Abstract
Zscan4 is an early embryonic gene cluster expressed in mouse embryonic stem and induced pluripotent stem cells where it plays critical roles in genomic stability, telomere maintenance, and pluripotency. Zscan4 expression is transient, and characterized by infrequent high expression peaks that are quickly down-regulated, suggesting its expression is tightly controlled. However, little is known about the protein degradation pathway responsible for regulating the human ZSCAN4 protein levels. In this study we determine for the first time the ZSCAN4 protein half-life and degradation pathway, including key factors involved in the process, responsible for the regulation of ZSCAN4 stability. We demonstrate lysine 48 specific polyubiquitination and subsequent proteasome dependent degradation of ZSCAN4, which may explain how this key factor is efficiently cleared from the cells. Importantly, our data indicate an interaction between ZSCAN4 and the E3 ubiquitin ligase RNF20. Moreover, our results show that RNF20 depletion by gene knockdown does not affect ZSCAN4 transcription levels, but instead results in increased ZSCAN4 protein levels. Further, RNF20 depletion stabilizes the ZSCAN4 protein half-life, suggesting that RNF20 negatively regulates ZSCAN4 stability. Due to the significant cellular functions of ZSCAN4, our results have important implications in telomere regulation, stem cell biology, and cancer.
Keywords: ZSCAN4, RNF20, Proteasome, Lysine-48 ubiquitination, Cancer, Stem cells, Telomeres
1. Introduction
The embryonic gene Zscan4 (Zinc finger and SCAN domain containing 4) promotes telomere and genomic stability in mouse embryonic stem (ES) cells [1]. Knockdown of Zscan4 in mouse ES cells results in telomere shortening and karyotype abnormalities, slowing cell proliferation until reaching culture crisis. Zscan4 is highly but transiently expressed [1], with protein expression bursts that facilitate chromatin remodeling [2,3] and transcriptional reprogramming during the generation of induced pluripotent stem (iPS) cells [4–6]. A short expression burst of Zscan4 was further demonstrated to replace Myc and enhance the efficiency of mouse iPS cell formation through activation of early embryonic genes [4]. The human ZSCAN4 has been shown to interact with factors important for telomere maintenance [7,8], and has been suggested to play a role in cancer [1,8]. Given the important role of ZSCAN4 and its transitory nature in the cell [1,9], maintaining the delicate balance between its protein synthesis and degradation is critical for stem cell and potentially cancer cell function.
Concentrations and spatial gradients of specific proteins must be able to rapidly change in response to extracellular cues and according to current cell state [10]. Small protein imbalances can drastically impact such important cellular processes. Therefore, intracellular protein degradation and turnover play a significant role in cell life cycle [11]. Two major pathways responsible for the degradation of proteins in cells are autophagy and the ubiquitin proteasome system (UPS). Autophagy is the process responsible for degradation of longer lived, structural proteins and organelles. This process depends on the formation of a double membrane autophagosome, which takes up its cargo and subsequently fuses with lysosomes, leading to degradation [12,13]. The canonical UPS is an ATP-dependent degradation pathway [14]. Proteins are marked for proteasomal degradation by ubiquitin, a small 8.5 kDa regulatory protein, which is added to lysine residues of the target protein. Polyubiquitination, or formation of an ubiquitin side chain, specifically on lysine residue 48 of the ubiquitin moieties, targets proteins to the 26S proteasome for degradation. The ubiquitination process involves three steps: activation of the ubiquitin molecule by E1 ubiquitin enzymes, conjugation of ubiquitin to an E2 ubiquitin ligase, and ligation of the ubiquitin molecule to substrate. E3 ubiquitin ligases play a particularly important role as they connote substrate specificity and facilitate the ligation of the ubiquitin molecule to the target protein [15].
Transient expression of high levels of Zscan4 [1] leads to drastic changes in stem cell properties and potency [2,3]. Therefore, stringent regulation of the ZSCAN4 protein is required to effectively control its cellular functions. However, the regulation of human ZSCAN4 protein, and more specifically its turnover dynamics, remains obscure. As a growing body of evidence suggests a significant role for ZSCAN4 in stem cells and cancer, knowledge of its protein regulation is critical. In this study, we demonstrate for the first time that ZSCAN4 protein degradation is regulated by the ubiquitin-proteasome system. Further, we identify the E3 ubiquitin ligase RNF20 as an important negative regulator of ZSCAN4 protein stability.
2. Materials and methods
2.1. Cell culture
Tu167 cells were obtained from the University of Texas MD Anderson Cancer Center (Houston, TX, USA). All cell lines used in this study were grown in complete DMEM medium (Invitrogen) supplemented with 10% fetal bovine serum (Sigma), 2 mM Gluta-MAX, penicillin (100 U/mL), streptomycin (100 µg/mL) and were tested free of mycoplasma.
2.2. RNF20 knockdown by siRNA
Wild type (WT) Tu167 cells were grown in monolayer to 70% confluence and transfected with 25 nM of either ON-TARGETplus individual siRNAs (targeting sequences GCUAAACAGUGGAGAUAAU and GUAUCAUCCUUAAACGUUA) or SMARTpool siRNA reagent targeting human RNF20 (Dharmacon). For non-targeting control conditions, Tu167 cells were transfected with 25 nM MISSION siRNA Fluorescent Universal Negative Control siRNA (Sigma). Cells were transfected using DharmaFECT reagent (Dharmacon) according to manufacturer's instructions. Knockdown was confirmed after 48 h incubation by immunoblot.
2.3. Determination of ZSCAN4 half-life
WT and doxycycline (Dox) inducible tet-ZSCAN4 Tu167 cells were treated with 1 µg/mL Dox (or kept untreated) for 24 h to induce ZSCAN4. For RNF20 knockdown, WT Tu167 cells were transfected for 48 h with either NTC-siRNA, pool RNF20 siRNA or RFN20 siRNA1–2 as described above. Cells were treated with 25 µg/ml CHX (Sigma), for the indicated time points. Total cell lysate in RIPA buffer was loaded on 10% SDS PAGE gel and immunoblotted with ZSCAN4 antibodies (1:1000; Origene) or controls, β-actin (1:1000; Millipore) and Lamin B antibodies (1:2000; Santa Cruz). Band intensities of ZSCAN4 were quantified using ImageJ software [16] and normalized to controls. The relative levels of ZSCAN4 in sample not treated with CHX was considered as initial level of ZSCAN4 and considered as 1 unit. The half-life of ZSCAN4 was determined using formula t1/2 = ln2/k (k is the slope of the degradation curve).
2.4. Autophagy pathway assay
tet-ZSCAN4 Tu167 cells were treated for 24 h with Dox and then autophagy inhibitors: 5 nM of Bafilomycin A1 or 25 µM of Chloroquine (Sigma) were added for 24 h. Whole cell lysate (50 µg) in RIPA buffer was used on 8% SDS-PAGE analyzed by immunoblot to visualize the following antigens: anti-ZSCAN4 (1:1000; Origene), anti-p62 (1:5000; Sigma), anti-LC3 (1:1000; CellSignaling Technology), Anti-Beta Actin (1:10,000; Millipore). All data shown represent at least 3 independent experiments.
2.5. Immunoblot analysis
Nuclear proteins were fractionated using Nuclear Extraction Kit following manufacture's protocol (Active Motif). Total cell lysate was prepared in RIPA buffer and sonicated. For the detection of endogenous ZSCAN4 in Tu167 cells, cells were harvested by accutase (Millipore) and Cytoskeleton buffer (10 mM PIPES, 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2, 1 mM EGTA and 0.5% Triton X100) was used to fractionate cytosolic proteins. Then, pellets were lysed in urea solution (8 M Urea in 0.01 Tris pH 8 + 0.1 M NaH2PO4) and sonicated. Nuclear proteins were electrophoresed in 8% polyacrylamide gels and transferred to a PVDF membrane. Immunoblot was performed using the following primary antibodies: ZSCAN4 (Origene; 1:1000), GAPDH (Santa Cruz; 1:5000), Actin (Sigma; 1:500), Lamin B (Santa Cruz; 1:2000) and with HRP (horseradish peroxidase) conjugated secondary antibodies (Millipore; 1:5000). Protein bands were detected using Pierce ECL Western Blotting Substrate (Thermo Scientific). SuperSignal West Femto (Thermo Scientific) was used to detect endogenous ZSCAN4 in Tu167 cells. All immunoblots shown represent at least 3 independent experiments.
2.6. Ubiquitination assay
WT Tu167 cells were treated with indicated concentration of MG132 for 3–12 h. Cells treated with vehicle only were used as controls. Nuclear protein was isolated using urea extraction buffer. Then, 100 µg of nuclear lysate was diluted in IP-RIPA buffer with 0.1% SDS and denatured by heating to 90 °C for 10 min. Samples were taken for co-immunoprecipitation and immunoblot analyses.
2.7. Co-immunoprecipitation
WT Tu167 were lysed in RIPA buffer and sonicated. Protein A/G beads (Invitrogen) were incubated with 1 µg of anti-ZSCAN4 in 100 µL of RIPA buffer without SDS for 1 h at room temperature. Then, the beads were washed twice with RIPA buffer and crosslinked to the beads with 5 mM BS3 solution (ThermoFisher Scientific) according to manufacturer's protocols. Then the beads were pre-cleared with 100 µg of nuclear lysate in 100 µL IP buffer without SDS (Cell Signaling Technology) overnight at 4 °C. Cell lysates were loaded and bound antigens were eluted with 25 µL RIPA with 0.1% SDS and 25 µL of 2× loading dye. Proteins were separated in 10% SDS PAGE and immunoblotted with corresponding antibodies.
2.8. Immunofluorescence confocal microscopy
Cells were fixed in ice cold methanol/acetic acid (3:1). The fixed cells were dropped on microscope slides. Antigen retrieval was performed followed by blocking for 10 min at room temperature. Primary antibody was incubated overnight at 4 °C for the following antigens: ZSCAN4 (Origene, 1:1000), RNF20 (Cell Signaling, 1:2000), and Ubiquitin Lysine 48 (Millipore 1: 2000). The slides were washed and incubated with secondary antibodies conjugated with Alexa-488 or 568 at room temperature for 1 h and then treated with DAPI and To-Pro-3 to stain the nuclei. Slides were mounted and visualized by a Zeiss 510-confocal microscope. Co-localization analyses were performed by ImageJ software [16].
2.9. Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)
RNA was isolated by Trizol (invitrogene) and 1 µg of total RNA was reverse transcribed by Superscript III (Invitrogen) following the manufacturer's protocol. Then, 10 ng cDNA was used per well in triplicates using SYBR green (Roche) following the manufacturer's protocol. Real time PCR was performed on LightCycler 480 (Roche). Fold induction was calculated by the absolute quantification method. Primers used: ZSCAN4 forward 5′-ATCCACCTGCCTTAGTCCAC-3′ and ZSCAN4 reverse 5′-TCGAAGAACTGTTCCAGCCA-3′, and RPLP0 reverse 5′-CCCATTCTATCATCAACGGGTACAA-3’.
2.10. Statistical analyses
All data and representative images shown are the result of at least three independent experiments, with biological replicates. Student's t-test, one-way or two-way ANOVAs with repeated measures were followed by Tukey's posthoc comparison tests (when appropriate) for statistical analyses. Statistical analyses were performed with STATISTICA-12 and GraphPad-Prism7 software. All P values < 0.05 were considered as statistically significant.
3. Results
3.1. ZSCAN4 protein turnover and half-life
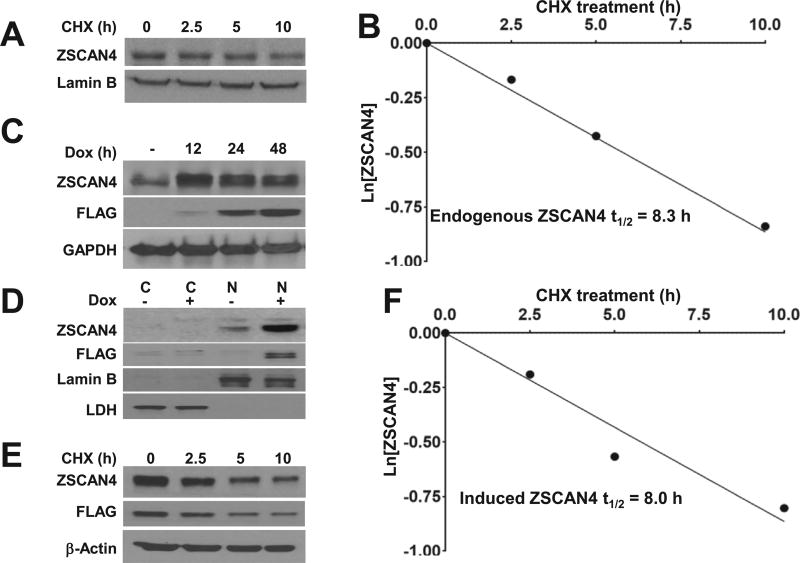
Zscan4 expression is transient and limited to a small fraction of cells, suggesting it requires tight regulation at the protein level. Interestingly, ZSCAN4 has been found to be relatively stable in mouse ES cells, with a half-life of up to 6 h [17]. While normal physiological expression is limited to early stage embryonic cells, ZSCAN4 has been shown to be aberrantly reactivated in cancer [8]. Therefore, we chose to study ZSCAN4 protein stability in the human head and neck cancer cell line Tu167. To determine ZSCAN4 half-life, Tu167 cells were incubated with the protein translation inhibitor cycloheximide (CHX) and ZSCAN4 concentration was monitored by immunoblot over 10 h. The CHX chase assay indicates that ZSCAN4 protein half-life is 8.3 h (Fig. 1A and B).
Fig. 1. Human ZSCAN4 protein half-life.
A. Immunoblot analysis of ZSCAN4 in wild type (WT) Tu167 cells after treatment with cycloheximide (CHX) results in decreased ZSCAN4 protein. B. Protein half-life analysis indicates that endogenous ZSCAN4 protein half-life is 8.3 h. C. Immunoblot in tet-ZSCAN4 cell lines show addition of doxycycline (Dox+) to medium for 0–48 h results in ZSCAN4-FLAG induction in as early as 12 h. D. Cells fractionation to cytoplasmic (C) and nuclear (N) proteins, show the endogenous ZSCAN4 (anti-ZSCAN4) and FLAG-tagged ZSCAN4 (anti-FLAG), are localized to the nucleus. E. Immunoblot in isogenic (Dox+) tet-ZSCAN4 cells after treatment with CHX and F. Protein half-life analyses indicate that the ectopic ZSCAN4 half-life is 8.0 h. Images and results represent data of at least three independent experiments.
ZSCAN4 is weakly expressed in the human cell line Tu167. Therefore, in order to further study ZSCAN4 in these cells, we developed a doxycycline (Dox) inducible ZSCAN4 lentiviral vector. We used this vector to generate inducible ZSCAN4 cells (Tu167) and named them tet-ZSCAN4. Addition of Dox, a tetracycline analog, to the media results in ZSCAN4 induction. Our immunoblot analysis reveals that ZSCAN4 is induced in these cells as early as 12 h Fig.1C) and as expected, is localized to the nucleus (Fig. 1D). Using our tet-ZSCAN4 cell line, we then analyzed the stability of the ZSCAN4 protein. Cells were treated for 24 h with Dox and then incubated with CHX. ZSCAN4 concentration was again monitored by immunoblot over 10 h. Our CHX chase assay indicates that the induced ectopic ZSCAN4 protein half-life of 8.0 h is similar to the endogenous ZSCAN4, suggesting its utility as a tool to further study ZSCAN4 stability (Fig. 1E and F). These data together suggest that the stability of the human ZSCAN4 in human cancer cells is similar to that of mouse ES cells [17].
3.2. ZSCAN4 degradation is proteasome dependent
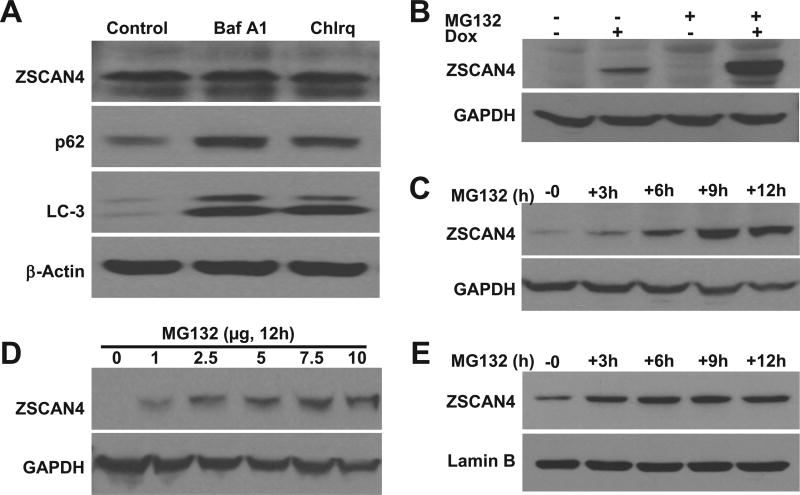
We next sought to determine the pathway responsible for ZSCAN4 turnover and degradation. We first examined the possible clearance of ZSCAN4 by autophagy. tet-ZSCAN4 cells (Tu167) were incubated for 12 h with autophagy inhibitors Bafilomycin A1 and Chloroquine [12]. Our immunoblot analysis shows that both p62 and LC-3, two proteins specifically degraded by autophagy, accumulate in the cells (Fig. 2A). These results indicate that the autophagy pathway was successfully inhibited. However, ZSCAN4 levels remained unchanged (Fig. 2A), excluding autophagy as the mechanism responsible for ZSCAN4 turnover.
Fig. 2. ZSCAN4 degradation is proteasome dependent.
A. Immunoblot analyses indicate that inhibition of autophagy with Bafilomycin A1 and Chloroquine results in accumulation of autophagy targets p62 and LC-3. However, ZSCAN4 was not affected. B. Induction of ZSCAN4 by Dox followed by treatment with the proteasomal inhibitor MG132 demonstrates ZSCAN4 accumulates in the cells. C–D. Kinetics experiments show that accumulation of ZSCAN4 following by MG132 treatment is time and dose dependent. E. Similar studies performed in WT cells show the accumulation of ZSCAN4 following MG132 treatment.
This prompted us to examine the UPS. We incubated tet-ZSCAN4 cells with the proteasome inhibitor MG132 at multiple time points. Our data indicate a time dependent accumulation of ZSCAN4 through the 12 h incubation (Fig. 2B). Furthermore, we show a time dependence (Fig. 2C) and a dose response (Fig. 2D) to proteasome inhibition by MG132. These data show that UPS inhibition results in ZSCAN4 accumulation and suggests it is the pathway by which ZSCAN4 protein is degraded.
To exclude that the response was specific to the exogenous ZSCAN4 induction, we treated wild type (WT) Tu167 cells with 5uM MG132 for 0–12 h. Our results validate that endogenous ZSCAN4 accumulates with proteasome inhibition (Fig. 2E). These data indicate that both the endogenous and the induced ZSCAN4 proteins are not degraded by autophagy but instead in a proteasome dependent manner.
3.3. ZSCAN4 is marked for proteasomal degradation by lysine 48 ubiquitination
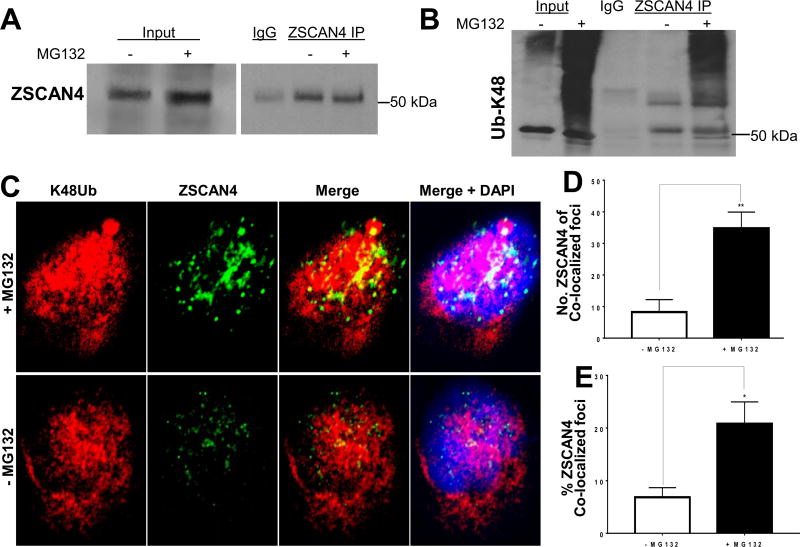
Proteins turnover allows for clearance of non-functional proteins. Proteins destined for degradation by the proteasome are tagged by ubiquitin side chains [18]. Lysine 48 ubiquitin side chains, also known as K48 polyubiquitination, connote specificity to proteasomal degradation [15]. Therefore, we next examined if ZSCAN4 is lysine 48 polyubiquitinated by inhibiting the proteasome with MG132 for 12 h in WT Tu167 cells. Then, we selected for covalently bound K48 ubiquitin chains by performing ZSCAN4 IP experiments in denaturing conditions. Following ZSCAN4 IP, we immunoblotted with a lysine 48 ubiquitin (K48-Ub) antibody and observed a strong ubiquitin signal at the 50 kDa molecular weight of ZSCAN4 (Fig. 3A). Moreover, a significant smear above the 50 kDA band indicates ZSCAN4 with bound ubiquitin side chains of varying lengths (Fig. 3B).
Fig. 3. ZSCAN4 is Lysine 48 (K48Ub) polyubiquitinated.
A. Endogenous ZSCAN4 immunoprecipitation (IP) in treated (+MG132) or untreated (−MG132) WT cells followed by ZSCAN4 immunoblot. B. Denaturing conditions where used followed by ZSCAN4-IP and immunoblot with anti-Lysine 48 (K48) ubiquitin (anti-K48Ub). MG132 treated cells indicate that ZSCAN4 is polyubiquitinated. Untreated cells were used as controls. C. Co-immunostaining and confocal microscopy analyses in WT Tu167 cells treated with MG132 (+MG132) or untreated (−MG132) using anti-ZSCAN4 (green) and anti-K48 ubiquitin (red) indicates the ZSCAN4 overlaps with K48Ub. D. Colocalization analyses with ImageJ show an increase in the number of ZSCAN4 foci colocalized with K48Ub and E. the percent (%) of K48Ub colocalized ZSCAN4 foci (n = 6 per group and average of >300 foci per group). Asterisks indicate * p < 0.05, **p < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To further validate the lysine 48 polyubiquitination of ZSCAN4 protein, WT Tu167 cells were treated with or without MG132 for 5 h and co-immunostained for anti-K48-Ub and anti-ZSCAN4 (Fig. 3C). Co-localization analysis indicates that about 5% of ZSCAN4 co-localizes with K48-Ub in the untreated conditions, whereas this fraction is 4 fold higher following 5 h inhibition of the proteasome with MG132 (Fig. 3D and E). Our data suggest that ZSCAN4 is targeted to the proteasome via canonical lysine 48 polyubiquitination.
3.4. The E3 ubiquitin ligase RNF20 negative regulates ZSCAN4 protein stability
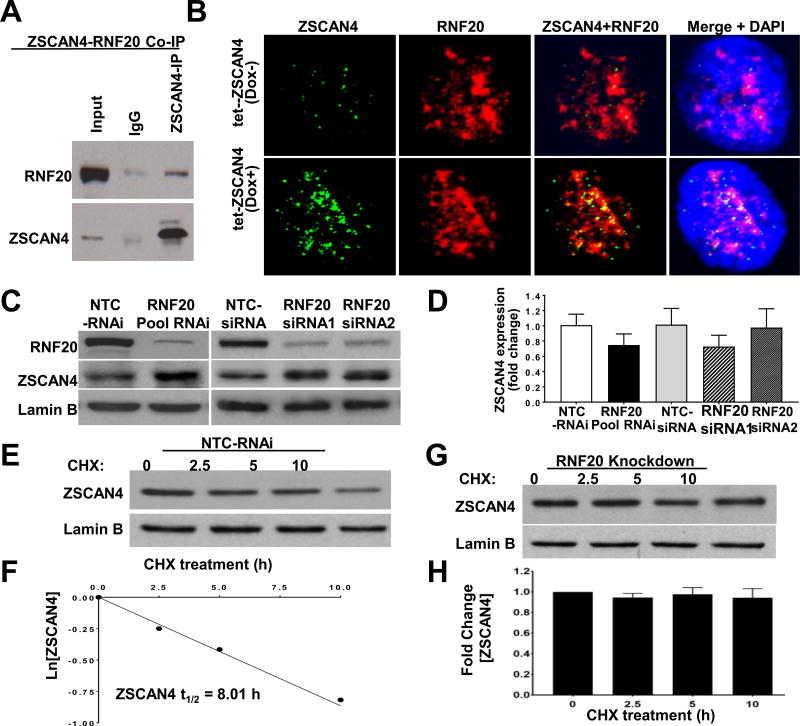
To further characterize the ZSCAN4 ubiquitination mechanism, we performed ZSCAN4 immunoprecipitation (IP) followed by mass spectrometry. Ranking atop the list of interacting proteins, we found the E3 ubiquitin ligase RNF20. Co-IP for ZSCAN4 followed by RNF20 immunoblot validates that ZSCAN4 associates with RNF20 (Fig. 4A). Immunofluorescence staining followed by confocal microscopy confirm the co-localization between ZSCAN4 and RNF20, which also increases after ZSCAN4 induction (Fig. 4B).
Fig. 4. The E3 ubiquitin ligase RNF20 negatively regulates and interacts with ZSCAN4.
A. Co-IP of ZSCAN4 and immunoblot with anti-RNF20 displays interaction. B. Confocal microscope images in untreated tet-ZSCAN4 cells (Dox−) show co-localization of RNF20 (red) and ZSCAN4 (green). DNA is stained with DAPI (blue). Furthermore, co-localization increases following ZSCAN4 induction (Dox+). C. Immunoblot analyses show successful RNF20 knockdown by SMARTpool siRNA and two different siRNA (RNAi 1 and RNAi 2) compared to non-template controls (NTC). Conversely, RNF20 depletion results in a significant increase in ZSCAN4. D. qRT-PCR analysis in RNF20 depleted by the indicated siRNAs show no significant change in ZSCAN4 expression compared to NTC-siRNA controls. E–F. Protein half-life analyses after treatment with CHX and immunoblot with corresponding antibodies indicate that NTC-siRNA do not alter the ZSCAN4 protein half-life which remains about 8 h, yet, as shown in G–H. Knockdown of RNF20 (pool siRNA) leads to stabilization of ZSCAN4 protein. All data shown represent three independent experiments. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
RNF20, also known as BRE1, is an E3 ubiquitin ligase that has been implicated in protein degradation [19]. Therefore, we next sought t to study the effect of RNF20 depletion on ZSCAN4 protein stability. WT Tu167 cells were transfected with RNF20 RNAi (siRNA) for 48 h and compared to isogenic cells transfected with non-targeting control (NTC-siRNA). Interestingly, our immunoblot analyses reveal that depletion of RNF20 by SMARTpool siRNA, or independently by two different RNF20 siRNA sequences, results in a significant increase in ZSCAN4 protein when compared to NTC-siRNA (Fig. 4C). Conversely, our quantitative reverse transcription PCR (qRT-PCR) analysis in the RNF20 knockdown cells excludes transcriptional regulation of ZSCAN4 by RNF20 (Fig. 4D). These results were validated in multiple independent experiments in triplicatesm and suggest that RNF20 regulates ZSCAN4 levels in a post-translational manner.
To determine the effect of RNF20 depletion on ZSCAN4 half-life, we preformed protein half-life analysis by CHX following RNF20 knockdown (pool siRNA) in WT Tu167 cells. As expected, the half-life of ZSCAN4 remains at 8.1 h in NTC-siRNA transfected cells (Fig. 4E and F). However, our data indicate that RNF20 knockdown stabilizes ZSCAN4, as protein levels remained stable for the entire duration of the assay (10 h) (Fig. 4G and H). Data was reproduced in three independent experiments (n = 3; p < 0.001). These results suggest that RNF20 depletion leads to accumulation of ZSCAN4 protein and indicate that RNF20 negatively regulates ZSCAN4 protein stability.
4. Discussion
ZSCAN4 plays a significant role in ES telomere regulation and genomic stability. ZSCAN4 has been indicated as a nuclear reprogramming factor [2], and only a short induction restores the developmental potency of ES cells. Previous reports have suggested that the human ZSCAN4 is aberrantly activated in cancer [1], and interacts with the shelterin components important for telomere stability [7,8]. However, to date, little is known about ZSCAN4 protein turnover or regulation.
Previous studies have shown that transient ZSCAN4 expression must be tightly controlled in order to exert its significant role in preservation of stem cell lifespan. This suggests the importance of regulating ZSCAN4 not just transcriptionally, but more importantly at the protein level. Here we show for the first time, the specific regulation of ZSCAN4 protein turnover and the pathway through which this is achieved. Our ZSCAN4 degradation assay demonstrated that the half-life of ZSCAN4 is 8 h. We show that ZSCAN4 is not degraded by autophagy, but instead is regulated by a proteasome dependent pathway. Our data reveal that inhibition of the proteasome results in accumulation of ZSCAN4 in a time and dose dependent manner. Proteins are targeted to the proteasome for degradation by ligation of ubiquitin side chains [18,20]. Specifically, chains linked through lysine 48 ubiquitination serve as the signal for proteasomal degradation. Our Co-IP assays confirm that ZSCAN4 is lysine 48 polyubiquitinated. These data are further validated by co-localization studies between ZSCAN4 and lysine 48 specific ubiquitin. Thus, we infer that the UPS plays a significant role in modulating ZSCAN4 levels and activity in the cell.
The enzymes responsible for specific ubiquitination of proteins are termed E3 ubiquitin ligases. E3 ubiquitin ligases facilitate the transfer and of ubiquitin moieties by bringing E2 ubiquitin conjugating enzymes and substrate into close contact [20]. Our ZSCAN4 pull down assay followed by mass spectrometry identifies RNF20 as an interacting E3 ubiquitin ligase. RNF20, also known as BRE1, has been implicated in epigenetic regulation [21,22] and as a putative tumor suppressor in cancer [23]. RNF20 was also shown to promote the polyubiquitination and proteasomal degradation of proteins [19]. Our data confirm the interaction between ZSCAN4 and RNF20 through co-immunostaining and co-IP. Importantly, our results show that depletion of RNF20 does not affect ZSCAN4 RNA transcription, yet it leads to the accumulation and stabilization of ZSCAN4 protein, suggesting it as a novel negative regulator of ZSCAN4 protein stability. Due to the significant role of ZSCAN4 in the generation of iPS cells, our results have important implications in telomere and genomic stability regulation, stem cell biology, and cancer.
Supplementary Material
Acknowledgments
This work was supported in part by the Maryland Stem Cell Research Fund (MSCRF) grant 2013-MSCRFII-0165 and the National Institutes of Health/ NINDS grant 1R21 NS095088-01A1 (MZ). We thank the Weitzman family for their contribution and support of this work. RK was partially supported by the MSCRF grant 2014-MSCRF-0719.
Abbreviations
- UPS
ubiquitin-proteasome system
- ES cells
embryonic stem cells
- iPS cells
induced pluripotent stem cells
- Dox
doxycycline
- CHX
cycloheximide
- co-IP
co-immunoprecipitaiton
- kDA
kilo Dalton
- K48Ub
lysine-48 Ubiquitination
- DAPI
diamidino-2-phenylindole
- Cyto
cytoplasmic cell fraction
- Nuc
nuclear cell fraction
Footnotes
Author's contributions
RK, BAP and MZ conceived the project and designed the experiments. MZ supervised the project and secured funding. BAP, RK, WAM, JMM conducted the experiments, analyzed the data and generated the figures. BAP drafted the initial manuscript and all authors were involved in reviewing and editing the manuscript.
Transparency document
Transparency document related to this article can be found online at https://doi.org/10.1016/j.bbrc.2018.02.155.
References
- 1.Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, Wersto RP, Ko MSH. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–U866. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akiyama T, Xin L, Oda M, Sharov AA, Amano M, Piao Y, Cadet JS, Dudekula DB, Qian Y, Wang W, Ko SB, Ko MS. Transient bursts of Zscan4 expression are accompanied by the rapid derepression of heterochromatin in mouse embryonic stem cells. DNA Res. 2015;5:307–318. doi: 10.1093/dnares/dsv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amano T, Hirata T, Falco G, Monti M, Sharova LV, Amano M, Sheer S, Hoang HG, Piao Y, Stagg CA, Yamamizu K, Akiyama T, Ko MS. Zscan4 restores the developmental potency of embryonic stem cells. Nat. Commun. 2013;4:1966. doi: 10.1038/ncomms2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirata T, Amano T, Nakatake Y, Amano M, Piao Y, Hoang HG, Ko MS. Zscan4 transiently reactivates early embryonic genes during the generation of induced pluripotent stem cells. Sci. Rep. 2012;2:208. doi: 10.1038/srep00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang J, Lv W, Ye X, Wang L, Zhang M, Yang H, Okuka M, Zhou C, Zhang X, Liu L, Li J. Zscan4 promotes genomic stability during reprogramming and dramatically improves the quality of iPS cells as demonstrated by tetraploid complementation. Cell Res. 2013;23:92–106. doi: 10.1038/cr.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park HS, Hwang I, Choi KA, Jeong H, Lee JY, Hong S. Generation of induced pluripotent stem cells without genetic defects by small molecules. Biomaterials. 2015;39:47–58. doi: 10.1016/j.biomaterials.2014.10.055. [DOI] [PubMed] [Google Scholar]
- 7.Lee K, Gollahon LS. ZSCAN4 and TRF1: a functionally indirect interaction in cancer cells independent of telomerase activity. Biochem. Biophys. Res. Commun. 2015;466:644–649. doi: 10.1016/j.bbrc.2015.09.107. [DOI] [PubMed] [Google Scholar]
- 8.Lee K, Gollahon LS. Zscan4 interacts directly with human Rap1 in cancer cells regardless of telomerase status. Canc. Biol. Ther. 2014;15:1094–1105. doi: 10.4161/cbt.29220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T, Ko MS. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev. Biol. 2007;307:539–550. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010;584:1393–1398. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- 11.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nature reviews, Mol. Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 12.Eskelinen EL, Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim. Biophys. Acta. 2009;1793:664–673. doi: 10.1016/j.bbamcr.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Q, Figueiredo-Pereira ME. Ubiquitin/proteasome pathway impairment in neurodegeneration: therapeutic implications, Apoptosis. Int. J. Programmed Cell Death. 2010;15:1292–1311. doi: 10.1007/s10495-010-0466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 16.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Storm MP, Kumpfmueller B, Bone HK, Buchholz M, Sanchez Ripoll Y, Chaudhuri JB, Niwa H, Tosh D, Welham MJ. Zscan4 is regulated by PI3-kinase and DNA-damaging agents and directly interacts with the transcriptional repressors LSD1 and CtBP2 in mouse embryonic stem cells. PLoS One. 2014;9:e89821. doi: 10.1371/journal.pone.0089821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W, Ye Y. Polyubiquitin chains: functions, structures, and mechanisms, Cellular and molecular life sciences. CMLS. 2008;65:2397–2406. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren P, Sheng Z, Wang Y, Yi X, Zhou Q, Zhou J, Xiang S, Hu X, Zhang J. RNF20 promotes the polyubiquitination and proteasome-dependent degradation of AP-2alpha protein. Acta Biochimica et Biophysica Sinica. 2014;46:136–140. doi: 10.1093/abbs/gmt136. [DOI] [PubMed] [Google Scholar]
- 20.Nandi D, Tahiliani P, Kumar A, Chandu D. The ubiquitin-proteasome system. J. Biosci. 2006;31:137–155. doi: 10.1007/BF02705243. [DOI] [PubMed] [Google Scholar]
- 21.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 22.Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 23.Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, Hublarova P, Moyal L, Gana-Weisz M, Shiloh Y, Yarden Y, Johnsen SA, Vojtesek B, Berger SL, Oren M. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 2008;22:2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.