Abstract
A cDNA clone encoding the Prx7 peroxidase from barley (Hordeum vulgare L.) predicted a 341-amino acid protein with a molecular weight of 36,515. N- and C-terminal putative signal peptides were present, suggesting a vacuolar location of the peroxidase. Immunoblotting and reverse-transcriptase polymerase chain reaction showed that the Prx7 protein and mRNA accumulated abundantly in barley coleoptiles and in leaf epidermis inoculated with powdery mildew fungus (Blumeria graminis). Two isoperoxidases with isoelectric points of 9.3 and 7.3 (P9.3 and P7.3, respectively) were purified to homogeneity from barley coleoptiles. P9.3 and P7.3 had Reinheitszahl values of 3.31 and 2.85 and specific activities (with 2,2′-azino-di-[3-ethyl-benzothiazoline-6-sulfonic acid], pH 5.5, as the substrate) of 11 and 79 units/mg, respectively. N-terminal amino acid sequencing and matrix-assisted laser desorption/ionization time-of-flight mass-spectrometry peptide analysis identified the P9.3 peroxidase activity as due to Prx7. Tissue and subcellular accumulation of Prx7 was studied using activity-stained isoelectric focusing gels and immunoblotting. The peroxidase activity due to Prx7 accumulated in barley leaves 24 h after inoculation with powdery mildew spores or by wounding of epidermal cells. Prx7 accumulated predominantly in the epidermis, apparently in the vacuole, and appeared to be the only pathogen-induced vacuolar peroxidase expressed in barley tissues. The data presented here suggest that Prx7 is responsible for the biosynthesis of antifungal compounds known as hordatines, which accumulate abundantly in barley coleoptiles.
The structurally diverse secretory plant peroxidases constitute a class within the superfamily of peroxidases (EC 1.11.1.7; H2O2 oxidoreductase) (Welinder, 1992). Each group of structural relatives in the superfamily is likely to be involved in specific and distinct biosynthetic pathways. Plant development and environmental changes, including biotic stress, are often followed by dramatic changes in peroxidase activity and in the number of isoperoxidases present in specific tissues. Peroxidases participate in a variety of plant defense mechanisms (Moerschbacher, 1992) in which H2O2 is often supplied by an oxidative burst, a common event in defense responses (Lamb and Dixon, 1997). The cell wall appears to be a major site for defense-related peroxidase polymerization reactions such as lignification (Hammerschmidt and Kuc, 1982), suberization (Espelie et al., 1986), cross-linking of structural cell wall proteins (Bradley et al., 1992), and dimerization of ferulate esters (Ikegawa et al., 1996).
Appositions at the cell wall known as papillae are induced in the pathogenic interaction between barley (Hordeum vulgare L.) and the biotrophic powdery mildew fungus (Blumeria [syn. Erysiphe] graminis f.sp. hordei) (Aist and Bushnell, 1991; Collinge et al., 1997) and are a race-nonspecific defense component of major importance. The papilla response is well described at the cellular and biochemical levels, with active phenylpropanoid biosynthesis (Carver et al., 1994) and early accumulation of peroxidase mRNA (Boyd et al., 1994), apparently important processes for inhibition of the fungus.
Several proteins, such as peroxidases (Scott-Craig et al., 1995), accumulate in the papillae and in the leaf (Kerby and Somerville, 1989; Thordal-Christensen et al., 1992). The papillae are surrounded by an autofluorescent halo, which is most likely due to the presence of phenolic compounds. There is also evidence suggesting that these phenolics are polymerized p-coumarylagmatine and p-coumarylhydroxyagmatine (Wei et al., 1994; von Röpenack et al., 1998). Hordatines, soluble dimers of the hydroxycinnamic acid amides p-coumarylagmatine and ferulylagmatine, have antifungal activity and can be isolated from barley coleoptiles (Stoessl, 1967) or dark-grown seedlings (Stoessl and Unwin, 1970). Coleoptiles are resistant to invasion by Cochliobulus sativus ([Ito and Kurib.] Drechs. ex Dastur), allowing the seedling to remain free from infection for the first few days after emergence. There is correlative evidence that hordatines are the mediator of this resistance (Stoessl, 1967, and refs. therein). Hordatines also accumulate slowly but substantially in the barley leaf in response to powdery mildew fungus attack (Smith and Best, 1978). Stoessl (1967) showed that hordatines could be generated in vitro by horseradish peroxidase from the appropriate p-coumarylagmatines, suggesting that a peroxidase mediates the last step in the biosynthetic pathway to hordatine.
The barley Prx7 peroxidase was first cloned as a partial cDNA from dark-grown barley (Rasmussen et al., 1991a). The gene mapped tightly to the MlLa powdery mildew resistance locus on chromosome 2 (Giese et al., 1993). Because it was the seventh peroxidase gene to be mapped in barley, the gene was designated Prx7 and the encoded protein Prx7. Prx7 mRNA has been shown to accumulate in response to pathogens in barley leaves (Thordal-Christensen et al., 1992) and roots (Valé et al., 1994) and also in the barley cell-elongation mutant Slender (Schünmann et al., 1994). In the present study we describe the cDNA cloning, purification, and characterization of Prx7.
MATERIALS AND METHODS
Plant and Fungal Material
Barley (Hordeum vulgare L. cv Pallas near-isogenic line P-02; Kølster et al., 1986) plants were grown in a 16-h photoperiod at 220 μE m−2 s−1, at 18°C day/16°C night temperatures, and at a constant (65%) RH. Coleoptiles were cut from 7-d-old barley seedlings. The barley powdery mildew fungus isolates C15 and A6 were used to inoculate plants and were maintained on cv Manchuria. The primary leaves of 7-d-old barley plants were held down on a horizontal plexiglass plate to expose the abaxial epidermis for inoculation. Inoculation was performed in a settling tower using pressurized air to release spores from the conidial chains. Wounding of 7-d-old primary leaves was done by gently rubbing powdered silicon carbide (no. 2FE coarse, Carborundum Company, Manchester, UK) onto the leaves. Epidermal strips and the remaining mesophyll from primary barley leaves were obtained by removing spores from the abaxial epidermis with moist cotton balls. An incision was then made through the adaxial epidermis and mesophyll. The cut side of the epidermis was immobilized on electrical tape, and the intact inoculated epidermis was stripped off. Epidermal strips and mesophyll were immediately frozen in liquid nitrogen.
Cloning of pcD1311BE
A barley leaf cDNA library in λ phage (λZAP-XR, Stratagene) (Scott-Craig et al., 1995) was screened under low-stringency conditions with 5× SSPE (20× SSPE is 3 m NaCl, 0.2 m NaH2PO4, and 0.02 m EDTA), by probing with the partial clone (pcD1311, accession no. X62438) as described previously (Rasmussen et al., 1991a). One positive plaque was purified after a wash at high stringency with 0.1× SSPE. The 1316-bp BlnI/EcoRI fragment of the insert was blunt-ended and subcloned into the SmaI site of pUC13. This clone was designated pcD1311BE (accession no. AJ003141).
DNA Sequencing and RNA and DNA Manipulations
Sequencing was carried out on a DNA sequencer (model ABI 373, Perkin-Elmer) using a dye-terminator cycle-sequencing kit (Thermo Sequenase, Amersham). Both strands were sequenced using internal primers. Sequences were proofread using Sequencher software (version 3.0, Gene Codes Corporation, Ann Arbor, MI). Plasmids for sequencing were prepared using the Wizard kit (Promega). Restriction digests and other standard procedures were as instructed by the manufacturers or by using the method of Sambrook et al. (1989). Total RNA was prepared from frozen tissue samples by the method of Chirgwin et al. (1979). RT-PCR using beads (Ready-to-Go, Amersham-Pharmacia Biotech, Allerød, Denmark) was essentially as described by the manufacturer.
Before reverse transcription, 0.8 μg of total RNA was denatured in the presence of 0.5 μg of oligo-T and 1 unit/μL RNasin (Promega) in a volume of 33 μL for 5 min at 95°C. The RNA/oligo-T mixture was then divided and transferred to three PCR tubes for separate amplification of GAPDH, Prx7, and Prx8 mRNA, each in a final volume of 50 μL. Negative controls in which the murine leukemia virus RT had been heat inactivated were used to check for DNA contamination, and no fragments were amplified in these controls. Reverse transcription was carried out without amplification primers at 42°C for 20 min. After the addition of the relevant primers and initial denaturing at 95°C for 5 min, PCR was carried out for 25 cycles using the following protocol: denaturing at 94°C for 45 s, annealing at 58°C for 45 s, and elongation at 72°C for 1 min. The primers TH1 (5′-TATCTCTCACATGTCAGCGGC-3′) and TH2 (5′-TACTACTTCGACCTGATCGCG-3′) were used to amplify a 277-bp fragment from Prx7 mRNA. The primers RT-Prx8F (5′-TGTTCAACAACGACACCACC-3′) and RT-Prx8R (5′-CATTCACGTGTCGTGCTAGC-3′) were used for Prx8 mRNA to generate a 222-bp fragment. GAPDH1 (5′-CAAGGACTGGAGRGGTGG-3′) and GAPDH2 (5′-CCCACTCGTTGTCRTACC-3′) were used to amplify a 376-bp fragment from mRNA corresponding to the barley GAPDH gene as an internal positive control.
Construction of Recombinant Prx7 Plasmid and Expression in Escherichia coli
The blunt-ended BglI-DraI fragment of pcD1311BE was ligated into the blunt-ended BamHI site of the pET15b expression vector (Novagen, Madison, WI), and the orientation was confirmed by sequencing. E. coli BL21 (DE3) pLysS harboring the recombinant Prx7 (rPrx7) construct was grown in superbroth (32 g of tryptone, 20 g of yeast extract, and 5 g/L NaCl, pH 7.0) at 37°C. Induction of rPrx7 expression involved the addition of isopropyl-β-d-thiogalactoside to 0.5 mm at an A600 of 10 to 12. Cells were harvested after an additional 4 h of incubation and lysed by two freeze-thaw cycles, followed by sonication in the presence of denaturing buffer I (8 m urea, 100 mm NaH2PO4, and 10 mm Tris-HCl, pH 8.0). Solubilized His-tagged rPrx7 was purified from the supernatant by Ni+-nitrilotriacetic acid agarose (Qiagen, Hilden, Germany) affinity chromatography as described by the manufacturer. His-tagged protein was desalted on a P-6 Bio-Gel column (Bio-Rad) and bound on a Resource-Q column (Amersham-Pharmacia Biotech). Protein was eluted with a linear gradient of 20 mm Tris-HCl, pH 8.6, 4 m urea, and 1 m NaCl in the loading buffer. The purity of the rPrx7 fractions was verified by SDS-PAGE.
Production of Polyclonal Antibodies
Anti-rPrx7 antibodies were raised in Danish White rabbits (Dako, Glostrup, Denmark). Immunoglobulins in blood serum were purified on protein A-agarose beads (KemEnTec, Copenhagen, Denmark) as described by Harlow and Lane (1988).
Purification of Peroxidases from Coleoptiles
Coleoptiles (75 g fresh weight) were ground to a fine powder in liquid nitrogen using a mortar and pestle. The powder was extracted for 2 h with 500 mL of buffer II (50 mm Tris-HCl, pH 8.0, 0.5 m NaCl, and 0.2 mm PMSF) at 4°C, filtered, and centrifuged for 30 min at 16,000g. Protein in the cleared crude extract was precipitated by the addition of saturated (NH4)2SO4 to 60% saturation and incubated for 4 h. The precipitate was collected by centrifugation at 15,000g for 40 min, redissolved in 60 mL of deionized water, and dialyzed overnight against 2× 5 L of dialysis buffer C (10 mm Na-acetate, pH 5.2, and 1 mm CaCl2). The remaining peroxidase in the (NH4)2SO4 supernatant was batch adsorbed to 20 mL of Phenyl-Sepharose CL4B medium (Amersham-Pharmacia Biotech). Bound protein was eluted stepwise using buffer III (15 mm Na-acetate, pH 4.75). The first 120 mL eluted was dialyzed against 10 mm Na-acetate, pH 5.2, and 1 mm CaCl2, and the volume was adjusted to 200 mL. All steps were performed at 4°C or on ice unless indicated otherwise.
Column-chromatography purification steps were performed on a fast-protein liquid chromatography system (Amersham-Pharmacia Biotech) at room temperature. Dialyzed protein was loaded separately onto a 6-mL Resource-S column, and bound proteins were eluted with a linear gradient of buffer IV (15 mm Na-acetate, pH 4.75, and 1.0 m NaCl). Unbound activities were purified on a 1-mL Resource-Q column in 50 mm Tris-HCl, pH 8.0, and eluted with a linear gradient of buffer V (50 mm Tris-HCl, pH 8.0, and 1.0 m NaCl). Fractions with peroxidase activity were pooled and prepared for HIC by adding (NH4)2SO4 to a final concentration of 1.5 m. HIC was performed on a 1.7-mL phenyl ether column (POROS-PE, PerSeptive Biosystems, Framingham, MA) equilibrated in 50 mm Tris-HCl, pH 8.0, and 1.5 m (NH4)2SO4, and the bound peroxidases were eluted with a linear gradient of 50 mm Tris-HCl, pH 8.0. Fractions were analyzed by electrophoresis (IEF, SDS-PAGE, and immunoblotting).
N-Terminal Amino Acid Sequencing
Purified P9.3 and P7.3 were run on SDS-PAGE, blotted onto a PVDF membrane, and stained with Coomassie Brilliant Blue R-250. Bands were cut from the membrane and the proteins were deblocked by cleavage of the N-terminal pyroglutamic acid with pyroglutamyl amino peptidase (EC 3.4.19.3) (Unizyme Laboratories, Hørsholm, Denmark) as described previously (Hirano et al., 1993). Amino acid sequencing was performed on a sequencer (model 477A, Applied Biosystems) with an on-line phenylthiohydantoin amino acid analyzer.
MS and Tryptic in-Gel Digestion
After SDS-PAGE, the protein band from the Coomassie Blue-stained gel was excised, washed in 200 μL of 100 mm NH4HCO3, and treated with 3 mm DTT (Sigma) and 100 mm NH4HCO3 in 150 μL at 60°C for 30 min. The protein was alkylated by the addition of 10 μL of 100 mm iodoacetamide (Merck, Darmstadt, Germany) and incubation for 30 min in the dark at room temperature. The solvent was discarded and the gel piece was washed in 500 μL of 50% (v/v) CH3CN (Merck) and 100 mm NH4HCO3 for 1 h. The gel pieces were dehydrated in 50 μL of CH3CN for 10 min, dried under vacuum, and reswelled for 10 min with 10 μL of 25 mm NH4HCO3 containing 0.2 μg of trypsin (EC 3.4.21.4) (Sigma). After the addition of 20 μL of 25 mm NH4HCO3, the digestion was carried out overnight at 37°C. The supernatant was saved and the peptides were extracted from the gel slices twice with 50 μL of 60% CH3CN and 0.1% (v/v) trifluoroacetic acid (Sigma) for 20 min. The supernatant and extracts were combined and dried under vacuum. Peptides were reconstituted in 0.1% (v/v) trifluoroacetic acid and mixed 1:1 (v/v) with the matrix (33 mm α-cyano-4-hydroxy-trans-cinnamic acid [MALDI grade] from Hewlett-Packard). The mass spectra of the peptides were recorded in the range up to 104 D in a linear positive mode with an acceleration voltage of 28 kV using a MALDI-TOF MS system (model G2025A, Hewlett-Packard) with a sampling rate of 200 MHz. All spectra were recorded at a tube pressure of less than 9 × 10−6 torr. To analyze the whole protein, sinapinic acid (Hewlett-Packard) was used as the matrix and the mass spectra were recorded in the range up to 105 D.
Enzyme Assays
A rapid spot test for peroxidase activity based on a guaiacol substrate (80 mm guaiacol [Sigma], 80 mm H2O2 [Merck], and 50 mm Na-acetate, pH 5.5) was used for visual monitoring of column fractions. Substrate (10 μL) was spotted onto Parafilm (American National Can, Greenwich, CT), followed by 5 μL of the sample fraction. Activity was estimated visually. Specific activities were measured at 20°C on an ELISA plate scanner (Titertek Multiscan MKII, Flow Laboratories, Lugano, Switzerland) equipped with a 405-nm filter using 100 mm Na-acetate, pH 5.5, 0.36 mm ABTS (Sigma), and 6 mm H2O2 as substrate. The increase in absorbance was followed for 90 s at 10-s intervals. In each reaction, 0.035 to 4.5 μg of protein (based on A280) was analyzed. The absorption coefficient for the ABTS product used was 36.0 mm−1 cm−1 (Childs and Bardsley, 1975). Absorption spectra and Reinheitszahl (A403–408/A280) values were determined using a spectrophotometer (model UV2101PC, Shimadzu Scientific Instruments, Columbia, MD, or model M4QII, Zeiss).
Protein Sample Preparation
Total protein was prepared from barley tissues by homogenizing frozen samples in liquid nitrogen using a mortar and pestle. The frozen tissue powder was extracted with 2 volumes (v/w) of ice-cold buffer (50 mm K-acetate, pH 5.2, 100 mm KCl, 1 m NaCl, 1 mm CaCl2, 1 mm ascorbic acid, 0.1% Triton X-100, and 0.2 mm PMSF) followed by centrifugation at 15,000g for 10 min at 4°C and measurement of protein concentration. For IEF the supernatant was dialyzed against 3 mm Na-acetate, pH 5.2, containing 1 mm CaCl2 followed by lyophilization and measurement of protein concentration. Extraction of extracellular proteins was performed by vacuum infiltration and low-speed centrifugation as described previously (Kerby and Somerville, 1989) using 50 mm K-acetate, pH 5.2, and 100 mm KCl as an infiltration buffer. This extraction method does not lead to leakage of intracellular protein into the washing fluid (Kristensen et al., 1997). The extracted washing fluid and all other protein samples were adjusted to 1 mm CaCl2 and 1 mm ascorbic acid before storage at −20°C.
Protein Determination
Protein was quantified in duplicate by spectrophotometry using A2800.1% (w/v) = 1 cm−1 or according to the method of Bradford (1976), with IgG as the standard protein (Bio-Rad).
Electrophoretic Protein Analyses and Immunoblotting
Proteins separated by SDS-PAGE (Laemmli, 1970) were stained with Coomassie Blue G-250 or electroblotted using a semidry apparatus (Kyhse-Andersen, 1984) onto 0.45-μm PVDF membranes (Millipore) for sequencing or onto nitrocellulose membranes (Amersham) for immunodetection. Membranes were probed with anti-rPrx7 polyclonal rabbit antibodies diluted 1:10,000 in TBST (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 0.05% [v/v] Tween 20). The secondary antibody was alkaline phosphatase-conjugated anti-rabbit goat antibody (Stratagene) diluted 1:2,000 in TBST. The blots were washed and developed (Harlow and Lane, 1988) with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate as substrates (Sigma). IEF was performed as described previously (Kristensen et al., 1997) and stained for peroxidase activity using 3-amino-9-ethylcarbazole (Sigma) as the substrate (Kerby and Somerville, 1989).
RESULTS
Cloning and Sequencing of a Full-Length Prx7 cDNA
A partial cDNA clone, pcD1311, had previously been obtained by heterologous probing with the cDNA clone pcR7 corresponding to barley grain peroxidase BP1 (Rasmussen et al., 1991a). In the present study we used this partial clone as a probe on a λ-phage cDNA library prepared from barley leaves inoculated with powdery mildew (Scott-Craig et al., 1995). One full-length clone, pcD1311BE, was isolated and the 700-bp sequence in the 3′ end of the DNA was found to be identical to pcD1311. The open reading frame starts 93 bases from the 5′ end of the cDNA clone and encodes a 341-amino acid protein with a calculated Mr of 36,515. We designated this protein Prx7 in accordance with the restriction fragment-length polymorphism mapping history of the partial clone (Giese et al., 1993). The C-terminal amino acid sequence of the protein derived from pcD1311 (Rasmussen et al., 1991a) does not correspond to the C-terminal sequence of Prx7 derived from pcD1311BE because of an artifact in the sequencing of the pcD1311 clone, in which a base was inserted, resulting in a +1 reading frame shift.
All of the domains characteristic of secretory plant peroxidases (Welinder, 1992) are highly conserved in Prx7. The functions of these domains are consistent with the properties of purified Prx7. In addition to these domains, there are two consensus putative Asn-glycosylation sites at residues 91 and 176 (Fig. 1). Like all plant peroxidase precursors, the uncleaved form of Prx7 has an N-terminal putative ER-targeting peptide. The cleavage site between residue 24 (G) and 25 (Q) can be deduced from alignment with HRP-C (Welinder, 1979) and Prx8 (Kristensen et al., 1997), for which the first N-terminal residues in the mature protein are known (Fig. 1). Aligning Prx7 with HRP-C (Fujiyama et al., 1988), the barley peroxidases Prx8 (Thordal-Christensen et al., 1992), BP1 (Rasmussen et al., 1991b), and BP2 (Theilade and Rasmussen, 1992) revealed that Prx7 carries a C-terminal elongation similar to those in HRP-C, BP1, and BP2, although there was low sequence similarity among these elongations.
Figure 1.
Amino acid sequence of Prx7 and alignment to selected peroxidases. Alignment to peroxidases from barley: Prx8 (pBH6-301; Thordal-Christensen et al., 1992), BP1 (Rasmussen et al., 1991b), and horseradish HRP-C gene A (Welinder, 1979; Fujiyama et al., 1988). The lower line shows invariant residues, and variable residues are indicated by dots. The eight Cys residues forming disulfide bridges and the two His residues essential for catalysis are indicated in bold and are underlined. Gaps (-) are introduced to maximize the alignment. N- and C-terminal sequences shown to be absent from the mature proteins are in lowercase italics. Residues shown in bold in Prx7 were determined by amino acid sequencing. Underlined regions in the Prx7 sequence correspond to peptides identified by MALDI-TOF MS tryptic fingerprinting. The underlined and bold Asn residues at positions 91 and 176 in the Prx7 sequence are putative N-glycosylation sites.
Nakamura and Matsuoka (1993) suggested that a hydrophobic/acidic motif structure, rather than any specific amino acid sequence, forms the core of the C-terminal vacuolar sorting signal. The hydrophobic/acidic motif typically consists of three to four hydrophobic amino acids followed by one or two acidic residues. Such a motif is also present in the peroxidase HRP-C and in the vacuolar BP2 (Theilade et al., 1993). The motif is repeated once in the C terminus of the Prx7 preprotein, AVAGDEGIAADM, suggesting that Prx7 is also located in the vacuole. For both vacuolar HRP-C (Welinder, 1979; Fujiyama et al., 1988) and BP1 (Rasmussen et al., 1991b), it has been shown that these C-terminal domains are absent from the mature protein, whereas BP2 has been localized by immunogold electron microscopy to the vacuole of scutellum cells in barley grains (Theilade et al., 1993). These results suggest that the C terminus of Prx7 is a vacuole-targeting signal and that mature Prx7 is located in the vacuole.
Purification of Peroxidases from Barley Coleoptiles
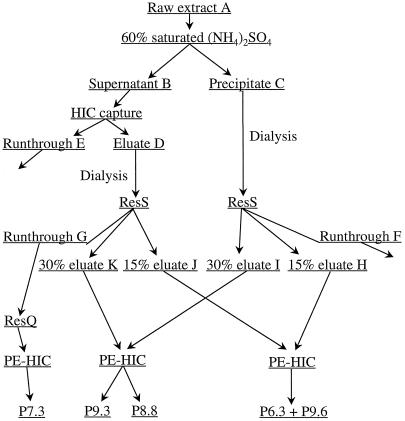
Seven-day-old coleoptiles were chosen as the starting material for the purification of Prx7, because anti-rPrx7 antibodies reacted with substantial amounts of the protein in this tissue (see below). Initial trials using variations on the purification method showed a substantial peroxidase precipitation in coleoptile extracts at pH 8.0 and 60% (NH4)2SO4 saturation. Approximately one-fourth of the peroxidase activity in the coleoptile extract (Table I) was precipitated by (NH4)2SO4. Figure 2 provides an overview of the purification scheme. The remaining peroxidase activity in the (NH4)2SO4 supernatant was completely adsorbed to HIC medium.
Table I.
Purification of barley coleoptile peroxidases
| Purification Step | Total Protein | Specific Activity | Total Activity | RZ |
|---|---|---|---|---|
| mg | units/mg | units | ||
| Cleared crude extract (A) | 270 | 0.97 | 263 | n.d.a |
| (NH4)2SO4 supernatant (B) | 194 | 1.03 | 199 | n.d. |
| (NH4)2SO4 precipitate (C) | 42 | 1.73 | 73.0 | 0.11 |
| HIC capture of B, eluate (D) | 45 | 2.73 | 123 | 0.13 |
| Cation exchange of C, runthrough (F) | 9.0 | 0.03 | 0.3 | n.d. |
| Cation exchange of D, runthrough (G) | 16 | 5.16 | 81.5 | 0.79 |
| Cation exchange of C, 15% eluate (H) | 1.8 | 1.46 | 2.7 | n.d. |
| Cation exchange of C, 30% eluate (I) | 6.8 | 5.27 | 36.1 | 0.56 |
| Cation exchange of D, 15% eluate (J) | 0.1 | 50.1 | 6.0 | n.d. |
| Cation exchange of D, 30% eluate (K) | 0.5 | 1.97 | 0.9 | n.d. |
| P7.3 | 0.1 | 78.9 | 5.5 | 2.85 |
| P9.3 | 0.8 | 11.1 | 7.3 | 3.31 |
Specific activity was measured as ABTS oxidation at pH 5.5. Total activity is given as units of ABTS oxidation. The Reinheitszahl unit (RZ) is the ratio of the absorbance of the heme group (A403–408) to the absorbance of the aromatic amino acids (A280) and is a measure of peroxidase purity.
n.d., Not determined.
Figure 2.
Flow chart for purification of barley coleoptile peroxidases P7.3, P8.8, P9.3, and P9.6. ResS, Cation-exchange chromatography; ResQ, anion-exchange chromatography. Details of the purification scheme are described in Methods.
Because it was not determined at this point whether an isoform-specific or a nonspecific, incomplete precipitation had taken place, the two protein pools were kept separate for cation-exchange purification. Two distinct peaks of peroxidase activity were eluted by cation-exchange chromatography of the extracts from both the (NH4)2SO4 precipitate and the HIC-captured (NH4)2SO4 supernatant, one at approximately 15% and the other at approximately 30% elution buffer. Activity-stained IEF showed that the approximately 15% elution-buffer activity from the (NH4)2SO4 precipitate and the HIC-captured (NH4)2SO4 supernatant was a mixture of P6.3 and P9.6. The peroxidase eluted at a gradient buffer strength of approximately 30% was P8.8 and P9.3. Most of the peroxidase that did not bind to the cation exchanger also did not bind to the anion exchanger.
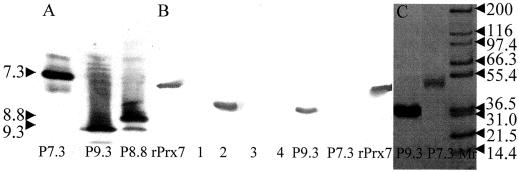
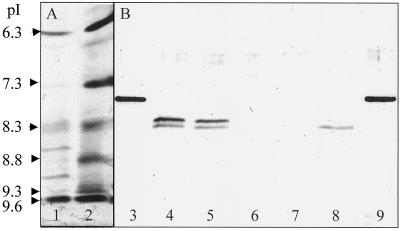
One peak of activity was eluted from the anion-exchange column, but this peroxidase was too diluted for further analysis and was discarded. The peroxidase in the anion-exchange unbound fraction was bound on the phenyl ether-HIC column and was characterized as a P7.3 (Fig. 3A). P7.3 is immunologically unrelated to Prx7 (Fig. 3B) and has several molecular forms with apparent molecular masses of about 53 to 55 kD (Fig. 3C). The diffuse bands on the SDS-PAGE gel indicate a highly glycosylated peroxidase. The high Reinheitszahl value (2.85) of the P7.3 preparation makes it unlikely that the heterogeneity was the result of contamination by other proteins. P9.6 and P6.3 activities were lost on the phenyl ether-HIC column under the conditions used; the activity from P8.8 and P9.3 was partially resolved.
Figure 3.
Analysis of peroxidase purification fractions. A, Activity staining of purified peroxidases P7.3, P8.8, and P9.3 after IEF. pH values calculated using pI markers are shown on the left. B, Immunoblot analysis. Protein was detected using polyclonal anti-rPrx7 antibodies. Analysis of rPrx7. Lane 1, Protein not bound to cation-exchange chromatography (ResS) or anion-exchange chromatography (ResQ), captured on phenyl ether-HIC, and containing P7.3; lane 2, pooled 30% elution buffer eluate from ResS; lane 3, pooled 15% elution buffer eluate from ResS; and lane 4, activity bound on and eluted from ResQ. C, SDS-PAGE of P9.3 and P7.3. The gel was stained with Coomassie Blue. Molecular mass markers (×103) are shown on the right.
IEF analysis of the fractions revealed that four fractions (PE16–PE19) contained pure P9.3, whereas the next four fractions (PE20–PE23) contained predominantly P8.8 (Fig. 3A). SDS-PAGE analysis showed that the P8.8-containing fractions contained several polypeptides (not shown), but only a trace amount of protein with a size identical to P9.3. None of the dominant polypeptides, which were all larger than P9.3, cross-reacted with the antibodies (Fig. 3B, lane 2), showing that P8.8 is immunologically unrelated to Prx7. P9.3 was judged to be highly pure on the basis of its Reinheitszahl value (3.31). The protein responsible for P9.3 activity cross-reacted strongly with anti-rPrx7 antibodies (Fig. 3B). The apparent molecular mass was about 33 kD (Fig. 3C), and this plus the sharpness of the P9.3 band indicated that P9.3 was nonglycosylated. Immunoblotting of crude coleoptile extract (see below) and pooled activities before phenyl ether-HIC revealed that only P9.3 cross-reacted with the anti-rPrx7 antibodies (Fig. 3B), indicating that P9.3 is the only product of the mRNAs corresponding to pcD1311BE. For the oxidation of ABTS, P7.3 had a specific activity of 78.9 units/mg, whereas P9.3 had a specific activity of 11.1 units/mg (Table I).
N-Terminal Sequencing of P9.3 and P7.3
To verify the identity of P9.3 and its N-terminal signal-peptide cleavage site, P9.3 was blotted onto PVDF membranes and subjected to Edman degradation. Unexpectedly, the peroxidase yielded a sequence without pyroglutamyl deblocking. The 10-residue sequence ATXPDLERIV, which was identical to residues 8 to 18 in the deduced mature protein (residues 33–43 using pre-Prx7 numbering; Fig. 1), verified the identity of P9.3 as Prx7. No alternative sequence was obtained after treatment with pyroglutamyl amino peptidase, showing that mature Prx7 starts at residue 33. The mobility of Prx7 on immunoblots was identical when freshly prepared coleoptile extract (see below) and purified Prx7 were analyzed. This shows that the N-terminal sequence determined here was truly for native Prx7 and was not the result of contaminating proteases. The deblocking yield from P7.3 was very low, and no sequence was obtained.
MALDI-TOF MS Analysis of P9.3 and P7.3
The barley peroxidases P9.3 and P7.3 were digested by trypsin in both polyacrylamide gel pieces and solution. Self-digested trypsin and trypsin-digested recombinant Prx7 from E. coli were used as internal and positive controls for the analyses of P9.3 and P7.3. The digests were analyzed by MALDI-TOF MS. For each run, 20 to 40 spectra were summarized into one and the peptide masses were determined.
Only peptides detected in two or more individual runs were considered for analysis. Peaks from self-digested trypsin were subtracted from the spectra manually and mean peptide masses were calculated for the remaining peaks. Seventeen peptides were found for the P9.3 sample, whereas 13 peptides were detected from P7.3 (Table II) and 11 peptides were detected from rPrx7. The calculated average peptide masses were entered in the PeptideSearch program(www.mann.embl-heidelberg.de/Services/PeptideSearch/FR_PeptideSearchForm.html).Thedatabase searches were performed with the following restrictions: (a) calculated masses should be average masses; (b) the scored protein should be in the 30- to 38-kD range; (c) Met residues were unmodified and Cys residues were carbamidomethylated; (d) peptides were protonated; and (e) the accuracy of the peptide masses should be 0.1%. This accuracy was the same as the sd among the measured masses.
Table II.
Peptide masses from tryptic digests of P9.3 and P7.3 determined by MALDI-TOF MS
| Residuesa | P9.3 (M + H) | P7.3 (M + H) |
|---|---|---|
| 826.2 | ||
| 861.55 | ||
| 963.15 | ||
| 984.9 | ||
| 1012.12 | 1031.9 | |
| 252 –259 | 1061.3 | |
| 1103.4 | 1054.65 | |
| 134 –145 | 1147.97 | |
| 1170.16 | ||
| 265 –276 | 1311.8 | |
| 1334.35 | 1075.95 | |
| 1350.0 | 1095.7 | |
| 41 –52 | 1506.20 | |
| 311 –323 | 1571.42 | |
| 282 –295 | 1663.25 | |
| 1684.65 | 1816.8 | |
| 1794.96 | 1854.9 | |
| 1803.2 | 1872.7 | |
| 244 –295 | 1978.75 | 1978.9 |
| 81 –108 | 2605.25 | |
| 2870.9 | 3077.45 |
Calculated from Prx7 cDNA (Fig. 1).
Searching the database with the 17-peptide masses from P9.3 gave 47 matches. The best of these matches involved the recognition of three peptides more than the second-best match. The best match was the cDNA clone pcD1311BE encoding Prx7, which we had previously deposited in the database. Nine of the 17 peptides matched tryptic fragments derived from pcD1311BE. A second-pass search on the cDNA revealed that one peptide matched Prx7 more closely if it was assumed to be modified covalently by acrylamide. In total, these 10 peptides covered 147 of the 341 amino acids in the preprotein. These peptides are marked in Figure 1, showing the amino acid sequence of Prx7. The C-terminal peptide (residues 311–323 using pre-Prx7 numbering) extends precisely to the deduced cleavage site for the C-terminal putative signal peptide. The C-terminal cleavage site was estimated by alignment with other peroxidase proteins with known C termini and by comparison with the empirical pI of the mature peroxidase. One of the detected peptides carried one of the two N-glycosylation sites. The mass of this peptide confirmed that at least some of the Prx7 molecules were not glycosylated at this residue, supporting our previous notion that Prx7 from coleoptiles does not appear to be glycosylated, based on the sharpness of the Prx7 band obtained by SDS-PAGE.
Eleven tryptic peptide masses were determined from rPrx7, and seven of these had a counterpart in the P9.3 digest (overlapping sd values), showing the identity between the two proteins.
As expected, the search with the 13-peptide masses from P7.3 did not give any matches. Although P7.3 has been characterized as being constitutively expressed in the epidermis (Kogel et al., 1994), it has not yet been cloned.
The molecular mass of nondigested P9.3 was determined with the aim of discovering whether the protein was glycosylated and whether P9.3 was also processed in the C terminus. The determined mass did not include the heme group and structural Ca2+ ions that are lost during desorption. From seven individual spectra the mean Mr was determined to be 31,521 D (sd = 374 D), which is in close correspondence to the calculated mass of Prx7-containing residues 33 to 323 (31,587 D), as indicated by peptide fingerprinting and N-terminal protein sequencing (Table III). From this analysis we conclude that the C terminus is processed, that Prx7 is nonglycosylated in coleoptile tissue, and that residue 323 is the C-terminal residue in the mature protein (because the tryptic peptide covering residues 311 to 323 was found in the peptide analysis). On the 31,587 m/z peak in one of the seven mass spectra, weak shoulders on the higher m/z side could be seen (not shown), indicating the presence of small amounts of alternatively processed Prx7 in the sample. Wobble in the processing of C-terminal peptides has been described for BP1 (Rasmussen et al., 1991b), and this may have been responsible for the microheterogeneity seen here.
Table III.
Posttranslational modification of Prx7
| Calculated Mass of Prx7 Preprotein | Residues (Peptide Length) | N- and C-Terminal Amino Acids | Polypeptide Mass of P9.3 by MALDI-TOF MS |
|---|---|---|---|
| D | D | ||
| 31,431 | 33–322 (290) | AT-NR | |
| 31,587 | 33–323 (291) | AT-NRR | 31,521 ± 374 |
| 32,481 | 24–323 (299) | QL-NRR | |
| 33,372 | 33–341 (309) | AT-DM | |
| 34,266 | 24–341 (317) | QL-DM |
Calculated masses using the cDNA and measured mass of P9.3 (Prx7).
Induction of Peroxidase by Powdery Mildew Inoculation and Wounding
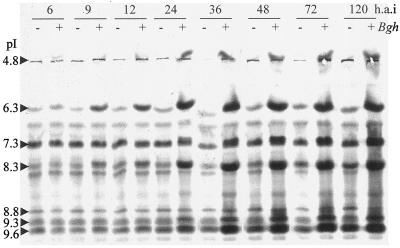
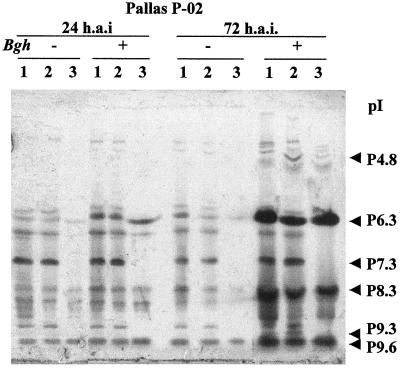
Time-course studies using barley plants inoculated with powdery mildew isolates were designed to investigate the accumulation pattern of P9.3 in response to inoculation with a pathogen and in relation to other isoperoxidases (Fig. 4). Accumulation of P6.3, P8.3 (Prx8), and P9.6 in total leaf extract was evident from 9 h after inoculation. Both P9.3 and P4.8 had accumulated significantly at 24 h after inoculation.
Figure 4.
Time course (h.a.i, h after inoculation) of peroxidase accumulation in barley leaves after inoculation with powdery mildew. IEF total protein was activity stained for peroxidase. Shown are protein extracts (20 μg) from P-02 leaves after inoculation with powdery mildew fungus A6 spores (incompatible interaction) (+) and from noninoculated control leaves (−). The pI values of the isozymes are shown on the left.
The race-specific resistance in the isogenic line used, cv Pallas P-02 (mla3), manifested itself as a multicell hypersensitive response at approximately 72 h after inoculation against the incompatible powdery mildew fungus A6 isolate. From 24 h after inoculation until the end of the time course (5 d after inoculation), no new peroxidase isoforms accumulated and there appeared to be no difference in the timing of accumulation of isoperoxidases between the compatible (cv Pallas P-02/powdery mildew fungus C15) and the incompatible (cv Pallas P-02/powdery mildew fungus A6) interaction (data not shown). This is in agreement with a study by Kerby and Somerville (1989) using a different combination of powdery mildew isolates and barley isolines. Wounding of epidermal cells by rubbing with silicon carbide powder induced the same peroxidases and with the same timing of accumulation as powdery mildew inoculation (data not shown).
Tissue-Specific Peroxidase Expression
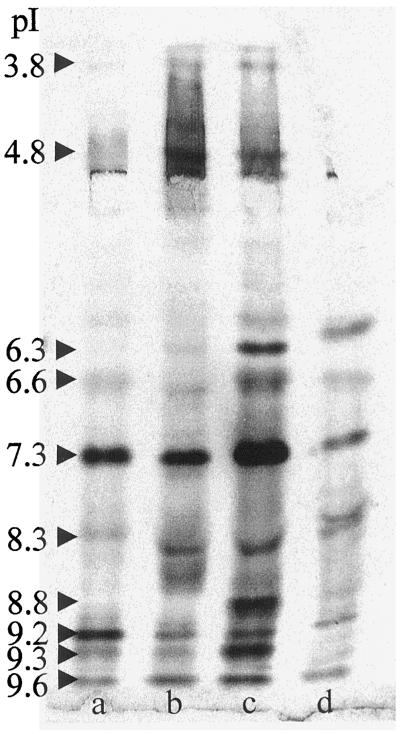
Each of the four tested organs had its own characteristic isoperoxidase profile (Fig. 5). A total of 12 isoenzymes could be distinguished in the various organs. On the basis of peroxidase-band intensity, primary leaves contained the least peroxidase on a fresh-weight basis. The coleoptile contained the most peroxidase and also had the largest abundance of each of the peroxidase isoenzymes among the four tissues. P9.3 was particularly abundant in the epidermis-rich coleoptile, whereas it was present in low amounts in the other tissues. This difference was also suggested by immunoblotting (see below). Activity due to P7.3 was the most dominant peroxidase activity in all tissues of the seedling. A similar constitutive expression has also been noted for the Arabidopsis ATP1 and ATP2 peroxidases (Kjærsgård et al., 1997), suggesting basic metabolic functions for these peroxidases.
Figure 5.
Isoperoxidase expression in various barley tissues. IEF and peroxidase activity staining of total protein extracts (20 μg) derived from the basal part of the primary leaf sheath covered by the coleoptile from the apical meristem (a), root (b), coleoptile (c), and primary leaf (d) of 7-d-old P-02 seedlings. The pI values of the isozymes are shown on the left.
Intracellular Versus Extracellular Accumulation of Barley Peroxidases
IEF and activity staining of peroxidases in extracellular fluid from leaves showed that three isoperoxidases, P6.3, P8.3, and P9.6, accumulated in the extracellular space 24 h after inoculation in both the compatible and the incompatible interaction (Fig. 6). Analysis at later times (72 h after inoculation) showed that P3.8, P4.8, and P8.8 also accumulated in the extracellular space after inoculation.
Figure 6.
Peroxidase accumulation in total protein extracts, intercellular washing fluid, and residual protein extracted from leaves after extraction of intercellular proteins. Activity-stained IEF of protein corresponding to extracts from a half leaf. Total protein (lanes 1), residual protein (lanes 2), and intercellular protein (lanes 3) were analyzed. Protein was extracted from P-02 leaves 24 and 72 h after inoculation with powdery mildew fungus A6 spores (+), and from noninoculated control leaves (−) at the same times.
Four isoperoxidases, P6.6, P7.3, P9.2, and P9.3, were never detected in the extracellular fluid, indicating their intracellular localization. None of the intracellular isoperoxidases appeared to increase in abundance as strongly as the extracellular isoperoxidases in response to inoculation or infection.
Accumulation of Peroxidases and Peroxidase Gene Transcripts in Epidermis and Mesophyll in Response to Powdery Mildew Inoculation
Immunoblotting of SDS-PAGE-separated proteins using antibodies raised against rPrx7 showed the presence of Prx7 in noninoculated epidermis and a slight enhancement upon inoculation (Fig. 7B). No immunologically reacting peroxidases could be detected in either inoculated or noninoculated mesophyll. The immunoblot of epidermal extract had an extra band compared with that for the coleoptile extract. This extra band may have represented a glycosylated form of Prx7, as is the case with Prx8 (Kerby and Somerville, 1992), or it may have been a distinct but immunologically related peroxidase present in the leaf but not in the coleoptile (Fig. 7B, lane 8). However, all of the isoforms present in the leaf were also present in the coleoptile (Fig. 5), and because anti-rPrx7 antibodies did not recognize proteins in fractions from the purification other than those containing P9.3, the antibodies were specific for Prx7 and the alternative explanation given above is unlikely. We also observed that storage of the epidermal samples at 4°C followed by western analysis at later times led to increasing abundance of the low-Mr band (not shown). This band likely represented a nonglycosylated protein. Van Huystee and McManus (1998) made similar observations by showing that secreted β-galactosidase present in crude extract was able to alter the size and lectin-binding properties of the glycosylated anionic peanut peroxidase.
Figure 7.
Peroxidase accumulation in coleoptiles, leaf epidermis, and leaf mesophyll. A, IEF of protein extracts from 7-d-old barley leaves (P-02) stained for peroxidase activity. Shown are extracts from mesophyll (lane 1) and epidermis (lane 2) obtained from leaves 24 h after inoculation with powdery mildew fungus A6 spores. Total protein (20 μg) was applied in each lane. pI values calculated using markers are shown on the left. B, Immunoblot of rPrx7 and protein extracts obtained as in A using polyclonal anti-rPrx7 antibodies for detection of Prx7. Extracts from epidermis (lanes 4 and 5) and mesophyll (lanes 6 and 7) were obtained from barley leaves inoculated with powdery mildew fungus A6 spores 24 h after inoculation (lanes 4 and 6) and noninoculated leaves (lanes 5 and 7). Extracts from 7-d-old coleoptiles (lane 8) were also analyzed. In lanes 4 to 8, approximately 20 μg of total protein was analyzed. In lanes 3 and 9, 200 ng of rPrx7 was analyzed. Protein was separated on 12% SDS-PAGE gels.
Isoperoxidase analysis of protein from epidermis obtained 24 h after inoculation showed the location of six isoenzymes (Fig. 7A): P6.3, P7.3, P8.3, P8.8, P9.3, and P9.6. In mesophyll, P9.3 and P8.8 were not detected, whereas P6.3, P8.3, P9.2, and P9.6 were detected.
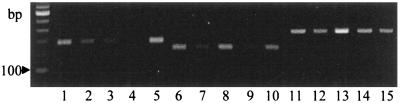
RT-PCR analyses of mRNA from epidermis and mesophyll using Prx7-specific primers showed enhancement in inoculated epidermis and mesophyll at 24 h after inoculation, but less accumulation in the mesophyll than in the epidermis (Fig. 8). These results indicated that Prx7 was expressed preferentially in epidermal cells and was present in the leaf as two molecular forms, one of which was likely to be glycosylated.
Figure 8.
RT-PCR analysis of transcript accumulation in epidermis and mesophyll in response to powdery mildew inoculation. RNA was extracted from 8-d-old barley (P-02) primary leaves 24 h after inoculation with powdery mildew fungus A6 spores or from noninoculated control leaves. Prx7 primers were used in lanes 1 to 5, Prx8 primers in lanes 6 to 10, and GAPDH primers in lanes 11 to 15. RNA was obtained from inoculated epidermis (lanes 1, 6, and 11), noninoculated epidermis (lanes 2, 7, and 12), inoculated mesophyll (lanes 3, 8, and 13), noninoculated mesophyll (lanes 4, 9, and 14), and 7-d-old P-02 coleoptiles (lanes 5, 10, and 15). A 100-bp DNA ladder is shown in the far left lane. Two independent inoculations were performed, and extracted RNA was analyzed twice. All four PCR experiments gave the same banding-intensity pattern.
RT-PCR analysis of mRNA from stripped mesophyll and epidermis showed that the Prx8 messenger accumulated preferentially in the mesophyll (Fig. 8). Gregersen et al. (1997) obtained similar results. Prx8 protein can be obtained from extracellular spaces in barley after inoculation with powdery mildew by a gentle vacuum-infiltration procedure (Kerby and Somerville, 1989). These data indicate that Prx8 was transcribed preferentially in mesophyll cells and that Prx8 accumulated in the extracellular space surrounding mesophyll cells and in vascular bundles, from which it was easily extracted.
DISCUSSION
The coleoptile is a rapidly growing protective tissue that shields the emerging primary leaf (and later the basal part of the primary leaf sheath) from both physical damage and infection by antagonistic soil organisms. The coleoptile is rich in epidermal tissue and contains no chlorophyll. Because of their rapid growth and linear cell pattern, coleoptiles from monocots are one of the preferred experimental tissues for studies of cell elongation and auxin action. For more than 30 years, coleoptiles have also been used widely in microscopy-based physiological studies of the barley-powdery mildew fungus interaction (Bushnell et al., 1967).
We chose barley coleoptiles as the starting material for Prx7 purification because immunoblotting of coleoptile proteins and RT-PCR showed substantial accumulation of Prx7 in this tissue. After purification of several isoperoxidases from coleoptiles, we were able to identify the P9.3 isozyme as Prx7, and were then able to follow the accumulation of Prx7 in barley leaves in response to powdery mildew inoculation and to investigate the tissue-specific and subcellular accumulation pattern of Prx7.
The C-terminal vacuolar-targeting hydrophobic/acidic motif (Nakamura and Matsuoka, 1993) is repeated once in the C terminus of the Prx7 preprotein, suggesting that Prx7 is located in the vacuole or in endosomes. We showed that Prx7 is targeted to the secretory pathway, because the putative N-terminal targeting peptide is absent from the mature protein. However, in spite of its abundance in total extracts the protein was never recovered in the extracellular washing fluid. Furthermore, the molecular mass determined by MALDI-TOF MS of pure Prx7 (31.5 kD) showed that the C terminus was processed like that of vacuolar BP1 (Rasmussen et al., 1991b). These results strongly suggest that Prx7 is a vacuolar peroxidase. The unusual processing of the Prx7 N terminus may not be related to a distinct intracellular route, but could be the result of nonspecific carboxypeptidase activity in the final destination of the putative vacuolar peroxidase.
Prx7 was expressed preferentially in epidermal cells and was present in the leaf as two molecular forms, one of which was likely to be glycosylated. In the coleoptile only one nonglycosylated form of Prx7 was present, as shown by MS of purified Prx7 and by immunoblotting of crude extract.
The accumulation patterns of Prx7 and Prx8 mRNAs in barley leaves inoculated with powdery mildew fungus were first reported by Thordal-Christensen et al. (1992). The mRNA corresponding to Prx8 showed a two-peak accumulation pattern at the time of penetration attempts of the fungus, but the Prx7 mRNA always accumulated later and to a much lower extent. The delay in the appearance of Prx7 compared with Prx8 seen in this study corresponded to the delay in the increase of the transcript, suggesting that translational regulation is not important in the regulation of these genes.
The general notion that the cell wall is the major target for peroxidase-mediated modifications is also supported by our data, because only the levels of Prx7 and P7.3 were enhanced intracellularly, whereas the levels of five isoenzymes were enhanced in the apoplast after inoculation with powdery mildew. This finding suggests a special role of the putative vacuolar Prx7 in biochemical pathways involving peroxidases.
Hordatines are antifungal compounds found in the young barley seedling. Hordatine A is a dimer of two p-coumarylagmatines, and hordatine B is a dimer of a p-coumarylagmatine and a ferulylagmatine (Stoessl, 1967).
Stoessl (1967) showed that hordatines could be generated in vitro from the appropriate hydroxycinnamic acid amides by horseradish peroxidase, suggesting that a peroxidase may mediate the last step in the biosynthetic pathway to hordatine. Hordatine M, a mixture of glycoconjugates of hordatine A and B, was found by Stoessl (1967) to be present in substantial amounts (277 mg/kg fresh weight) in 6-d-old barley coleoptiles, along with small amounts of hordatine A and B. Transfer of dark-grown barley seedlings to the light rapidly reduced the biosynthesis of the hordatines compared with that in etiolated seedlings, especially that of hordatine M (Smith and Best, 1978). The subcellular locations of the production and storage of these compounds are unknown, but like the majority of glycosylated secondary plant metabolites (Boller and Wiemken, 1986), hordatine M is likely to accumulate in the vacuole. Stoessl (1967) showed that hordatines could be generated in vitro by horseradish peroxidase; however, unlike the hordatines isolated from coleoptiles, the in vitro products were not optically active.
Lignans are optically active dimers of hydroxycinnamic acids and are likely to be synthesized by peroxidases (Nose et al., 1995; Dinkova-Kostova et al., 1996) and the novel stereospecific dirigent protein in concert (Davin et al., 1997). Lignans are believed to be stored as glycoconjugates in the vacuole (Lewis and Yamamoto, 1990; Lewis and Davin, 1994) and to have antimicrobial properties, in addition to having a role as lignification nuclei in the wall (Nose et al., 1995). Stoessl (1967) also speculated that hordatine A and B have a role in the biogenesis of the barley cell wall. The enhanced expression of Prx7 in the leaf-elongation zone in the Slender mutant of barley (Schünmann et al., 1994) may indicate that this peroxidase does not restrain cell wall expansion, but rather provides building blocks for the wall.
Prx7 is abundant in the coleoptile, where hordatine M also is present, and the slow accumulation of Prx7 in response to inoculation with powdery mildew fungus parallels the accumulation of hordatine (Smith and Best, 1978). Likewise, Prx7 is present at low levels in the noninoculated, light-grown, green primary leaf, where the biosynthesis of hordatine is down-regulated (Smith and Best, 1978). These observations strongly suggest that Prx7 is the peroxidase responsible for the last step in the biosynthesis of hordatine. A protein similar to the dirigent protein may also be involved.
Because Prx7 and the constitutive P7.3 differ significantly in their specific activities, as measured by ABTS oxidation at pH 5.5, Prx7 is likely to have a function distinct from that of the constitutive putative vacuolar peroxidase P7.3. Even though both enzymes appear to be vacuolar or endosomal, their preferred substrates, pH optima, and functions in vivo are likely to be different. It must be noted that the artificial substrate ABTS has very little similarity to p-coumarylagmatine or hydroxycinnamic acids and alcohols, so the specific activities determined in the present study may not reflect activities measured with more physiologically relevant substrates.
ACKNOWLEDGMENTS
We thank Dr. J.S. Scott-Craig and Dr. S. Somerville for the barley leaf cDNA library and Dr. Thomas H. Roberts for constructive reading of the manuscript. Charlotte Koutras and Stanko Djudjevic are thanked for skilled technical assistance. Yvonne Berger and Arne Jensen of the University of Copenhagen kindly performed the amino acid sequencing. Unizyme A/S (Hørsholm, Denmark) generously donated pyroglutamyl amino peptidase.
Abbreviations:
- ABTS
2,2′-azino-di-(3-ethyl-benzothiazoline-6-sulfonic acid)
- GAPDH
glyceraldehyde-3-P dehydrogenase
- HIC
hydrophobic interaction chromatography
- MALDI-TOF
matrix-assisted laser desorption/ionization time of flight
- P#
peroxidase with a pI of #
- RT
reverse transcriptase
Footnotes
This study was supported by the Danish Agricultural and Veterinary Research Council (grant no. 5.23.26.10), Molecular Strategies for Crop Improvements, the Danish Research Academy, and the Danish Cereal Network, which is supported by the Ministry of Food, Agriculture, and Fisheries.
LITERATURE CITED
- Aist JR, Bushnell WR (1991) Invasion of plants by powdery mildew fungi, and cellular mechanisms of the resistance. In GT Cole, HC Hoch, eds, The Fungal Spore and Disease Initiation in Plants and Animals. Plenum Press, New York, pp 299–321
- Boller T, Wiemken A. Dynamics of vacuolar compartmentation. Annu Rev Plant Physiol. 1986;37:137–164. [Google Scholar]
- Boyd LA, Smith PH, Brown JKM. Molecular and cellular expression of quantitative resistance in barley to powdery mildew. Physiol Mol Plant Pathol. 1994;45:47–58. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bradley DJ, Kjellborn P, Lamb CJ. Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defense response. Cell. 1992;70:21–30. doi: 10.1016/0092-8674(92)90530-p. [DOI] [PubMed] [Google Scholar]
- Bushnell WR, Dueck J, Rowell JB. Living haustoria and hyphae of Erysiphe graminis f.sp. hordei with intact and partly dissected host cells of Hordeum vulgare. Can J Bot. 1967;45:1719–1732. [Google Scholar]
- Carver TLW, Zeyen RJ, Bushnell WR, Robbins MP. Inhibition of phenylalanine ammonia lyase and cinnamyl alcohol dehydrogenase increases quantitative susceptibility of barley to powdery mildew (Erysiphe graminis DC.) Physiol Mol Plant Pathol. 1994;44:261–272. [Google Scholar]
- Childs RE, Bardsley WG. The steady-state kinetics of peroxidase with 2,2′-azino-di-(3-ethyl-benzthiazole-6-sulfonic acid) as chromogen. Biochem J. 1975;145:93–103. doi: 10.1042/bj1450093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin JJ, Przbyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Collinge DB, Bryngelsson T, Gregersen PL, Smedegaard-Petersen V, Thordal-Christensen H (1997) Resistance against fungal pathogens: its nature and regulation. In AS Basra, RK Basra, eds, Mechanisms of Environmental Stress Resistance in Plants. Harwood Academic Publishers, London, pp 335–372
- Davin LB, Wang HB, Crowell AL, Bedgar DL, Martin DM, Sarkanen S, Lewis NG. Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science. 1997;275:362–366. doi: 10.1126/science.275.5298.362. [DOI] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Gang DR, Davin LB, Bedgar DL, Chu A, Lewis NG. (+)-Pinoresinol/(+)-lariciresinol reductase from Forsythia intermedia: protein purification, cDNA cloning, heterologous expression and comparison to isoflavone reductase. J Biol Chem. 1996;271:29473–29482. doi: 10.1074/jbc.271.46.29473. [DOI] [PubMed] [Google Scholar]
- Espelie KE, Franceschi VR, Kolattukudy PE. Immunocytochemical localization and time course of appearance of an anionic peroxidase associated with suberization in wound-healing potato tuber tissue. Plant Physiol. 1986;81:487–492. doi: 10.1104/pp.81.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama K, Takemura H, Shibayama S, Kobayashi K, Choi J-K, Shinmyo A, Takano M, Yamada Y, Okada H. Structure of the horseradish peroxidase C genes. Eur J Biochem. 1988;173:681–687. doi: 10.1111/j.1432-1033.1988.tb14052.x. [DOI] [PubMed] [Google Scholar]
- Giese H, Holm-Jensen AG, Jensen HP, Jensen J. Localization of the Laevigatum powdery mildew resistance gene to barley chromosome 2 by the use of RFLP markers. Theor Appl Genet. 1993;85:897–900. doi: 10.1007/BF00225035. [DOI] [PubMed] [Google Scholar]
- Gregersen PL, Thordal-Christensen H, Forster H, Collinge DB. Differential gene transcript accumulation in barley leaf epidermis and mesophyll in response to attack by Blumeria graminis f.sp. hordei (syn. Erysiphe graminis f.sp. hordei) Physiol Mol Plant Pathol. 1997;51:85–97. [Google Scholar]
- Hammerschmidt R, Kuc J. Lignification as a mechanism for induced systemic resistance in cucumber. Physiol Plant Pathol. 1982;20:61–71. [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Hirano H, Komatsu S, Kajiwara H, Takagi Y, Tsunasawa S. Microsequence analysis of the N-terminally blocked proteins immobilized on polyvinylidene difluoride membrane by western blotting. Electrophoresis. 1993;14:839–846. doi: 10.1002/elps.11501401134. [DOI] [PubMed] [Google Scholar]
- Ikegawa T, Mayama S, Nakayashiki H, Kato H. Accumulation of diferulic acid during the hypersensitive response of oat leaves to Puccinia coronata f. sp. avenae and its role in the resistance of oat tissues to cell wall degrading enzymes. Physiol Mol Plant Pathol. 1996;48:245–255. [Google Scholar]
- Kerby K, Somerville S. Enhancement of specific intercellular peroxidases following inoculation of barley with Erysiphe graminis f. sp. hordei. Physiol Mol Plant Pathol. 1989;35:323–337. [Google Scholar]
- Kerby K, Somerville SC. Purification of an infection-related, extracellular peroxidase from barley. Plant Physiol. 1992;100:397–402. doi: 10.1104/pp.100.1.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjærsgård, Jespersen HM, Rasmussen SK, Welinder KG. Sequence and RT-PCR expression analysis of two peroxidases from Arabidopsis thaliana belonging to a novel evolutionary branch of plant peroxidases. Plant Mol Biol. 1997;33:699–708. doi: 10.1023/a:1005707813801. [DOI] [PubMed] [Google Scholar]
- Kogel KH, Beckhove U, Dreschers J, Munch S, Romme Y. Acquired resistance in barley. The resistance mechanism induced by 2,6-dichloroisonicotinic acid is a phenocopy of a genetically based mechanism governing race-specific powdery mildew resistance. Plant Physiol. 1994;106:1269–1277. doi: 10.1104/pp.106.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kølster P, Munk L, Stølen O, Løhde J. Near-isogenic barley lines with genes for resistance to powdery mildew. Crop Sci. 1986;26:903–907. [Google Scholar]
- Kristensen BK, Brandt J, Bojsen K, Thordal-Christensen H, Kerby KB, Collinge DB, Mikkelsen JD, Rasmussen SK. Expression of a defense-related intercellular barley peroxidase in transgenic tobacco. Plant Sci. 1997;122:173–182. [Google Scholar]
- Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- Laemmli JJ. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamb C, Dixon RA. The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- Lewis NG, Davin LB. Evolution of lignan and neolignan biochemical pathways. In: Nes WD, editor. Isopentenoids and Other Natural Products: Evolution and Function. Symposium Series 562. Washington, DC: American Chemical Society; 1994. pp. 202–246. [Google Scholar]
- Lewis NG, Yamamoto E. Lignin: occurrence, biogenesis and biodegradation. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:455–496. doi: 10.1146/annurev.pp.41.060190.002323. [DOI] [PubMed] [Google Scholar]
- Moerschbacher BM (1992) Plant peroxidases: involvement in response to pathogens. In C Penel, T Gaspar, H Greppin, eds, Plant Peroxidases 1980–1990: Topics and Detailed Literature on Molecular, Biochemical, and Physiological Aspects. University of Geneva, Switzerland, pp 91–99
- Nakamura K, Matsuoka K. Protein targeting to the vacuole in plant cells. Plant Physiol. 1993;101:1–5. doi: 10.1104/pp.101.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nose M, Bernards MA, Furlan M, Zajicek J, Eberhardt TL, Lewis NG. Towards the specification of consecutive steps in macromolecular lignin assembly. Phytochemistry. 1995;39:71–79. doi: 10.1016/0031-9422(95)95268-y. [DOI] [PubMed] [Google Scholar]
- Rasmussen SK, Johansson A, Rasmussen HN, Theilade B (1991a) Molecular analysis and cloning of barley peroxidase genes. In J Lobarzewski, H Greppin, C Penel, T Gaspar, eds, Biochemical, Molecular and Physiological Aspects of Plant Peroxidases. University of Geneva, Switzerland, pp 21–29
- Rasmussen SK, Welinder KG, Hejgaard J. cDNA cloning, characterization and expression of an endosperm-specific barley peroxidase. Plant Mol Biol. 1991b;16:317–327. doi: 10.1007/BF00020562. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schünmann PHD, Harrison J, Ougham HJ. Slender barley, an extension growth mutant. J Exp Bot. 1994;45:1753–1760. [Google Scholar]
- Scott-Craig JS, Kerby K, Stein BD, Somerville SC. Expression of an extracellular peroxidase that is induced in barley (Hordeum vulgare) by the powdery mildew pathogen (Erysiphe graminis f.sp. hordei) Physiol Mol Plant Pathol. 1995;47:407–418. [Google Scholar]
- Smith TA, Best G. Distribution of the hordatines in barley. Phytochemistry. 1978;17:1093–1098. [Google Scholar]
- Stoessl A. The antifungal factors in barley. IV. Isolation, structure, and synthesis of the hordatines. Can J Chem. 1967;45:1745–1760. [Google Scholar]
- Stoessl A, Unwin CH. The antifungal factors in barley. V. Antifungal activity of the hordatines. Can J Bot. 1970;48:465–470. [Google Scholar]
- Theilade B, Rasmussen SK. Structure and chromosomal localization of the gene encoding barley seed peroxidase BP 2A. Gene. 1992;118:261–266. doi: 10.1016/0378-1119(92)90197-w. [DOI] [PubMed] [Google Scholar]
- Theilade B, Rasmussen SK, Rosenkrands I, Frøkier H, Hejgaard J, Theilade J, Pihakaski-Maunsbach K, Maunsbach AB (1993) Subcellular localization of barley grain peroxidase by immuno-electron microscopy. In KG Welinder, SK Rasmussen, C Penel, H Greppin, eds, Plant Peroxidases: Biochemistry and Physiology. University of Geneva, Switzerland, pp 321–324
- Thordal-Christensen H, Brandt J, Cho BH, Rasmussen SK, Gregersen PL, Smedegaard-Petersen V, Collinge DB. cDNA cloning and characterization of two barley peroxidase transcripts induced differentially by the powdery mildew fungus Erysiphe graminis. Physiol Mol Plant Pathol. 1992;40:395–409. [Google Scholar]
- Valé GP, Torrigiani E, Gatti A, Delogu G, Potta-Puglia A, Vannacci G, Cattivelli L. Activation of genes in barley roots in response to infection by two Drechslera graminea isolates. Physiol Mol Plant Pathol. 1994;44:207–215. [Google Scholar]
- Van Huystee RB, McManus MT. Glycans of higher plant peroxidases: recent observations and future speculations. Glycoconjugate J. 1998;15:101–106. doi: 10.1023/a:1006955903531. [DOI] [PubMed] [Google Scholar]
- von Röpenack E, Parr A, Schulze-Lefert P. Structural analyses and dynamics of soluble and cell wall-bound phenolics in a broad spectrum resistance to the powdery mildew fungus in barley. J Biol Chem. 1998;273:9013–9022. doi: 10.1074/jbc.273.15.9013. [DOI] [PubMed] [Google Scholar]
- Wei YD, de Neergaard E, Thordal-Christensen H, Collinge DB, Smedegaard-Petersen V. Accumulation of a putative guanidine compound in relation to other early defense reactions in epidermal cells of barley and wheat exhibiting resistance to Erysiphe graminis f.sp. hordei. Physiol Mol Plant Pathol. 1994;45:469–484. [Google Scholar]
- Welinder KG. Amino acid sequence studies of horseradish peroxidase: amino and carboxyl termini, cyanogen bromide and tryptic fragments, the complete sequence, and some structural characteristics of horseradish peroxidase C. Eur J Biochem. 1979;96:483–502. doi: 10.1111/j.1432-1033.1979.tb13061.x. [DOI] [PubMed] [Google Scholar]
- Welinder KG. Superfamily of plant, fungal and bacterial peroxidases. Curr Opin Struct Biol. 1992;2:388–393. [Google Scholar]