Abstract
Mucosal-associated invariant T cells (MAIT cells) recognize bacterial metabolites as antigen and are found in blood and tissues, where they are poised to contribute to barrier immunity. Recent data demonstrate that MAIT cells located in mucosal barrier tissues are functionally distinct from their blood counterparts, but the relationship and circulation of MAIT cells between blood and different tissue compartments remains poorly understood. Previous studies raised the possibility that MAIT cells do not leave tissue and may either be retained or undergo apoptosis. To directly address if human MAIT cells exit tissues, we collected human donor–matched thoracic duct lymph and blood and analyzed MAIT cell phenotype, transcriptome, and T cell receptor (TCR) diversity by flow cytometry and RNA sequencing. We found that MAIT cells were present in the lymph, despite being largely CCR7– in the blood, thus indicating that MAIT cells in the lymph migrated from tissues and were capable of exiting tissues to recirculate. Importantly, MAIT cells in the lymph and blood had highly overlapping clonotype usage but distinct transcriptome signatures, indicative of differential activation states.
Keywords: Immunology
Keywords: Cell migration/adhesion, Cellular immune response, Lymph
Human mucosal-associated invariant T (MAIT) cells circulate from tissues to the lymph and exhibit distinct functional responses across different tissue compartments.
Introduction
Mucosal-associated invariant T cells (MAIT cells) recognize bacterially derived metabolites in the context of MHC class I–related protein (MR1) (1). MAIT cells typically make up 1%–8% of T cells in human blood and mucosal tissues, and they appear most abundant in the liver, where as many as 20%–45% of T cells are MAIT cells (2, 3). Once activated, human MAIT cells express IFN-γ and granzyme B (4, 5), but a small subset can also secrete IL-17 when stimulated ex vivo (6, 7). A decrease in MAIT cell frequencies in the blood has been described in a series of studies including acute and chronic infections, cancer, and autoimmune disorders, indicating that MAIT cells respond and are thus relevant in a wide array of conditions (8–14). Interestingly, a common theme of these studies is an irreversible decrease or loss of MAIT cells in the blood in chronic infections (HIV, hepatitis C virus [HCV], and hepatitis B virus [HBV]) and autoimmune diseases (11–15). It is unclear if this is due to a systemic loss of MAIT cells or one-directional migration into tissues, leading to a decrease in the periphery. MAIT cells have an effector memory–like phenotype in the sense that they respond quickly to restimulation but do not express C-C chemokine receptor type 7 (CCR7) and L-selectin (CD62L) (2). This effector memory–like designation refers to a conventional memory T cell subset that has, in part, been defined based on the ability of cells to enter the lymph node. A landmark study by Sallusto and colleagues nearly 20 years ago demonstrated unique functional properties in these memory subsets and coined the terms central memory T cells and effector memory T cells (16). Expression of CCR7 and CD62L by a T cell is required for entering a lymph node from the blood via high endothelial venules (HEV). The expression patterns of CCR7 and its chemokine ligands CCL19 and CCL21 have been defined in mice and humans and indicate that the same principal mechanisms apply in both cases (17). In general, only naive and central memory T cells express both CCR7 and CD62L, but not effector memory cells (18, 19) or MAIT cells.
The trafficking pattern and relationship of conventional T cells in nonlymphoid tissues, lymph/lymph nodes, and blood has been extensively studied in animal model systems (20, 21), but little data are available from human studies. Animal studies have also demonstrated that some memory T cells reside permanently in tissues (tissue-resident memory T cells [Trm]), where they can get directly reactivated by antigen-presenting cells upon antigen reencounter (22, 23). Elegant studies using human tissue obtained from biopsies and organ donors have further provided evidence for the existence of human Trm within the conventional T cell compartment (24–28). Given the decrease of MAIT cells from the periphery in various inflammatory disease scenarios and data showing that MAIT cells in tissues express CRTAM (6), a gene that confers retention in tissues (29), we wanted to address if human MAIT cells are, in principle, capable of leaving tissues, which is a critical piece of information needed to understand the role of MAIT cells in tissue-based immune responses.
Results
MAIT cell chemokine receptor expression pattern suggests that MAIT cells in the lymph migrated from the tissue.
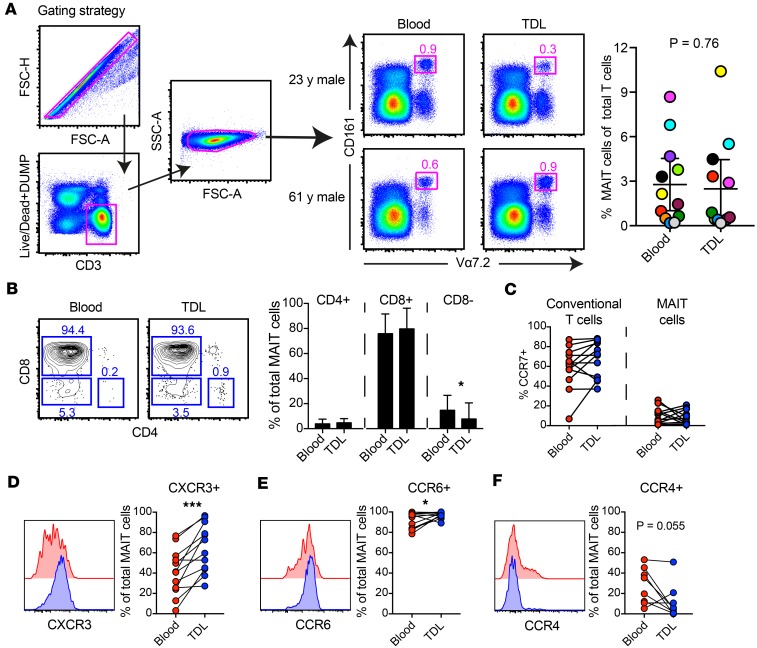
We measured the frequency of MAIT cells (CD161hiVα7.2+) within the T cell (CD3+) compartment in blood and thoracic duct lymph and found that MAIT cells are present in both (Figure 1, A and B). Most blood and lymph MAIT cells were CD8+ (Figure 1B), which is also the most abundant MAIT cell subset in tissues (13).
Figure 1. Characterization of MAIT cells in human lymph compared with peripheral blood.
(A) Gating strategy, frequencies and (B) representative flow plots and subsets of CD161hiVα7.2+ MAIT cells gated on live CD3+ cells from human lymph and matched peripheral blood (n = 12 donors). (C) Comparison of CCR7 expression between total CD3+ cells and MAIT cells. Surface expression of (D) CXCR3, (E) CCR6, and (F) CCR4 on MAIT cells in human lymph and peripheral blood. Each point corresponds to 1 patient, and lines connect matched samples. Wilcoxon matched-pairs signed rank tests were performed. *P ≤ 0.05 , ***P ≤ 0.001.
We compared CCR7 expression of MAIT cells and other T cells and found that most conventional T cells in the lymph are CCR7+, while most MAIT cells in blood and lymph are CCR7– (Figure 1C). We interrogated expression patterns of CXCR3, CCR6, and CCR4. We found that more MAIT cells in the lymph expressed CXCR3 and CCR6 compared with their blood counterparts (Figure 1, D and E), while more MAIT cells in the blood expressed CCR4 compared with MAIT cells in the lymph (Figure 1F). Together, these data demonstrate that MAIT cells in the lymph are equipped to respond to proinflammatory chemotactic cues sensed by CXCR3 (CXCL9, -10, and -11) (30) and CCR6-mediated tissue homing.
Selection of a MAIT cell subset for RNA-seq analysis.
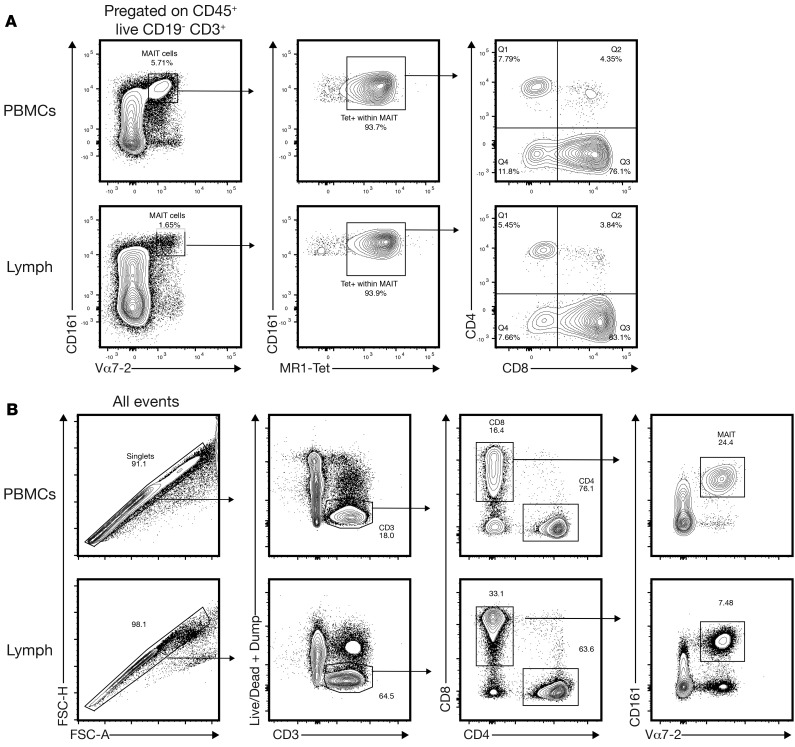
MAIT cells were initially defined as CD161hiVα7.2+ cells. The recent development of MR1 tetramers also allows for an alternative identification of MAIT cells (and independently of CD161 expression levels, which may change; ref. 31). Importantly, these 2 populations have been reported to be nearly, but not fully, congruent (32, 33). We found that almost all CD161hiVα7.2+ cells were MR1-tetramer+ (Figure 2A, left and middle panel) and CD8+ (Figure 2A, right panel). The same was true when the gating scheme was reversed (almost all MR1-tetramer+ cells are CD161hiVα7.2+ cells; data not shown) indicating that these 2 populations are nearly identical in the lymph, as has been previously shown in blood. We next wanted to interrogate MAIT cells in lymph and blood in an unbiased manner by RNA sequencing (RNA-seq) analysis. Importantly, when designing the RNA-seq experiment, we considered that CD8– and CD8+ MAIT cells isolated from human blood and mucosal tissues have distinct transcriptional profiles (6). We therefore focused our analysis on the more abundant CD8+ MAIT cell subset, which we identified as CD3+CD8+CD161hiVα7.2+ (Figure 2B). We sorted ~200 CD8+ MAIT cells from blood and lymph from 4 donors by FACS for subsequent RNA-seq analysis. Importantly, we had technical replicates for 6 of 8 samples (lymph and blood for each of the 4 donors), which all clustered when analyzed in a multidimensional scaling plot, demonstrating that there is little to no technical noise or variability in our samples (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.98487DS1).
Figure 2. CD161hiVα7.2+ are nearly uniformly MR1-tetramer+ (A) Donor-matched human lymph and peripheral blood were analyzed for CD161, Vα7.2, and MR1-tetramer staining (n = 4 donors).
CD161hiVα7.2+ MAIT cells (gated on live CD3+ cells) stain nearly uniformly positive with MR1-tetramer. (B) MAIT sorting scheme for the RNA-seq experiments.
Equivalent TCR usage of MAIT cells in the blood and lymph.
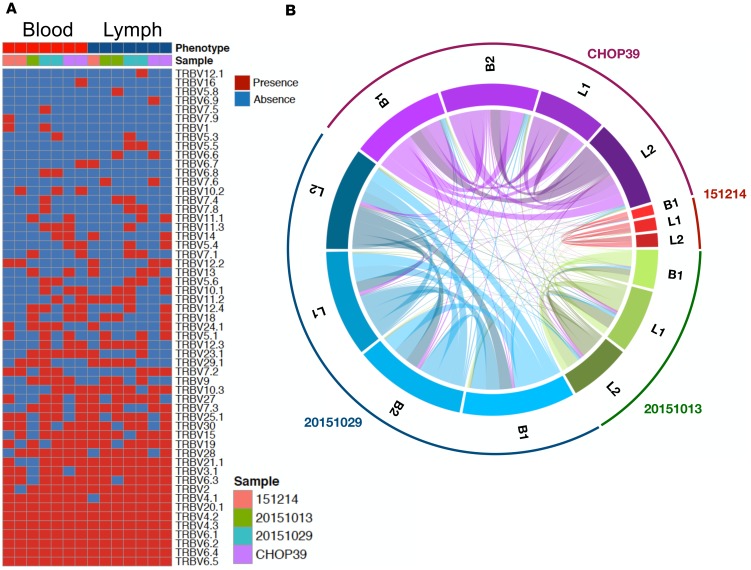
To gain a much more detailed understanding of the relationship of MAIT cells in lymph and blood, we used an RNA-seq approach to determine whether the MAIT cell populations in these 2 compartments are comparable since a lack of recirculation would result in disparate MAIT T cell receptor (TCR) repertoires in blood and lymph. We first mined the RNA-seq dataset for TCRα and -β expression to understand the clonal relationship between these populations. We found that MAIT cells in blood and lymph had a very similar TCRα repertoire, as is expected given the nature of the semi-invariant TCR (data not shown). The TCRβ repertoire for each donor and tissue is shown in Figure 3A. Importantly, since the cDNA is amplified as part of the RNA-seq protocol, we cannot confidently compare the frequencies of TCRα and -β usage in blood and lymph, but we can examine diversity by comparing the number of overlapping clonotypes (i.e., not just shared β usage, but also shared CDR3 region) in the 2 tissue compartments. We found that TCRβ clonotypes were heavily shared between blood and lymph for each donor (Figure 3B). Matching clonotypes are connected by a line, which grows in thickness when the number of matching clonotypes increases (Figure 3B). These data suggest that, in principle, most clonotypes have the ability to exit tissue and leave via the lymphatics.
Figure 3. MAIT cells in blood and lymph have equivalent TCR diversity.
(A) TCRβ usage in each of the n = 4 donors and tissue is listed. The Vβ chains found in the donors are listed on the right side. Red (present) and blue (absent) squares indicate whether a particular Vβ was found in blood and lymph of each donor. Each donor is color coded and listed at the top of the graph. When available, both technical replicates are shown (3 of 4 samples in each, blood and lymph). (B) The clonotype overlap between blood (when available, technical replicates for each donor are listed as B1 and B2) and lymph samples (when available, technical replicates for each donor are listed as L1 and L2) is illustrated by visualizing the number of shared clonotypes. Clonotypes are connected by a line, which increases in thickness with the number of shared clonotypes.
Differential activation signature of MAIT cells in the blood and lymph.
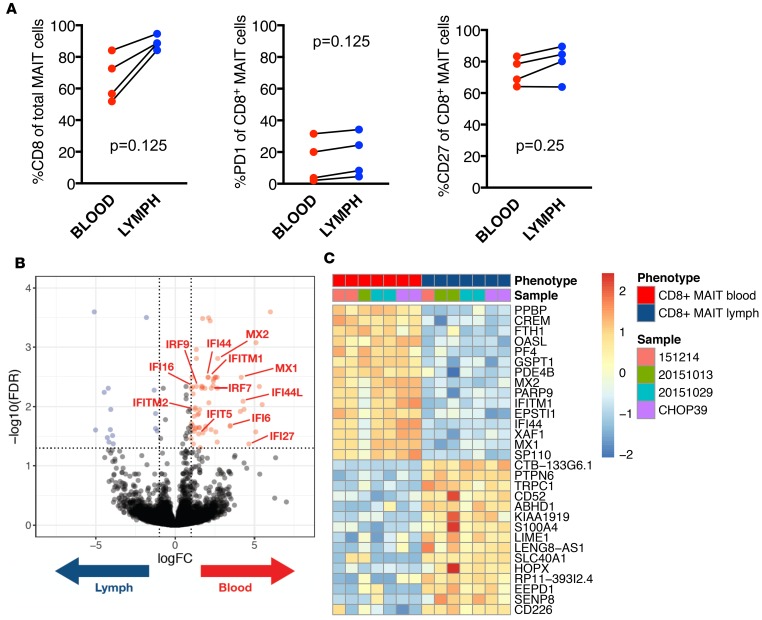
CD8+ MAIT cells in blood and lymph were present at typical frequencies (Figure 4A, left panel) and had similar PD-1 expression patterns (Figure 4A, middle panel), indicating that MAIT cells in lymph do not have exacerbated immune activation or dysfunction. MAIT cells had similar CD27 expression in lymph and blood (Figure 4A, right panel), which was comparable to conventional T cell populations in the lymph (data not shown). CD27 is a member of the tumor necrosis factor receptor superfamily and provides costimulatory function. A decrease in CD27 expression is used as a biomarker for senescence and can also be used as an indicator of exposure to inflammation (34). Given the similar PD-1 and CD27 expression levels, these data could indicate a similar activation status of MAIT cells in blood and lymph. However, when we next compared the transcriptome of MAIT cells in blood and lymph in our 4 donors, we found 62 transcripts that were more highly expressed in CD8+ MAIT cells isolated from the blood versus lymph and 15 transcripts that were upregulated in lymph versus blood (Figure 4B). A substantial number of transcripts upregulated in the blood belong to the IFN-inducible gene family (Figure 4B), which were also part of the top differentially expressed genes (DEGs) (Figure 4C).
Figure 4. MAIT cells in blood and lymph have distinct transcriptional profiles.
(A) The frequency of PD-1 and CD27 expressing MAIT cells in blood (red) and lymph (blue) is shown for each of the 4 donors analyzed by RNA-seq. Wilcoxon matched-pairs signed rank tests were performed to determine statistical significance. (B) A Volcano plot reporting FDR as a function of log2 fold change between the samples from n = 4 donors is shown. We found 77 differentially expressed genes (FDR 5% and |log2FC| ≥ 1), 62 are upregulated in blood (colored in red), and 15 are upregulated in lymph (colored in blue). (C) The top 10 differentially expressed genes upregulated in blood or lymph MAIT cells are listed (20 total).
Protein expression analysis of 17 biomarkers shows differential clustering of MAIT cells isolated from the blood and lymph.
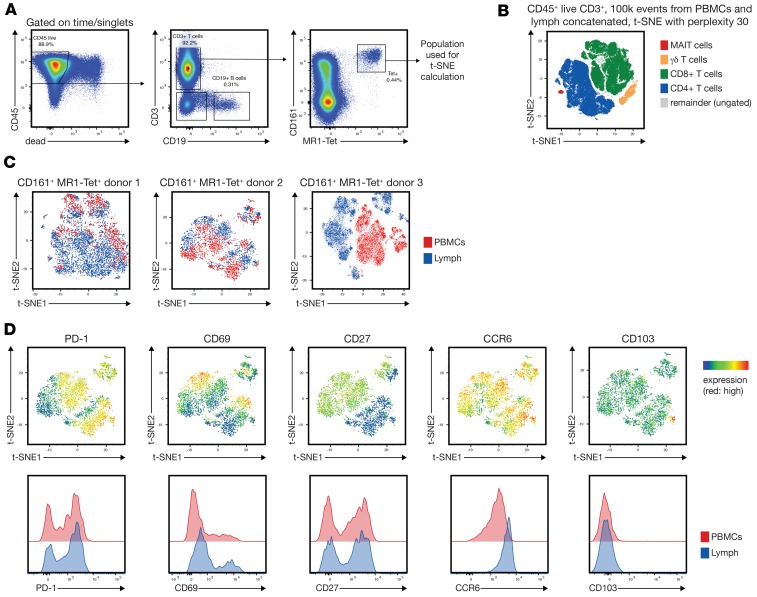
To further support the notion that MAIT cells in blood and lymph are distinct, we developed a 27-color flow cytometry panel to analyze MAIT cells in blood and lymph of 3 additional donors (Figure 5A). We recently described the development of such a high-parameter flow cytometry panel (35). Dimensionality reduction using t-stochastic neighbor embedding (t-SNE) clearly separated all canonical T cell populations, including MAIT cells (Figure 5B), and revealed phenotypic heterogeneity in MAIT cells isolated from blood and lymph (Figure 5C) based on the expression of 17 surface biomarkers (Supplemental Figure 2). For all 3 donors, the MAIT cells in blood versus lymph showed distinct phenotypic separation (Figure 5C). PD-1, CD27, and CCR6 expression patterns were similar to the donors shown in Figures 1 and 4 (Figure 5D); CD103 expression was only observed on very few MAIT cells in the lymph (Figure 5D).
Figure 5. MAIT cells in donor-matched blood and lymph differ phenotypically.
(A) The gating strategy to identify MAIT cells is shown. (B) Two hundred thousand total concatenated CD45+CD3+ events from blood (100,000) and lymph (100,000) were overlaid with manually gated T cell populations in a t-SNE plot as indicated. (C) t-SNE analysis of the blood and lymph-derived MAIT cells from 3 donors using the biomarkers listed in Supplemental Figure 2 demonstrates phenotypic differences based on tissue origin (blood vs. lymph). (D) A heatmap of selected markers from donor 2 (C, middle panel).
Discussion
Recent reports provide compelling evidence that MAIT cells located in mucosal barrier tissues are functionally distinct from their blood counterparts in regard to phenotype, activation status (6, 36), and functional properties, including IL-22 production in mucosal tissue–derived MAIT cells (36). However, the relationship and circulation of MAIT cells between blood and different tissue compartments remains poorly understood. We collected human donor–matched thoracic duct lymph and blood to analyze the MAIT cell population and determine if MAIT cells exit tissues and recirculate. The lymphocytes that are present in the thoracic duct lymph represent those cells that have egressed from nearly all tissues in the body, with the exception of the head/neck and right upper quadrant of the body (which drain via the lymphatic duct). We found that MAIT cells were present in the lymph (Figure 1A), indicating that they must have exited from tissues — either lymphoid tissue or other tissues. Mouse parabiosis experiments would be necessary to formally demonstrate that MAIT cells exit tissues and recirculate, which may be challenging given that MAIT cells in mice have a naive phenotype and, thus, likely different steady state trafficking. To address these questions with human MAIT cells, we used a combination of flow cytometry and RNA-seq analysis approaches to infer if MAIT cells can exit tissues and recirculate.
Since MAIT cells are largely CCR7– in the blood (Figure 1, B and C), this suggests that MAIT cells in the lymph exited from nonlymphoid tissues (as opposed to entering the lymph node from the blood via HEVs in a CCR7- and CD62L-dependent manner). We acknowledge that this conclusion rests on the assumption that MAIT cells enter the HEVs (from the blood) in a CCR7-depedent manner, similar to conventional T cells.
If MAIT cells in the lymph entered by tissue egress, one would also expect other phenotypic differences. We found that significantly more MAIT cells isolated from the lymph expressed CXCR3 and CCR6 compared with their blood counterparts (Figure 1, D and E), indicating that MAIT cells from blood and lymph differ in their ability to migrate to tissues (with MAIT cells in the lymph being ready to respond to these chemotactic cues). We used a broad 27-color immunophenotyping approach to further compare MAIT cells from blood and lymph via t-SNE analysis and found distinct clustering patterns based on their tissue origin. We only found a very limited number of CD103+ MAIT cells in the lymph (Figure 5D and data not shown), which could indicate that CD103+ MAIT cells found in mucosal tissue (37) are indeed tissue resident. Together, these data are consistent with the notion that MAIT cells in blood and lymph are phenotypically distinct and further suggest that MAIT cells in the lymph entered by tissue egress.
To further address the potential origin of MAIT cells in the lymph, we also compared MAIT cells from blood and lymph by RNA-seq analysis (Figures 3 and 4). A lack of recirculation would result in disparate MAIT TCR repertoires in blood and lymph, but we found that MAIT cells in the lymph and blood had highly overlapping clonotype usage (Figure 3), suggesting that MAIT cells recirculate between blood and tissues. Furthermore, our RNA-seq data revealed an increased IFN signature in MAIT cells isolated from the blood compared with lymph (Figure 4, B and C). This could be simply interpreted as MAIT cells in the blood receiving more IFN signals compared with the MAIT cells in the lymph, but this interpretation fails to explain the lack of induction of downstream effectors and the similarity in PD-1 and CD27 expression patterns. Interestingly, we observed higher expression levels of PTPN6 (SHP1) and HOPX in MAIT cells isolated from lymph (Figure 4C; the 2nd and 11th gene more highly expressed in lymph, respectively). SHP1 is a phosphatase and negative regulator of TCR signaling but is also involved in other signaling pathways (38). HOPX is a transcription cofactor that downregulates AP-1 (39), which is a key transcription factor complex downstream of cytokine and other signaling pathways. Since both HOPX and SHP1 decrease T cell responsiveness, we favor the interpretation that the increased IFN signature observed in the blood (relative to lymph) reflects a decrease in overall responsiveness to (possibly tonic) signals received by MAIT cells in the lymph. This change could be due to different signals that MAIT cells receive in tissue or lymph compared with blood. Alternatively, MAIT cells with such a phenotype may preferentially leave tissues. Single-cell data of MAIT cells isolated from human mucosal tissue indeed show such heterogeneity in regard to their activation status (6). Future studies will need to address whether MAIT cell functional responses are finely tuned across different tissue compartments. The notion of a differential tissue–dependent activation status would be in line with our previous observation that a population of MAIT cells in mucosal barrier tissues is poised to respond rapidly compared with MAIT cells in the blood (6).
Overall, our data clearly show that human MAIT cells are present in the lymph, thus capable of exiting (either lymphoid or other) tissues. Our flow cytometry and RNA-seq data support a model where MAIT cells can enter the lymph by tissue egress and recirculate via the lymphatics during steady state conditions. Finally, it will be interesting to address if the MAIT migration patterns change in conditions where a permanent decrease of MAIT cells in the blood has been observed, such as HIV+ patients despite antiretroviral treatment (11, 14, 15).
Methods
Human samples.
Thoracic duct lymph was collected from patients with chylopericardium and chylothorax (trauma or idiopathic disease). Sample size (n = 12) was based on the availability of biological samples rather than a prespecified effect size. The age of the participants varied from 4–67 years of age (mean, 30 years of age). Lymph was accessed as described previously (40, 41). Aspirated lymph was stored in heparin tubes and centrifuged over a Ficoll-Hypaque density gradient on the same day to remove contrast agents. Lymph samples were cryopreserved in 90% FBS/10% DMSO or used directly in flow cytometry experiments. Matched peripheral blood mononuclear cells (PBMCs) were collected from venous blood during the surgical intervention and, similar to above, centrifuged over a Ficoll-Hypaque density gradient and either cryopreserved or used directly.
Flow cytometry.
Fresh PBMCs and lymph samples were used for all phenotype experiments. Briefly, cells were washed with PBS and prestained for chemokine receptors for 10 minutes at 37°C. Cells were then stained with LIVE/DEAD Aqua (Invitrogen) for 10 minutes, and after that, they were stained with an optimized antibody cocktail for further 20 minutes to detect additional surface markers. Cells were then washed with FACS buffer (PBS containing 0.1% sodium azide and 1% BSA), fixed in PBS containing 1% paraformaldehyde (MilliporeSigma) and stored at 4°C. All samples were acquired within 3 days using an LSRII (BD Biosciences). Data were analyzed with FlowJo software (version 9.8.8 or higher, TreeStar Inc.). The gating strategies are depicted in Figure 1A. Cryopreserved cells were used for sorting. Cells were thawed and rested overnight in complete media; they were stained in 15-ml conical tubes following the procedure described above but with higher concentrations of antibodies (i.e., not diluted 1:50 in FACS buffer). Cells were then washed with PBS and suspended in complete media. All sorting experiments were carried out on a FACSAriaII (BD Biosciences).
For the 27-color phenotypic analysis, cryopreserved samples were thawed according to standard procedures, treated with Fc-Block (Human TruStain FcX, BioLegend) and UV Blue Live/Dead reagent (Thermo Fisher), stained with a Phycoerythrin-labelled MR1-Tetramer in FACS buffer for 30 minutes at room temperature (RT), and then with the remaining antibodies in Brilliant Staining buffer (BD Biosciences) for 20 minutes at RT, followed by fixation using 4.2% PFA (Cytofix/Cytoperm, BD Biosciences). MR1 tetramer was provided by the NIH Tetramer Core facility as permitted to be distributed by the University of Melbourne (Melbourne, Australia) (32). The samples were acquired on a BD FACSymphony instrument equipped with 5 laser lines (355 nm, 405 nm, 488 nm, 532 nm, and 628 nm). Manual analysis as well as t-SNE dimensionality reduction was performed using FlowJo 10.4.x. Specific clone information is provided in Supplemental Figure 2.
RNA-seq.
MAIT cells from cryopreserved PBMCs and lymph of 5 patients were sorted and processed with the SMARTseq v4 kit (Clontech). After cDNA amplification, sequencing libraries were prepared using the Nextera XT DNA Library Preparation Kit (Illumina). Barcoded libraries were pooled and quantified using a Qubit Fluorometer (Invitrogen). Single-read sequencing of the pooled libraries was carried out on a HiSeq2500 sequencer (Illumina) with 58-base reads, either using TruSeq v4 or Rapid Run v2 Cluster and SBS kits (Illumina).
The RNA-seq data were aligned to the human genome (UCSC Human Genome Assembly [http://hgdownload.cse.ucsc.edu/downloads.html#human], reference sequence GRCh38) using STAR (v2.4.2a) (42), and gene expression quantification was performed using RSEM (v1.2.22) (43). Genes with less than 5 nonzero read counts were discarded, leaving 15,719 expressed genes for the analysis. Libraries (samples) with less than 200,000 reads; 12,000 detected genes; and an exon rate <60% were also removed. Fourteen of the 18 prepared libraries (from 4 of 5 patients) passed these quality criteria.
TCR repertoire data analysis.
Analysis of TCRα and TCRβ was performed by mixCR software (v2.1.1) (44). Deeper TCRα and -β repertoire analysis was done using VDJtools software (v1.1.4) (45). To assess differences in TCR diversity between blood and lymph samples, the number of overlapping clonotypes (TCRs) was quantified.
Data availability.
All sequencing data has been deposited to the GEO under the accession GSE106288.
Statistics.
Raw count data from RSEM was imported into R (v3.4.0). The edgeR Bioconductor package was used to calculate normalization factors to scale the raw library sizes (46), followed by a normalization using the voom transformation from the limma Bioconductor package (47, 48). It transforms count data to log2 counts per million and estimates the mean-variance relationship to compute appropriate observation-level weights. For differential gene expression analysis, we used the linear models for microarray data (limma Bioconductor package) (48, 49). A linear model was fitted to each gene, and empirical Bayes moderated t-statistics (2-tailed) were used to assess differences in expression (49). A contrast comparing blood and lymph samples was tested. Intraclass correlations were estimated to account for measures originating from the same patients (50). An absolute log2-fold change cutoff of 1 and an FDR cutoff of 5% were used to determine DEGs.
To compare phenotypes between MAIT cells in the blood and lymph, Wilcoxon matched-pairs signed rank test were performed. P > 0.05 was considered not significant (ns), and values denoted with asterisks reflect significance levels as follows: *P ≤ 0.05 and ***P ≤ 0.001. RNA-seq analysis methods are described in detail in the supplemental methods. RNA-seq data are in NCBI’s Gene Expression Omnibus database (GSE106288).
Study approval.
All participants signed for informed consent, and the protocols were approved by IRBs at the University of Pennsylvania (822686) and Children’s Hospital of Philadelphia (15-012159).
Author contributions
VV, MB, CKS, FM, RG, MRB, and MP conceived and designed experiments; VV, MB, CKS, JDB, FM, RG, MRB, and MP wrote the paper; VV, MB, CKS, JDB, FM, RG, MRB, and MP performed experiments and analyzed data; and MMA, YD, GN, and MGI provided manuscript feedback, advice, enrolled human subjects, and collected blood and lymph.
Supplementary Material
Acknowledgments
This work was supported by NIH grants DP2 DE023321 (to MP), R01 AI118694 (to MRB), T32 AI007509-16 (to CKS), and T32 GM007270 (JDB). MB is funded through the Swedish Research Council (Dnr 537-2014-6829), Karolinska Institutet, Magnus Bergvall foundation, and Lars Hiertas foundation. FM is an ISAC scholar. We thank the James B. Pendleton Charitable Trust for their equipment donation and Aaron Tyznik and Suraj Saksena (BD Biosciences) for providing reagents and advice. This research was funded in part through the NIH/NCI Cancer Center support grant P30 CA015704.
Version 1. 04/05/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(7):e98487. https://doi.org/10.1172/jci.insight.98487.
Contributor Information
Valentin Voillet, Email: vvoillet@fredhutch.org.
Marcus Buggert, Email: marcus.buggert@ki.se.
Florian Mair, Email: fmair@fredhutch.org.
Yoav Dori, Email: doriy@email.chop.edu.
Gregory Nadolski, Email: Gregory.Nadolski@uphs.upenn.edu.
Raphael Gottardo, Email: rgottard@fhcrc.org.
Martin Prlic, Email: mprlic@fhcrc.org.
References
- 1.Chandra S, Kronenberg M. Activation and Function of iNKT and MAIT Cells. Adv Immunol. 2015;127:145–201. doi: 10.1016/bs.ai.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Dusseaux M, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. 2011;117(4):1250–1259. doi: 10.1182/blood-2010-08-303339. [DOI] [PubMed] [Google Scholar]
- 3.Jo J, et al. Toll-like receptor 8 agonist and bacteria trigger potent activation of innate immune cells in human liver. PLoS Pathog. 2014;10(6):e1004210. doi: 10.1371/journal.ppat.1004210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ussher JE, et al. CD161++ CD8+ T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. Eur J Immunol. 2014;44(1):195–203. doi: 10.1002/eji.201343509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finak G, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slichter CK, et al. Distinct activation thresholds of human conventional and innate-like memory T cells. JCI Insight. 2016;1(8):e86292. doi: 10.1172/jci.insight.86292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mpina M, et al. Controlled Human Malaria Infection Leads to Long-Lasting Changes in Innate and Innate-like Lymphocyte Populations. J Immunol. 2017;199(1):107–118. doi: 10.4049/jimmunol.1601989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gapin L. Check MAIT. J Immunol. 2014;192(10):4475–4480. doi: 10.4049/jimmunol.1400119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magalhaes I, Kiaf B, Lehuen A. iNKT and MAIT Cell Alterations in Diabetes. Front Immunol. 2015;6:341. doi: 10.3389/fimmu.2015.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salou M, Franciszkiewicz K, Lantz O. MAIT cells in infectious diseases. Curr Opin Immunol. 2017;48:7–14. doi: 10.1016/j.coi.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 11.Wong EB, Ndung’u T, Kasprowicz VO. The role of mucosal-associated invariant T cells in infectious diseases. Immunology. 2017;150(1):45–54. doi: 10.1111/imm.12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bianchini E, et al. Invariant natural killer T cells and mucosal-associated invariant T cells in multiple sclerosis. Immunol Lett. 2017;183:1–7. doi: 10.1016/j.imlet.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 13.Berkson JD, Prlic M. The MAIT conundrum - how human MAIT cells distinguish bacterial colonization from infection in mucosal barrier tissues. Immunol Lett. 2017;192:7–11. doi: 10.1016/j.imlet.2017.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saeidi A, et al. Functional role of mucosal-associated invariant T cells in HIV infection. J Leukoc Biol. 2016;100(2):305–314. doi: 10.1189/jlb.4RU0216-084R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leeansyah E, et al. Activation, exhaustion, and persistent decline of the antimicrobial MR1-restricted MAIT-cell population in chronic HIV-1 infection. Blood. 2013;121(7):1124–1135. doi: 10.1182/blood-2012-07-445429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401(6754):708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 17.Förster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol. 2008;8(5):362–371. doi: 10.1038/nri2297. [DOI] [PubMed] [Google Scholar]
- 18.Gunn MD, Tangemann K, Tam C, Cyster JG, Rosen SD, Williams LT. A chemokine expressed in lymphoid high endothelial venules promotes the adhesion and chemotaxis of naive T lymphocytes. Proc Natl Acad Sci USA. 1998;95(1):258–263. doi: 10.1073/pnas.95.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Förster R, et al. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99(1):23–33. doi: 10.1016/S0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- 20.Klonowski KD, Williams KJ, Marzo AL, Blair DA, Lingenheld EG, Lefrançois L. Dynamics of blood-borne CD8 memory T cell migration in vivo. Immunity. 2004;20(5):551–562. doi: 10.1016/S1074-7613(04)00103-7. [DOI] [PubMed] [Google Scholar]
- 21.Schenkel JM, Masopust D. Tissue-resident memory T cells. Immunity. 2014;41(6):886–897. doi: 10.1016/j.immuni.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carbone FR. Tissue-Resident Memory T Cells and Fixed Immune Surveillance in Nonlymphoid Organs. J Immunol. 2015;195(1):17–22. doi: 10.4049/jimmunol.1500515. [DOI] [PubMed] [Google Scholar]
- 23.Wakim LM, Waithman J, van Rooijen N, Heath WR, Carbone FR. Dendritic cell-induced memory T cell activation in nonlymphoid tissues. Science. 2008;319(5860):198–202. doi: 10.1126/science.1151869. [DOI] [PubMed] [Google Scholar]
- 24.Clark RA, et al. Skin effector memory T cells do not recirculate and provide immune protection in alemtuzumab-treated CTCL patients. Sci Transl Med. 2012;4(117):117ra7. doi: 10.1126/scitranslmed.3003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar BV, et al. Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites. Cell Rep. 2017;20(12):2921–2934. doi: 10.1016/j.celrep.2017.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sathaliyawala T, et al. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity. 2013;38(1):187–197. doi: 10.1016/j.immuni.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thome JJ, Farber DL. Emerging concepts in tissue-resident T cells: lessons from humans. Trends Immunol. 2015;36(7):428–435. doi: 10.1016/j.it.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe R, et al. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci Transl Med. 2015;7(279):279ra39. doi: 10.1126/scitranslmed.3010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cortez VS, et al. CRTAM controls residency of gut CD4+CD8+ T cells in the steady state and maintenance of gut CD4+ Th17 during parasitic infection. J Exp Med. 2014;211(4):623–633. doi: 10.1084/jem.20130904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groom JR, Luster AD. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol Cell Biol. 2011;89(2):207–215. doi: 10.1038/icb.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma PK, et al. High expression of CD26 accurately identifies human bacteria-reactive MR1-restricted MAIT cells. Immunology. 2015;145(3):443–453. doi: 10.1111/imm.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corbett AJ, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509(7500):361–365. doi: 10.1038/nature13160. [DOI] [PubMed] [Google Scholar]
- 33.Reantragoon R, et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J Exp Med. 2013;210(11):2305–2320. doi: 10.1084/jem.20130958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prlic M, Sacks JA, Bevan MJ. Dissociating markers of senescence and protective ability in memory T cells. PLoS ONE. 2012;7(3):e32576. doi: 10.1371/journal.pone.0032576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mair F, Prlic M. OMIP-044: 28-color immunophenotyping of the human dendritic cell compartment. Cytometry A. doi: 10.1002/cyto.a. [published online ahead of print January 22, 2018]. https://doi.org/10.1002/cyto.a.23331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibbs A, et al. MAIT cells reside in the female genital mucosa and are biased towards IL-17 and IL-22 production in response to bacterial stimulation. Mucosal Immunol. 2017;10(1):35–45. doi: 10.1038/mi.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Booth JS, et al. Mucosal-Associated Invariant T Cells in the Human Gastric Mucosa and Blood: Role in Helicobacter pylori Infection. Front Immunol. 2015;6:466. doi: 10.3389/fimmu.2015.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorenz U. SHP-1 and SHP-2 in T cells: two phosphatases functioning at many levels. Immunol Rev. 2009;228(1):342–359. doi: 10.1111/j.1600-065X.2008.00760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hawiger D, Wan YY, Eynon EE, Flavell RA. The transcription cofactor Hopx is required for regulatory T cell function in dendritic cell-mediated peripheral T cell unresponsiveness. Nat Immunol. 2010;11(10):962–968. doi: 10.1038/ni.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nadolski G, Itkin M. Thoracic duct embolization for the management of chylothoraces. Curr Opin Pulm Med. 2013;19(4):380–386. doi: 10.1097/MCP.0b013e3283610df2. [DOI] [PubMed] [Google Scholar]
- 41.Nadolski GJ, Itkin M. Feasibility of ultrasound-guided intranodal lymphangiogram for thoracic duct embolization. J Vasc Interv Radiol. 2012;23(5):613–616. doi: 10.1016/j.jvir.2012.01.078. [DOI] [PubMed] [Google Scholar]
- 42.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolotin DA, et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12(5):380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 45.Shugay M, et al. VDJtools: Unifying Post-analysis of T Cell Receptor Repertoires. PLoS Comput Biol. 2015;11(11):e1004503. doi: 10.1371/journal.pcbi.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 50.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21(9):2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data has been deposited to the GEO under the accession GSE106288.