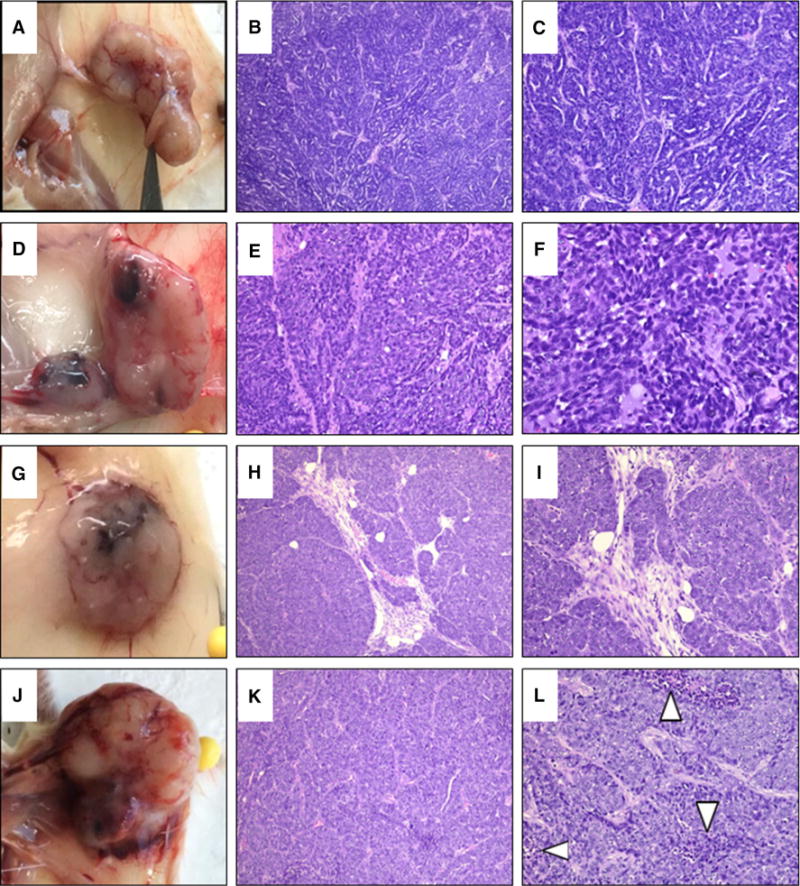
Mammary hyperplasia, dysplasia, and tumorigenesis in NSD3 transgenic females. a Tumor isolated from the left inguinal mammary gland of an NSD3 transgenic female. b–c H&E staining of histological sections of the tumor shown in panel A displaying a trabecular growth pattern. d Tumor isolated from the left inguinal gland of an NSD3 transgenic female. e–f H&E staining of histological sections of the tumor shown in panel D. g Tumor isolated from the right thoracic gland of an NSD3 transgenic female. h–i H&E staining of histological sections of the tumor shown in panel G. H&E staining shows precursor trabecular growth as well as invasive ductal carcinoma. j Tumor isolated from the cervical gland of an NSD3 transgenic female. k–l H&E staining of the tumor shown in panel J, showing invasive ductal carcinoma with trabecular growth patterns coupled with areas of necrosis
It is noteworthy that in the course of these studies, we also generated KATA6A (Myst3) transgenic mice, as we have previously implicated this gene from the 8p11-p12 region as a breast cancer oncogene [14]. In contrast to the results obtained with the NSD3 transgenic mice, KATA6 mice did not exhibit any altered morphologic phenotypes or lactation defects, and never developed mammary pre-neoplastic or neoplastic lesions.
To confirm transgene overexpression in the mammary glands of transgenic mice-bearing tumors or hyperplasias/dysplasias, immunohistochemical analysis was performed on the histological sections from these mammary glands. Figure 5a shows that indeed, these mammary gland lesions and tumors stained positively for NSD3, as well as ERa, consistent with our recently published association between NSD3 and ER expression in human breast cancer cell lines [16]. Finally, when tumors were harvested from transgenic female mice, pieces of the tumor were subjected to enzymatic dissociation and plated into primary culture. After one to two weeks in culture, cells were harvested and assessed for expression of NSD3 by western blot analysis. Figure 5b confirms that these mouse mammary tumor cells express high levels of NSD3 protein.
Fig. 5.
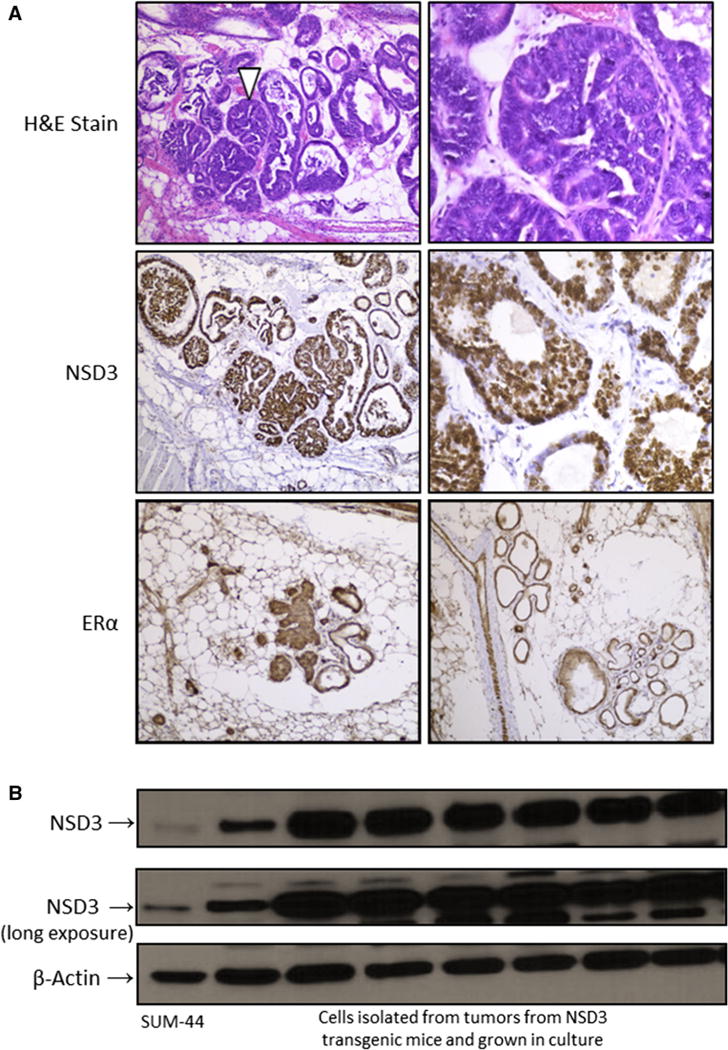
Overexpression of NSD3 in mammary tumors from transgenic females. a The upper two panels show H&E stained mammary glands from an NSD3 transgenic female exhibiting areas of hyperplasia and dysplasia. The middle two panels display positive nuclear staining for NSD3 by immunohistochemistry (IHC) in the mammary gland of an NSD3 female. The bottom two panels show positive ERa staining by IHC in mammary glands from a tumor-bearing NSD3 transgenic female. b. Western blot of NSD3 expression in cells isolated from tumors in NSD3 females and grown in culture under various conditions. Because they have been shown to express significant levels of NSD3, whole cell lysate from SUM-44 are included for comparison [16]
In summary, these studies demonstrate the oncogenic potential of NSD3 when expression is targeted to the mammary gland of FVB mice and support the hypothesis that NSD3 is a driving breast cancer oncogene from the 8p11 amplicon.
Discussion
8p11-p12 amplification is associated with poor survival and distant recurrence in luminal breast cancer [1, 3, 4, 17]. We and others have shown that overexpression of several genes within the 8p11-p12 region can provide a pathological gain-of-function to breast cells [15, 16, 22]. Within focal regions of gene amplification, it is often difficult to determine which genes are most important and act as driving genes. Driving genes provide a necessary growth advantage that is essential for tumor survival, whereas while passenger genes are often amplified with driving genes, they are not necessary for proliferation or survival. Previous published data are consistent with the notion that the 8p11-p12 amplicon contains both driver and passenger genes, and studies from other laboratories have proposed different drivers [2, 4, 15, 25, 26]. Predictions based on Cancer Genome Atlas data, including GISTIC analyses, have helped to narrow the number of putative 8p11 amplicon driving genes, and have identified NSD3 as a driving gene in multiple tumor types, including breast cancer [23]. In the present study, we demonstrated that targeted expression of NSD3 in the mammary epithelium of FVB mice resulted in drastic morphological alterations in the mammary gland, inhibited functional differentiation, and resulted in the development of hyperplasias, dysplasias, carcinoma in situ, and mammary carcinoma development.
We first observed that targeted expression of NSD3 to the mammary epithelium blocked functional differentiation of the alveolar units in the mammary gland resulting in stunted developmental growth of the offspring when compared to pups born to WT females. The developmental lag was evident by postpartum day 9; pups nursed by NSD3 females had delayed hair growth compared to pups nursed by WT females. This phenotype persisted through weaning age, resulting in significantly smaller NSD3 transgenic female-nursed pups. In contrast, pups nursed by WT females displayed normal developmental patterns. Interestingly, the developmental lag was evident in all pups nursed by NSD3 transgenic females regardless of the genotype of the pups. Indeed, carmine staining of mammary glands from lactating NSD3 transgenic females showed areas where alveoli had failed to undergo full functional differentiation.
In addition to the lactation deficiency seen in NSD3 females, these mice developed mammary gland hyperplasias, dysplasias, and carcinomas. Tumors from NSD3 transgenic females presented as infiltrating ductal carcinomas with high nuclear grade. These tumors were not well circumscribed and displayed irregular borders. The phenotypes of these mouse mammary carcinomas are significant because they are similar to the phenotypic characteristics observed in human breast cancers. Furthermore, the majority of glands in tumor-bearing NSD3 transgenic females showed areas of hyperplasia, dysplasia, and carcinoma in situ.
Conclusions
Previous studies from our lab and others have demonstrated the potent transforming properties of NSD3, which is consistent with its role as an important driving oncogene on the 8p11-p12 amplicon [15, 16, 22]. Here, we provide direct evidence for NSD3 as a breast cancer oncogene by showing that targeted expression of NSD3 in the mouse mammary gland prevents alveolar differentiation and results in development of pre-malignant lesions and mammary gland ductal carcinomas. At the present time, the only clinical studies that have been performed in patients exhibiting the 8p11 amplicon have focused on targeting the FGFR1 gene, which is present on the amplicon and is often co-amplified with NSD3. The results of these clinical studies have been disappointing in both breast and lung cancer patients bearing the amplicon [27, 28]. These clinical findings suggest that FGFR1 is not a major driving oncogene in 8p11 amplified breast and lung cancers. Thus, it is now important to consider NSD3 as an important driving oncogene and a potential druggable target in breast cancer patients, and perhaps lung cancer patients, bearing the 8p11 amplicon and exhibiting amplification and overexpression of NSD3.
Acknowledgments
We would like to specifically thank Michael J. Kern, Ph.D. of the Gene Function Core at the Medical University of South Carolina and all of the technicians for their assistance in generating the NSD3 transgenic mice. We would also like to thank Kiwana Gibbs and Yuan Shao, Ph.D. of the Biorepository and Tissue Analysis Shared Resource at the Medical University of South Carolina for providing histological sections and staining of all mammary glands and tumors.
Funding This study was funded by the 2RO1CA100724, NCI 2P30CA138313, and the Chalsty Breast Cancer Research Fund 1K12CA457688-O1A1.
Footnotes
Authors’ contribution
Conception and design of the study was conducted by SE. Transgenic mice were handled by ES for ear tagging/tail snips, euthanizing, and harvesting of all mammary glands for whole mount preparation. Genotyping and transgenic mouse record keeping for the NSD3 colony was performed by BI. Both ES and BI performed carmine staining of whole mounts, cultured tumor-derived mammary cells, and performed western blotting on lysates obtained from NSD3-derived mammary tumor cells. AR organized mammary glands and generated images for scoring hyperplastic lesions seen in whole mounts. LS is the breast cancer pathologist for the Hollings Cancer Center and along with JM performed histopathologic analysis of mammary lesions and tumors. The manuscript was written by SE, BI, ES, AR, and LS.
Compliance with ethical standards
Conflict of interest The authors declare that they have no conflict of interest.
Ethical approval All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. The Institutional Animal Care and Use Committee (IACUC) at the Medical University of SC (MUSC) approved all animal experiments, AR#3397. Male and female WT FVB/N mice were bred under typical conditions with unlimited access to food and water. Animals were euthanized per IACUC approved protocol during all experiments. This article does not contain any studies with human participants performed by any of the authors.
References
- 1.Adelaide J, Chaffanet M, Imbert A, Allione F, Geneix J, Popovici C, van Alewijk D, Trapman J, Zeillinger R, Borresen-Dale AL, et al. Chromosome region 8p11-p21: refined mapping and molecular alterations in breast cancer. Genes Chromosomes Cancer. 1998;22(3):186–199. doi: 10.1002/(sici)1098-2264(199807)22:3<186::aid-gcc4>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 2.Ray ME, Yang ZQ, Albertson D, Kleer CG, Washburn JG, Macoska JA, Ethier SP. Genomic and expression analysis of the 8p11–12 amplicon in human breast cancer cell lines. Cancer Res. 2004;64(1):40–47. doi: 10.1158/0008-5472.can-03-1022. [DOI] [PubMed] [Google Scholar]
- 3.Bilal E, Vassallo K, Toppmeyer D, Barnard N, Rye IH, Almendro V, Russnes H, Borresen-Dale AL, Levine AJ, Bhanot G, et al. Amplified loci on chromosomes 8 and 17 predict early relapse in ER-positive breast cancers. PLoS ONE. 2012;7(6):e38575. doi: 10.1371/journal.pone.0038575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gelsi-Boyer V, Orsetti B, Cervera N, Finetti P, Sircoulomb F, Rouge C, Lasorsa L, Letessier A, Ginestier C, Monville F, et al. Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol Cancer Res. 2005;3(12):655–667. doi: 10.1158/1541-7786.MCR-05-0128. [DOI] [PubMed] [Google Scholar]
- 5.Carapeti M, Aguiar RC, Watmore AE, Goldman JM, Cross NC. Consistent fusion of MOZ and TIF2 in AML with inv (8) (p11q13) Cancer Genet Cytogenet. 1999;113(1):70–72. doi: 10.1016/s0165-4608(99)00007-2. [DOI] [PubMed] [Google Scholar]
- 6.Stec I, den Dunnen JT. WHSC1L1, on human chromosome 8p11.2, closely resembles WHSC1 and maps to a duplicated region shared with 4p16.3. Genomics. 2001;76(1–3):5–8. doi: 10.1006/geno.2001.6581. [DOI] [PubMed] [Google Scholar]
- 7.Cully M, Shiu J, Piekorz RP, Muller WJ, Done SJ, Mak TW. Transforming acidic coiled coil 1 promotes transformation and mammary tumorigenesis. Cancer Res. 2005;65(22):10363–10370. doi: 10.1158/0008-5472.CAN-05-1633. [DOI] [PubMed] [Google Scholar]
- 8.Elbauomy Elsheikh S, Green AR, Lambros MB, Turner NC, Grainge MJ, Powe D, Ellis IO, Reis-Filho JS. FGFR1 amplification in breast carcinomas: a chromogenic in situ hybridisation analysis. Breast Cancer Res. 2007;9(2):R23. doi: 10.1186/bcr1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Streicher KL, Yang ZQ, Draghici S, Ethier SP. Transforming function of the LSM1 oncogene in human breast cancers with the 8p11-12 amplicon. Oncogene. 2007;26(14):2104–2114. doi: 10.1038/sj.onc.1210002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernard-Pierrot I, Gruel N, Stransky N, Vincent-Salomon A, Reyal F, Raynal V, Vallot C, Pierron G, Radvanyi F, Delattre O. Characterization of the recurrent 8p11-12 amplicon identifies PPAPDC1B, a phosphatase protein, as a new therapeutic target in breast cancer. Cancer Res. 2008;68(17):7165–7175. doi: 10.1158/0008-5472.CAN-08-1360. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Liu X, Datta A, Govindarajan K, Tam WL, Han J, George J, Wong C, Ramnarayanan K, Phua TY, et al. RCP is a human breast cancer-promoting gene with Ras-activating function. J Clin Invest. 2009;119(8):2171–2183. doi: 10.1172/JCI37622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slorach EM, Chou J, Werb Z. Zeppo1 is a novel metastasis promoter that represses E-cadherin expression and regulates p120-catenin isoform expression and localization. Genes Dev. 2011;25(5):471–484. doi: 10.1101/gad.1998111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G, Liu G, Wang X, Sethi S, Ali-Fehmi R, Abrams J, Zheng Z, Zhang K, Ethier S, Yang ZQ. ERLIN2 promotes breast cancer cell survival by modulating endoplasmic reticulum stress pathways. BMC Cancer. 2012;12:225. doi: 10.1186/1471-2407-12-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner-Ivey B, Guest ST, Irish JC, Kappler CS, Garrett-Mayer E, Wilson RC, Ethier SP. KAT6A, a chromatin modifier from the 8p11-p12 amplicon is a candidate oncogene in luminal breast cancer. Neoplasia. 2014;16(8):644–655. doi: 10.1016/j.neo.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang ZQ, Liu G, Bollig-Fischer A, Giroux CN, Ethier SP. Transforming properties of 8p 11-12 amplified genes in human breast cancer. Cancer Res. 2010;70(21):8487–8497. doi: 10.1158/0008-5472.CAN-10-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irish JC, Mills JN, Turner-Ivey B, Wilson RC, Guest ST, Rutkovsky A, Dombkowski A, Kappler CS, Hardiman G, Ethier SP. Amplification of WHSC1L1 regulates expression and estrogen-independent activation of ERalpha in SUM-44 breast cancer cells and is associated with ERalpha over-expression in breast cancer. Mol Oncol. 2016;10:850–865. doi: 10.1016/j.molonc.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Angrand PO, Apiou F, Stewart AF, Dutrillaux B, Losson R, Chambon P. NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics. 2001;74(1):79–88. doi: 10.1006/geno.2001.6524. [DOI] [PubMed] [Google Scholar]
- 18.Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, Khatry DB, Protopopov A, You MJ, Aguirre AJ, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci USA. 2005;102(27):9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taketani T, Taki T, Nakamura H, Taniwaki M, Masuda J, Hayashi Y. NUP98-NSD3 fusion gene in radiation-associated myelodysplastic syndrome with t(8;11) (p11;p15) and expression pattern of NSD family genes. Cancer Genet Cytogenet. 2009;190(2):108–112. doi: 10.1016/j.cancergencyto.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Z, Thomsen R, Kahns S, Nielsen AL. The NSD3L histone methyltransferase regulates cell cycle and cell invasion in breast cancer cells. Biochem Biophys Res Commun. 2010;398(3):565–570. doi: 10.1016/j.bbrc.2010.06.119. [DOI] [PubMed] [Google Scholar]
- 21.He C, Li F, Zhang J, Wu J, Shi Y. The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition. J Biol Chem. 2013;288(7):4692–4703. doi: 10.1074/jbc.M112.426148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, Roe JS, Suzuki Y, Pappin DJ, Joshua-Tor L, Vakoc CR. NSD3-short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol Cell. 2015;60(6):847–859. doi: 10.1016/j.molcel.2015.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, McGee J, Chen X, Doman TN, Gong X, Zhang Y, Hamm N, Ma X, Higgs RE, Bhagwat SV, et al. Identification of druggable cancer driver genes amplified across TCGA datasets. PLoS ONE. 2014;9(5):e98293. doi: 10.1371/journal.pone.0098293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo J, Liu S, Leung S, Gru AA, Tao Y, Hoog J, Ho J, Davies SR, Allred DC, Salavaggione AL, et al. An mRNA gene expression-based signature to identify FGFR1-amplified estrogen receptor-positive breast tumors. J Mol Diagn. 2017;19(1):147–161. doi: 10.1016/j.jmoldx.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang ZQ, Streicher KL, Ray ME, Abrams J, Ethier SP. Multiple interacting oncogenes on the 8p11-p12 amplicon in human breast cancer. Cancer Res. 2006;66(24):11632–11643. doi: 10.1158/0008-5472.CAN-06-2946. [DOI] [PubMed] [Google Scholar]
- 26.Cornen S, Guille A, Adelaide J, Addou-Klouche L, Finetti P, Saade MR, Manai M, Carbuccia N, Bekhouche I, Letessier A, et al. Candidate luminal B breast cancer genes identified by genome, gene expression and DNA methylation profiling. PLoS ONE. 2014;9(1):e81843. doi: 10.1371/journal.pone.0081843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andre F, Bachelot T, Campone M, Dalenc F, Perez-Garcia JM, Hurvitz SA, Turner N, Rugo H, Smith JW, Deudon S, et al. Targeting FGFR with dovitinib (TKI258): preclinical and clinical data in breast cancer. Clin Cancer Res. 2013;19(13):3693–3702. doi: 10.1158/1078-0432.CCR-13-0190. [DOI] [PubMed] [Google Scholar]
- 28.Pearson A, Smyth E, Babina IS, Herrera-Abreu MT, Tarazona N, Peckitt C, Kilgour E, Smith NR, Geh C, Rooney C, et al. High-level clonal FGFR amplification and response to FGFR inhibition in a translational clinical trial. Cancer Discov. 2016;6(8):838–851. doi: 10.1158/2159-8290.CD-15-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]


