Abstract.
Between 2012 and 2016, over 80% of registered malaria cases in Anhui province were Plasmodium falciparum returned from Africa. However, drug-resistance marker polymorphisms in imported P. falciparum cases have not been assessed. This study looked at the distribution of antimalarial–drug resistance by evaluating K13-propeller, pfmdr1, and pfcrt gene mutations. Fourteen synonymous and 15 nonsynonymous mutations in the K13-propeller gene were detected in samples from nine African countries, yet no candidate and validated K13 resistance mutations were found. The prevalence of pfcrt K76T and pfmdr1 N86Y mutants was 27.7% and 19.9%, respectively. Six different pfcrt genotypes were found, with C72V73M74N75T76 being the most common (89.2%). The pfcrt 76-pfmdr1 86 haplotype combination was evaluated in 173 isolates, and the N86T76 genotype was the most prevalent (50.3%). Notably, the prevalence of the N86Y mutation in Africa marked a decline from 31.0% in 2012 to 8.2% in 2016. Our findings suggest that there is no immediate threat to artemisinin efficacy in imported P. falciparum infections returned from Africa to Anhui province. Nevertheless, pfcrt K76T and pfmdr1 N86Y mutations were modestly prevalent, suggesting the presence of chloroquine resistance in these cases. Accordingly, dihydroartemisinin + piperaquine may be a better choice than artesunate + amodiaquine for the treatment of uncomplicated P. falciparum infections in Anhui province. In addition to, artemether–lumefantrine can be introduced as an alternative measure.
INTRODUCTION
Malaria is a serious global public health problem. At the beginning of 2016, malaria was considered to be endemic in 91 countries and territories. Updated estimates point to 212 million cases globally in 2015, leading to 429,000 deaths.1 In China, the action plan for the elimination of malaria aims to eliminate local malaria transmission nationwide. The plan was initiated in 2010 on a national and local scale, including in Anhui province, in eastern China. Indigenous malaria in Anhui has been effectively controlled through comprehensive intervention, and no indigenous cases were reported in 2014. Nevertheless, imported malaria has been a great public health challenge because of frequent travel worldwide. Plasmodium falciparum accounted for more than 80% of registered malaria cases have been identified as the main agent responsible for the increasing impact.
Artemisinin-based combination therapies (ACTs) are well tolerated and highly effective, and have been recommended as first-line treatment against uncomplicated P. falciparum infection by the World Health Organization.2 Morbidity and mortality of malaria in highly prevalent areas of the world have fallen dramatically because of the use of ACTs.3 However, the curative effect of ACTs has declined gradually along with its use. The emergence of P. falciparum resistance to artemisinin and its derivatives was first reported in 2008 in western Cambodia.4 Then, ACT–resistance became prevalent in the Greater Mekong Subregion.5–7 K13-propeller was identified as a molecular marker of artemisinin resistance in 2014 by Ariey et al.8 Since then, numerous studies have looked at K13-propeller mutations. K13-propeller mutations associated with artemisinin resistance have been found mainly in Southeast Asia. A major concern nowadays is whether artemisinin resistance has spread westward to Africa. Some studies reported that no mutations associated with artemisinin resistance were detected in Africa.9,10 However, recently, Lu et al.11 described a previously unreported nonsynonymous single-nucleotide polymorphism (SNP) at amino acid position 579 (M579I), which displayed a substantially higher survival rate than control P. falciparum in an in vitro ring-stage survival assay. Feng et al.12 reported that polymorphism C580Y was associated with delayed clearance in samples from Ghana. To understand discrepancies between different studies, further dynamic surveillance will be needed. To gather new evidence, we have evaluated the prevalence of K13-propeller gene polymorphisms in imported P. falciparum cases returned from Africa between 2012 and 2016. It should be noted that outside the Greater Mekong Subregion, treatment failure with ACTs has occurred in the absence of artemisinin resistance and can be ascribed mainly to partner drug resistance.13 Previous studies have confirmed that mutation K76T in the pfcrt gene can reduce the parasite’s susceptibility to antimalarial drugs, such as amodiaquine, piperaquine (PPQ),14 and quinine.15 Similarly, mutation N86Y in the pfmdr1 gene was associated with treatment failure in P. falciparum malaria cases given amodiaquine (odds ratio = 5.4, 95% confidence interval = 2.6–11.2).16 Therefore, the present study reports the assessment of antimalarial resistance marker polymorphisms in pfmdr1, pfcrt, and K13-propeller genes in P. falciparum samples originating from Africa. The results might provide rational evidence for further molecular surveillance of drug-resistant P. falciparum in Anhui province, and will be useful for developing and updating antimalarial guidelines.
METHODS
Sample collection and DNA extraction.
Blood samples were obtained from patients who returned from Africa to Anhui Province between 2012 and 2016 before initiating antimalarial treatment. The final diagnosis was confirmed by microscopic examination of Giemsa-stained thick blood films and fluorescent–polymerase chain reaction (PCR). Approximately 200 μL of finger-prick blood was spotted on a piece of Whatman filter paper (3MM) and air-dried. The samples were labeled with study numbers, names, and dates and stored at −20°C until DNA extraction. DNA was extracted using the QIAamp DNA Mini Kit (QIAGEN Inc., Hilden, Germany) according to the manufacturer’s instructions and stored at −20°C for use in PCR assays.
Genotyping.
Primers and PCR conditions used to amplify the K13-propeller gene were as reported in the Artemisinin Resistance Multicenter Assessment study.17 Nested PCR was performed to amplify the fragments of the pfmdr1 and pfcrt gene. First-round PCR was performed in a 30-μL reaction volume containing 12 μL of ddH2O, 1 μL each of the forward and reverse primers (10 μmol/L; Tianyi Huiyuan Biotech Co., Ltd., Wuhan, China), 15 μL of 2 × Power Taq PCR MasterMix (Tianyi Huiyuan Biotech Co., Ltd.), and 1 μL of DNA template. PCR reaction conditions were: 95°C for 15 minutes; followed by 30 cycles at 95°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds; and a final extension at 72°C for 5 minutes. The second round contained 1 μL of the first-round product as the template. Except for the number of cycles, which was increased to 35, all other conditions were as in the first round. The amplified products were sequenced by Tianyi Huiyuan Biotech Co., Ltd.
First-round primers were as follows: pfcrt-f: CCCTTGTCGACCTTAACAGATG, pfcrt-r: AAAATGACTGAACAGGCATCTAAC; pfmdr1-f: AGAGCAGAAAGAGAAAAAAGATGGT, pfmdr1-r: TTAATATAGTCTTTTCTCCACAATA; Second-round primers were as follows: pfcrt-f: TCTTGGTAAATGTGCTCATGTG, pfcrt-r: AAA-GTTGTGAGTTTCGGATGTT; pfmdr1-f: TTTTTACCGTTTAAATGTTTACCTG, pfmdr1-r: AAACCTAAAAAGGAACTGGCAT.
Sequencing alignments and data analysis.
Sequences were analyzed using the Basic Local Alignment Search Tool program (http://blast.ncbi.nlm.nih.gov/). Reference sequences were downloaded from PlasmoDB (http://www.plasmodb.org, Pfcrt: PF3D7_0709000, Pfmdr1: PF3D7_0523000, K13-propeller: PF3D7_1343700). Multiple nucleotide sequence alignments and analysis were performed using the BioEdit 7.2 tool (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Data were analyzed using Microsoft Excel (Microsoft, Redmond, WA) and SPSS 17.0 (IBM, Chicago, IL). The map was created by MapInfo 15.0 (Pitney Bowes, Troy, NY). The χ2 test or Fisher’s exact test was used to assess differences. All reported P values were two sided, and a P value < 0.05 was considered statistically significant.
RESULTS
Patients’ epidemiological characteristics.
A total of 460 samples were collected from P. falciparum cases returned from 26 countries in Africa to Anhui Province during 2012–2016. Of these, 433 samples were successfully sequenced and included in the study. The annual number of cases were 29 (2012), 121 (2013), 107 (2014), 78 (2015), and 98 (2016). The male:female ratio was 47:1 (424/9), and mean age was 42.0 ± 8.7 years. Most patients recovered from malaria, except for four patients who died; K13-propeller mutations were observed in two samples from these four patients.
K13-propeller point mutations.
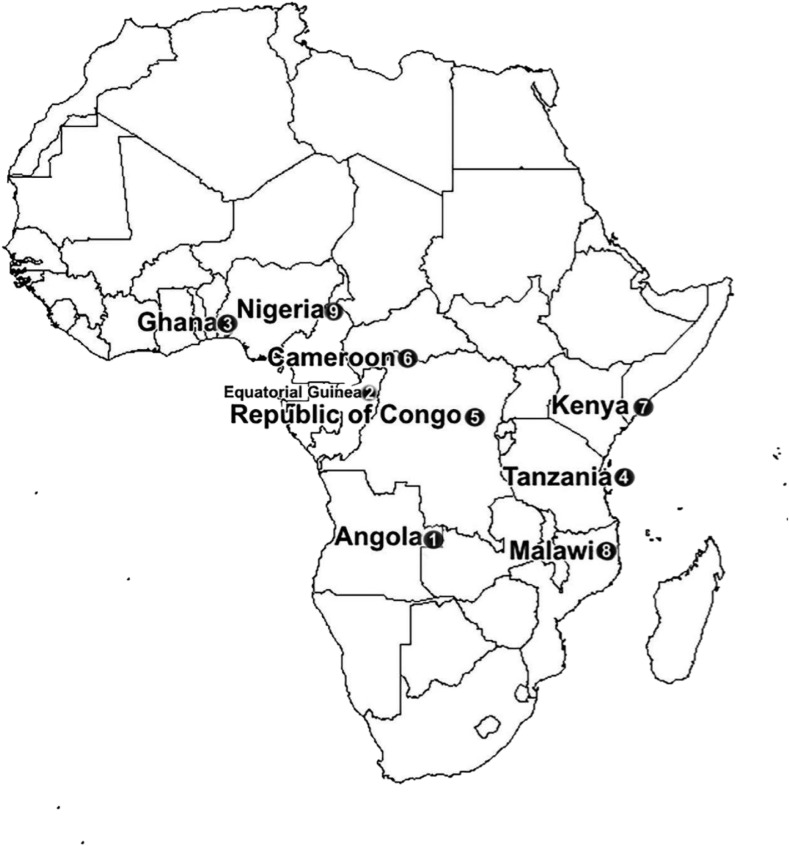
An 809-bp fragment was amplified by nested PCR for each of the 433 samples. All products were successfully sequenced, and 7.4% (32/433) contained SNPs at 24 locations. Fourteen synonymous and 15 nonsynonymous mutations were detected. The synonymous mutations were C469C, A676A, G454G, R417R, V589V, P443P, R471R, V510V, Y493Y, G533G, V666V, R575R, S477S, and T685T. Except for C469C (0.5%, N = 2), the others were detected only once. Of the 15 nonsynonymous mutations, the most prevalent was A578S (1.2%, N = 5), whereas the rest were observed only once. At position 613, we observed mutations Q613E and Q613H. No candidate and validated K13-resistance mutations were found in our study. However, we detected a C469F mutation. At this point, previous studies confirmed that C469Y mutation was reported to be associated with artemisinin-resistant in vivo or in vitro tests.13 Furthermore, two patients were found to have mixed genotypes: E619S621 and Y434N437, respectively (Table 1). Notably, the latter was found in one of the reported deaths. Details of K13 genotyping by site and year are provided in Table 1 and Figure 1.
Table 1.
Nonsynonymous polymorphisms observed in the K13-propeller gene in Plasmodium falciparum isolates
| Country (n) | Codon position | Amino acid reference | Nucleotide reference | Amino acid mutation | Nucleotide mutation* | Prevalence of mutation (%) |
|---|---|---|---|---|---|---|
| Angola (139) | 443 | P | CCA | R | cGT | 0.71 (N = 1) |
| 589 | V | GTC | I | gtT | 0.71 (N = 1) | |
| 613 | Q | CAA | E | Gaa | 0.71 (N = 1) | |
| 434† | F | TTT | Y | tAt | 0.71 (N = 1) | |
| 437† | T | ATT | N | aAt | 0.71 (N = 1) | |
| Equatorial Guinea (82) | 578 | A | GCT | S | Tct | 3.66 (N = 3) |
| 469 | C | TGC | F | tTT | 1.21 (N = 1) | |
| 462 | D | GAT | N | Aat | 1.21 (N = 1) | |
| Republic of Congo (16) | 634 | I | ATA | T | aCa | 6.25 (N = 1) |
| 578 | A | GCT | S | Tct | 6.25 (N = 1) | |
| Ghana (22) | 619† | L | TTA | E | GAa | 4.55 (N = 1) |
| 621† | A | GCT | S | AGC | 4.55 (N = 1) | |
| 578 | A | GCT | S | Tct | 4.55 (N = 1) | |
| Cameroon (26) | 457 | L | TTA | S | tCa | 3.84 (N = 1) |
| Kenya (2) | 683 | I | ATA | R | aGa | 50.00 (N = 1) |
| Tanzania (3) | 613 | Q | CAA | H | caT | 33.33 (N = 1) |
| 488 | L | TTG | V | Gtg | 33.33 (N = 1) |
Mutations are in boldface.
Two mixed genotypes (E619S621 and Y434N437) were found in two patients, respectively.
Figure 1.
Geographical distribution of K13-propeller mutations in Africa during 2012–2016.
Mutational analysis of pfcrt and pfmdr1.
The fragment of the pfcrt gene covering codons 72–76 and that of the pfmdr1 gene covering codon 86 were successfully sequenced in 433 samples. The prevalence of pfcrt mutations C72S, M74I, N75K, and K76T was 0.7% (3/433), 5.5% (24/433), 1.9% (8/433), and 27.7% (120/433), respectively (Table 2). Mutations of the pfcrt gene in codons 72–76 were found in 120 isolates. Six different pfcrt genotypes were found, among which C72V73M74N75T76 was the most common (89.2%) (Table 3). Mutation N86Y was detected in 86 isolates from 13 countries in Africa (Table 2) with 19.9% (86/433) prevalence. Notably, the prevalence of N86Y in Africa exhibited a decline from 31.03% in 2012 to 8.2% in 2016 (Table 4). The pfcrt 76-pfmdr1 86 haplotype combination was detected in 173 isolates, and the N86T76 genotype was the most prevalent (50.3%, 87/173) (Table 3).
Table 2.
Geographical distribution of pfmdr1 and pfcrt mutants based on the country of origin
| Nation | Cases | pfmdr1 | pfcrt | |||
|---|---|---|---|---|---|---|
| N86Y | C72S | M74I | N75K | K76T | ||
| Angola | 139 | 20 | 1 | 10 | 3 | 53 |
| Equatorial Guinea | 82 | 35 | 1 | 4 | 1 | 16 |
| Nigeria | 46 | 6 | 1 | 1 | 1 | 17 |
| Cameroon | 26 | 7 | 0 | 1 | 1 | 5 |
| Ghana | 22 | 0 | 0 | 1 | 1 | 1 |
| Democratic Republic of the Congo | 18 | 4 | 0 | 0 | 0 | 3 |
| Republic of Congo | 16 | 2 | 0 | 0 | 0 | 8 |
| Zambia | 16 | 0 | 0 | 0 | 0 | 0 |
| Guinea | 10 | 2 | 0 | 1 | 0 | 3 |
| Ivory Coast | 8 | 1 | 0 | 1 | 1 | 1 |
| Gabon | 7 | 3 | 0 | 1 | 0 | 3 |
| Malawi | 7 | 0 | 0 | 0 | 0 | 0 |
| Mozambique | 5 | 0 | 0 | 0 | 0 | 0 |
| Ethiopia | 4 | 2 | 0 | 0 | 0 | 3 |
| Benin | 4 | 0 | 0 | 1 | 0 | 3 |
| Uganda | 4 | 0 | 0 | 2 | 0 | 0 |
| Sierra Leone | 3 | 1 | 0 | 0 | 0 | 1 |
| Tanzania | 3 | 0 | 0 | 0 | 0 | 0 |
| Togo | 2 | 0 | 0 | 0 | 0 | 0 |
| Kenya | 2 | 0 | 0 | 0 | 0 | 0 |
| Madagascar | 2 | 2 | 0 | 0 | 0 | 0 |
| Niger | 2 | 0 | 0 | 0 | 0 | 0 |
| Chad | 2 | 1 | 0 | 0 | 0 | 1 |
| Algeria | 1 | 0 | 0 | 0 | 0 | 1 |
| Unknown* | 1 | 0 | 0 | 1 | 0 | 1 |
| South Sudan | 1 | 0 | 0 | 0 | 0 | 0 |
| Total | 433 | 86 | 3 | 24 | 8 | 120 |
According to our investigation records, the patient was infected in Egypt. However, Egypt had already eradicated malaria in the 1990s. The patient should have been infected in other African countries. But, the exact place of the infection can not be confirmed now, because we lost contact with the patient in 2013.
Table 3.
Prevalence of genotypes of candidate genes
| Gene | Genotype* | Prevalence of mutation (%) |
|---|---|---|
| pfmdr1–pfcrt | N86T76 (N = 87) | 50.29 (87/173) |
| Y86K76 (N = 53) | 30.64 (53/173) | |
| Y86T76 (N = 33) | 19.08 (33/173) | |
| pfcrt | S72V73I74N75K76 (N = 1) | 0.83 (1/120) |
| C72V73I74K75T76 (N = 1) | 0.83 (1/120) | |
| S72V73M74N75T76 (N = 2) | 1.67 (2/120) | |
| C72V73I74K75K76 (N = 4) | 3.33 (4/120) | |
| C72V73I74N75T76 (N = 5) | 4.17 (5/120) | |
| C72V73M74N75T76 (N = 107) | 89.17 (107/120) |
Mutated amino acids are in boldface.
Table 4.
Annual prevalence of K76T and N86Y mutations in Angola, Nigeria, Equatorial Guinea, and Africa as a whole
| Mutation | Region | Prevalence (%) of mutation | χ2 | P | ||||
|---|---|---|---|---|---|---|---|---|
| 2012 | 2013 | 2014 | 2015 | 2016 | ||||
| K76T | Africa | 27.58 (8/29) | 28 (34/121) | 29.9 (31/107) | 28.21 (22/78) | 25.51 (25/98) | 0.34 | 0.987 |
| Angola | 23.08 (3/13) | 40.6 (13/32) | 38.46 (10/26) | 38.71 (12/31) | 40.54 (15/37) | 1.43 | 0.839 | |
| Nigeria | 33.33 (1/3) | 42.86 (6/14) | 41.67 (5/12) | 28.57 (2/7) | 30.00 (3/10) | 0.99 | 0.941 | |
| Equatorial Guinea | 33.33 (2/6) | 13.79 (4/29) | 23.33 (7/30) | 8.33 (1/12) | 40.00 (2/5) | 4.15 | 0.348 | |
| N86Y | Africa | 31.03 (9/29) | 30.58 (37/121) | 22.4 (24/107) | 10.26 (8/78) | 8.16 (8/98) | 24.40 | 0.000 |
| Angola | 30.77 (4/13) | 25.00 (8/32) | 11.54 (3/26) | 12.9 (4/31) | 2.7 (1/37) | 10.32 | 0.024 | |
| Nigeria | 0.00 (0/3) | 28.57 (4/14) | 16.67 (2/12) | 0.00 (0/7) | 0.00 (0/10) | 4.62 | 0.267 | |
| Equatorial Guinea | 33.33 (2/6) | 44.8 (13/29) | 43.33 (13/30) | 25.00 (3/12) | 80.00 (4/5) | 4.41 | 0.364 | |
DISCUSSION
As recommended by the WHO in China artesunate + amodiaquine (AS + AQ) and dihydroartemisinin + piperaquine (DHA–PPQ) have been used against uncomplicated P. falciparum infections in Anhui province since 2009.18,19 However, little is known about current drug resistance, especially among imported P. falciparum cases. Here, we report an evaluation of antimalarial resistance marker polymorphisms of the K13-propeller, pfcrt, and pfmdr1 genes. The results could provide useful suggestions for a more rational administration of antimalarial drugs.
In 2014, using whole-genome sequencing of an artemisinin-resistant parasite line from Africa and clinical parasite isolates from Cambodia, Ariey et al.8 were the first to report an association between K13-propeller and artemisinin resistance. To date, more than 200 nonsynonymous mutations have been reported in the K13-propeller gene.13 N458Y, Y493H, R539T, R561H, I543T, and C580Y have been validated as associated with artemisinin resistance. These mutations are found mainly in Southeast Asia, with C580Y being the most prevalent. Results of the therapeutic efficacy studies conducted in 2015 in Cambodia, Lao PDR, and Vietnam have shown that C580Y has become the dominant mutation ranging from 48.8% in Lao PDR to 92.6% in Cambodia.13 In recent years, the possibility that artemisinin resistance has spread westward to Africa has attracted worldwide attention. To address such concern, extensive studies have been carried out in Africa, but the results have not always been consistent. Here, we evaluated the prevalence of K13-propeller gene polymorphisms in imported P. falciparum cases returned from Africa between 2012 and 2016. In this study, the gene encoding the K13-propeller domain was successfully sequenced in 433 samples. We identified 29 point mutations in 32 samples, of which 14 were synonymous and 15 were nonsynonymous. No candidate and validated K13-resistance mutations were found among nonsynonymous mutations. The most prevalent nonsynonymous mutation was A578S, which was found to be susceptible to artemisinin in vitro using a ring-stage survival assay.17 Notably, two of the deceased patients presented mutations F434Y, T437N, and I683R, and one exhibited a mixed Y434N437 genotype. Unfortunately, no further clinical studies were carried out at this time; therefore, it remains to be determined whether mutations F434Y, T437N, and I683R are associated with altered drug sensitivity. Surprisingly, we could not detect mutation C580Y in the 23 isolates from Ghana, which is in contrast with the study by Feng et al.,12 but could be possibly explained by the small sample pool. Similarly, we could not detect mutations R539T, P574L, and M579I, which had been previously associated with artemisinin resistance in samples returned to China from Africa.11,20 Our data indicate that the most prevalent African mutation was A578S, whereas other mutations had low prevalence. This puts our findings in line with those reported by Ménard et al.,17 whereby many K13-propeller mutations appeared to be neutral and failed to undergo clonal expansion in Africa. Such state of the facts would suggest no immediate threat to artemisinin efficacy.
In Africa, treatment failure with ACTs has occurred mainly because of partner drug resistance.13 Two genes in P. falciparum have been identified as candidates for chloroquine resistance: the multidrug-resistance gene pfmdr1, and the chloroquine resistance transporter gene pfcrt. The pfcrt K76T mutation is the primary determinant of resistance21,22 and can be enhanced by the pfmdr1 N86Y mutation.23 These two mutations that present strong linkage disequilibrium,24 have been reported to be associated also with in vitro responses to chloroquine, mefloquine, lumefantrine, quinine, monodesethylamodiaquine, and DHA.25 This study reports a modest prevalence of pfcrt K76T and pfmdr1 N86Y mutations. Pfcrt, which is located on chromosome 7, was identified as the candidate gene for chloroquine resistance by genetic cross studies.26 Mutation K76T in pfcrt may result in altered chloroquine flux or reduced drug binding to hematin through an effect on digestive vacuole pH. In our study, K76T was detected in 120 isolates form 16 countries, and its prevalence was 27.7%. Annual mutation rates were 27.6%, 28.1%, 30.0%, 28.2%, and 25.5% from 2012 to 2016, respectively. Recently, a study carried out in Zambia found the disappearance of chloroquine-resistant malaria after the removal of chloroquine drug pressure.27 Similarly, Kiarie et al.28 reported that the prevalence of pfcrt K76T diminished 13 years after cessation of chloroquine use in Kenya. These results are consistent with our study; we did not detect the K76T mutation in samples from Kenya and Zambia. However, the prevalence of K76T was still existent in Angola (38.1%, 53/139) and Nigeria (37.0%, 17/46). Our results showed that the return of chloroquine-susceptible malaria to the entire African region was not synchronous. Chloroquine pressure was still present in Angola and Nigeria, explaining the maintenance of the K76T phenotype. Thus, use of chloroquine for P. falciparum treatment in these areas should be kept to a minimum. PFMDR1 belongs to the ATP-binding cassette transporter superfamily. The mutation in codon 86 of PFMDR1 is the most important because it alters the transport activity of the protein.29,30 In a meta-analysis, the N86Y mutation was found to be associated with amodiaquine failure, with an odds ratio of 5.4.16 Another report showed that the N86 allele decreased susceptibility to DHA, whereas the 86Y mutation was significantly associated with increased susceptibility to DHA (P = 0.0387).31 The result was confirmed by Veiga et al.,32 who demonstrated that the N86Y mutation enhanced susceptibility to lumefantrine, mefloquine, and DHA. In contrast, N86Y increases resistance to chloroquine and amodiaquine. In the present study, N86Y was detected in 86 isolates from 13 countries, with 19.86% prevalence. Notably, we observed a decline in the prevalence of N86Y in Africa: from 31.03% in 2012 to 8.2% in 2016. In this study, more than half of overall cases came back from Angola (139), Equatorial Guinea (82), and Nigeria (46). The decrease was observed in Angola (from 30.8% to 2.7%) and Nigeria (from 28.6% to 0%), but not in Equatorial Guinea. The decrease in the prevalence of the N86Y mutation might be caused by drug pressure,33 however, more evidence is required to confirm it.
In conclusion, no candidate or validated K13 resistance mutations were found in our study, which suggests that there is no immediate threat to artemisinin efficacy. However, the prevalence of pfcrt K76T and pfmdr1 N86Y was still present, which indicates the presence of imported chloroquine-resistant P. falciparum cases. In summary, present results and other published evidence indicate that: 1) Increasing use of amodiaquine in Africa threatens to cause a selective sweep of highly amodiaquine-resistant and chloroquine-resistant parasites with pfcrt and pfmdr1 mutations34; 2) The pfmdr1 N86Y mutation enhances susceptibility to lumefantrine, mefloquine, and DHA, but increases resistance to chloroquine and amodiaquine32; 3) The pfcrt K76T mutation is a drug-specific contributor to enhanced P. falciparum susceptibility to lumefantrine in vivo and in vitro35; 4) DHA–PPQ is recommended for use in settings where chloroquine resistance is high.29 We suggest that DHA–PPQ is a better choice than AS + AQ, and artemether–lumefantrine can also be introduced for the treatment of uncomplicated P. falciparum infections in Anhui province.
Acknowledgment:
We thank Jun Feng, He Yan, National Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention Shanghai, China, for technological guidance.
Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Anhui Provincial Center for Disease Control and Prevention.
REFERENCES
- 1.World Health Organization , 2016. World Malaria Report 2016. Available at: http://apps.who.int/iris/handle/10665/252038. Accessed October 1, 2017.
- 2.World Health Organization , 2015. Guidelines for the Treatment of Malaria, 3rd edition. Available at: http://www.portal.pmnch.org/malaria/publications/atoz/9789241549127/en/. Accessed October 1, 2017.
- 3.Wang LQ, Liu YH, Zhao SX, Yang ZQ, 2014. Advances in the study of artemisinin resistance in Plasmodium falciparum and methods of its detection. J Pathog Biol 9: 1142–1144. [Google Scholar]
- 4.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM, 2008. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med 359: 2619–2620. [DOI] [PubMed] [Google Scholar]
- 5.Müller O, Sié A, Meissner P, Schirmer RH, Kouyaté B, 2009. Artemisinin resistance on the Thai–Cambodian border. Lancet 374: 1418–1419. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, Shrestha S, Li X, Miao J, Yuan L, Cabrera M, Grube C, Yang Z, Cui L, 2015. Prevalence of K13-propeller polymorphisms in Plasmodium falciparum from China–Myanmar border in 2007–2012. Malar J 14: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takala-Harrison S, et al. 2015. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in southeast Asia. J Infect Dis 211: 670–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ariey F, et al. 2014. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505: 50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor SM, et al. 2015. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis 211: 680–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamau E, et al. 2015. K13-propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J Infect Dis 211: 1352–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu F, et al. 2017. Emergence of indigenous artemisinin-resistant Plasmodium falciparum in Africa. N Engl J Med 376: 991–993. [DOI] [PubMed] [Google Scholar]
- 12.Feng J, Li J, Yan H, Feng X, Xia Z, 2015. Evaluation of antimalarial resistance marker polymorphism in returned migrant workers in China. Antimicrob Agents Chemother 59: 326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Health Organization , 2017. Artemisinin and Artemisinin-Based Combination Therapy Resistance Available at: http://apps.who.int/iris/handle/10665/255213. Accessed October 1, 2017.
- 14.Eastman RT, Dharia NV, Winzeler EA, Fidock DA, 2011. Piperaquine resistance is associated with a copy number variation on chromosome 5 in drug-pressured Plasmodium falciparum parasites. Antimicrob Agents Chemother 55: 3908–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper RA, Lane KD, Deng B, Mu J, Patel JJ, Wellems TE, Su X, Ferdig MT, 2010. Mutations in transmembrane domains 1, 4 and 9 of the Plasmodium falciparum chloroquine resistance transporter alter susceptibility to chloroquine, quinine and quinidine. Mol Microbiol 63: 270–282. [DOI] [PubMed] [Google Scholar]
- 16.Picot S, Olliaro P, de Monbrison F, Bienvenu AL, Price RN, Ringwald P, 2009. A systematic review and meta-analysis of evidence for correlation between molecular markers of parasite resistance and treatment outcome in falciparum malaria. Malar J 8: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ménard D, et al. 2016. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N Engl J Med 374: 2453–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization , 2006. The application of artemisinin monotherapies for malaria treatment was demanded to end by WHO [In Chinese] ADRJ 8: 152. [Google Scholar]
- 19.National Health and Family Planning Commission of the People’s Republic of China , 2009. Antimalarial Drug Policy in China Available at: http://www.nhfpc.gov.cn/zwgkzt/wsbysj/200907/41610.shtml. Accessed October 1, 2017.
- 20.Yang C, Zhang H, Zhou R, Qian D, Liu Y, Zhao Y, Li S, Xu B, 2017. Polymorphisms of Plasmodium falciparum k13-propeller gene among migrant workers returning to Henan Province, China from Africa. BMC Infect Dis 17: 560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fidock DA, et al. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell 6: 861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sidhu ABS, Verdierpinard D, Fidock DA, 2002. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science 298: 210–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duraisingh MT, Cowman AF, 2005. Contribution of the pfmdr1 gene to antimalarial drug-resistance. Acta Trop 94: 181–190. [DOI] [PubMed] [Google Scholar]
- 24.Awasthi G, Das A, 2013. Genetics of chloroquine-resistant malaria: a haplotypic view. Mem Inst Oswaldo Cruz 108: 947–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wurtz N, et al. 2014. Role of pfmdr1 in vitro Plasmodium falciparum susceptibility to chloroquine, quinine, monodesethylamodiaquine, mefloquine, lumefantrine, and dihydroartemisinin. Antimicrob Agents Chemother 58: 7032–7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fidock DA, et al. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell 6: 861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mwanza S, Joshi S, Nambozi M, Chileshe J, Malunga P, Kabuya JB, Hachizovu S, Manyando C, Mulenga M, Laufer M, 2016. The return of chloroquine-susceptible Plasmodium falciparum malaria in Zambia. Malar J 15: 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiarie WC, Wangai L, Agola E, Kimani FT, Hungu C, 2015. Chloroquine sensitivity: diminished prevalence of chloroquine-resistant gene marker pfcrt-76 13 years after cessation of chloroquine use in Msambweni, Kenya. Malar J 14: 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Danquah I, Coulibaly B, Meissner P, Petruschke I, Müller O, Mockenhaupt FP, 2010. Selection of pfmdr1 and pfcrt alleles in amodiaquine treatment failure in north-western Burkina Faso. Acta Trop 114: 63–66. [DOI] [PubMed] [Google Scholar]
- 30.Eastman RT, Dharia NV, Winzeler EA, Fidock DA, 2011. Piperaquine resistance is associated with a copy number variation on chromosome 5 in drug-pressured Plasmodium falciparum parasites. Antimicrob Agents Chemother 55: 3908–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wurtz N, et al. 2014. Role of pfmdr1 in in vitro Plasmodium falciparum susceptibility to chloroquine, quinine, monodesethylamodiaquine, mefloquine, lumefantrine, and dihydroartemisinin. Antimicrob Agents Chemother 58: 7032–7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veiga MI, Dhingra SK, Henrich PP, Straimer J, Gnädig N, Uhlemann AC, Martin RE, Lehane AM, Fidock DA, 2016. Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat Commun 7: 11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carrara VI, et al. 2009. Changes in the treatment responses to artesunate-mefloquine on the northwestern border of Thailand during 13 years of continuous deployment. PLoS One 4: e4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sá JM, Twu O, Hayton K, Reyes S, Fay MP, Ringwald P, Wellems TE, 2009. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci USA 6: 18883–18889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sisowath C, Petersen I, Veiga MI, Mårtensson A, Premji Z, Björkman A, Fidock DA, Gil JP, 2009. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. J Infect Dis 199: 750–757. [DOI] [PMC free article] [PubMed] [Google Scholar]