Abstract
Chlamydia trachomatis entry into host cells is mediated by pathogen-directed remodeling of the actin cytoskeleton. The chlamydial type III secreted effector, translocated actin recruiting phosphoprotein (Tarp), has been implicated in the recruitment of actin to the site of internalization. Tarp harbors G-actin binding and proline rich domains required for Tarp-mediated actin nucleation as well as unique F-actin binding domains implicated in the formation of actin bundles. Little is known about the mechanical properties of actin bundles generated by Tarp or the mechanism by which Tarp mediates actin bundle formation. In order to characterize the actin bundles and elucidate the role of different Tarp domains in the bundling process, purified Tarp effectors and Tarp truncation mutants were analyzed using Total Internal Reflection Fluorescence (TIRF) microscopy. Our data indicate that Tarp mediated actin bundling is independent of actin nucleation and the F-actin binding domains are sufficient to bundle actin filaments. Additionally, Tarp-mediated actin bundles demonstrate distinct bending stiffness compared to those crosslinked by the well characterized actin bundling proteins fascin and alpha-actinin, suggesting Tarp may employ a novel actin bundling strategy. The capacity of the Tarp effector to generate novel actin bundles likely contributes to chlamydia’s efficient mechanism of entry into human cells.
Keywords: Chlamydia trachomatis, Tarp, effector, actin bundles, bending persistence length, cytoskeleton
Introduction
The obligate intracellular bacterium, Chlamydia trachomatis is the most frequently reported sexually transmitted bacterial disease in the United States, with over 1 million cases reported annually to the Centers for Disease Control and Prevention (CDC) since 2006 [1]. C. trachomatis displays a unique biphasic developmental cycle consisting of two metabolically and morphologically distinct developmental forms [2]. The infectious extracellular form is called the elementary body (EB) whereas the vegetative intracellular form is called the reticulate body (RB) [3].
To facilitate the obligate intracellular lifestyle, Chlamydia trachomatis manipulates the host cell cytoskeleton to promote entry, development and exit [4]. Shortly after attachment of the EB to the host cell surface, C. trachomatis delivers several effector proteins into the host cell cytoplasm via a type III secretion system (T3SS) [5]. The translocated actin-recruiting phosphoprotein (Tarp) is one of the early translocated effectors and is spatially and temporally associated with the recruitment of actin to the site of EB invasion [6]. Tarp is a bacterial actin nucleating and bundling protein which harbors one G-actin binding domain (implicated in actin nucleation) as well as two F-actin binding domains (implicated in actin bundling) [7, 8].
The arrangement of actin filaments during entry of the EBs into the host cell is not known. One of the well characterized actin bundling proteins, fascin 1, co-localizes with filopodia on the leading edge of the growth cones of developing nerve cells and are implicated in the formation of actin bundles [9]. Likewise, Tarp may play a role in the creation of actin bundles located directly beneath the host-pathogen contact site to form pedestal-like structures that are important for chlamydial entry into host cells [8, 10]. Herein, we examined the biophysical properties of Tarp-generated actin bundles in vitro and thus demonstrate that Tarp-mediated actin bundle assembly is independent of actin nucleation and the F-actin binding domains are sufficient to bundle actin filaments. Additionally, Tarp-mediated actin bundles have distinct bending stiffness compared to that of known actin bundling proteins. To our knowledge, this is the first characterization of actin bundle flexibility engendered from a bacterial effector protein. Our findings indicate that Tarp employs a novel actin bundling strategy which may facilitate chlamydial invasion of human cells.
Materials and methods
Cloning, protein expression and purification
In-frame amino-terminal glutathione S-transferase (GST) and carboxyl-terminal polyhistindine fusion Tarp proteins were generated as previously described[8]. Two additional truncated Tarp effectors including the C-terminal domain of Tarp harboring the F-actin binding domain (FAB domain) (D761-G1005) and the N-terminal and central domains of Tarp excluding all known actin binding sites (N-terminal domain)(M1-P747) were generated by PCR amplifying the corresponding coding regions from C. trachomatis serovar L2 LGV 434 genomic DNA (Qiagen genomic purification kit, Valencia CA). PCR was performed with custom synthesized oligonucleotide primers (Integrated DNA technologies, Coralville, IA) engineered with BamHI and XhoI linkers. PCR products were purified, digested with restriction enzymes (New England Biolabs, Beverly, MA) and cloned into linearized pGEX-6P-1 vector (GE Health Sciences, Piscataway, NY) to generate the translation fusions. All clones were confirmed by restriction digest and Sanger sequencing. All Tarp containing pGEX-6P-1 plasmids were transformed into the BL21 strain of Escherichia coli (Novagen, Madison, WI). Protein expression and purification were performed according to the procedures outlined for Ni sepharose 6 Fast Flow and glutathione sepharose 4B in the bulk GST purification module (GE Health sciences, Chicago, IL). The GST tag was removed with PreScission Protease treatment according to the manufacturer’s recommendations (GE Health Sciences, Chicago, IL).
Actin nucleation pyrene assay
Pyrene actin polymerization assays were performed as previously described [7, 8, 11].
F-actin binding and bundling assay
Actin monomers (21 μM) were first polymerized to form filamentous actin (F-actin) in the presence of polymerization buffer (10 mM imidazole, pH 7.0, 50 mM KCl, 2 mM MgCl2) for 1 h at 25°C. To induce bundles, F-actin was then incubated with 35 nM Tarp proteins for one more hour at 25°C and spun at 10,000 × g for 30 min at 25°C in a Beckman Optima TLX Ultracentrifuge using a TLA 100.3 rotor (Beckman Coulter Inc., Fullerton, CA). The low-speed centrifugation was used to separate bundles from F- or G-actin in samples. α-actinin (16 μM, Cytoskeleton, Denver, CO) was used as a positive control. For the one step polymerization/bundling assay, 500 nM of G-actin was incubated with increasing concentrations of wild type Tarp (0 nM to 35 nM) for 2 h in presence of polymerization buffer at 25°C and spun at 10,000 × g for 30 min at 25°C. Equal volumes of supernatant and pellet were separated by 10% SDS-PAGE, stained with Coomassie blue for 1 h and destained overnight. Gels were analyzed by densitometry on a ChemiDoc MP Imaging System (Bio-Rad, Hercules, CA).
TIRF microscopy imaging and bending persistence length analysis
Rhodamine-labeled G-actin (Cytoskeleton, Denver, CO) was polymerized in polymerization buffer (10 mM Imidazole, pH 7.0, 50 mM KCl, 2 mM MgCl2, 1 mM ATP and 1 mM DTT) at the concentration of 8.3 μM for 1 h at room temperature (~22 °C) to form F-actin. Then the F-actin was incubated with Tarp, α-actinin (Cytoskeleton, Denver, CO) or fascin (Abcam, Cambridge, MA) at various molar ratio for 1 h at room temperature. Tarp-induced actin bundles were diluted with imaging buffer (10 mM imidazole, pH 7.0, 50 mM KCl, 2 mM MgCl2, 1 mM ATP, 1 mM DTT, 0.15 M glucose, 1 mg/ml catalase, 0.2 mg/ml glucose oxidase). Bundles were immobilized on poly-L-lysine (Sigma Aldrich, St. Louis, MO) coated microscope coverslips that were thoroughly cleaned with absolute ethanol and KOH followed by rinsing with ddH2O. Bundle images were taken using a Nikon Eclipse Ti TIRF microscope equipped with a Hamamatsu Image EM X2 CCD Camera, a 100X oil immersion objective (numerical aperture, 1.49), and Nikon LU-N4 Laser. At least 100 filaments and/or bundles were analyzed for each sample. Actin filament and/or bundle length and bending persistence length (Lp) were calculated from the two-dimensional average cosine correlation (<C(s)>) of the tangent angle (θ) along the segment lengths (s) of a filament and/or bundle by fitting to the following equation (where A is a scaling factor) as described [12] using ImageJ (NIH) and Persistence software:
| (1) |
Statistical analysis
Statistical analyses were performed using Graphpad Prism (version 7.04, Graphpad software, CA). Unless otherwise stated, data are given as mean ± 95% confidence intervals. Comparison between two groups were performed using Mann-Whitney t-test in case of non-parametric data and for multiple groups one-way analysis of variance (ANOVA) and Tukey’s post hoc comparisons were used.
Results
The Tarp FAB domain is sufficient for actin bundle formation
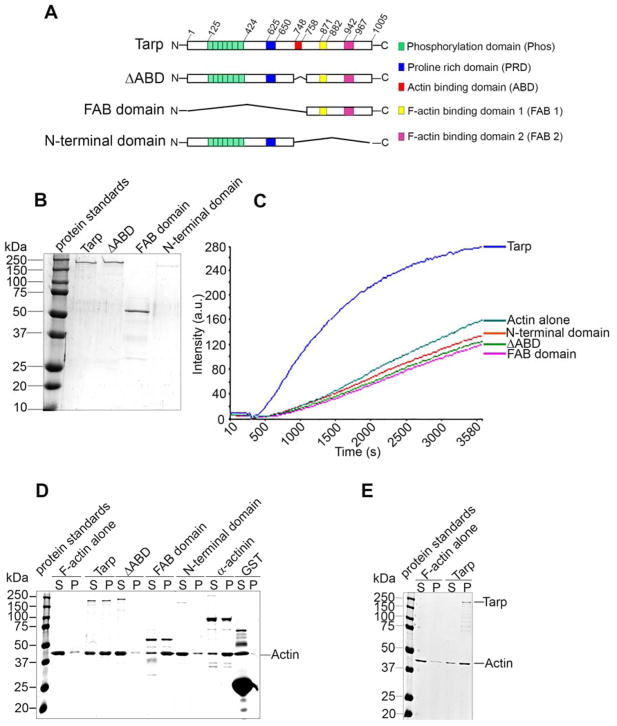
C. trachomatis Tarp containing one G-actin binding domain (ABD) and two filamentous actin binding domains (FAB) binds directly to both globular actin and filamentous actin in vitro respectively[8]. The ABD and proline rich domain (PRD) are required for Tarp-mediated actin nucleation [7]. However, the cohort of protein domains required for Tarp-mediated actin bundling has not been thoroughly examined. In order to determine which region(s) of Tarp is sufficient for bundling actin filaments we generated recombinant wild type Tarp and mutant Tarp proteins that harbor specific domain deletions (Figure 1A). Specifically, amino- and carboxyl- domain deletions were generated to create truncated Tarp proteins referred to as the FAB domain (deletion of amino acids M1 through P747) and the N-terminal domain (deletion of amino acids A748-G1005), respectively. A Tarp effector lacking the solitary G-actin binding domain (ΔABD) was also created (deletion of A748-K758) (Figure 1A). The purified proteins (Figure 1B) were analyzed for actin nucleation activity in pyrene actin polymerization assays (Figure 1C). In agreement with previous reports[8], all mutant Tarp proteins lacking the G-actin binding domain (ΔABD, FAB domain and N-terminal domain) failed to promote actin polymerization. In contrast, wild type Tarp demonstrated robust actin polymerization. These data reaffirm the requirement of the ABD for Tarp-mediated actin nucleation.
Figure 1.
The Tarp FAB domain is sufficient to bundle actin. (A) Schematics of Tarp proteins utilized in this study indicating the locations of the tyrosine rich repeat phosphorylation domain (green boxes), the proline rich domain (blue box), G-actin binding domain (red box), and F-actin binding domains 1 (yellow box) and 2 (pink box). The numbers indicate amino acid positions encoded within the C. trachomatis tarP gene. The “∧” indicates amino acids deleted from the wild type sequence. Mutant Tarp clones included Tarp lacking the G-actin binding domain (ΔABD) as well as Tarp fragments representing only the F-actin binding domains (FAB domains) or a Tarp truncation excluding all known actin (both G- and F-) binding domains (N-terminal domain). (B) Purified Tarp and Tarp mutants depicted in (A). (C) Tarp proteins described in panels A and B were assessed in pyrene actin polymerization assays. Tarp proteins were incubated with monomeric pyrene-labeled actin. An increase in actin polymerization after the addition of polymerization buffer at 300 s was measured as arbitrary fluorescence intensity [Intensity (a.u.)] over time [Time (s)]. The data are representative of three repeated experiments. (D) Purified recombinant Tarp proteins were incubated with preformed filamentous actin (F-actin) and isolated by low-speed centrifugation. Protein supernatants (S) and pellets (P) were resolved by SDS-PAGE and visualized by Coomassie blue staining. α-actinin and GST served as positive and negative controls, respectively. (E) Globular actin (G-actin) along with polymerization buffer was incubated with (Tarp) or without (F-actin alone) purified recombinant Tarp and isolated by low-speed centrifugation. Protein supernatants (S) and pellets (P) were resolved by SDS-PAGE and visualized by Coomassie blue staining.
In addition to actin nucleation, we demonstrated previously that Tarp harbors actin bundling activity[8]. In order to determine whether the Tarp FAB domains alone are sufficient to mediate this activity we examined the wild type and mutant proteins for the ability to assemble preformed actin filaments (Figure 1D). Actin bundles sediment at a higher rate compared to actin filaments and monomeric actin [13]. Therefore, proteins capable of bundling actin filaments will be present in the pellet upon low speed centrifugation. Interestingly, Tarp and FAB domain were detected in the pellet fraction with actin bundles, similar to the positive control protein α-actinin. Whereas, the ΔABD and N-terminal domain were detected exclusively in the supernatant fraction along with the negative control glutathione S-transferase (Figure 1D). These data demonstrate that the FAB domain alone is sufficient to bundle pre-formed actin filaments. However, when the FAB domain is associated with the N-terminal domain (but still lacking the G-actin binding domain in the ΔABD) bundle formation did not occur, probably suggesting ABD has an important role in maintaining functionality and structural conformation of Tarp.
Since, Tarp has the capacity to associate with G-actin to promote the development of actin filaments, we examined the cumulative effects of both Tarp-mediated actin nucleation and bundling in a one-step incubation reaction containing G-actin, Tarp and actin polymerization buffer. Subsequent low speed actin co-sedimentation revealed Tarp exclusively in the actin bundle pellet (Figure 1E).
Tarp and FAB domain-mediated actin structures are consistent with actin bundles
To gain more insight into the biophysical characteristics of actin bundles formed by wild type Tarp and the FAB domain mutant protein, we directly visualized the rhodamine-conjugated actin filaments and bundles with a Total Internal Reflection Fluorescence (TIRF) microscope (Figure 2). The N-terminal domain protein served as the experimental negative control and F-actin alone as the assay control. The actin cables detected in the images were quantitatively analyzed for cross-sectional fluorescence integrated density, an increase in which is known to correlate with actin bundle formation [14]. Representative TIRF images of the actin structures generated in the presence of the wild type and mutant Tarp proteins, as well as those for F-actin alone are shown in Figure 2A. The actin bundles formed in the presence of Tarp and FAB domain demonstrated significantly increased mean fluorescence integrated densities relative to F-actin filaments alone, whereas the N-terminal domain did not (Figure 2B). These data are consistent with the co-sedimentation assay that Tarp and FAB domain can assemble actin filaments to generate bundles. Therefore, our TIRF image analysis provide further support for the finding that, in vitro, the FAB domain of the Tarp protein is sufficient to form bundles from actin filaments independently without the involvement of additional host cell actin binding proteins.
Figure 2.
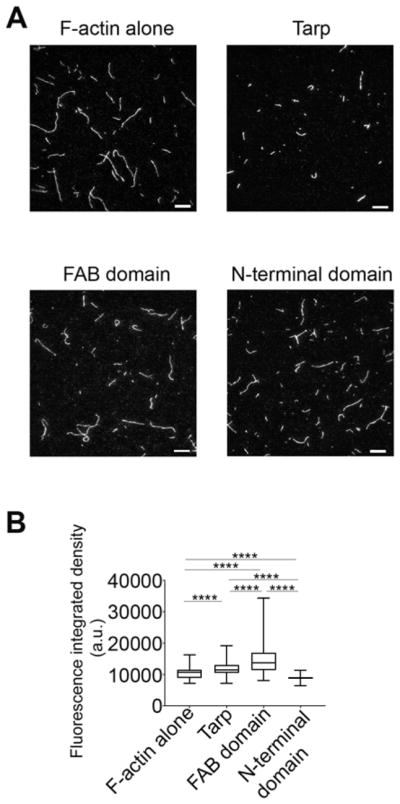
Tarp produces actin structures which have higher fluorescence intensities compared to actin filaments alone as determined by TIRF microscopy image analysis. (A) Representative TIRF images of rhodamine-labeled actin filaments or bundles assembled with Tarp or Tarp mutant proteins (Scale bar 10 μm). (B) Cumulative fluorescence integrated density of actin structures in the presence or absence of Tarp or mutant Tarp proteins plotted in a Box and Whiskers graph using GraphPad prism version 7.04 where the ends of the whiskers represent the minimum and maximum of the cumulative data. One-way ANOVA with Tukey’s multiple comparison test was used. **** represents p<0.0001.
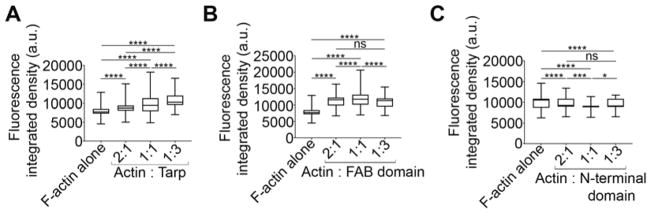
Increased molar concentrations of Tarp generate actin bundles with increased fluorescence intensity
In order to test whether actin bundle assembly was influenced by changes in Tarp protein concentration we examined TIRF images captured using increasing molar ratios of actin:Tarp (2:1, 1:1 and 1:3). (Figure 3). The fluorescent integrated density of the actin structures formed in the presence of wild type Tarp were found to increase significantly with increased Tarp concentration (Figure 3A). Interestingly, the FAB domain generated intense actin bundles in a 1:1 (actin to FAB domain) molar ratio (mean±SD being 11856±1858) compared to 2:1 or 1:3 (mean±SD being 11235±1588 and 10942±1540 respectively, Figure 3B). This increase in fluorescence intensity may indicate an increase in bundle thickness, length, or population, or all of these. Consistently, the N-terminal fragment which served as a negative control failed to make actin bundles even as higher concentrations of the mutant effector were tested (Figure 3C).
Figure 3.
Fluorescence integrated densities of the actin bundles increase in a Tarp concentration dependent manner. Rhodamine labeled F-actin (500 nM) was assembled with 250 nM, 500 nM and 1.5 μM of Tarp (A) or FAB domain (B) or N-terminal domain (C) to get 2:1, 1:1 and 1:3 molar ratio respectively. The fluorescence integrated densities were calculated by NIH ImageJ version 1.48 from the TIRF images. The Box and Whiskers graphs were generated using GraphPad prism version 7.04 where ends of the whiskers represent the minimum and maximum of the cumulative data. One-way ANOVA with Tukey’s multiple comparison test was used for this study where * = p<0.05, *** = p<0.001, **** = p<0.0001 and ns = not significant.
One-way ANOVA with Tukey’s multiple comparison test was used for this study. **** represents p<0.0001 and ‘ns’ is not significant.
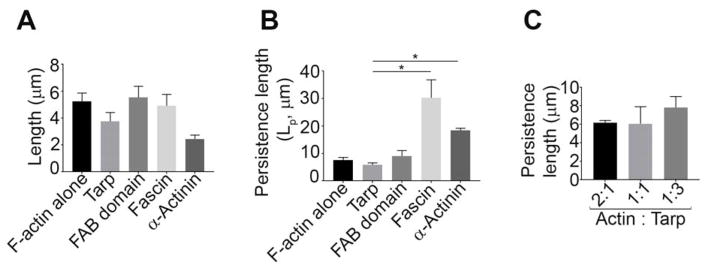
Tarp-mediated actin bundles have unique flexibility
According to the TIRF image analyses, the average length of the actin bundles formed in the presence of Tarp (3.75±1.60 μm) or α-actinin (2.44±0.50 μm), although not statistically significant, appeared to be shorter compared to F-actin alone (5.25±1.35 μm), as well as the actin bundles formed in the presence of the FAB domain (5.52±2.05 μm) and fascin (4.91±1.47 μm) (Figure 4A). To characterize the bending flexibility of the Tarp-mediated actin bundles relative to F-actin filaments and bundles generated by known bundling proteins, the bending persistence length (Lp) at constant temperature from two-dimensional angular correlation of the tangent angles were determined as shown in Supplemental Figure 1 [12]. The bending persistence length provides a useful quantitative measure of a polymer’s bending rigidity. Persistence length of a freely suspended thermally fluctuating polymer is the distance along the polymer over which the ‘memory’ of the tangent angle at a given point is maintained [15]. Our analysis demonstrated that both Tarp (Lp of 5.8±2.1 μm) and FAB domain (Lp of 9.0±5.0 μm) induced bundles were more flexible than those formed by fascin (Lp of 30.3±11.2 μm) and α-actinin (Lp of 18.4±1.4 μm) as indicated by persistence length values; though only the average persistent length of Tarp-mediated bundles was statistically significant relative to that of fascin and α-actinin (Figure 4B). Consistent with the increase in fluorescence intensity of actin bundles generated by increased molar concentrations of Tarp, an increasing trend in persistence length was also found to occur with increased molar concentrations of Tarp. (Figure 4C).
Figure 4.
Tarp generates flexible actin bundles. (A) The average length of the actin structures were measured from the TIRF microscopy images where 500 nM of rhodamine labelled F-actin was incubated with or without 500 nM of Tarp or the mutant Tarp proteins. (B) Persistence lengths (Lp) of actin filaments alone, and Tarp-, FAB domain-, α-actinin- or fascin-mediated actin bundles were analyzed ([actin] = 500 nM, [Tarp, FAB or fascin] = 500 nM, for α-actinin, [actin] = 5 μM and [α-actinin] = 1 μM). The data represent the average of three experiments. (C) Persistence length was measured for the actin bundles generated by co-incubation of F-actin and Tarp in three different molar ratios (2:1, 1:1 and 1:3) ([actin] = 500nM). Mann-Whitney test was performed between 2 non-parametric groups. The data plotted as mean ± SD. * = p<0.05.
Discussion
Many intracellular bacterial pathogens modulate actin dynamics of eukaryotic host cells to promote internalization, intracellular movement and cell-to-cell spread [16]. The Chlamydia trachomatis Tarp effector is known to be a candidate virulence factor, which manipulates actin dynamics of the host cell by nucleating G-actin directly [7] and signaling via SH2 domain containing host cell proteins that promote the activation of other host cell actin nucleators such as the Arp2/3 complex [17, 18]. Here, we have demonstrated that Tarp can generate actin bundles that are more flexible than those formed by previously well-characterized eukaryotic actin bundling proteins [15, 19]. Our previous studies revealed that Tarp harbors two distinct F-actin binding/bundling domains (FAB1 and 2)[8]. We now provide evidence that the C-terminal domain of Tarp containing only FAB1 and 2 is sufficient to bind to filamentous actin and generate bundles in the absence of both the actin nucleation and N-terminal phosphorylation domains (Figures 1 and 2). Interestingly, ΔABD (missing the G-actin binding/nucleation domain), was unable to bundle actin filaments compared to wild type Tarp even though ΔABD contains the FAB domains. It is possible that the ΔABD protein may not access actin filaments in the same way as the FAB domain alone because of steric interference from the amino terminal half of the protein. Additionally, a recent NMR study [20] suggests that the intrinsically disordered Tarp protein is stabilized following actin binding to the alpha helix which is missing in the ΔABD protein and this may contribute to the observed differential binding to preformed actin filaments.
Chlamydia trachomatis Tarp has the ability to bind G-actin as well as F-actin [8]. For the first time, herein, we demonstrate that Tarp can generate bundles from pre-formed actin filaments as well as from Tarp-derived actin monomers in a single process (Figure 1D and E), suggesting that actin nucleation by Tarp not only accelerates polymerization but also bundling kinetics.
Actin bundles predominantly play an important role in the formation and function of various protrusive and contractile cellular structures such as filopodia, microvilli, stereocillia, nerve growth cones and stress fibers [21]. The assembly of actin filaments into bundles is tightly regulated by a plethora of actin-binding proteins at spatial and temporal levels [22]. Actin bundling proteins facilitate the formation of bundles with fundamentally different mechanical properties and vastly different architectures [19, 23]. To gain insight into how cytoskeletal filaments and higher-order structures such as bundles provide mechanical responses in the presence of Tarp, we quantified the bending persistence length of Tarp-derived actin bundles. Interestingly, the actin bundles generated by Tarp were found to be more flexible as compared to bundles generated by eukaryotic actin bundling proteins such as α-actinin, fascin (Figure 4), scruin and vinculin tail domain [19, 24, 25]. Winkelman et al., (2016) demonstrated that interfilament spacing of a bundle can determine the number of filaments to be assembled in a bundle and also the flexibility of the bundle. Interestingly, the interfilament distance correlates with the size of the bundling proteins in question. Therefore, the formation of flexible bundles by Tarp could be attributed to the fact that Tarp forms large oligomers with itself generating more space between actin filaments and ultimately creating actin bundles that are more compliant than fascin or α-actinin [26].
Transverse arcs, one type of contractile fibers, are relatively flexible bundles of unbranched actin filaments of mixed polarity [27]. These arcs provide a starting block to push off from, for subsequent rounds of leading-edge protrusion. In contrast, sometimes the arcs accelerate actin disassembly via their contractility. One possible reason for the generation of the flexible bundles by Tarp could be the generation of transverse arcs and subsequent rapid dismantling of the actin structures associated with chlamydial entry.
Though further studies are needed to confirm the importance of the unique bundles generated by Tarp, we propose that the unique Tarp-mediated actin bundles contribute to the mechanisms of Chlamydia trachomatis entry of human host cells.
Supplementary Material
The average cosine correlation functions are plotted against segment length. Solid lines indicate best fits of data in accordance to Eq. (1), yielding bending persistence lengths of each filament/bundle.
The F-actin binding domain of Tarp is sufficient to bundle filamentous actin.
Actin bundles formed by Tarp are more flexible than that of α-actinin and fascin.
Acknowledgments
The authors thank members of the Mollie W. Jewett Laboratory for helpful discussions as well as acknowledge the technical assistance of Thomas Laux. This work is supported by the NIAID, NIH R21AI117013 awarded to T.J.J.
Footnotes
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Torrone E, Papp J, Weinstock H C. Centers for Disease, Prevention. Prevalence of Chlamydia trachomatis genital infection among persons aged 14–39 years--United States, 2007–2012. MMWR Morb Mortal Wkly Rep. 2014;63:834–838. [PMC free article] [PubMed] [Google Scholar]
- 2.Moulder JW. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Omsland A, Sager J, Nair V, Sturdevant DE, Hackstadt T. Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. Proc Natl Acad Sci U S A. 2012;109:19781–19785. doi: 10.1073/pnas.1212831109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wesolowski J, Paumet F. Taking control: reorganization of the host cytoskeleton by Chlamydia. F1000Res. 2017;6:2058. doi: 10.12688/f1000research.12316.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller KE, Plano GV, Fields KA. New frontiers in type III secretion biology: the Chlamydia perspective. Infect Immun. 2014;82:2–9. doi: 10.1128/IAI.00917-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci U S A. 2004;101:10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jewett TJ, Fischer ER, Mead DJ, Hackstadt T. Chlamydial TARP is a bacterial nucleator of actin. Proc Natl Acad Sci U S A. 2006;103:15599–15604. doi: 10.1073/pnas.0603044103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiwani S, Alvarado S, Ohr RJ, Romero A, Nguyen B, Jewett TJ. Chlamydia trachomatis Tarp harbors distinct G and F actin binding domains that bundle actin filaments. J Bacteriol. 2013;195:708–716. doi: 10.1128/JB.01768-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kliewe F, Scharf C, Rogge H, Darm K, Lindenmeyer MT, Amann K, Cohen CD, Endlich K, Endlich N. Studying the role of fascin-1 in mechanically stressed podocytes. Sci Rep. 2017;7:9916. doi: 10.1038/s41598-017-10116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carabeo RA, Grieshaber SS, Fischer E, Hackstadt T. Chlamydia trachomatis induces remodeling of the actin cytoskeleton during attachment and entry into HeLa cells. Infect Immun. 2002;70:3793–3803. doi: 10.1128/IAI.70.7.3793-3803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jewett TJ, Miller NJ, Dooley CA, Hackstadt T. The conserved Tarp actin binding domain is important for chlamydial invasion. PLoS Pathog. 2010;6:e1000997. doi: 10.1371/journal.ppat.1000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham JS, McCullough BR, Kang H, Elam WA, Cao W, De La Cruz EM. Multi-platform compatible software for analysis of polymer bending mechanics. PLoS One. 2014;9:e94766. doi: 10.1371/journal.pone.0094766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srivastava J, Barber D. Actin co-sedimentation assay; for the analysis of protein binding to F-actin. J Vis Exp. 2008 doi: 10.3791/690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann C, Moes D, Dieterle M, Neumann K, Moreau F, Tavares Furtado A, Dumas D, Steinmetz A, Thomas C. Live cell imaging reveals actin-cytoskeleton-induced self-association of the actin-bundling protein WLIM1. J Cell Sci. 2014;127:583–598. doi: 10.1242/jcs.134536. [DOI] [PubMed] [Google Scholar]
- 15.Takatsuki H, Bengtsson E, Mansson A. Persistence length of fascin-cross-linked actin filament bundles in solution and the in vitro motility assay. Biochim Biophys Acta. 2014;1840:1933–1942. doi: 10.1016/j.bbagen.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 16.Gouin E, Welch MD, Cossart P. Actin-based motility of intracellular pathogens. Curr Opin Microbiol. 2005;8:35–45. doi: 10.1016/j.mib.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 17.Lane BJ, Mutchler C, Al Khodor S, Grieshaber SS, Carabeo RA. Chlamydial entry involves TARP binding of guanine nucleotide exchange factors. PLoS Pathog. 2008;4:e1000014. doi: 10.1371/journal.ppat.1000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehlitz A, Banhart S, Maurer AP, Kaushansky A, Gordus AG, Zielecki J, Macbeath G, Meyer TF. Tarp regulates early Chlamydia-induced host cell survival through interactions with the human adaptor protein SHC1. J Cell Biol. 2010;190:143–157. doi: 10.1083/jcb.200909095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Claessens MM, Bathe M, Frey E, Bausch AR. Actin-binding proteins sensitively mediate F-actin bundle stiffness. Nat Mater. 2006;5:748–753. doi: 10.1038/nmat1718. [DOI] [PubMed] [Google Scholar]
- 20.Tolchard J, Walpole SJ, Miles AJ, Maytum R, Eaglen LA, Hackstadt T, Wallace BA, Blumenschein TMA. The intrinsically disordered Tarp protein from chlamydia binds actin with a partially preformed helix. Sci Rep. 2018;8:1960. doi: 10.1038/s41598-018-20290-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bathe M, Heussinger C, Claessens MM, Bausch AR, Frey E. Cytoskeletal bundle mechanics. Biophys J. 2008;94:2955–2964. doi: 10.1529/biophysj.107.119743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol Rev. 2003;83:433–473. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- 23.De La Cruz EM, Gardel ML. Actin Mechanics and Fragmentation. J Biol Chem. 2015;290:17137–17144. doi: 10.1074/jbc.R115.636472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin JH, Mahadevan L, So PT, Matsudaira P. Bending stiffness of a crystalline actin bundle. J Mol Biol. 2004;337:255–261. doi: 10.1016/j.jmb.2004.01.028. [DOI] [PubMed] [Google Scholar]
- 25.Oztug Durer ZA, McGillivary RM, Kang H, Elam WA, Vizcarra CL, Hanein D, De La Cruz EM, Reisler E, Quinlan ME. Metavinculin Tunes the Flexibility and the Architecture of Vinculin-Induced Bundles of Actin Filaments. J Mol Biol. 2015;427:2782–2798. doi: 10.1016/j.jmb.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasza KE, Broedersz CP, Koenderink GH, Lin YC, Messner W, Millman EA, Nakamura F, Stossel TP, Mackintosh FC, Weitz DA. Actin filament length tunes elasticity of flexibly cross-linked actin networks. Biophys J. 2010;99:1091–1100. doi: 10.1016/j.bpj.2010.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The average cosine correlation functions are plotted against segment length. Solid lines indicate best fits of data in accordance to Eq. (1), yielding bending persistence lengths of each filament/bundle.