Abstract
Transcription factor (TF) networks are a key determinant of cell fate decisions in mammalian development and adult tissue homeostasis, and are frequently corrupted in disease. However, our inability to experimentally resolve and interrogate the complexity of mammalian TF networks has hampered the progress in this field. Recent technological advances, in particular large-scale genome-wide approaches, single-cell methodologies, live-cell imaging, and genome editing, are emerging as important technologies in TF network biology. Several recent studies even suggest a need to re-evaluate established models of mammalian TF networks. Here, we provide a brief overview of current and emerging methods to define mammalian TF networks. We also discuss how these emerging technologies facilitate new ways to interrogate complex TF networks, consider the current open questions in the field, and comment on potential future directions and biomedical applications.
Introduction
During mammalian development, hundreds of unique cell types are specified in a complex spatio-temporal patterning process. In adults, stem and progenitor cell populations replenish mature cell types to maintain tissue homeostasis throughout life. Concerted gene expression programs are responsible for these fundamental biological processes and the underlying cell fate decisions. Transcription represents a major control point in gene expression (Figure 1A) and occurs within the context of chromatin. Precise spatial and temporal expression of combinations of a limited number of genes (~20,000 in humans) appears to be responsible for the intricate cellular processes of developmental specification and adult tissue homeostasis.
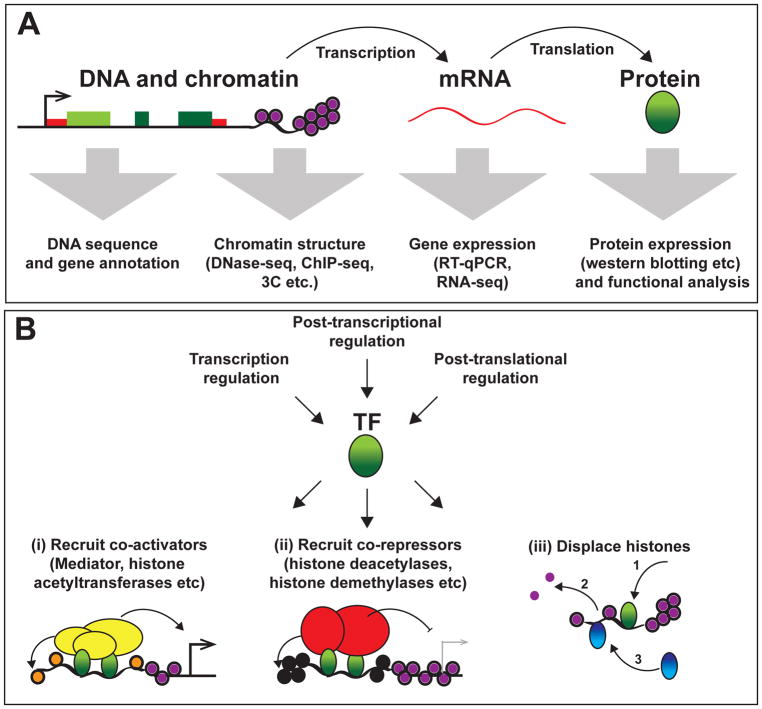
Figure 1. Central dogma of molecular biology and functions of transcription factors.
(A) Gene expression is the process of gene transcription into messenger (m)RNA followed by translation into protein. Genes are encoded within genomic DNA and packaged within the nucleus as chromatin. Genomic sequencing has allowed protein-coding genes to be identified and annotated. A range of techniques have been developed to investigate chromatin structure, including DNase I hypersensitivity assays (such as DNase-seq), chromatin immunoprecipitation (such as ChIP-seq for histone modifications and TF enrichment) and chromatin conformation capture (3C) methods. Gene products can be measured at both RNA and protein levels by a range of techniques.
(B) Regulation of TF expression, activity and function. TFs are regulated at transcriptional, post-transcriptional and post-translational levels. TFs (green) can function by multiple mechanisms including: (i) recruitment of co-activators (yellow) that may add activating histone modifications (H3K4me or H3K27Ac; denoted as orange histones) or recruit RNA pol II to promote gene transcription; (ii) recruitment of co-repressors (red) that apply repressive histone modifications (such as H3K29me; denoted by black histones) to promote histone compaction and gene silencing; or (iii) DNA binding that results in histone displacement, which allows other TFs (blue) to bind;. TFs usually bind cooperatively and regulation of TF expression levels (and post-translational modifications) may influence TF function and activities.
Sequence-specific transcription factors (TFs) are a large class of DNA binding protein that play central roles in regulating gene transcription, and account for almost 7% of genes (~1,400) in the human genome (Vaquerizas et al., 2009). TFs regulate gene promoter activity, but often act via interactions with other genomic locations that can be distant in primary DNA sequence. These are broadly defined as gene regulatory regions (Kellis et al., 2014), with an important subclass of positive regulatory regions being termed enhancers. Enhancers are composed of TF binding sites (TFBSs) or DNA motifs, which are are commonly short (4-12 nucleotides) (Jolma et al., 2013). Such motifs therefore frequently occur by chance in mammalian genomes and individual TF-DNA interactions can be weak. TF-DNA interactions must compete with histone-DNA interactions for stable and productive binding. Cooperativity in TF binding is therefore common, such as through protein-protein interactions with other TFs, co-activators, and/or co-repressors (Vaquerizas et al., 2009).
TFs can be thought of as “readers” of enhancers, with the combination (and spacing) of encoded TFBSs defining combinatorial binding capacity and stability. TF binding may directly activate or repress an enhancer and/or gene promoter, through recruitment of co-activators or co-repressors, or may act indirectly to influence gene expression such as through histone displacement (Figure 1B). The multi-protein complex Mediator is an important enhancer co-activator, which is thought to coordinate enhancer-promoter interactions and stimulate transcription (Malik and Roeder, 2010). TFs may also recruit other co-activators, such as histone methyltransferases, histone acetyltransferases, and chromatin-modifying complexes (Kouzarides, 2007). By contrast, enhancers and genes become repressed through TF recruitment of co-repressors such as histone demethylases (Whyte et al., 2012), histone deacetylases (HDACS), and polycomb complexes (Reynolds et al., 2013).
TFs have the ability to directly regulate their own expression through binding to enhancer(s) that control their own gene transcription. This can be thought of as a simple molecular circuit, a feedback loop. By understanding the concept that a TF can regulate its own expression, and expression of other TFs, it is possible to envisage the resulting TF circuits and networks that may be active within mammalian cells (Davidson, 2010). TF proteins, their genes and enhancers can be considered as the building blocks or constituents of a complex TF network (Alon, 2007). However, such a TF network is commonly not active in its entirety, but instead exists in various network states, comprising of active TF sub-networks. Of course, TFs not only regulate the transcription of TF genes, but also of genes involved in cellular structure/function. Therefore, the TF network state determines the global transcriptional program and the cell-type specific gene expression patterns that define cell identity and function.
Over the last thirty years, numerous experimental approaches have been used to define mammalian TF networks (Box 1). However, the inherent biological complexity (Box 2) has hampered these efforts. Recent technological innovations are now helping to define TF networks at unprecedented detail and accurately interrogate network logic and function, particularly in stem cell and cancer systems (Box 3). Several recent reports are even questioning established TF network models and suggest that some current paradigms may need updating. This Perspective aims to provide non-expert biologists with an overview of exciting recent developments in mammalian TF network biology, alongside a discussion of the open questions, the field’s future directions, and its potential applications in biomedicine.
Box 1. Defining TF networks in mammals.
To define mammalian TF networks, we must be able to identify its constituents and understand its underlying network logic. Numerous technological advances have helped us to define enhancer location and gene targets, measure enhancer activity, determine TF activity and function, and develop TF network models to predict logic. These are briefly summarized below, but further details can be found elsewhere (Brent, 2016, Blais and Dynlacht, 2005).
Defining enhancer location and gene targets
Putative enhancers were initially identified from conservation in non-coding elements within the genome, although we now know that sequence conservation is often a poor method to identify TFBSs (Villar et al., 2015). Numerous approaches have since been developed to identify putative enhancers, including DNase I hypersensitivity site (DHS) assays and chromatin immunoprecipitation (ChIP) (Noonan and McCallion, 2010), and more recently ATAC-seq (Buenrostro et al., 2013). Additionally, chromatin conformation capture (3C) methods (Dostie et al., 2006) and particularly the genome-wide adaptation of this method (Hi-C) are helping to better define enhancer-promoter interactions as well as spatial chromatin structure information including topologically-associated domains (Pombo and Dillon, 2015). Certain histone modifications, particularly histone 3 lysine 4 mono-methylation (H3K4me1) (Heintzman et al., 2007) and H3K27 acetylation, are commonly used to identify putative enhancers (Creyghton et al., 2010, Rada-Iglesias et al., 2011). Large (~10 kb) regions of H3K27Ac enrichment within the genome have recently been defined as “super-enhancers”, which appear to often contain numerous TFBSs (and perhaps multiple individual enhancers) (Hnisz et al., 2013, Whyte et al., 2013). However, the functional relevance of this classification is still unclear (Hay et al., 2016). While histone modification, TF enrichment, or open chromatin are often fairly good indicators of active enhancers, we do not yet have a universal active enhancer “mark” (Dogan et al., 2015).
Measuring enhancer activity
Enhancers are classically defined as functional DNA sequences with the ability to activate (enhance) the rate of transcription from a heterologous promoter, independent of location and orientation (Maniatis et al., 1987, Kim and Shiekhattar, 2015). This is the basis of the widely used enhancer assay: a putative enhancer is cloned up or downstream of a minimal promoter that drives expression of a reporter gene (Noonan and McCallion, 2010) (Figure 2A). However, recent technological advances are now allowing endogenous enhancers to be functionally interrogated (see the Emerging Technologies section). Synthetic TFs such as Zinc Fingers, Transcription Activator-Like Effectors (TALEs), and Cas9, can also be used to perturb endogenous enhancer activity and study TF networks. This is achieved by fusing genome-specific synthetic DNA binding domains to transcriptional effector domains such as the VP64 (activator) and KRAB (repressor) domains (Gao et al., 2013, Wilkinson et al., 2014). These approaches provide an opportunity to inducibly activate or silence enhancers, or synthetically engineer more complex transcriptional circuitry.
Determining TF function
The genome-editing revolution associated with CRISPR/Cas9 technologies (Cho et al., 2013, Cong et al., 2013, Mali et al., 2013) now allows TF genes, as well as enhancer regions, to be deleted and mutated easily. Importantly, these methods also allow for the first time, large-scale analysis within human cells, where efficiencies of homologous recombination are normally too low for traditional targeted genetic manipulation (see Emerging Technologies section).
Defining TFBSs
TF DNA-binding specificity can be determined by in vitro assays such as electrophoretic mobility shift assays (EMSAs) or systematic evolution of ligands by exponential enrichment (SELEX) (Figure 2B). DNase-seq is also being used to describe the binding patterns of TF-DNA interactions through deep sequencing, which allows TF binding motifs or “footprints” within DHSs to be resolved (Hesselberth et al., 2009). Although such DNase footprinting only generates candidate assignments for a class of TF (not a specific TF), this methodology has provided fundamental insights into TF network topology, and its conservation within mammals (Stergachis et al., 2014, Boyle et al., 2014).
Developing TF network models
By combining our knowledge of TF binding events within TF gene loci with enhancer assays and functional analysis, we can build models of TF networks to predict biological behavior in silico (Figure 2C). To date, most network models are relatively simple, often consisting of simple diagrams of TF sub-networks annotated with nodes and edges. However, Boolean (Xu et al., 2014, Dunn et al., 2014, Moignard et al., 2015) and Bayesian (Dowell et al., 2014) modeling approaches have been built from such information to provide dynamic and executable network models. PetriNets, a mathematical modeling approach to graphically model networks, have also been successfully used to computationally encode TF networks (Bonzanni et al., 2013). Alternative methods have been used to “reverse engineer” networks from gene expression data, such as by using mutual information or partial correlation analysis including the ARACNE algorithm (Margolin et al., 2006), and have been recently applied to single cell gene expression data (Wilkinson et al., 2014, Moignard et al., 2015). We refer readers to recent reviews of TF network modeling for further information (Woodhouse et al., 2016, Le Novère, 2015).
Box 2. Emerging concepts in the regulation of TF networks.
TF network states are not static but dynamic and often unstable. It is well understood that phenotypically different cells will contain different TF network states. However, phenotypically identical cells also have considerable functional heterogeneity, even with highly purified cell populations (Yamamoto et al., 2013), and differ in their response to extracellular signals (Satija and Shalek, 2014). Such functional heterogeneity highlights that TF network states can also differ between individual phenotypically similar (identical) cells. Extracellular signaling plays a major role in influencing the TF network state in mammalian cells, although numerous intrinsic mechanisms also influence TF network state stability and transitions between them, including intrinsic dynamics within the TF network (and its constituents), indirect TF interactions, cell cycle progression, and metabolic state (Figure 3). It is worth highlighting that different TF sub-networks (regulating particular biological functions such as cell cycle or response to stress) may be overlapping or independent of each other. Recent examples of these mechanisms of TF network regulation are briefly summarized below, and can be found in more detail elsewhere (Long et al., 2016, Davidson, 2010).
Extracellular signaling
All multicellular organisms require intercellular signaling pathways to allow the coordinated formation and maintenance of complex tissues. Signaling pathways often have multiple functions at different stages of development and in different cell types (Massagué, 2012). This can at least partially be explained by the transcriptional response to a stimulus being dependent on the TF network state (Trompouki et al., 2011, Mullen et al., 2011). Many signaling pathway effectors are themselves TFs, and therefore directly integrate with the TF network. The downstream transcriptional targets of these signaling effectors also often include TF genes (Kageyama et al., 2007), which go on to influence future TF network states.
TF network states are dynamic and heterogeneous
Positive feedback loops help to reinforce TF expression, and can thereby stabilize a TF network state. By contrast, TF antagonism causes inherent instability and appears to play important roles in cell fate decisions. One of the best-described examples is the antagonism between the TFs Gata1 and PU.1 in erythroid-myeloid lineage specification during hematopoietic differentiation (Burda et al., 2010) (although this TF antagonism has recently been questioned – see the Emerging Technologies section). Of course, TF antagonism can occur by various mechanisms, for example, competition for binding to the same TFBS (Bresnick et al., 2010).
TF stability and dose-dependence
The stability of the TF network as a whole depends on the stability of its constituents, particularly protein stability, rates of transcription and mRNA stability. While the DNA encoding TF genes and enhancers is permanent (although copy number can change, as in polyploidy and aneuploidy), relative accessibility of these genomic regions can be modulated by epigenetic modification. TF protein concentration appears to be particularly important for their function, with several TFs having dosage-dependent functions in development and homeostasis (Sigvardsson, 2012).
Indirect interactions and regulation of TF activity
Considering only TFs, enhancers and gene loci as the TF regulatory networks oversimplifies the biological complexity. Numerous levels of regulation overlay each other to “fine tune” gene expression. For example, numerous microRNAs post-transcriptionally regulate TF genes and mediate indirect TF network interactions (Martinez and Walhout, 2009). The function of other noncoding RNAs such as enhancer RNAs (eRNAs), RNA transcripts that originate from active enhancers, are yet to be fully understood (Kim and Shiekhattar, 2015). It is important to also remember that TFs are frequently post-translationally modified, which can profoundly influence TF activity and localization (Filtz et al., 2014).
Cell cycle
A major cause of intrinsic destabilization is the cell cycle. Cell cycle progression and division directly impacts on DNA accessibility (Ma et al., 2015) as well as TF protein (and mRNA) concentration. It is important to remember that the TF network plays a key role in regulating cell cycle progression (Müller, 1995), while cell cycle stage itself influences rates of gene expression (Bertoli et al., 2013) and cell fate decisions (Pauklin and Vallier, 2013).
Metabolic status and intracellular signaling
A cell must be able to adapt to its intracellular and extracellular metabolic status. There are several highly conserved metabolic signaling pathways that regulate such cellular adaption (Efeyan et al., 2015). While these pathways, in particular the Integrated Stress Response (ISR) pathway and mTOR signaling, can alter TF networks through altering rates of global translation, these pathways also more directly influence transcriptional states (de Nadal et al., 2011). For example, activation of the ISR pathway suppresses global translation, it acts to increase translation of certain TFs, notably its canonical effector ATF4 (Wek et al., 2006).
Box 3. Using stem cells and cancer models to understand TF network regulation and dysregulation.
While TF networks have been investigated in numerous mammalian cell types, such networks have been most intensively studied in pluripotent stem cells (Ng and Surani, 2011, Orkin et al., 2008), adult muscle stem cells (Buckingham and Rigby, 2014, Tapscott, 2005), and adult hematopoietic stem cells (Göttgens, 2015) (Figure 4). Additionally, TF network dysregulation is a common theme in cancer, particularly leukemia (Sive and Gottgens, 2014).
Pluripotent stem cells
Pluripotent stem cells (PSCs) have the capacity to form any embryonic type (Figure 4B) (Murry and Keller, 2008). In vitro PSC self-renewal and differentiation provides an important and widely used tractable model of early developmental cell fate decisions. Induced pluripotent stem (iPS) cell reprogramming experiments have highlighted the importance of TFs (all four Yamanaka factors are TFs) in the acquisition and maintenance of the pluripotent state (Takahashi and Yamanaka, 2006). Live cell imaging and single cell RNA-seq methods have recently revealed unexpected heterogeneity of the TF network associated with pluripotency (Filipczyk et al., 2015, Kolodziejczyk et al., 2015), suggesting we still do not fully understand this TF network state. Consistent with this, several pluripotency TF network models recently built from detailed knowledge of the key TFs regulating pluripotency (Xu et al., 2014, Dunn et al., 2014, Dowell et al., 2014) were unable to fully predict cellular behavior.
Adult stem cells
Adult stem cells are thought to provide life-long homeostasis of several adult mammalian tissues. TF interactions regulating the generation, self-renewal, and differentiation of unipotent muscle stem cells and multipotent hematopoietic stem cells (HSCs) are arguably the best understood (Figure 4B). The TF MyoD is a central regulator of muscle formation; it is upregulated in differentiating muscle stem cells and its overexpression in a number of cell types can induce trans-differentiation to muscle (Tapscott et al., 1988, Davis et al., 1987). Numerous TFs have been found to regulate HSC function, and can be found reviewed elsewhere (Wilkinson and Gottgens, 2013). Several laboratories have recently developed methods to reprogram, trans-differentiate and forward program cells into hematopoietic stem and progenitor cells (Riddell et al., 2014, Xie et al., 2004, Sandler et al., 2014). All the methods published so far have used TF overexpression (usually in multi-TF combinations), highlighting the instructive role of TFs, and their combinatorial interactions, in initiating and maintaining hematopoietic cell identity.
Dysregulation in cancer
A diverse set of molecular mechanisms has so far been described to interfere with normal TF network logic in cancer including the mutation of TFs, co-activators/co-repressors, and enhancer regions. Two novel mechanism by which enhancer regions are mutated are particularly noteworthy. First, a chromosomal inversion event has been shown to cause spatial rearrangement of a GATA2 enhancer to be proximal to the EVI1. This alteration in the TF regulatory network logic results in oncogenic EVI1 overexpression, which results in leukemogenesis (Groschel et al., 2014, Yamazaki et al., 2014). Second, somatic mutation of an enhancer upstream of the TAL1 gene has been shown to introduce a novel Myb DNA motif, which drives oncogenic TAL1 overexpression and acute lymphoblastic leukaemia (Mansour et al., 2014).
Recent insights from emerging technologies
New technologies remain major drivers in advancing our understanding of TF network biology. Most recently, single cell transcriptomics, live-cell imaging, and CRISPR/Cas9 genome editing have been applied to the field. These technologies are providing new angles from which to approach TF network biology, and alongside ongoing large-scale annotation approaches, including epigenome annotation, are helping to shed new light on mammalian TF networks.
Large-scale and high-throughput annotation
While TF ChIP-seq experiments have been possible for several years (Johnson et al., 2007) (Box 1), the reduction in next generation sequencing (NGS) costs are now allowing large-scale ChIP-seq studies. From assessing the binding of multiple TFs within the same and different cell types, it is possible to build a comprehensive annotation of TF network interactions. Several large-scale TF ChIP-seq studies have been undertaken, and have identified highly interconnected TF networks in several cell types (Tsankov et al., 2015, Wilson et al., 2016, Goode et al., 2016). Just as sequencing of mammalian genomes has provided a blueprint to study mammalian genomics, these studies provide a central resource to investigate TF binding events within multiple cell types. By undertaking ChIP-seq experiments in similar cell types from different mammals, insights into evolutionary conservation of TF binding events and enhancer function are also being gained (Boyle et al., 2014, Cheng et al., 2014, Villar et al., 2015).
Recently large-scale efforts, including those by the Human Epigenome, ENCODE, and BLUEPRINT consortiums, have provided unprecedented resolution of the epigenetic state and conformation of chromatin in numerous cell types. For example, recent large-scale Hi-C analysis of almost 40 human cell types have helped to define topologically-associated domains (TADs) and assign enhancer-promoter interactions (both constitutive and cell-type specific) (Schmitt et al., 2016, Javierre et al., 2016). Such approaches are providing an important and data-rich annotation of mammalian epigenomes that is necessary for comprehensive TF network assignment. Interestingly, single cell DNase-seq, ATAC-seq, Hi-C and ChIP-seq protocols have also recently been published (Jin et al., 2015, Buenrostro et al., 2015, Cusanovich et al., 2015, Nagano et al., 2013, Rotem et al., 2015), suggesting single cell level chromatin structure and accessibility can also be employed to investigate TF network states in single cells.
Large-scale approaches have also been applied to understand cooperative TF binding. For example, Consecutive Affinity-Purification Systematic Evolution of Ligands by Exponential Enrichment (CAP-SELEX) (Jolma et al., 2015) has been developed to provide high-throughput TF pair binding site characterization, which have yielded important insights into cooperative TF binding events. Additionally, Fuxman Bass and co-workers recently provided a yeast one-hybrid based method to quantify human TF binding to enhancers, and probe the effects of genetic variation on TF interactions (Fuxman Bass et al., 2015). Such methods afford high-throughput analysis from which general principles of mammalian TF cooperativity may be extracted.
High-throughput genome engineering
While CRISPR/Cas9 technologies can be used to make single genetic mutations, these methods are amenable to high-throughput studies, which allows for genome-wide coverage (Parnas et al., 2015) or saturation of a single genomic region (Canver et al., 2015). Several CRISPR gRNA libraries have recently been published (Sanjana et al., 2014, Tzelepis et al., 2016, Horlbeck et al., 2016), for both genetic deletion and transcriptional activation/repression. As with any screening method, it will be important to develop appropriate readouts and/or reporters for these assays. However, such tools are poised to provide significant insights into enhancer regulation and TF network interactions within mammalian cell types.
Canver et al. (2015) provided an elegant demonstration of the application of CRISPR/Cas9 for saturating mutagenesis of a single enhancer of BCL11A, allowing functional element mapping over a 12kb enhancer region (Canver et al., 2015). Importantly, such an approach allows for mutational analysis of endogenous enhancers, rather than the traditional reliance on heterologous enhancer reporter assays. Large-scale application of these CRISPR/Cas9 methods is likely to provide important fundamental insights into TF network architecture and principles.
Single cell transcriptomics
Fluorescent-Activated Cell Sorting (FACS) has long been used to purify single cells (Osawa et al., 1996), and has highlighted functional variability in highly purified cell populations (Yamamoto et al., 2013). In combination with microfluidics technologies, FACS has recently afforded single cell gene expression analysis. Multiplexed qPCR initially allowed expression of 10–100 genes to be quantified in 100s of FACS-isolated single cells (Sanchez-Freire et al., 2012, White et al., 2011). Such methods have been particularly useful to investigate TF networks during embryogenesis and in adult stem cell populations, where limited cell numbers have often prevented population level analysis (Moignard et al., 2013, Moignard et al., 2015, Wilson et al., 2015). However, given the limited number of genes that could be simultaneously quantified by such methods, TF networks could not be comprehensively studied.
The advent of single-cell whole transcriptome RNA-seq methodologies (Tang et al., 2010, Picelli et al., 2013, Macosko et al., 2015) has provided important new opportunities. Several recent reports have demonstrated the potential of single cell RNA-seq to provide new resolution of TF network architecture (Kolodziejczyk et al., 2015, Olsson et al., 2016, Scialdone et al., 2016). It is of course important to remember the experimental caveats. First, mRNA levels do not always correspond to TF protein level (or activity). Second, single cell RNA-seq alone cannot distinguish indirect vs. direct TF interactions, although here, its combination with genetic deletion of specific TFs has yielded important resolution (Olsson et al., 2016, Scialdone et al., 2016). Third, current single cell RNA-seq approaches generally have lower sequencing coverage than bulk cell analyses, which may influence the transcript detection and/or observed intercellular heterogeneity.
For example, single cell RNA-seq has recently been used to re-evaluate the role of the TF Tal1 (also known as Scl) in mesoderm specification into hematopoietic and cardiac fates (Scialdone et al., 2016). Tal1 has been thought to autonomously activate (and stabilize) a hematopoietic TF network state while actively repressing a cardiac state, as its deletion blocks developmental blood cell formation and induces cardiomyocyte formation (Van Handel et al., 2012, Ismailoglu et al., 2008). While single cell RNA-seq analysis confirmed that loss of Tal1 inhibited activation of blood-associated TFs, it failed to identity a corresponding immediate upregulation of a cardiac transcriptional program. These results suggest aberrant cardiac formation is likely a slower, and perhaps secondary consequence of Tal1 deletion, rather than a direct lineage fate switch.
Recently, CRISPR/Cas9 screening has been combined with single cell RNA-seq in an approach that promises to provide detailed resolution of TF network circuitry in single cells (Dixit et al., 2016, Jaitin et al., 2016). Given that TF network states are heterogeneous at single cell level, the approaches by Dixit et al. (2016) and Jaitin et al. (2016) promise to provide a data-rich method to analyze CRISPR screening. Importantly, the approaches provide high-throughput and quantitative analysis of the direct transcriptomic consequences of genetic mutations. Through such methods, it should be possible to derive a comprehensive TF network map, and through profiling many single cells, infer fundamental principles of network state dynamics. However, to do so we will need to develop new bioinformatics methods to analyze and integrate these large and multidimensional datasets.
Live-cell imaging
While single cell transcriptomics determines the expression of many TFs, these technologies provide only a snapshot of gene expression. By lysing the cell for such gene expression studies, its future fate and potential cannot be simultaneously assessed (Hoppe et al., 2014). Live-cell imaging provides a powerful method to study the dynamics of TFs and allow TF network states to be correlated with cell fate decisions (within future generations of the cell). By tracking fluorescent reporters linked to TF expression (often directly fused to the TF of interest), live-cell imaging has provided important insights into the relationship between TF expression and the cell cycle (Kueh et al., 2013), TF network plasticity (Filipczyk et al., 2015), TF antagonism (Hoppe et al., 2016), and extracellular signaling interactions (Kueh et al., 2016).
An early application of live-cell imaging was provided by Kueh et al. (2013), who focused on the transcription factor PU.1 and its interaction with the cell cycle. PU.1 is known to play a dose-dependent and context-dependent role in driving proliferation vs. terminal differentiation within hematopoietic cells (Mak et al., 2011). By tracking PU.1 expression over multiple cell divisions during differentiation into lymphoid and myeloid cell commitment, Kueh and colleagues were able to demonstrate that cell cycle kinetics could directly influence accumulation or loss of PU.1 over several generations, and thereby alter cell fate decisions. By regulating cell cycle progression, PU.1 could itself influence its own accumulation, outside of its regulation of positive auto-feedback at the transcriptional level (Kueh et al., 2013). One caveat of this study was that PU.1 levels were indirectly measured using a PU.1-IRES-GFP reporter, rather than a directly fused PU.1-fluorescent reporter. However, directly fused TF-fluorescent reporters hold their own caveats, such as potentially altering TF function and dynamics (TF-DNA or TF-TF interactions), as well as protein half-life.
More recently, quantitative live-cell imaging has been further used to investigate PU.1 within the context of PU.1-Gata1 antagonism in the erythroid vs. myeloid cell fate decision during hematopoietic stem cell (HSC) differentiation (Hoppe et al., 2016). Bulk cell analyses have implicated a direct cross-antagonism of Gata1 and PU.1 proteins, and this is widely used as an example of mammalian TF circuit interactions (Burda et al., 2010). However, live-cell imaging of Gata1-mCherry and PU.1-eYFP expression during the differentiation of individual HSCs questioned these long-held assumptions as few cells co-express Gata1 and PU.1 simultaneously, a requirement for cross-antagonism. Instead of being responsible for the cell fate decision, upregulation of Gata1 or PU.1 appears to only occur following cell fate decisions, and therefore more likely acts to reinforce a pre-determined cell fate decision. While it is currently unclear what is upstream, and thereby responsible for the erythroid vs. myeloid fate decision, these data question the simplistic TF network models for mammalian cell fate decisions.
An alternative to tagging TFs with fluorescent proteins is the use of HaloTag ligand-based technology to tag TFs with organic dyes, which even affords subcellular TF localization in live cells using light-sheet microscopy. For example, Liu et al (2014) used this powerful approach to track Sox2-DNA binding in human ESCs and 3D diffusion within the nucleus (Liu et al., 2014). However, it is worth noting that live cell imaging methods are currently limited in the number of TFs that can be simultaneous detected (due to fluorescent spectral overlap). However, it is clear such powerful approaches are providing important quantitative insights into TF network biology.
Open questions and future directions
As described above, recent technical advances are helping to drive forward our characterization of mammalian TF regulatory networks. However, many of these technologies are yet to reach their full potential. Several key questions remain open. We hope that these technologies and others will be able to provide answers in the future.
Moving towards protein level quantification
Cell fate decisions are made at the single cell level. Important biological understanding is therefore being yielded from the single cell approaches described above. Single cell RNA-seq technologies certainly provide unprecedented single cell resolution of transcriptional programs, but we must remember that TF proteins are the functional products that determine TF network activity. While current technical limitations prevent single cell proteomics, new technologies are moving towards this goal, including mass cytometry (Bendall et al., 2011) and single cell protein expression using microfluidic systems and protein-PCR based quantification (Macaulay and Voet, 2014). Ultimately however, we need to develop in vivo live cell imaging systems to measure multiple endogenous proteins simultaneously.
Dynamic modeling of a comprehensive TF network
While several in silico computational modeling methods have been used to study TF networks, these have so far largely failed to accurately predict biological systems. More complex modeling approaches are needed to better predict and extract the biological logic of TF networks. Here, it will be important to move from static models to dynamic models of TF regulatory networks, which better reflect biological complexity. In silico models of mammalian TF networks have also so far tended to investigate limited sub-networks within the larger TF network. By moving towards larger-scale comprehensive analysis of enhancer activity and TF network interactions, such as methods used in other organisms (Arnold et al., 2013, MacNeil et al., 2015), modeling of entire mammalian TF regulatory networks should be possible. For example, Arnold et al. recently developed a powerful genome-wide assay called STARR-seq to quantitiate enhancer activity genome-wide in Drosophila (Arnold et al., 2013). Additionally, by integrating data from various experimental sources, greater predictive power is possible. For example, by integrating data from ChIP-seq and enhancer assays into dynamic Bayesian computational modeling, accurate TF sub-network modeling has been possible (Schütte et al., 2016).
General principles in mammalian TF network biology
While many general principles that govern mammalian gene expression, enhancer activity, and TF interactions, have been described, we still have few general principles that hold for explaining mammalian TF networks. We hope that through integrating the above technologies, along with others, we will soon start to be able to develop meaningful principles that govern this key biological decision-making process.
Applications of TF network biology
Understanding the human TF regulatory network has important implications for health and disease. These include improving disease diagnosis and developing new therapeutic strategies, as outlined below.
Disease diagnosis
Central to the realization of personalized medicine is the ability to discriminate whether a patient will respond to a particular treatment or develop resistance, will relapse or go into remission, etc. Accurate biomarkers are key to this predict power. TF network components are often mutated in cancers (see Box 3), but genetic mutations alone often fail to accurately predict disease progression. Understanding the TF network states associated with a certain disease, and unique output (e.g. gene expression profile), may help to accurately predict clinical response and outcome. For example, regression analysis has recently been applied to large gene expression datasets from leukemia patients to identify a highly predictive and clinically informative 17-gene signature for therapy resistance (Ng et al., 2016). The application of our understanding of TF networks associated with (and specific to) human diseases also has significant potential in identifying novel therapeutic targets.
Cellular engineering
Reprogramming, forward programming, and trans-differentiation approaches hold important promise for regenerative medicine (Graf and Enver, 2009). However, the generation of transplantation-grade cells is a major hurdle for the clinical application of these approaches. TFs are most commonly used in these approaches, with the best combinations of TFs being identified from experimental screening. However, such approaches are often expensive, time-consuming and fail to generate fully functional cell types. Several network biology-based bioinformatics tools have been developed to predict TF combinations optimal for reprogramming and trans-differentiation, such as the CellNet platform (Cahan et al., 2014, Morris et al., 2014). Recently, a more comprehensive bioinformatics platform has been developed, called Mogrify, which combines gene expression datasets from over 300 cell types with TF network information to predict TFs for cellular reprogramming and trans-differentiation (Rackham et al., 2016). Given the importance of TF network state for cellular function and potential, such approaches and in silico modeling methodologies will likely play an increasingly important role in future translational research efforts.
Conclusion
TF regulatory network biology is an inter-disciplinary research field, with molecular, cellular, genetic, genomic, and computational approaches currently driving forward our understanding in a range of mammalian cell lineages and developmental stages. It is particularly exciting that human TF regulatory network biology is becoming an increasing research focus within the field, with its significance in understanding human health and disease. Using recent technological advances, we can now quantify global gene expression at single cell resolution, track TF dynamics within live cells, and investigate the impact of multiple mutations on cellular (and molecular) function. These new approaches are questioning some of the long-held assumptions in TF network biology. We expect that future advances will further drive forwards our understanding of mammalian TF networks.
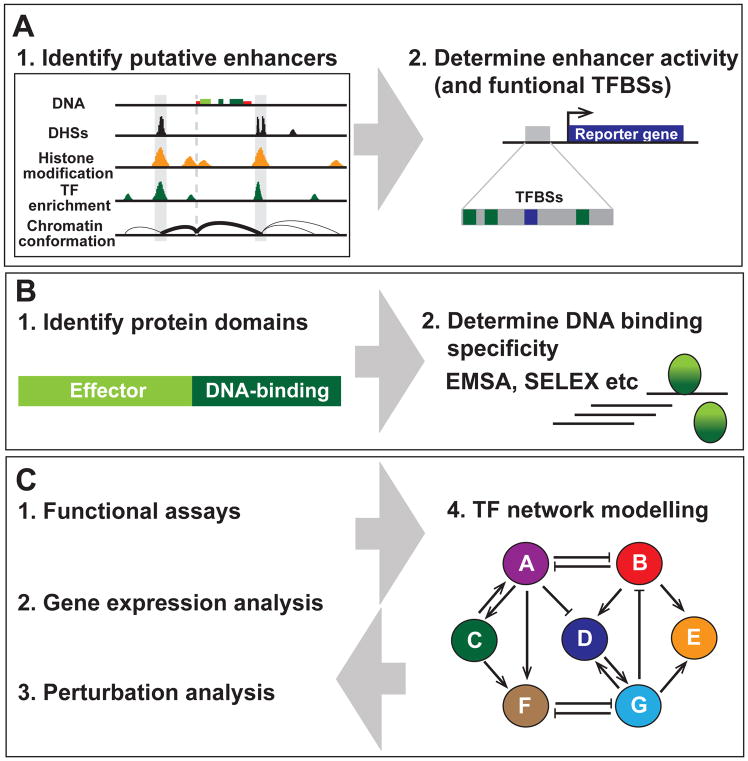
Figure 2. Approaches to build TF regulatory network models.
(A) Enhancers. Putative enhancers can be identified by a number features including DNase I hypersensitivity sites (DHSs), histone modifications (such as H3K4me), TF enrichment and DNA looping (measured by chromatin conformation capture methods such as Hi-C). Enhancer activity can be assessed using in vivo or in vitro enhancer assays, and the function of the TFBSs (DNA motifs) identified within such enhancers can assessed by mutational analysis.
(B) Transcription factors. TFs can be identified by their DNA binding domains. TFs also contain effector domains, which are responsible for protein-protein interactions. A range of methods including electrophoretic mobility shift assays (EMSAs) and systematic evolution of ligands by exponential enrichment (SELEX) have been used to determine individual and cooperative TF DNA binding specificities.
(C) Building TF network models. Methods in (A) and (B) can be combined with functional assays (such as enhancer mutagenesis), gene expression analysis and/or TF pertubation analysis to build and train TF regulatory network models that can be “executed” in silico. These models provide important insights into the biological logic underpinning mammalian cell fate decisions, which feedback into experimental research.
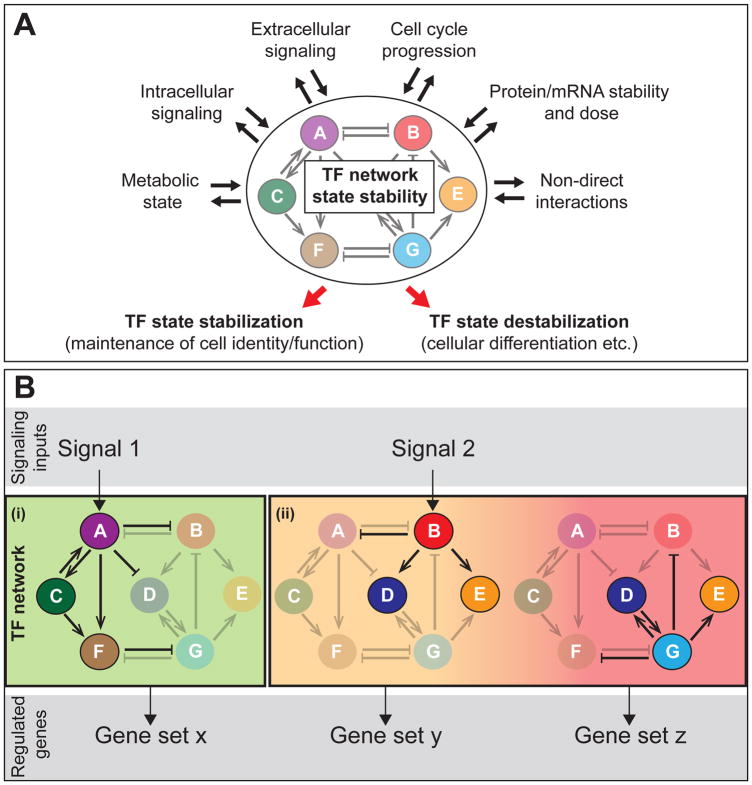
Figure 3. Mechanism of TF network regulation.
(A) A summary of mechanisms that influence TF network state stability. Numerous extrinsic and intrinsic mechanisms regulate TF network stability. These mechanisms are also often influenced by TF network state). TF network stabilization results in maintenance of a cell identity/function, such as stem cell self-renewal, while TF network destabilization induces TF network state transitions can lead to changing cellular identity/function and cellular differentiation.
(B) A schematic of how different signalling pathways activate certain sub-networks or states of a TF network. Depending on the logic of TF interactions and signalling inputs, states may be (i) stabilized or (ii) destabilized (resulting in state transitions). The set of TFs expressed determines the selection of genes regulated/expressed, which influences cellular identity and function. This review focuses on the TF networks, rather than upstream signaling inputs or downstream regulated genes/patterns of expression. For simplicity, the TF protein, its enhancer(s) and gene are represented as a single circle (A-G). As described in the main text, TF proteins regulate the activity of enhancers of other TFs, to activate or repress gene transcription (and may also be involved in auto-feedback regulation).
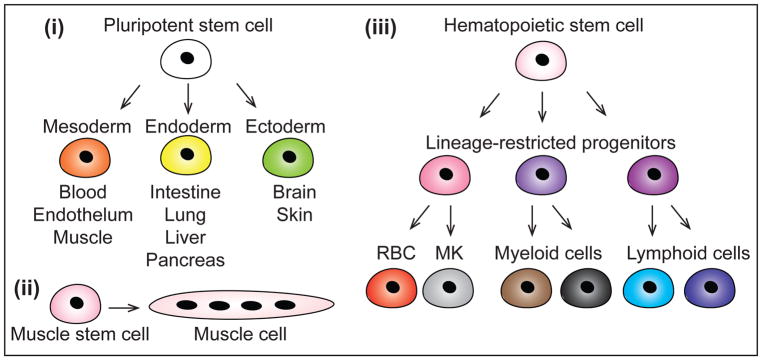
Figure 4. Commonly used mammalian systems to study TF networks.
Mammalian TF networks have been commonly investigated in the context of pluripotency, muscle formation and hematopoiesis. (i) Pluripotent stem cells (embryonic stem cells or induced pluripotent stem cells) can self-renew or differentiate into any embryonic cell type through commitment to mesoderm, endoderm or ectoderm germ layers. (ii) Muscle stem cells (or satellite cells) can self-renew or differentiate into muscle cells. (iii) Hematopoietic stem cells (HSCs) have the ability to self-renew or differentiate into any mature blood cell type, through increasingly lineage-restricted haematopoietic progenitor cells. Red blood cells (RBCs), megakaryocytes (MKs), myeloid cells and lymphoid cells can be specified from HSCs.
Acknowledgments
We apologize to those authors whose work could not be cited due to space constraints. We thank Dr. Hannah Long and Dr. Craig Mak for helpful discussion and feedback on the manuscript. ACW is funded by a Bloodwise Visiting Fellowship. HN is funded by the Japan Science and Technology Agency, the California Institute of Regenerative Medicine and Ludwig Foundation. BG is funded by Bloodwise, Cancer Research UK, the Wellcome Trust, the MRC, NIH-NIDDK and core funding from the Wellcome Trust to the Cambridge Stem Cell Institute.
Glossary
- Sequence-specific transcription factor (TF)
A sequence-specific DNA binding protein that regulates gene expression. These differ from other classes of transcription factors, such as general transcription factors, that regulate gene expression but do not bind DNA with sequence specificity.
- TF network
The entire collection of TFs, their genes, and enhancer regions, within a cell that directly or indirectly interact to form a complex, interconnected molecular circuit.
- Network state
The subset of the TF network, including the TFs and genes/enhancers that are expressed/active at a given moment.
- Enhancer region
A genomic region (often composed of multiple DNA elements) that positively regulates (enhances) transcription from a gene promoter. An enhancer may be proximal to or distal from its target sequence in primary DNA sequence.
- Transcription factor binding site (TFBS)
A specific (usually short) DNA sequence or motif to which a TF binds, a functional DNA element within an enhancer region.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8(6):450–61. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Stelzer C, Boryñ Ł, Rath M, Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339(6123):1074–7. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- Bendall SC, Simonds EF, Qiu P, Amir e-A, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, Balderas RS, Plevritis SK, Sachs K, Pe’er D, Tanner SD, Nolan GP. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–96. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013;14(8):518–28. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais A, Dynlacht BD. Constructing transcriptional regulatory networks. Genes Dev. 2005;19(13):1499–511. doi: 10.1101/gad.1325605. [DOI] [PubMed] [Google Scholar]
- Bonzanni N, Garg A, Feenstra KA, Schütte J, Kinston S, Miranda-Saavedra D, Heringa J, Xenarios I, Göttgens B. Hard-wired heterogeneity in blood stem cells revealed using a dynamic regulatory network model. Bioinformatics. 2013;29(13):i80–8. doi: 10.1093/bioinformatics/btt243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Araya CL, Brdlik C, Cayting P, Cheng C, Cheng Y, Gardner K, Hillier LW, Janette J, Jiang L, Kasper D, Kawli T, Kheradpour P, Kundaje A, Li JJ, Ma L, Niu W, Rehm EJ, Rozowsky J, Slattery M, Spokony R, Terrell R, Vafeados D, Wang D, Weisdepp P, Wu YC, Xie D, Yan KK, Feingold EA, Good PJ, Pazin MJ, Huang H, Bickel PJ, Brenner SE, Reinke V, Waterston RH, Gerstein M, White KP, Kellis M, Snyder M. Comparative analysis of regulatory information and circuits across distant species. Nature. 2014;512(7515):453–6. doi: 10.1038/nature13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent MR. Past Roadblocks and New Opportunities in Transcription Factor Network Mapping. Trends Genet. 2016;32(11):736–750. doi: 10.1016/j.tig.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S. GATA switches as developmental drivers. J Biol Chem. 2010;285(41):31087–93. doi: 10.1074/jbc.R110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M, Rigby PW. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev Cell. 2014;28(3):225–38. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523(7561):486–90. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda P, Laslo P, Stopka T. The role of PU.1 and GATA-1 transcription factors during normal and leukemogenic hematopoiesis. Leukemia. 2010;24(7):1249–57. doi: 10.1038/leu.2010.104. [DOI] [PubMed] [Google Scholar]
- Cahan P, Li H, Morris SA, Lummertz da Rocha E, Daley GQ, Collins JJ. CellNet: network biology applied to stem cell engineering. Cell. 2014;158(4):903–15. doi: 10.1016/j.cell.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, Luc S, Kurita R, Nakamura Y, Fujiwara Y, Maeda T, Yuan GC, Zhang F, Orkin SH, Bauer DE. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527(7577):192–7. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Ma Z, Kim BH, Wu W, Cayting P, Boyle AP, Sundaram V, Xing X, Dogan N, Li J, Euskirchen G, Lin S, Lin Y, Visel A, Kawli T, Yang X, Patacsil D, Keller CA, Giardine B, Kundaje A, Wang T, Pennacchio LA, Weng Z, Hardison RC, Snyder MP Consortium ME. Principles of regulatory information conservation between mouse and human. Nature. 2014;515(7527):371–5. doi: 10.1038/nature13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–2. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107(50):21931–6. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, Steemers FJ, Trapnell C, Shendure J. Epigenetics. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348(6237):910–4. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468(7326):911–20. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- de Nadal E, Ammerer G, Posas F. Controlling gene expression in response to stress. Nat Rev Genet. 2011;12(12):833–45. doi: 10.1038/nrg3055. [DOI] [PubMed] [Google Scholar]
- Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, Adamson B, Norman TM, Lander ES, Weissman JS, Friedman N, Regev A. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell. 2016;167(7):1853–1866.e17. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan N, Wu W, Morrissey CS, Chen KB, Stonestrom A, Long M, Keller CA, Cheng Y, Jain D, Visel A, Pennacchio LA, Weiss MJ, Blobel GA, Hardison RC. Occupancy by key transcription factors is a more accurate predictor of enhancer activity than histone modifications or chromatin accessibility. Epigenetics Chromatin. 2015;8:16. doi: 10.1186/s13072-015-0009-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16(10):1299–309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell KG, Simons AK, Bai H, Kell B, Wang ZZ, Yun K, Hibbs MA. Novel insights into embryonic stem cell self-renewal revealed through comparative human and mouse systems biology networks. Stem Cells. 2014;32(5):1161–72. doi: 10.1002/stem.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn SJ, Martello G, Yordanov B, Emmott S, Smith AG. Defining an essential transcription factor program for naïve pluripotency. Science. 2014;344(6188):1156–60. doi: 10.1126/science.1248882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517(7534):302–10. doi: 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipczyk A, Marr C, Hastreiter S, Feigelman J, Schwarzfischer M, Hoppe PS, Loeffler D, Kokkaliaris KD, Endele M, Schauberger B, Hilsenbeck O, Skylaki S, Hasenauer J, Anastassiadis K, Theis FJ, Schroeder T. Network plasticity of pluripotency transcription factors in embryonic stem cells. Nat Cell Biol. 2015;17(10):1235–46. doi: 10.1038/ncb3237. [DOI] [PubMed] [Google Scholar]
- Filtz TM, Vogel WK, Leid M. Regulation of transcription factor activity by interconnected post-translational modifications. Trends Pharmacol Sci. 2014;35(2):76–85. doi: 10.1016/j.tips.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxman Bass JI, Sahni N, Shrestha S, Garcia-Gonzalez A, Mori A, Bhat N, Yi S, Hill DE, Vidal M, Walhout AJ. Human gene-centered transcription factor networks for enhancers and disease variants. Cell. 2015;161(3):661–73. doi: 10.1016/j.cell.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Yang J, Tsang JC, Ooi J, Wu D, Liu P. Reprogramming to Pluripotency Using Designer TALE Transcription Factors Targeting Enhancers. Stem Cell Reports. 2013;1(2):183–97. doi: 10.1016/j.stemcr.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode DK, Obier N, Vijayabaskar MS, Lie-A-Ling M, Lilly AJ, Hannah R, Lichtinger M, Batta K, Florkowska M, Patel R, Challinor M, Wallace K, Gilmour J, Assi SA, Cauchy P, Hoogenkamp M, Westhead DR, Lacaud G, Kouskoff V, Göttgens B, Bonifer C. Dynamic Gene Regulatory Networks Drive Hematopoietic Specification and Differentiation. Dev Cell. 2016;36(5):572–87. doi: 10.1016/j.devcel.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462(7273):587–94. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Groschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BA, Erpelinck C, van der Velden VH, Havermans M, Avellino R, van Lom K, Rombouts EJ, van Duin M, Dohner K, Beverloo HB, Bradner JE, Dohner H, Lowenberg B, Valk PJ, Bindels EM, de Laat W, Delwel R. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157(2):369–81. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- Göttgens B. Regulatory network control of blood stem cells. Blood. 2015;125(17):2614–20. doi: 10.1182/blood-2014-08-570226. [DOI] [PubMed] [Google Scholar]
- Hay D, Hughes JR, Babbs C, Davies JO, Graham BJ, Hanssen LL, Kassouf MT, Oudelaar AM, Sharpe JA, Suciu MC, Telenius J, Williams R, Rode C, Li PS, Pennacchio LA, Sloane-Stanley JA, Ayyub H, Butler S, Sauka-Spengler T, Gibbons RJ, Smith AJ, Wood WG, Higgs DR. Genetic dissection of the α-globin super-enhancer in vivo. Nat Genet. 2016;48(8):895–903. doi: 10.1038/ng.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, Fields S, Stamatoyannopoulos JA. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6(4):283–9. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe PS, Coutu DL, Schroeder T. Single-cell technologies sharpen up mammalian stem cell research. Nat Cell Biol. 2014;16(10):919–27. doi: 10.1038/ncb3042. [DOI] [PubMed] [Google Scholar]
- Hoppe PS, Schwarzfischer M, Loeffler D, Kokkaliaris KD, Hilsenbeck O, Moritz N, Endele M, Filipczyk A, Gambardella A, Ahmed N, Etzrodt M, Coutu DL, Rieger MA, Marr C, Strasser MK, Schauberger B, Burtscher I, Ermakova O, Bürger A, Lickert H, Nerlov C, Theis FJ, Schroeder T. Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature. 2016;535(7611):299–302. doi: 10.1038/nature18320. [DOI] [PubMed] [Google Scholar]
- Horlbeck MA, Gilbert LA, Villalta JE, Adamson B, Pak RA, Chen Y, Fields AP, Park CY, Corn JE, Kampmann M, Weissman JS. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. Elife. 2016:5. doi: 10.7554/eLife.19760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismailoglu I, Yeamans G, Daley GQ, Perlingeiro RCR, Kyba M. Mesodermal patterning activity of SCL. Experimental Hematology. 2008;36(12):1593–1603. doi: 10.1016/j.exphem.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Jaitin DA, Weiner A, Yofe I, Lara-Astiaso D, Keren-Shaul H, David E, Salame TM, Tanay A, van Oudenaarden A, Amit I. Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell. 2016;167(7):1883–1896.e15. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, Cairns J, Wingett SW, Várnai C, Thiecke MJ, Burden F, Farrow S, Cutler AJ, Rehnström K, Downes K, Grassi L, Kostadima M, Freire-Pritchett P, Wang F, Stunnenberg HG, Todd JA, Zerbino DR, Stegle O, Ouwehand WH, Frontini M, Wallace C, Spivakov M, Fraser P Consortium B. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell. 2016;167(5):1369–1384.e19. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Tang Q, Wan M, Cui K, Zhang Y, Ren G, Ni B, Sklar J, Przytycka TM, Childs R, Levens D, Zhao K. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature. 2015;528(7580):142–6. doi: 10.1038/nature15740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316(5830):1497–502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, Palin K, Vaquerizas JM, Vincentelli R, Luscombe NM, Hughes TR, Lemaire P, Ukkonen E, Kivioja T, Taipale J. DNA-binding specificities of human transcription factors. Cell. 2013;152(1–2):327–39. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature. 2015;527(7578):384–8. doi: 10.1038/nature15518. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007;134(7):1243–51. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Marinov GK, Ward LD, Birney E, Crawford GE, Dekker J, Dunham I, Elnitski LL, Farnham PJ, Feingold EA, Gerstein M, Giddings MC, Gilbert DM, Gingeras TR, Green ED, Guigo R, Hubbard T, Kent J, Lieb JD, Myers RM, Pazin MJ, Ren B, Stamatoyannopoulos JA, Weng Z, White KP, Hardison RC. Defining functional DNA elements in the human genome. Proc Natl Acad Sci U S A. 2014;111(17):6131–8. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Shiekhattar R. Architectural and Functional Commonalities between Enhancers and Promoters. Cell. 2015;162(5):948–59. doi: 10.1016/j.cell.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejczyk AA, Kim JK, Tsang JC, Ilicic T, Henriksson J, Natarajan KN, Tuck AC, Gao X, Bühler M, Liu P, Marioni JC, Teichmann SA. Single Cell RNA-Sequencing of Pluripotent States Unlocks Modular Transcriptional Variation. Cell Stem Cell. 2015;17(4):471–85. doi: 10.1016/j.stem.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kueh HY, Champhekhar A, Nutt SL, Elowitz MB, Rothenberg EV. Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science. 2013;341(6146):670–3. doi: 10.1126/science.1240831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueh HY, Yui MA, Ng KK, Pease SS, Zhang JA, Damle SS, Freedman G, Siu S, Bernstein ID, Elowitz MB, Rothenberg EV. Asynchronous combinatorial action of four regulatory factors activates Bcl11b for T cell commitment. Nat Immunol. 2016;17(8):956–65. doi: 10.1038/ni.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Novère N. Quantitative and logic modelling of molecular and gene networks. Nat Rev Genet. 2015;16(3):146–58. doi: 10.1038/nrg3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Legant WR, Chen BC, Li L, Grimm JB, Lavis LD, Betzig E, Tjian R. 3D imaging of Sox2 enhancer clusters in embryonic stem cells. Elife. 2014;3:e04236. doi: 10.7554/eLife.04236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Prescott SL, Wysocka J. Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell. 2016;167(5):1170–1187. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Kanakousaki K, Buttitta L. How the cell cycle impacts chromatin architecture and influences cell fate. Front Genet. 2015;6:19. doi: 10.3389/fgene.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay IC, Voet T. Single cell genomics: advances and future perspectives. PLoS Genet. 2014;10(1):e1004126. doi: 10.1371/journal.pgen.1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil LT, Pons C, Arda HE, Giese GE, Myers CL, Walhout AJ. Transcription Factor Activity Mapping of a Tissue-Specific in vivo Gene Regulatory Network. Cell Syst. 2015;1(2):152–162. doi: 10.1016/j.cels.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161(5):1202–14. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak KS, Funnell AP, Pearson RC, Crossley M. PU.1 and Haematopoietic Cell Fate: Dosage Matters. Int J Cell Biol. 2011;2011:808524. doi: 10.1155/2011/808524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11(11):761–72. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Goodbourn S, Fischer JA. Regulation of inducible and tissue-specific gene expression. Science. 1987;236(4806):1237–45. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, Etchin J, Lawton L, Sallan SE, Silverman LB, Loh ML, Hunger SP, Sanda T, Young RA, Look AT. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346(6215):1373–7. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Walhout AJ. The interplay between transcription factors and microRNAs in genome-scale regulatory networks. Bioessays. 2009;31(4):435–45. doi: 10.1002/bies.200800212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13(10):616–30. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moignard V, Macaulay IC, Swiers G, Buettner F, Schutte J, Calero-Nieto FJ, Kinston S, Joshi A, Hannah R, Theis FJ, Jacobsen SE, de Bruijn MF, Gottgens B. Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis. Nat Cell Biol. 2013;15(4):363–72. doi: 10.1038/ncb2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moignard V, Woodhouse S, Haghverdi L, Lilly AJ, Tanaka Y, Wilkinson AC, Buettner F, Macaulay IC, Jawaid W, Diamanti E, Nishikawa S, Piterman N, Kouskoff V, Theis FJ, Fisher J, Göttgens B. Decoding the regulatory network of early blood development from single-cell gene expression measurements. Nat Biotechnol. 2015;33(3):269–76. doi: 10.1038/nbt.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Cahan P, Li H, Zhao AM, San Roman AK, Shivdasani RA, Collins JJ, Daley GQ. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell. 2014;158(4):889–902. doi: 10.1016/j.cell.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Lovén J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell. 2011;147(3):565–76. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132(4):661–80. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Müller R. Transcriptional regulation during the mammalian cell cycle. Trends Genet. 1995;11(5):173–8. doi: 10.1016/S0168-9525(00)89039-3. [DOI] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502(7469):59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Surani MA. The transcriptional and signalling networks of pluripotency. Nat Cell Biol. 2011;13(5):490–6. doi: 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, Arruda A, Popescu A, Gupta V, Schimmer AD, Schuh AC, Yee KW, Bullinger L, Herold T, Görlich D, Büchner T, Hiddemann W, Berdel WE, Wörmann B, Cheok M, Preudhomme C, Dombret H, Metzeler K, Buske C, Löwenberg B, Valk PJ, Zandstra PW, Minden MD, Dick JE, Wang JC. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540(7633):433–437. doi: 10.1038/nature20598. [DOI] [PubMed] [Google Scholar]
- Noonan JP, McCallion AS. Genomics of long-range regulatory elements. Annu Rev Genomics Hum Genet. 2010;11:1–23. doi: 10.1146/annurev-genom-082509-141651. [DOI] [PubMed] [Google Scholar]
- Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, Grimes HL. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 2016;537(7622):698–702. doi: 10.1038/nature19348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Wang J, Kim J, Chu J, Rao S, Theunissen TW, Shen X, Levasseur DN. The transcriptional network controlling pluripotency in ES cells. Cold Spring Harb Symp Quant Biol. 2008;73:195–202. doi: 10.1101/sqb.2008.72.001. [DOI] [PubMed] [Google Scholar]
- Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273(5272):242–5. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I, Sanjana NE, Shalem O, Satija R, Raychowdhury R, Mertins P, Carr SA, Zhang F, Hacohen N, Regev A. A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell. 2015;162(3):675–86. doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S, Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155(1):135–47. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Björklund Å, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10(11):1096–8. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- Pombo A, Dillon N. Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol. 2015;16(4):245–57. doi: 10.1038/nrm3965. [DOI] [PubMed] [Google Scholar]
- Rackham OJ, Firas J, Fang H, Oates ME, Holmes ML, Knaupp AS, Suzuki H, Nefzger CM, Daub CO, Shin JW, Petretto E, Forrest AR, Hayashizaki Y, Polo JM, Gough J Consortium F. A predictive computational framework for direct reprogramming between human cell types. Nat Genet. 2016;48(3):331–5. doi: 10.1038/ng.3487. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–83. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, O’Shaughnessy A, Hendrich B. Transcriptional repressors: multifaceted regulators of gene expression. Development. 2013;140(3):505–12. doi: 10.1242/dev.083105. [DOI] [PubMed] [Google Scholar]
- Riddell J, Gazit R, Garrison BS, Guo G, Saadatpour A, Mandal PK, Ebina W, Volchkov P, Yuan GC, Orkin SH, Rossi DJ. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell. 2014;157(3):549–64. doi: 10.1016/j.cell.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol. 2015;33(11):1165–72. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Freire V, Ebert AD, Kalisky T, Quake SR, Wu JC. Microfluidic single-cell real-time PCR for comparative analysis of gene expression patterns. Nat Protoc. 2012;7(5):829–38. doi: 10.1038/nprot.2012.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler VM, Lis R, Liu Y, Kedem A, James D, Elemento O, Butler JM, Scandura JM, Rafii S. Reprogramming human endothelial cells to haematopoietic cells requires vascular induction. Nature. 2014;511(7509):312–8. doi: 10.1038/nature13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–4. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Shalek AK. Heterogeneity in immune responses: from populations to single cells. Trends Immunol. 2014;35(5):219–29. doi: 10.1016/j.it.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AD, Hu M, Jung I, Xu Z, Qiu Y, Tan CL, Li Y, Lin S, Lin Y, Barr CL, Ren B. A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome. Cell Rep. 2016;17(8):2042–2059. doi: 10.1016/j.celrep.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütte J, Wang H, Antoniou S, Jarratt A, Wilson NK, Riepsaame J, Calero-Nieto FJ, Moignard V, Basilico S, Kinston SJ, Hannah RL, Chan MC, Nürnberg ST, Ouwehand WH, Bonzanni N, de Bruijn MF, Göttgens B. An experimentally validated network of nine haematopoietic transcription factors reveals mechanisms of cell state stability. Elife. 2016;5:e11469. doi: 10.7554/eLife.11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scialdone A, Tanaka Y, Jawaid W, Moignard V, Wilson NK, Macaulay IC, Marioni JC, Göttgens B. Resolving early mesoderm diversification through single-cell expression profiling. Nature. 2016;535(7611):289–93. doi: 10.1038/nature18633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigvardsson M. Transcription factor dose links development to disease. Blood. 2012;120(18):3630–1. doi: 10.1182/blood-2012-09-455113. [DOI] [PubMed] [Google Scholar]
- Sive J, Gottgens B. Transcriptional network control of normal and leukaemic haematopoiesis. Exp Cell Res. 2014 doi: 10.1016/j.yexcr.2014.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Sandstrom R, Haugen E, Reynolds AP, Zhang M, Byron R, Canfield T, Stelhing-Sun S, Lee K, Thurman RE, Vong S, Bates D, Neri F, Diegel M, Giste E, Dunn D, Vierstra J, Hansen RS, Johnson AK, Sabo PJ, Wilken MS, Reh TA, Treuting PM, Kaul R, Groudine M, Bender MA, Borenstein E, Stamatoyannopoulos JA. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature. 2014;515(7527):365–70. doi: 10.1038/nature13972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Nordman E, Li B, Xu N, Bashkirov VI, Lao K, Surani MA. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc. 2010;5(3):516–35. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132(12):2685–95. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988;242(4877):405–11. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, Li P, Durand EM, Mosimann C, Heffner GC, Daley GQ, Paulson RF, Young RA, Zon LI. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147(3):577–89. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov AM, Gu H, Akopian V, Ziller MJ, Donaghey J, Amit I, Gnirke A, Meissner A. Transcription factor binding dynamics during human ES cell differentiation. Nature. 2015;518(7539):344–9. doi: 10.1038/nature14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzelepis K, Koike-Yusa H, De Braekeleer E, Li Y, Metzakopian E, Dovey OM, Mupo A, Grinkevich V, Li M, Mazan M, Gozdecka M, Ohnishi S, Cooper J, Patel M, McKerrell T, Chen B, Domingues AF, Gallipoli P, Teichmann S, Ponstingl H, McDermott U, Saez-Rodriguez J, Huntly BJ, Iorio F, Pina C, Vassiliou GS, Yusa K. A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia. Cell Rep. 2016;17(4):1193–1205. doi: 10.1016/j.celrep.2016.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Handel B, Montel-Hagen A, Sasidharan R, Nakano H, Ferrari R, Boogerd CJ, Schredelseker J, Wang YL, Hunter S, Org T, Zhou J, Li XM, Pellegrini M, Chen JN, Orkin SH, Kurdistani SK, Evans SM, Nakano A, Mikkola HKA. Scl Represses Cardiomyogenesis in Prospective Hemogenic Endothelium and Endocardium. Cell. 2012;150(3):590–605. doi: 10.1016/j.cell.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10(4):252–63. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- Villar D, Berthelot C, Aldridge S, Rayner TF, Lukk M, Pignatelli M, Park TJ, Deaville R, Erichsen JT, Jasinska AJ, Turner JM, Bertelsen MF, Murchison EP, Flicek P, Odom DT. Enhancer evolution across 20 mammalian species. Cell. 2015;160(3):554–66. doi: 10.1016/j.cell.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34(Pt 1):7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- White AK, VanInsberghe M, Petriv OI, Hamidi M, Sikorski D, Marra MA, Piret J, Aparicio S, Hansen CL. High-throughput microfluidic single-cell RT-qPCR. Proc Natl Acad Sci U S A. 2011;108(34):13999–4004. doi: 10.1073/pnas.1019446108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482(7384):221–5. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–19. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson AC, Gottgens B. Transcriptional regulation of haematopoietic stem cells. Adv Exp Med Biol. 2013;786:187–212. doi: 10.1007/978-94-007-6621-1_11. [DOI] [PubMed] [Google Scholar]
- Wilkinson AC, Kawata VK, Schütte J, Gao X, Antoniou S, Baumann C, Woodhouse S, Hannah R, Tanaka Y, Swiers G, Moignard V, Fisher J, Hidetoshi S, Tijssen MR, de Bruijn MF, Liu P, Göttgens B. Single-cell analyses of regulatory network perturbations using enhancer-targeting TALEs suggest novel roles for PU.1 during haematopoietic specification. Development. 2014;141(20):4018–30. doi: 10.1242/dev.115709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Kent DG, Buettner F, Shehata M, Macaulay IC, Calero-Nieto FJ, Sánchez Castillo M, Oedekoven CA, Diamanti E, Schulte R, Ponting CP, Voet T, Caldas C, Stingl J, Green AR, Theis FJ, Göttgens B. Combined Single-Cell Functional and Gene Expression Analysis Resolves Heterogeneity within Stem Cell Populations. Cell Stem Cell. 2015;16(6):712–24. doi: 10.1016/j.stem.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Schoenfelder S, Hannah R, Sánchez Castillo M, Schütte J, Ladopoulos V, Mitchelmore J, Goode DK, Calero-Nieto FJ, Moignard V, Wilkinson AC, Jimenez-Madrid I, Kinston S, Spivakov M, Fraser P, Göttgens B. Integrated genome-scale analysis of the transcriptional regulatory landscape in a blood stem/progenitor cell model. Blood. 2016;127(13):e12–23. doi: 10.1182/blood-2015-10-677393. [DOI] [PubMed] [Google Scholar]
- Woodhouse S, Moignard V, Göttgens B, Fisher J. Processing, visualising and reconstructing network models from single-cell data. Immunol Cell Biol. 2016;94(3):256–65. doi: 10.1038/icb.2015.102. [DOI] [PubMed] [Google Scholar]
- Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117(5):663–76. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- Xu H, Ang YS, Sevilla A, Lemischka IR, Ma’ayan A. Construction and validation of a regulatory network for pluripotency and self-renewal of mouse embryonic stem cells. PLoS Comput Biol. 2014;10(8):e1003777. doi: 10.1371/journal.pcbi.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R, Morita Y, Ooehara J, Hamanaka S, Onodera M, Rudolph KL, Ema H, Nakauchi H. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 2013;154(5):1112–26. doi: 10.1016/j.cell.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Suzuki M, Otsuki A, Shimizu R, Bresnick EH, Engel JD, Yamamoto M. A remote GATA2 hematopoietic enhancer drives leukemogenesis in inv(3)(q21;q26) by activating EVI1 expression. Cancer Cell. 2014;25(4):415–27. doi: 10.1016/j.ccr.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]