Abstract
BACKGROUND. The effect of gene expression data on diagnosis remains limited. Here, we show how diagnosis and classification of growth hormone deficiency (GHD) can be achieved from a single blood sample using a combination of transcriptomics and random forest analysis.
METHODS. Prepubertal treatment-naive children with GHD (n = 98) were enrolled from the PREDICT study, and controls (n = 26) were acquired from online data sets. Whole blood gene expression was correlated with peak growth hormone (GH) using rank regression and a random forest algorithm tested for prediction of the presence of GHD and in classification of GHD as severe (peak GH <4 μg/l) and nonsevere (peak ≥4 μg/l). Performance was assessed using area under the receiver operating characteristic curve (AUC-ROC).
RESULTS. Rank regression identified 347 probe sets in which gene expression correlated with peak GH concentrations (r = ± 0.28, P < 0.01). These 347 probe sets yielded an AUC-ROC of 0.95 for prediction of GHD status versus controls and an AUC-ROC of 0.93 for prediction of GHD severity.
CONCLUSION. This study demonstrates highly accurate diagnosis and disease classification for GHD using a combination of transcriptomics and random forest analysis.
TRIAL REGISTRATION. NCT00256126 and NCT00699855.
FUNDING. Merck and the National Institute for Health Research (CL-2012-06-005).
Keywords: Endocrinology
Keywords: growth factors
Diagnosis and disease classification for growth hormone deficiency was achieved using a combination of transcriptomics and Random Forest analysis.
Introduction
High-throughput technologies, including next-generation sequencing, microarrays, mass spectrometry, and protein chips, now allow measurement of many thousands of biological variables at relatively low cost. While next-generation-based DNA sequencing panels are now affecting clinical practice (1, 2) for diagnosis of genetic disease, gene expression data are far less frequently utilized in routine clinical practice. A large number of studies have examined the utility of gene expression data in prognosis and classification of tumors (3, 4); however, in other fields in which affected tissue is of more limited availability, the clinical effect of transcriptomics has been minimal. One of the main challenges in utilizing these technologies is identifying the useful biological signal in such complex data. Combining transcriptomics with machine learning approaches has proven useful in disease classification in autism (5). Here, we show how the utility of transcriptomic data for diagnosis can be refined using a combination of machine learning and network based prioritization.
Many diagnostic tests in endocrinology require administration of pharmacological agents, multiple blood sampling, and hospital admission, making them expensive and unpleasant for patients. This study, using growth hormone deficiency (GHD) as an exemplar, demonstrates how a single blood test with extracted mRNA applied to a microarray could replace endocrine stimulation tests.
GHD is a rare but important cause of short stature in childhood, with a prevalence of approximately 1:4,000 (6). Consensus guidelines recommend an approach to the diagnosis of childhood GHD that integrates clinical, biochemical, and auxological data (7). Biochemical investigations are key to the diagnosis, particularly pharmacological growth hormone (GH) stimulation testing where a cutoff level is used, below which children are diagnosed with GHD. There are, however, many problems associated with these stimulation tests — they display poor reproducibility (8) and, in addition, the peak GH level achieved varies with body composition (9, 10), pharmacological stimuli (11), and assay (12) used. The first cutoff level proposed for the diagnosis of GHD was 5 μg/l (13) in 1968 on the basis that this seemed to best identify children with a GHD phenotype. With the increased availability of GH, this cutoff was subsequently increased to 7 μg/l and then 10 μg/l based on very limited evidence. Dependent upon the assay used, recent studies classifying children as GHD or not-GHD, based on auxological criteria, have suggested cutoff levels between 4 and 7 μg/l (12). Clearly, there remains uncertainty as to the optimal cutoff level for the diagnosis or GHD. Given the multiple problems associated with pharmacologic stimulation tests, there is also no clear cutoff for the differentiation between “mild” and “severe” GHD. A peak GH cutoff of 10 μg /l was used to define GHD in the PREDICT study, and, in the present analysis, we have chosen a cutoff of 4 μg /l to define “severe” GHD.
There is, therefore, a need to develop new tools that are not susceptible to the many problems associated with pharmacological stimulation tests to aid with the diagnosis and classification of childhood GHD. The PREDICT study (14) was a 1-month, phase IV, open-label, prospective multicenter study in GH treatment–naive children with GHD (NCT00256126) that aimed to identify genetic and transcriptomic markers of response to GH therapy. Children enrolled in the study all had a peak GH level of <10 μg/l on two stimulation tests, blood samples taken for whole-genome gene expression analysis, and candidate SNP genotyping prior to starting treatment.
Using this cohort along with gene expression data from healthy control children, this exploratory study aimed to (a) define the set of genes whose expression correlates with peak GH levels; (b) determine the usefulness of these gene expression data in the diagnosis and classification of GHD; (c) identify the biological function and regulators of these genes; and (d) identify SNPs associated with peak GH levels and examine the utility of these SNPs, either alone or in combination with the gene expression data and/or demographic/biochemical data, for classification of GHD subjects.
Results
SNPs associated with peak GH.
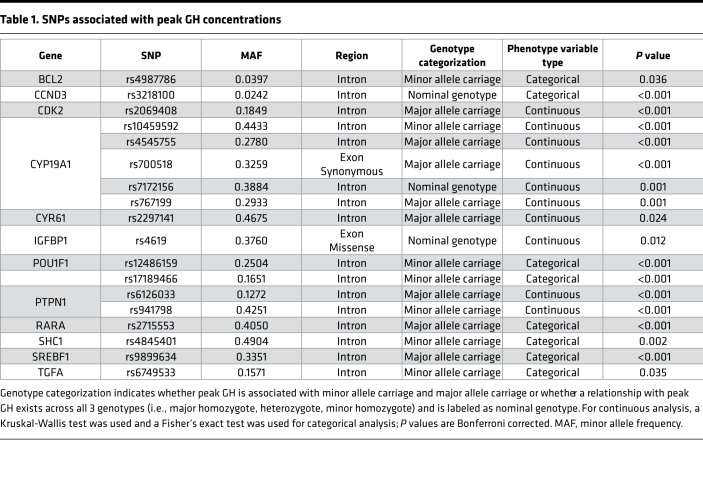
Eighteen SNPs in twelve genes were associated with peak GH concentrations (see Table 1). Sixteen of the eighteen SNPs are intronic, with one synonymous exonic SNP and one missense exonic SNP. None of the SNPs were rare (defined as a minor allele frequency <1%), with 16 of 18 SNPs having a minor allele frequency >10%. The function of the genes associated with the SNPs included pituitary developmental transcription (POU1F1), generation of estrogen (CYP19A1), IGF binding (IGFBP1), apoptosis (BCL2 and SHC1), cell cycle (CCND3 and CDK2), angiogenesis (CYR61), growth (TGFA), transcription (SREBF1), and signal transduction (PTPN1, RARA).
Table 1. SNPs associated with peak GH concentrations.
Principal component analysis and gene expression profiling in GHD and control children.
No differences were observed in the overall distribution of gene expression between GHD and control subjects using unsupervised principal component analysis (PCA) on the transcriptomic data from the different studies described (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.93247DS1). This determined that there was no overall effect of study or associated covariates on the data sets and therefore that further direct comparison was meaningful.
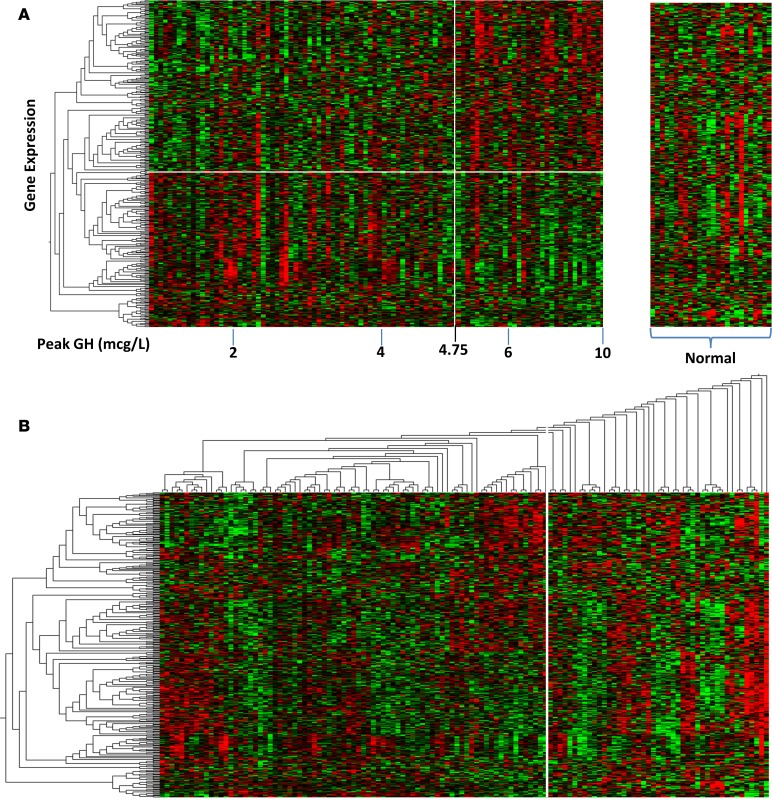
Rank regression identified 347 probe sets (representing 271 unique genes) in which expression correlated with peak GH concentrations in the 98 GHD children (188 probe sets correlated positively and 159 correlated negatively, r ± 0.28, P < 0.01), and these 347 probe sets were also present in control arrays (see Supplemental Table 1). Of these 347 probe sets, 65 were identified as also being expressed in the human growth plate. The gene expression of the 347 probe sets is displayed on a heatmap for both children with GHD and normal children (assigned to a peak GH of 10 μg/l) in Figure 1A. A clear distinction can be seen between the normal subjects and the subjects with GHD, and, in addition, a point of inflexion can be seen in the GHD subjects at a peak GH of 4.75 μg/l (Figure 1A).
Figure 1. Heatmap of gene expression for those probe sets whose expression correlated with peak GH levels.
(A) Normal children (n = 26) were combined with GHD patients (n = 98), rank regression analysis was adjusted for sex and age as covariates, and clusters of similar gene expression were identified using the Euclidean metric and marked using a dendrogram and white boxes (347 probe sets, 271 unique genes). The distinction between normal subjects is marked by the break in the heatmap; GHD is defined by a cutoff level of 10 μ/l growth hormone, as measured by provocation testing. The vertical white line demarcates the point of inflexion for gene expression at a peak GH level of 4.75 μ/l, while the horizontal white line demarcates those probe sets positively and negatively associated with peak GH levels (< or >4.75 μ/l). (B) Two-way cluster analysis of gene expression in GHD and control subjects. Four distinct clusters of GHD subgroups can be seen from the dendrogram on the horizontal axis derived via a Euclidian metric. There is, however, a large number of subjects it was not possible to classify (right of white line). This group contained all but 1 of the normal control subjects and 20 GHD subjects.
These 347 probe sets were then displayed on a heatmap with 2-way hierarchical clustering using both the control and GHD subjects (Figure 1B). On the x axis of the dendrogram, 4 clusters of GHD subjects are shown, and, in addition, there were 45 subjects (20 GHD, 25 controls) who could not be classified into any cluster (Figure 1B). Only one control subject was classified into one of the 4 clusters, with the remaining being GHD subjects. There were no significant differences in age, sex, height standard deviation score (SDS), weight SDS, or BMI SDS between the 20 GHD subjects not classified into the 4 GHD clusters and the 78 GHD subjects classified into one of those 4 clusters.
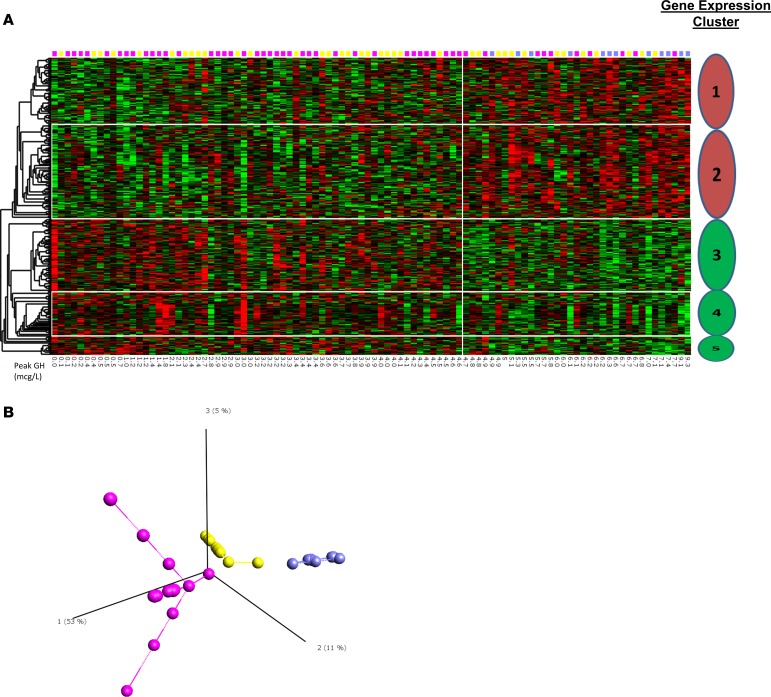
An additional heatmap of the 347 probe sets identified by rank regression using peak GH as a continuous variable in the GHD group only was generated (Figure 2A). In this heatmap, 5 clusters of gene expression — 2 related to genes where there was a positive correlation with peak GH and 3 related to genes where there was a negative correlation with peak GH — were identified. In this heatmap, all subjects could be classified via a Euclidian metric into one of the 5 clusters.
Figure 2. Identification of clusters of variation of gene expression related to GHD severity.
(A) Heatmap for the probe sets identified by correlation with peak GH (347 probe sets, 271 unique genes). Five distinct clusters of gene expression are identified via the dendrogram — two positively correlated (red) with peak GH and three negatively correlated (green). Pink, yellow, and blue squares indicate the principal component analysis group for each patient (see Figure 1B). (B) Isomap supervised principal component analysis using only those probe sets whose expression correlated to peak GH identified 3 distinct groups of GHD subjects (n = 98; pink n = 49, yellow n = 37, and blue n = 12).
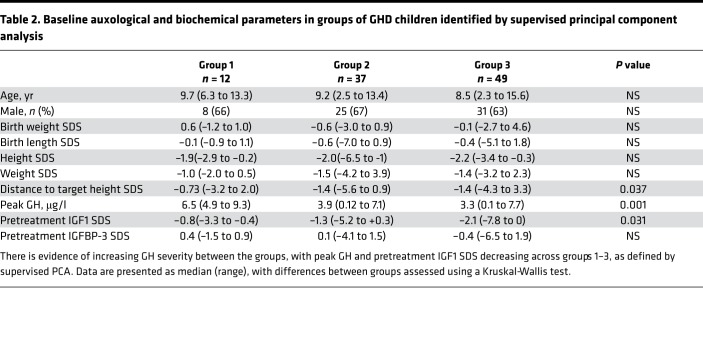
Supervised principal component and isomap multidimensional scaling identified 3 distinct groups of GHD patients (Figure 2B). There were significant differences between groups in peak GH levels, distance to target height SDS, and baseline insulin-like growth factor 1 (IGF1) SDS (see Table 2).
Table 2. Baseline auxological and biochemical parameters in groups of GHD children identified by supervised principal component analysis.
Network analysis.
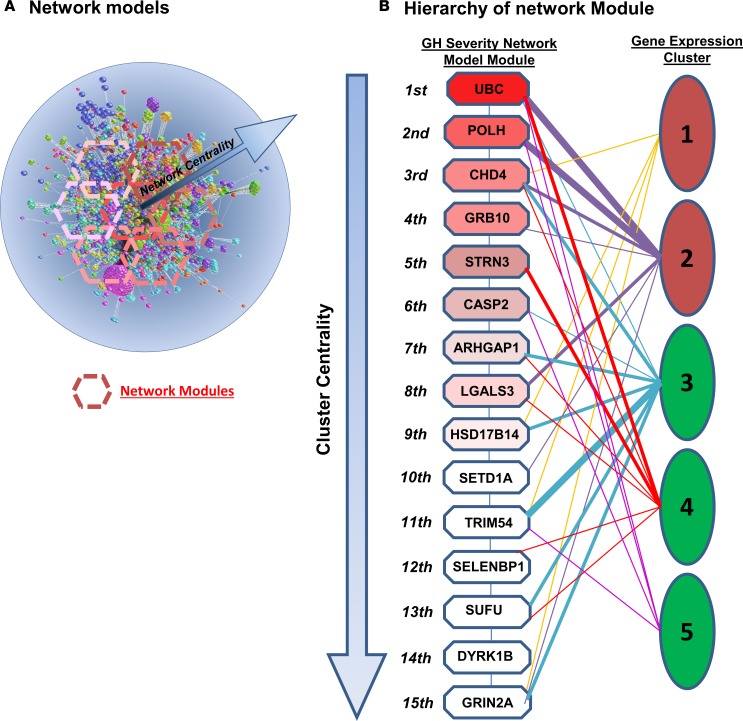
Using the genes identified by the rank regression, a network with 2,427 nodes and 3,604 links was generated. Decomposition into a hierarchical modular structure revealed 43 network modules. Functionality was assessed on the top 15 modules as ranked by network centrality (Figure 3). Of the 15 modules, 5 were related to circadian clock, 4 related to growth factor signaling, and 3 related to DNA replication.
Figure 3. Network modeling of the overlap of gene expression between clinical markers.
(A) Network models generated using BioGRID (version 3.2.117) were analyzed to define modules of functionally related genes. The “community structure” of these modules was assessed and ranked by their “centrality” score to form a hierarchy related to the biological action of the network. (B) Community structure of modules within the network was assessed using the ModuLand algorithm in Cytoscape 2.8.3. Hierarchy of the first 15 network modules in each of the network models of gene expression overlap between clinical markers. Modules are shown as octagons labeled with the most central gene in the cluster and ranked by network centrality (1st through 15th).
The gene expression clusters (Figure 2) were overlapped with the network modules (see Figure 3 and Supplemental Figure 2). Overlapping simply involved comparing the list of genes present in the gene expression clusters and network modules (those with ≥3 shared genes considered to be linked). Gene cluster 1 linked to only 1 network module (HSD17B14) related to cell cycle, while gene cluster 5 also linked to only 1 network module (CASP2) related to apoptosis pathways. Gene cluster 2 associated with the second, third, fourth, and tenth network modules related to circadian clock, chromatin organization, and growth factor signaling. Gene clusters 3 and 4 each linked to 4 network modules covering the whole spectrum of pathways identified, except for apoptosis. SSX2IP, STRN3, and PTGDS contained within the first and second (SSX2IP), third (STRN3), and fifth (PTGDS), clusters as determined by centrality hierarchy (Supplemental Figure 2), had previously been identified in the top 10 genes with variable importance in the random forest model.
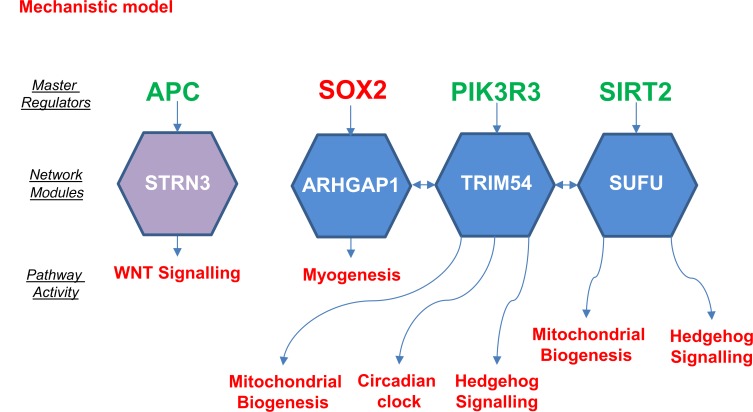
Causal network analysis (CNA) (15) identified 4 causal elements within the network model, which mapped to the 15 network modules (see Figure 4). This provides robust supporting evidence for the functional pathways described by the network modules. Master regulators identified included APC2, which regulated the STRN3 network module related to apoptosis and gene cluster 4. SOX2, PI3KR3, and SIRT2 were identified as regulators of the ARHGAP1, TRIM54, and SUFU network modules linked to gene clusters 3 and 4, and they affected Hedgehog signaling, circadian clock, mitochondrial biogenesis, and myogenesis pathways.
Figure 4. Summary of predicted activity and regulators derived via causal network analysis for the network modules.
The hierarchy of clusters of gene expression shown in Figure 2 were mapped onto identified causal networks. Activity of pathways and master regulators (colored red) show a positive correlation with the GHD severity or (colored green) show negatively correlated activity. Pathway ontology of all modules in the hierarchy is shown in Supplemental Figure 2.
Prediction of GHD status and classification of GH severity.
Random forest analysis for predicting GHD versus control subjects yielded an area under the receiver operating characteristic curve (AUC-ROC) of 0.95 (95%CI 0.91–0.99), with sensitivity of 96%, specificity of 100%, and an out-of-box AUC-ROC (OOB-AUC) of 0.99. 53 probe sets representing 40 named and 13 unnamed genes were confirmed by Boruta as containing predictive capacity (see Supplemental Table 2). Of the 53 probe sets identified by Boruta, 10 of these were also expressed in the growth plate. This represents an enrichment of growth plate genes in comparison to those selected just by the rank regression model (hypogeometric P = 1.14 × 10–12). Although the predictive capacity given by the transcriptomic data was excellent, we also assessed the ability of network biology to improve prediction by selecting probe sets ranked by network centrality. Selecting the top 10 probe sets ranked by network centrality yielded an AUC-ROC of 0.94 (95% CI 0.91–0.95), while 4 different combinations of 10 probe sets (randomly selected from the probe sets where expression was correlated with peak GH) yielded an inferior AUC-ROC of 0.84 (95% CI 0.78–0.90).
Demographic, biochemical, genomic, and transcriptomic data were used with random forest analysis to assess their predictive value in determining severe GHD (defined by peak GH <4 μg/l). Each of these data categories were assessed separately and then in combination (see Table 3). The transcriptomic data (AUC-ROC of 0.93) performed better than the genomic (AUC-ROC of 0.85) or biochemical/demographic data (AUC-ROC of 0.88). The addition of the genomic or biochemical/demographic data (either alone or in combination) to the transcriptomic data did not improve the AUC-ROC (all 0.93). In the model using all data categories, of the top 10 variables of greatest importance (as ranked by mean decrease in accuracy), 9 were gene expression probe sets (NRXN1, SSX2IP, STRN3, RNF43, SUZ12P, RAB7A, PROC, GATSL3 and PTGDS) and 1 was a SNP (rs2715553). The functions of the encoded proteins were diverse but included several that were clearly linked to growth: STRN3 is a WD40 domain containing protein that enhances cancer cell survival and activating AKT (16), RAB7A is an oncogene involved in the RAS pathway, RNF43 is a tumor suppressor involved in ubiquitination, and SSX2IP is known to bind to a synovial sarcoma-associated protein that promotes growth (17). Other genes and their encoded proteins did not have a clear role in GH secretion or growth — PROC is a coagulation factor, SUZ12P is a pseudogene, GATSL3 is associated with rheumatoid arthritis (18) but has no known function, NRXN1 is a neuroligin synapse receptor (19), and PTGDS is involved in prostaglandin production.
Table 3. Prediction of GH severity (peak GH ≤4 μg/l or >4 μg/l) via random forest model.
Discussion
This exploratory study aimed to identify whether gene expression profiling could aid with the diagnosis of GHD or in our classification and understanding of the factors influencing the severity of GHD. Despite the use of GH as a therapeutic agent since 1958 (20) and the ability to measure serum GH levels since 1963 (21), the diagnosis of GHD remains challenging; there is no “gold standard” test for diagnosis. In this study, we examined whether gene expression profiling could distinguish children from the PREDICT study with GHD from healthy controls. The development of a test based on gene expression would be a significant advance for patients, potentially avoiding the need for hospital admission and the use of pharmacological stimulation tests. The AUC-ROC of the random forest–based algorithm from our gene expression data was excellent at 0.98 and, with a specificity of 100% and a sensitivity of 96%, clearly distinguished GHD subjects from controls, thus suggesting that this could be developed into a useful test for diagnosing of GHD.
In addition to assessing the predictive ability of random forest analysis, we also assessed whether network prioritization of input genes improved prediction. Limiting a prediction algorithm to a small number of parameters may be helpful in developing a cost-effective test that can easily be applied to large numbers of patients. The predictive ability of 10 probe sets was increased by selecting them based on their network centrality. This combination of network analysis with a machine learning approach (in this case random forest analysis) may be particularly effective in developing “omic-based” approaches to diagnosis. Selecting probe sets based on the Boruta algorithm, which is designed to identify the probe sets most likely to contain true predictive capacity, resulted in an enrichment of selected probe sets for those expressed in the growth plate.
These data support the use potential use of a gene expression–based test, but there are significant limitations to our study. First, the patients and control children were assembled from different studies, although extensive work was undertaken to normalize both between and within batches of arrays. Second, rather than comparing children with GHD to normal healthy children, it would be better to compare GHD children to short children without GHD, as this is the distinction that is required of a clinical test. In addition, as a multicenter international study with the GHD diagnosis made at each study center, GH stimulation tests and assays were not standardized in the PREDICT study. PREDICT aimed to recruit a cohort of children with GHD diagnosed according to international practice, encompassing the variations in diagnostic criteria between centers and countries. In general, there will be reasonable correlations between peak GH levels after different stimuli, such that a low peak GH after arginine will also be low after an insulin tolerance test or a glucagon test and higher levels in a test will be mirrored in a second test. Within KIGS, which collected “real world” data on GHD patients, the correlation between the first and second GH stimulation tests in >3,000 patients was reasonable at r = +0.515 (22). A third limitation is that we did not have a peak GH level for the control subjects; due to the inaccuracy of GH stimulation tests, we cannot be sure if they had been tested that they would indeed have had a peak GH level >10 μg/, although as healthy controls it is highly unlikely that any of them had GHD. Future studies should concentrate on prospective recruitment of children undergoing GH stimulation testing using standardized stimulation tests and GH assays to determine in that cohort whether there is any evidence of a change in pattern of gene expression at any particular cutoff level for peak GH. A small number of subjects in this study received additional hormone supplementation with hydrocortisone or thyroxine; given that this was physiological replacement, we do not expect this to have significantly affected gene expression.
In addition, this study has demonstrated the utility of gene expression profiling and SNP genotyping in identifying a cohort of children with more severe GHD. A cutoff of 4 μg/l was selected, as this allowed us to create two groups (one with more severe GHD) with sufficient numbers for prediction. The most accurate tool for identifying GH severity status was the transcriptomic data, which performed better than the genomic data, clinical data, or genomic and clinical data combined. This is highly suggestive of the possibility of accurately identifying, on the basis of a basal gene expression, a child with severe GHD from among a cohort of subjects with short stature and a range of peak GH concentrations classified as GHD.
Unsupervised PCA did not identify clinically distinct groups of GHD patients, and we therefore undertook a supervised PCA using those genes where expression correlated with severity of GHD, as defined by peak GH concentration to stimulation testing. This supervised analysis identified 3 groups of GHD patients. There was a clear clinical separation between group 1 and groups 2 and 3, with group 1 representing a less severe cohort of patients with a higher peak GH level, higher IGF1 SDS, and lower distance to target height SDS (Table 2). There was clear separation of groups 2 and 3 in the PCA, but no clear auxological/biochemical differences between these groups.
In addition to identifying a gene expression profile associated with peak GH concentrations, we also identified 18 SNPs from 12 different genes where genotype was associated with peak GH concentrations. Five of these twelve genes (SHC1, CCND3, BCL2, CDK2, and RARA) were also present in the network. Of those five genes, two each are involved with apoptosis and cell cycle. For many patients with GH deficiency, anterior pituitary hypoplasia is also present, and these SNPs may mediate their effects by affecting somatotrope differentiation and survival. One of the SNPs was within POU1F1, a pituitary transcription factor essential for differentiation of somatotropes, lactotropes, and thyrotropes (23). Five SNPs were identified within CYP19A1, the gene that encodes the enzyme aromatase, which is responsible for the generation of oestrogen. Sex steroids augment GH peak concentrations during stimulation tests and mediate pituitary growth during puberty, and oestrogen inhibits GH signal transduction by stimulating expression of SOCS-2 (24). Although all children enrolled in PREDICT were prepubertal, it is possible that very low prepubertal oestrogen concentrations can be influenced by these SNPs and hence effect GH levels.
To explore the function of the genes with expression linked to peak GH levels, we generated a network model and ranked functional modules of genes according to the network centrality. Gene clusters 2, 3, and 4 all mapped to network modules involved in growth factor signaling, including WNT and hedgehog signaling, while gene cluster 5 mapped to a module involved in apoptosis. It is perhaps not surprising that a strong signature for growth factor signaling and apoptosis would be identified in the genes related to severity of GHD. Both GH and its downstream effector hormone IGF1 are known inhibitors of apoptosis (25). Clearly, with increasing severity of GHD, we would expect reduced growth factor signaling and increased apoptosis. This study, however, defines the distinct gene expression clusters that differentially link to growth factor signaling and apoptosis. It was interesting to find a strong signature for the circadian clock. GH is secreted in pulses mainly overnight (26), and this finding may reflect either disturbances of the circadian clock, leading to reduced secretion of GH, or perhaps an acceleration of the circadian clock rhythm in an attempt to maximize GH pulse frequency where pulse amplitude has been limited by somatotroph hypoplasia. GH secretion has not only circadian but also ultradian rhythms (27), and disturbance of these can lead to a form of GHD termed neurosecretory dysfunction. This is a disorder in which the child presents with GH deficiency with a normal pharmacological GH stimulation test but abnormal spontaneous GH secretion, with reduced frequency and amplitude of GH pulses (28).
CNA allowed us to identify 4 master regulators — APC2, SOX2, PIK3R3, and SIRT2. Loss-of-function mutations in anaphase-promoting complex 2 (APC2) have been associated with a Sotos syndrome–like phenotype of overgrowth and neurodevelopmental delay (29), and it is a negative regulator of WNT signaling through its targeting of β-catenin for ubiquitin-mediated proteolysis (30). WNT signaling is known to be involved in pituitary development, promoting the expression of PITX2 and proliferation of pituitary precursors (31). Sex determining region Y box 2 (SOX2) is a member of the SRY-related HMG box B1 (SOXB1) subfamily of transcription factors and is expressed in the developing brain and posterior neural tube, including Rathke’s pouch and hypothalamus (32). In humans, heterozygous loss-of-function mutations in SOX2 lead to eye abnormalities (microphthalmia and anophthalmia) and hypopituitarism (hypogonadotropic hypogonadism and variable GHD). SOX2 expression in the postnatal and adult pituitary marks a subpopulation of hormone-negative cells, which are pituitary progenitor stem cells capable of differentiating into endocrine-producing cells (31). SOX2 may therefore be regulating developmental processes, such as pituitary stem cell proliferation, in addition to myogenesis. PIK3R3 encodes a regulatory subunit of phosphoinositide-3-kinase, a component of both the GH and IGF1 signal transduction systems, as well as many other cell signal transduction cascades. PI3K is involved in a diverse range of functions, including proliferation, cell survival, degranulation, vesicular trafficking, and cell migration. SIRT2 is one of a class of NAD(+)-dependent deacetylases with antiaging activity in model organisms, an effect increased by caloric restriction (33). The sirtuins induce mitochondrial biogenesis (the generation of new mitochondria) to reduce the accumulation of toxic reactive oxygen species seen in caloric restriction (33). In addition, SIRT2-regulated adipocyte differentiation inhibits p53 accumulation and is regulated by Src tyrosine kinase (a component of the GH signal transduction system) (34).
This study has demonstrated the potential for gene expression profiling to aid in both the diagnosis and classification of GHD and, in addition, has identified the functions of the networks of genes related to peak GH concentrations along with their master regulators. Moving from a diagnosis requiring the use of pharmacological stimulation tests to a single blood sample would be a major advance, particularly for pediatric patients. This work could potentially be extended from GHD to other hormone deficiencies, allowing the full assessment of pituitary function with a single blood test.
Methods
Patients.
The PREDICT study was a phase IV, open-label, prospective pharmacogenomic study examining response to GH therapy; it enrolled 125 prepubertal children (78 male, 47 female), aged 2–15 years, with a diagnosis of GH deficiency, reached after two pharmacological stimulation tests according to local protocols, with a peak GH concentration of <10 μg/l. Details of the inclusion and exclusion criteria have previously been reported (14, 35). In brief, prior to enrollment in the study, none of the children had received GH therapy; children with GHD due to central nervous system tumors or radiotherapy were excluded, but children born small for gestational age were not. Of the 125 children in the PREDICT study, baseline gene expression data were available on 98 subjects aged 2–15 years (34 female, 64 male). Bone age was available for 92 patients. Mean bone age delay was 2.2 ± 1.5 years. A delay in bone age of ≥1 year was present in 72 patients. Birth weight was available for 82 patients, of whom 12 (15%) were born small for gestational age. In addition to GH therapy, 5 patients received both thyroxine and hydrocortisone replacement, 3 patients thyroxine alone, and 1 patient hydrocortisone alone.
The PREDICT study was conducted in compliance with ethical principles based on the Declaration of Helsinki, the International Conference on Harmonization Tripartite Guideline for Good Clinical Practice, and all applicable regulatory requirements.
Serum samples and assays.
Blood samples were drawn in the morning, after an overnight fast, and prior to and 1 month after start of treatment with recombinant human GH. Samples were centrally assayed at qLAB. Serum IGF1 and IGFBP-3 were measured using DPC chemiluminescent immunoassays (Immunolite 2000, Siemens Healthcare Diagnostics Inc.). Plasma insulin was measured with a 2-site immunoenzymometric assay (AIA-PACK IRI, Tosoh). Plasma glucose was determined by using the glucose oxidase method, and HDL cholesterol was measured with an enzymatic-calorimetric test.
Genotyping.
Genotyping of 1,536 SNPs, located on 103 candidate genes (related to the GH-IGF1 axis, bone and cell growth, and glucose and lipid metabolism), was performed as previously described (14) using the Illumina GoldenGate assays (Illumina). Before analysis, genotyping data were filtered to remove SNPs with a call rate <95% and those showing significant deviation from the Hardy-Weinberg equilibrium using a Bonferroni correction. After data cleaning, 1,171 SNPs in 97 genes remained for analysis.
Continuous analysis.
SNPs associated with peak GH were identified using the Kruskal-Wallis rank-sum test on the following models: genotype, presence of the major allele (dominant model), and presence of the minor allele (recessive model). For nonpseudoautosomal X-linked markers, boys were analyzed separately from girls. As a candidate gene rather than a whole-genome approach was being used, P values were adjusted for multiple testing using a Bonferroni correction, taking into account the number of linkage disequilibrium blocks present in the gene containing the SNP of interest and considered significant where adjusted P < 0.05.
Categorical analysis.
Markers were also tested in a categorical analysis, with patients classified by quartiles of peak GH; comparisons were made between those with low peak GH concentration versus those with intermediate and high peak GH and those with high peak GH concentration versus those with intermediate and low peak GH concentrations. All P values were calculated using Fisher’s exact test and adjusted for multiple testing using a Bonferroni correction, taking into account the number of linkage disequilibrium blocks within each candidate gene.
Transcriptome analysis for subjects in PREDICT study and gene expression data sets from normal childhood control subjects.
To assess transcriptomic relationships, gene expression profiling was carried out on whole blood RNA as previously described (14) and hybridized to Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays. These gene expression data have previously been uploaded to the NCBI Gene Expression Omnibus (GEO) database (GSE72439) (36). For control subjects, gene expression analysis was conducted on a library of gene expression data sets from normal children with age annotation collated from the publically available NCBI GEO and EBI ArrayExpress databases. The original Affymetrix CEL files from GSE9006 (37), GSE26440 (38), and TABM666 (39) were downloaded and combined into one group to form a main analysis data set following published guidelines (40). Details of the generation of this combined normative data set have previously been published (41). As the children in PREDICT were all above the age of 2 years, gene expression profiles from normal children were removed if they were aged <2 years old, which left 26 subjects (14 male, 12 female) aged 2–11 years. Mean age was significantly lower for control patients at 5.8 ± 2.7 years compared with PREDICT subjects at 8.8 ± 2.9 years (P < 0.01), but there was no significant difference in male sex (14 of 26, 54% in control subjects, and 61 of 98, 62% of PREDICT subjects, P = 0.58).
Normalization and quality control of gene expression data.
For background correction, the Robust Multichip Average (42) was applied to the combined CEL files (derived from GEO or PREDICT, see accession numbers above). The data set generated was subject to quality control to investigate the presence of outliers and further confounding effects. Dimensional scaling using PCA and isomap multidimensional scaling (MDS) (43, 44) was used to demonstrate data homogeneity (Qlucore Omics Explorer 2.2) and identify outliers using cross validation. Gene expression data were normalized for batch, age and sex.
Unsupervised and supervised PCA.
Unsupervised PCA was performed on the gene expression profiles to identify whether the variance in the data sets (GHD and controls) was consistent, a requirement for further statistical analysis between these groups. Supervised PCA was performed after initial statistical evaluation (rank regression, see below) to determine the presence of patient subgroups. All PCA was performed using Qlucore Omics Explorer (Qlucore). Quality control of all PCA was undertaken using cross-validation (sequential removal of all samples) to determine group stability. Unsupervised PCA was refined using variance filtering and a projection score (45). PCA was also confirmed using isomap multidimensional scaling.
Analysis of network models.
Network analysis allows the identification and prioritization of key functional elements within interactome models, which this study has used to prioritize genes for prediction and also to gain insights into biological function. To derive an interactome model, genes whose expression correlated with peak GH concentrations were used as “seeds,” and all known protein/protein interactions between the seeds and their inferred immediate neighbors were identified to generate a biological network using the output of the BioGRID model of the human interactome (3.3.122) (46). Network generation and processing was performed using Cytoscape 2.8.3 (47).
Clustering and “community structure” of modules within biological networks arise from variation in connectivity within the network and are known to be associated with function (48–50). To prioritize these functional components within interactome models, we used the ModuLand plug-in for Cytoscape 2.8.3 to determine overlapping modules and to identify hierarchical structure within the model, thus enabling the identification of key network elements (52). Network modules were prioritized for further investigation by their centrality property, and the most central set of 10 genes within each module was used to assess associated biological pathways using the Reactome database (52). The network structure observed with community modeling in ModuLand was confirmed by cluster analysis using the ClusterOne algorithm (53).
CNA.
CNA allows the identification and prioritization of regulatory system elements within transcriptomic models. CNA was performed within Ingenuity Pathways Analysis using the genes whose expression was correlated with peak GH concentrations.
CNA identifies upstream molecules, up to three steps distant, that control the expression of the genes in the data set (15). A prediction of the activation state for each regulatory factor (master regulator), based on the direction of change, was calculated (Z-score) using the gene expression patterns of the transcription factor and its downstream genes. An absolute Z-score of ≥|1.4| and a corrected P < 0.05 (Fisher’s exact test) was used to compare the regulators identified in each of the comparisons made using hierarchical clustering (Euclidean metric).
Regression and random forest analysis.
Rank regression of probe sets for association with peak GH levels was performed in Qlucore Omics Explorer (Qlucore). Differences in demographic characteristics were assessed using SPSS version 20 (IBM) via a Kruskal-Wallis test.
A random forest algorithm (54) was used to predict severity of GHD (<4 μg/l) based on demographic, biochemical, genomic, and transcriptomic data. A cutoff of 4 μg/l was chosen, as this divided the subjects into two groups of approximately equal size, maximizing accuracy of the random forest classification. Biochemical and demographic data included age, sex, and IGF1/BP3 before start of GH therapy. Genomic data comprised the SNPs identified as being associated with peak GH. For prediction of GHD versus control, the data were unbalanced (98 GHD subjects and 26 controls), and, with an unbalanced data set, random forest poorly predicts the minority class (in this case control subjects). To overcome this problem, a synthetic minority oversampling technique (55) was used to rebalance the data set prior to random forest prediction using age, sex, and transcriptomic data (final data used 114 subjects, 57 controls, and 57 patients). The predictions were assessed based on AUC-ROC and OOB-AUC as a validation set. Identifying those probe sets most likely to contain predictive capacity was achieved with the use of Boruta (56). The Boruta algorithm uses a 100-fold permutation to define the noise present in the data, the noise is modeled as shadow variables and used as a basis to assess confidence in the data. All statistical analyses were performed using R 2.15.3. Random forest analysis requires no explicit test and validation set, as the OOB-AUC functions as a validation data set. In developing the random forest algorithm, hundreds or thousands of decision trees (in our case 1,000 trees) were created. Each tree was generated using a random selection of input variables and randomly selected two-thirds of the subjects. Each tree produced a classification vote, and the majority vote across all trees determines final classification. For each tree, there is a random one-third of subjects whose data were not used in generating that tree — these data are then used to generate the OOB-AUC, which essentially functions as a validation data set.
Human growth plate gene expression and overlap with probe sets identified via random forest analysis.
Human gene expression data from growth plate–derived RNA was available for two subjects (1 male, 1 female) from the NCBI GEO database (GSE9160). For each subject, a sample of the distal femoral growth plate had been obtained, and populations of cells from reserve, proliferative, prehypertrophic, and hypertrophic zones were obtained by laser microdissection. RNA from each population of cells, corresponding to each zone of the growth plate, was amplified and hybridized to Affymetrix HU-133 2.0 arrays. Frozen robust multiarray analysis (57) was used to define absolute expression by comparison to publically available microarray data sets within R, and an expression barcode (58) was defined for each growth plate zone for each patient. Expression within the growth plate was defined by a gene expression barcode value of 1 in any zone of the growth plate in either patient.
Statistics.
The Kruskal-Wallis rank-sum test and Fisher’s exact test, both with Bonferroni corrections for multiple testing, were used to determine genetic associations with peak GH. The Kruskal-Wallis rank-sum test was also used to assess differences in clinical parameters between patient groups determined by multidimensional scaling. The significance of gene set overlaps was determined using the hypergeometric test. Analysis was performed in R or SPSS using a threshold of P < 0.05.
Gene expression was associated with peak GH levels using rank regression and a threshold of P < 0.01 in Qlucore Omics Explorer 2.2.
Study approval.
The PREDICT (NCT00256126) and PREDICT long-term follow-up (NCT00699855) studies were approved by the Scotland Medical Research and Ethics Committee (reference 05/MRE10/61) and the North West Research Ethics Committee (reference 08/H1010/77), respectively. Informed consent was obtained from parents for all study participants.
Author contributions
PEC, EK, and PC conceived and designed the PREDICT project and this study. Data analysis and methodology development were undertaken by PGM, AS, and CDL. The manuscript was written by PGM and AS and revised by EK, PC, and PEC.
Supplementary Material
Acknowledgments
Version 1. 04/05/2018
Electronic publication
Footnotes
Acknowledgments
The authors would like to thank all the PREDICT investigators and the patients who took part in the study. PGM thanks the National Institute for Health Research for their support (CL-2012-06-005). Members of the PREDICT investigator group are as follows: A. Belgorosky, Buenos Aires, Argentina; G. Ambler, Westmead, Australia; K. Kapelari, Innsbruck, Austria; C. Deal, Montreal, Canada; J. Hamilton, Toronto, Canada; J. Jääskeläinen, Kuopio, Finland; Y. Brusquet, Puyricard, France; P. Chatelain, Lyon, France; M. Colle, Bordeaux, France; R. Coutant, Angers, France; Y. Le Bouc, Paris, France; R. Reynaud, Marseille, France; J.-P. Salles, Toulouse, France; J. Weill, Lille, France; R. Pfäffle, Leipzig, Germany; M. Ranke, Tübingen, Germany; G. Binder, Tübingen, Germany; M. Bozzola, Pavia, Italy; F. Buzi, Brescia, Italy; M. Cappa, Rome, Italy; A. Cicognani, Bologna, Italy; M. Maghnie, Genova, Italy; L. Tato, Verona, Italy; F. Antoniazzi, Verona, Italy; D.H. Kim, Seoul, Korea; S.W. Yang, Seoul, Korea; H.W. Yoo, Seoul, Korea; E. Vangsøy Hansen, Bergen, Norway; D. Veimo, Bodø, Norway; E. Bashnina, St. Petersburg, Russia; V. Peterkova, Moscow, Russia; J. Skorodok, St. Petersburg, Russia; L. Sultanova, Kazan, Russia; A. Carrascosa, Barcelona, Spain; A. Ferrandez Longas, Zaragoza, Spain; R. Gracia Bouthellier, Madrid, Spain; J.P. Lopez Siguero, Malaga, Spain; S. Quinteiro, Las Palmas de Gran Canaria, Spain; M.D. Rodriguez-Arnao, Madrid, Spain; A. Rodriguez Sanchez, Madrid, Spain; J. Dahlgren, Göteborg, Sweden; L. Hagenäs, Stockholm, Sweden; J.W. Hou, Taoyuan, Taiwan; T.J. Wang, Kaohsiung County, Taiwan; P. Clayton, Manchester, UK; C. Kelnar, Edinburgh, UK.
Address all correspondence to: Peter Clayton, MRCP FRCPCH, Royal Manchester Children’s Hospital, 5th Floor Research, Oxford Road, Manchester M13 9WL, United Kingdom. Phone: 44.161.701.6949; Email: peter.clayton@manchester.ac.uk.
Conflict of interest: AS, PGM, CDL, PC, and PEC have received honoraria from Merck. EK is an employee of Merck.
Reference information: JCI Insight. 2018;3(7):e93247. https://doi.org/10.1172/jci.insight.93247.
The authors would like to thank all the PREDICT investigators and the patients who took part in the study. PGM thanks the National Institute for Health Research for their support (CL-2012-06-005). Members of the PREDICT investigator group are as follows: A. Belgorosky, Buenos Aires, Argentina; G. Ambler, Westmead, Australia; K. Kapelari, Innsbruck, Austria; C. Deal, Montreal, Canada; J. Hamilton, Toronto, Canada; J. Jääskeläinen, Kuopio, Finland; Y. Brusquet, Puyricard, France; P. Chatelain, Lyon, France; M. Colle, Bordeaux, France; R. Coutant, Angers, France; Y. Le Bouc, Paris, France; R. Reynaud, Marseille, France; J.-P. Salles, Toulouse, France; J. Weill, Lille, France; R. Pfäffle, Leipzig, Germany; M. Ranke, Tübingen, Germany; G. Binder, Tübingen, Germany; M. Bozzola, Pavia, Italy; F. Buzi, Brescia, Italy; M. Cappa, Rome, Italy; A. Cicognani, Bologna, Italy; M. Maghnie, Genova, Italy; L. Tato, Verona, Italy; F. Antoniazzi, Verona, Italy; D.H. Kim, Seoul, Korea; S.W. Yang, Seoul, Korea; H.W. Yoo, Seoul, Korea; E. Vangsøy Hansen, Bergen, Norway; D. Veimo, Bodø, Norway; E. Bashnina, St. Petersburg, Russia; V. Peterkova, Moscow, Russia; J. Skorodok, St. Petersburg, Russia; L. Sultanova, Kazan, Russia; A. Carrascosa, Barcelona, Spain; A. Ferrandez Longas, Zaragoza, Spain; R. Gracia Bouthellier, Madrid, Spain; J.P. Lopez Siguero, Malaga, Spain; S. Quinteiro, Las Palmas de Gran Canaria, Spain; M.D. Rodriguez-Arnao, Madrid, Spain; A. Rodriguez Sanchez, Madrid, Spain; J. Dahlgren, Göteborg, Sweden; L. Hagenäs, Stockholm, Sweden; J.W. Hou, Taoyuan, Taiwan; T.J. Wang, Kaohsiung County, Taiwan; P. Clayton, Manchester, UK; C. Kelnar, Edinburgh, UK.
Contributor Information
Chiara De Leonibus, Email: c.deleonibus@tigem.it.
Ekaterina Koledova, Email: ekaterina.koledova@merckgroup.com.
Pierre Chatelain, Email: pgechatelain@orange.fr.
References
- 1.Larsen CP, Durfee T, Wilson JD, Beggs ML. A custom targeted next-generation sequencing gene panel for the diagnosis of genetic nephropathies. Am J Kidney Dis. 2016;67(6):992–993. doi: 10.1053/j.ajkd.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weisschuh N, et al. Mutation detection in patients with retinal dystrophies using targeted next generation sequencing. PLoS ONE. 2016;11(1):e0145951. doi: 10.1371/journal.pone.0145951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinto R, De Summa S, Petriella D, Tudoran O, Danza K, Tommasi S. The value of new high-throughput technologies for diagnosis and prognosis in solid tumors. Cancer Biomark. 2014;14(2-3):103–117. doi: 10.3233/CBM-130328. [DOI] [PubMed] [Google Scholar]
- 4.Wang K, Huang C, Nice EC. Proteomics, genomics and transcriptomics: their emerging roles in the discovery and validation of colorectal cancer biomarkers. Expert Rev Proteomics. 2014;11(2):179–205. doi: 10.1586/14789450.2014.894466. [DOI] [PubMed] [Google Scholar]
- 5.Latkowski T, Osowski S. Computerized system for recognition of autism on the basis of gene expression microarray data. Comput Biol Med. 2015;56:82–88. doi: 10.1016/j.compbiomed.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Lindsay R, Feldkamp M, Harris D, Robertson J, Rallison M. Utah Growth Study: growth standards and the prevalence of growth hormone deficiency. J Pediatr. 1994;125(1):29–35. doi: 10.1016/S0022-3476(94)70117-2. [DOI] [PubMed] [Google Scholar]
- 7.Growth Hormone Research Society Consensus guidelines for the diagnosis and treatment of growth hormone (GH) deficiency in childhood and adolescence: summary statement of the GH Research Society. GH Research Society. J Clin Endocrinol Metab. 2000;85(11):3990–3993. doi: 10.1210/jcem.85.11.6984. [DOI] [PubMed] [Google Scholar]
- 8.Hilczer M, Smyczynska J, Stawerska R, Lewinski A. Stability of IGF-I concentration despite divergent results of repeated GH stimulating tests indicates poor reproducibility of test results. Endocr Regul. 2006;40(2):37–45. [PubMed] [Google Scholar]
- 9.Martha PM, Gorman KM, Blizzard RM, Rogol AD, Veldhuis JD. Endogenous growth hormone secretion and clearance rates in normal boys, as determined by deconvolution analysis: relationship to age, pubertal status, and body mass. J Clin Endocrinol Metab. 1992;74(2):336–344. doi: 10.1210/jcem.74.2.1730812. [DOI] [PubMed] [Google Scholar]
- 10.Stanley TL, Levitsky LL, Grinspoon SK, Misra M. Effect of body mass index on peak growth hormone response to provocative testing in children with short stature. J Clin Endocrinol Metab. 2009;94(12):4875–4881. doi: 10.1210/jc.2009-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghigo E, et al. Reliability of provocative tests to assess growth hormone secretory status. Study in 472 normally growing children. J Clin Endocrinol Metab. 1996;81(9):3323–3327. doi: 10.1210/jcem.81.9.8784091. [DOI] [PubMed] [Google Scholar]
- 12.Wagner, et al. Clinical evidence-based cutoff limits for GH stimulation tests in children with a backup of results with reference to mass spectrometry. Eur J Endocrinol. 2014;171(3):389–397. doi: 10.1530/EJE-14-0165. [DOI] [PubMed] [Google Scholar]
- 13.Kaplan SL, Abrams CA, Bell JJ, Conte FA, Grumbach MM. Growth and growth hormone. I. Changes in serum level of growth hormone following hypoglycemia in 134 children with growth retardation. Pediatr Res. 1968;2(1):43–63. doi: 10.1203/00006450-196801000-00005. [DOI] [PubMed] [Google Scholar]
- 14.Stevens A, et al. Pharmacogenomics of insulin-like growth factor-I generation during GH treatment in children with GH deficiency or Turner syndrome. Pharmacogenomics J. 2014;14(1):54–62. doi: 10.1038/tpj.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krämer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30(4):523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanti GK, Pandey S, Goswami SK. SG2NA enhances cancer cell survival by stabilizing DJ-1 and thus activating Akt. Biochem Biophys Res Commun. 2015;463(4):524–531. doi: 10.1016/j.bbrc.2015.05.069. [DOI] [PubMed] [Google Scholar]
- 17.de Bruijn DR, et al. The cancer-related protein SSX2 interacts with the human homologue of a Ras-like GTPase interactor, RAB3IP, and a novel nuclear protein, SSX2IP. Genes Chromosomes Cancer. 2002;34(3):285–298. doi: 10.1002/gcc.10073. [DOI] [PubMed] [Google Scholar]
- 18.Orozco G, et al. Novel rheumatoid arthritis susceptibility locus at 22q12 identified in an extended UK genome-wide association study. Arthritis Rheumatol. 2014;66(1):24–30. doi: 10.1002/art.38196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Araç D, et al. Structures of neuroligin-1 and the neuroligin-1/neurexin-1 beta complex reveal specific protein-protein and protein-Ca2+ interactions. Neuron. 2007;56(6):992–1003. doi: 10.1016/j.neuron.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 20.RABEN MS. Treatment of a pituitary dwarf with human growth hormone. J Clin Endocrinol Metab. 1958;18(8):901–903. doi: 10.1210/jcem-18-8-901. [DOI] [PubMed] [Google Scholar]
- 21.GLICK SM, ROTH J, YALOW RS, BERSON SA. IMMUNOASSAY OF HUMAN GROWTH HORMONE IN PLASMA. Nature. 1963;199:784–787. doi: 10.1038/199784a0. [DOI] [PubMed] [Google Scholar]
- 22. Rochiccioli P, Tauber M. Growth hormone deficiency: establishment of the diagnosis by means of growth hormone secretion tests in the Kabi international growth study. In: Ranke MB, Gunnarson Rm eds. Progress in growth hormone therapy–5 years of KIGS. Mannheim: Verlag; 69–76. [Google Scholar]
- 23.Kelberman D, Rizzoti K, Lovell-Badge R, Robinson IC, Dattani MT. Genetic regulation of pituitary gland development in human and mouse. Endocr Rev. 2009;30(7):790–829. doi: 10.1210/er.2009-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meinhardt UJ, Ho KK. Modulation of growth hormone action by sex steroids. Clin Endocrinol (Oxf) 2006;65(4):413–422. doi: 10.1111/j.1365-2265.2006.02676.x. [DOI] [PubMed] [Google Scholar]
- 25.Kiess W, Gallaher B. Hormonal control of programmed cell death/apoptosis. Eur J Endocrinol. 1998;138(5):482–491. doi: 10.1530/eje.0.1380482. [DOI] [PubMed] [Google Scholar]
- 26.Veldhuis JD, Keenan DM, Pincus SM. Motivations and methods for analyzing pulsatile hormone secretion. Endocr Rev. 2008;29(7):823–864. doi: 10.1210/er.2008-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thalange NK, et al. Infradian rhythms in urinary growth hormone excretion. J Clin Endocrinol Metab. 1996;81(1):100–106. doi: 10.1210/jcem.81.1.8550735. [DOI] [PubMed] [Google Scholar]
- 28.Spiliotis BE, August GP, Hung W, Sonis W, Mendelson W, Bercu BB. Growth hormone neurosecretory dysfunction. A treatable cause of short stature. JAMA. 1984;251(17):2223–2230. [PubMed] [Google Scholar]
- 29.Almuriekhi M, et al. Loss-of-function mutation in APC2 causes Sotos syndrome features. Cell Rep. 2015;10(9):1585–1598. doi: 10.1016/j.celrep.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 30.Yamulla RJ, et al. Testing models of the APC tumor suppressor/β-catenin interaction reshapes our view of the destruction complex in Wnt signaling. Genetics. 2014;197(4):1285–1302. doi: 10.1534/genetics.114.166496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kioussi C, et al. Identification of a Wnt/Dvl/beta-Catenin -- > Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111(5):673–685. doi: 10.1016/S0092-8674(02)01084-X. [DOI] [PubMed] [Google Scholar]
- 32.Kelberman D, et al. SOX2 plays a critical role in the pituitary, forebrain, and eye during human embryonic development. J Clin Endocrinol Metab. 2008;93(5):1865–1873. doi: 10.1210/jc.2007-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guarente L. Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol. 2007;72:483–488. doi: 10.1101/sqb.2007.72.024. [DOI] [PubMed] [Google Scholar]
- 34.Choi YH, Kim H, Lee SH, Jin YH, Lee KY. Src regulates the activity of SIRT2. Biochem Biophys Res Commun. 2014;450(2):1120–1125. doi: 10.1016/j.bbrc.2014.06.117. [DOI] [PubMed] [Google Scholar]
- 35.Clayton P, et al. A pharmacogenomic approach to the treatment of children with GH deficiency or Turner syndrome. Eur J Endocrinol. 2013;169(3):277–289. doi: 10.1530/EJE-13-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Leonibus C, Chatelain P, Knight C, Clayton P, Stevens A. Effect of summer daylight exposure and genetic background on growth in growth hormone-deficient children. Pharmacogenomics J. 2016;16(6):540–550. doi: 10.1038/tpj.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaizer EC, Glaser CL, Chaussabel D, Banchereau J, Pascual V, White PC. Gene expression in peripheral blood mononuclear cells from children with diabetes. J Clin Endocrinol Metab. 2007;92(9):3705–3711. doi: 10.1210/jc.2007-0979. [DOI] [PubMed] [Google Scholar]
- 38.Elo LL, et al. Early suppression of immune response pathways characterizes children with prediabetes in genome-wide gene expression profiling. J Autoimmun. 2010;35(1):70–76. doi: 10.1016/j.jaut.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 39.Wong HR, et al. Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Med. 2009;7:34. doi: 10.1186/1741-7015-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramasamy A, Mondry A, Holmes CC, Altman DG. Key issues in conducting a meta-analysis of gene expression microarray datasets. PLoS Med. 2008;5(9):e184. doi: 10.1371/journal.pmed.0050184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevens A, Hanson D, Whatmore A, Destenaves B, Chatelain P, Clayton P. Human growth is associated with distinct patterns of gene expression in evolutionarily conserved networks. BMC Genomics. 2013;14:547. doi: 10.1186/1471-2164-14-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 43.Nilsson J, Fioretos T, Höglund M, Fontes M. Approximate geodesic distances reveal biologically relevant structures in microarray data. Bioinformatics. 2004;20(6):874–880. doi: 10.1093/bioinformatics/btg496. [DOI] [PubMed] [Google Scholar]
- 44.Tenenbaum JB, de Silva V, Langford JC. A global geometric framework for nonlinear dimensionality reduction. Science. 2000;290(5500):2319–2323. doi: 10.1126/science.290.5500.2319. [DOI] [PubMed] [Google Scholar]
- 45.Fontes M, Soneson C. The projection score--an evaluation criterion for variable subset selection in PCA visualization. BMC Bioinformatics. 2011;12:307. doi: 10.1186/1471-2105-12-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chatr-Aryamontri A, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41(Database issue):D816–D823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ravasz E, Barabási AL. Hierarchical organization in complex networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2003;67(2 Pt 2):026112. doi: 10.1103/PhysRevE.67.026112. [DOI] [PubMed] [Google Scholar]
- 49.Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein M. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput Biol. 2007;3(4):e59. doi: 10.1371/journal.pcbi.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun J, Zhao Z. A comparative study of cancer proteins in the human protein-protein interaction network. BMC Genomics. 2010;11 Suppl 3:S5. doi: 10.1186/1471-2164-11-S3-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kovács IA, Palotai R, Szalay MS, Csermely P. Community landscapes: an integrative approach to determine overlapping network module hierarchy, identify key nodes and predict network dynamics. PLoS One. 2010;5(9):e12528. doi: 10.1371/journal.pone.0012528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haw R, Stein L. Using the reactome database. Curr Protoc Bioinformatics. 2012;Chapter 8:Unit8.7. doi: 10.1002/0471250953.bi0807s38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nepusz T, Yu H, Paccanaro A. Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods. 2012;9(5):471–472. doi: 10.1038/nmeth.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Svetnik V, Liaw A, Tong C, Culberson JC, Sheridan RP, Feuston BP. Random forest: a classification and regression tool for compound classification and QSAR modeling. J Chem Inf Comput Sci. 2003;43(6):1947–1958. doi: 10.1021/ci034160g. [DOI] [PubMed] [Google Scholar]
- 55.Nakamura M, Kajiwara Y, Otsuka A, Kimura H. LVQ-SMOTE - Learning vector quantization based synthetic minority over-sampling technique for biomedical data. BioData Min. 2013;6(1):16. doi: 10.1186/1756-0381-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kursa MB, Rudnicki WR. Feature selection with the Boruta package. J Stat Softw. 2010;36(11):1–13. [Google Scholar]
- 57.McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA) Biostatistics. 2010;11(2):242–253. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCall MN, et al. The Gene Expression Barcode 3.0: improved data processing and mining tools. Nucleic Acids Res. 2014;42(Database issue):D938–D943. doi: 10.1093/nar/gkt1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.