Abstract.
A phylogenetic analysis of Orientia tsutsugamushi was performed to elucidate its antigenic diversity in chiggers, small mammals, and patients. Between September 2014 and December 2016, a total of 3,816 chiggers were identified within nine species of four genera in the southwest region of Korea: Leptotrombidium scutellare (49.9%; 1,907/3,816), Leptotrombidium orientale (21.1%; 804/3,816), Leptotrombidium pallidum (12.4%; 474/3,816), Euchoengastia koreaensis (7.2%; 273/3,816), Leptotrombidium palpale (6.7%; 256/3,816), Neotrombicular gardellai (1.3%; 50/3,816), Leptotrombidium zetum (0.8%; 32/3,816), Walchia fragilis (0.5%; 18/3,816), and Neotrombicular japonica (> 0.1%; 2/3,816). Twelve chiggers (11 L. scutellare and one L. palpale) tested positive for O. tsutsugamushi by polymerase chain reaction and, except for 1 chigger (KY266830), were part of the Boryong strain cluster. Of the 413 small mammals that were analyzed for O. tsutsugamushi, Apodemus agrarius was the most common rodent species (89.5%; 370/413), followed by Crocidura lasiura (6.8%; 28/413) and Myodes regulus (3.6%; 15/413). The sequence identity of an O. tsutsugamushi sample obtained from the A. agrarius sample population belonged to the Saitama strain cluster. Furthermore, a phylogenetic analysis in 125 patients revealed four clusters (Boryong cluster: 82.4% [103/125], Karp: 13.6% [17/125], Kawasaki: 3.2% [4/125], and Saitama: 0.8% [1/125]). This study clarified the phylogenetic relationship for O. tsutsugamushi in chiggers, small mammals, and patients. The Boryong strain was the most common strain in chiggers and patients. In addition, various strains were identified, except for the Boryong strain, in the southwest region of Korea. Overall, the data presented here will be helpful for the establishment of prevention strategies for scrub typhus.
INTRODUCTION
Orientia tsutsugamushi is a rod-shaped, gram-negative, intracellular bacterium that belongs to the order Rickettsiales and is the causative agent of scrub typhus or Tsutsugamushi disease.1,2 Unlike other bacteria, the cell walls of O. tsutsugamushi lack a peptidoglycan and lipopolysaccharide layer. Scrub typhus has been recognized as a major cause of acute febrile disease that is encountered in rural regions such as bushes and abandoned grain field.3 This disease is endemic to the Western Pacific region, northern Australia, and Central Asia. Clinical characteristics of scrub typhus include eschar, fever, rash, meningitis, intracellular coagulation, lymphadenopathy, and multiple organ failure. Moving forward, if patients are not treated, the fatality rate will continue to increase.4
Furthermore, O. tsutsugamushi can be transmitted by the larval trombiculid mite (chigger) while feeding, which then can propagate to its progeny through transovarian transmission. The Leptotrombidium species are known to act as the primary vector for O. tsutsugamushi,5,6 where the major species, Leptotrombidium pallidum and Leptotrombidium scutellare, can be found in the central and southern regions of Korea, respectively.7,8
Wild rodents, such as Apodemus agrarius, are also natural hosts of scrub typhus as well as for chiggers.9,10 At the same regions where this study was performed, a serosurveillance for scrub typhus in small mammals was performed in 2014–2015 by Park, and a seropositive rate of 25% (37/145) in A. agrarius was observed.11
The 56-kDa type-specific antigen (TSA) is an immunodominant protein located on the surface of O. tsutsugamushi that is not expressed in other bacteria. Therefore, phylogenetic analysis of the 56-kDa TSA gene sequence is useful to clarify variations of scrub typhus.12–15
Worldwide, more than 20 variations of scrub typhus are caused by various prototype strains of O. tsutsugamushi. The Boryong strain is predominant in Korea. The Giliam, Karp, Kato, TA678, TA686, TA716, TA763, and TH1817 strains have been reported in Southeast Asia, Australia, and Taiwan. In addition, the Gilliam, Karp, Kato, Kawasaki, Kuroki, and Shimokoshi strains have been found in Japan.1 Furthermore, differences in virulence according to genotype variations of O. tsutsugamushi have been identified.16
Recently, climate change may have affected the life cycle of wild rodent and chiggers.17 Therefore, clarifying the relationship between the vector and environment may help predict future cases of scrub typhus.18–23 In northern China, a phylogenetic analysis for scrub typhus in chiggers, rodents, and patients was conducted, and consistency of circulating strains in the hosts was demonstrated.24
According to the Korea Centers for Disease Control and Prevention, 10,000 cases of scrub typhus were reported annually between 2013 and 2015, as well as 6,000 cases in 2005. The distribution of chiggers has also expanded from the southern region to the central region for the past decade.25 Therefore, scrub typhus is an increasingly important disease in Korea.26–29
For the development of strategies to control this disease, a better understanding of the vector’s life cycle is required as well as knowledge of the O. tsutsugamushi strain in the host. We surveyed a distribution of chiggers and performed a phylogenetic analysis of the 56-kDa TSA gene sequence to identify the genetic characteristics of O. tsutsugamushi in chiggers, small mammals, and patients in the southwest region of Korea.
MATERIALS AND METHODS
Collection of small mammals and chiggers.
All procedures were conducted under an animal use protocol approved by the Chosun University Animal Ethics Committee. From September 2014 to December 2016, small mammals were trapped monthly using Sherman live traps (3 in × 3.5 in × 9 in, USA) baited with peanut butter–covered biscuits in two southwest regions of Korea (Figure 1). After wild rodents were euthanized, their organs (spleen, kidney) were collected for detection of O. tsutsugamushi. Chiggers collected from the rodents were stored in 70% ethyl alcohol until used to acquire sample fluids for DNA extraction.
Figure 1.
Small mammals collection sites at A (35°13′51.7″ N, 126°54′23.8″ E) and B(35°09′19.2″ N, 126°45′05.4″ E) in the southwest area of Korea, from September 2014 to December 2016. This figure appears in color at www.ajtmh.org.
Patients’ blood sample.
From 2014 to 2015, blood samples were collected from patients who had visited Chosun University Hospital with suspected scrub typhus in the southwest region of Korea. Blood sample analysis was blinded. All experiments were conducted under approval of the Chosun University Medical College (IRB number: 2013-10-001).
Acquisition of fluids from chiggers and species identification.
The fluids from the engorged chiggers were acquired as previously described.30 The chigger’s internal contents (organ, fluid, and hemolymph) were squeezed out with two fine pins under a stereomicroscope (Carl Zeiss, Oberkochen, Germany). The chigger exoskeleton was identified by Ree’s fauna key.31
DNA extraction from chigger samples.
DNA from the chigger samples were purified using the G-spin Total DNA Extraction Kit (Intron Biotechnology, Seoul, Korea). The DNA from the rodent’s organs as well as the patients’ blood was purified using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. DNA was stored at −20°C until use.
Nested polymerase chain reaction (PCR) amplification for the 56-kDa TSA gene.
PCR was performed via the INNOPLEX TSUTSU detection kit (catalogue number: IPC10040; Intron Biotechnology). The kit was designed using primer sets (First F: 5ʹ-GCA ATA TTG CTA GTG CAA TGT CTG C-3ʹ, First R: 5ʹ-ATG CAT GCA TGR CGC TKC AAT TTA-3ʹ; Second F: 5ʹ-ATA GGC CTA TAA GTA TWG CKG ATC G-3ʹ, Second R: 5ʹ-CAT CTA GAY GCA CTA TTA GGC AAA-3ʹ) to detect the 56-kDa TSA gene of O. tsutsugamushi. The target was a 426–862-bp region that includes variable domain 2, 3 in the total TSA 56 gene (1,566 bp). The first PCR was amplified at 94°C for 5 minutes, followed by 40 cycles at 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 40 seconds, and a final elongation at 72°C for 5 minutes. The second PCR was amplified at 94°C for 5 minutes, followed by 30 cycles at 94°C for 30 seconds, 58°C for 30 seconds, 72°C for 40 seconds, and a final elongation at 72°C for 5 minutes using the GeneAmp 9700 Biosystem (ABI, California). The final 475-bp PCR products were evaluated by 1.5% agarose gel electrophoresis and visualized with ethidium bromide and an ultraviolet transilluminator.
Sequence and phylogenetic analysis.
The amplified PCR products were sent to Cosmogenetech (Daejeon, Korea) for sequencing using the ABI 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA). The nucleotide sequences were aligned using ClustalW to a reference sequence downloaded from the National Center for Biotechnology Information database, and phylogenetic analyses were conducted via the MEGA6 program. A neighbor-joining tree with 1,000 bootstrap replicates was constructed using the Kimura’s two-parameter model. Sequences obtained from chiggers, rodents, and human samples were submitted to GenBank (accession number: KY266824-KY266830, KX363954, and KY946003-KY946128, respectively).
RESULT
Small mammal collection and chigger infestation rate.
During the study period, 413 small mammals, including two species of rodents and one species of shrew, were captured. Apodemus agrarius were the most common small mammals (370; 89.5%), followed by Crocidura lasiura (28; 6.8%) and Myodes regulus (15; 3.6%). Furthermore, A. agrarius were captured year-round. The infestation rate of chiggers on the small mammals was 51.8% (214/413), A. agrarius 53.5% (198/370), M. regulus 100% (15/15), and C. lasiura 3.5% (1/28). Crocidura lasiura were infested with fewer chiggers than other small mammals. The infestation rate for M. regulus was 100% during the study period (Table 1).
Table 1.
Chigger infestation rates (%) on small mammals (number of chigger infested small mammals/number of captured small mammals) in the southwest area of Korea, from September 2014 to December 2016
| Month | Rodents | Shrew | Total (chigger infested/captured small mammals) | |
|---|---|---|---|---|
| Apodemus agrarus | Myodes regulus | Crocidura lasiura | ||
| September 2014 | 9/10 | – | – | 9/10 |
| October | 12/12 | – | – | 12/12 |
| November | 18/22 | – | 0/1 | 18/23 |
| December | 0/5 | 1/1 | 0/1 | 1/7 |
| January 2015 | 8/14 | 3/3 | 0/1 | 11/18 |
| February | 3/7 | 1/1 | – | 4/8 |
| March | 5/10 | 1/1 | – | 6/11 |
| April | 3/9 | 1/1 | – | 4/10 |
| May | 11/22 | 1/1 | 0/3 | 12/26 |
| June | 2/16 | – | – | 2/16 |
| July | 0/18 | – | – | 0/18 |
| August | 4/13 | – | – | 4/13 |
| September | 8/10 | – | 0/1 | 8/11 |
| October | 15/15 | – | 1/1 | 16/16 |
| November | 17/17 | 1/1 | – | 18/18 |
| December | 14/15 | – | – | 14/15 |
| January 2016 | 4/6 | 1/1 | – | 4/7 |
| February | 7/8 | – | – | 7/8 |
| March | 12/13 | 1/1 | 0/1 | 13/15 |
| April | 9/10 | 0/2 | 9/12 | |
| May | 6/25 | 1/1 | 0/1 | 7/27 |
| June | 0/22 | – | – | 0/22 |
| July | 0/13 | – | 0/1 | 0/14 |
| August | 0/7 | – | – | 0/7 |
| September | 6/12 | 1/1 | 0/1 | 7/14 |
| October | 5/5 | – | 1/5 | 6/10 |
| November | 12/12 | – | 0/4 | 12/16 |
| December | 8/25 | 2/2 | 0/5 | 10/32 |
| Total | 198/370 (53.5) | 15/15 (100) | 1/28 (3.5) | 214/413 (51.8) |
Monthly species distribution of chiggers.
A total of 3,816 chiggers were collected from 214 small mammals. Leptotrombidium scutellare was the most commonly collected species (1,907; 49.9%), followed by Leptotrombidium orientale (804; 21.1%), L. pallidum (474; 12.4%), Euchoengastia koreaensis (273; 7.2%), Leptotrombidium palpale (256; 6.7%), Neotrombicula gardellai (50; 1.3%), Leptotrombidium zetum (32; 0.8%), Walchia fragilis (18; 0.5%), and Neotrombicular japonica (2, < 0.1%). Regarding distribution, 3,512 (92.0%) chiggers were found on A. agrarius, followed by 301 on M. regulus (7.9%), and 3 (< 0.1%) on C. lasiura. Eight species of chiggers were observed on A. agrarius. Furthermore, W. fragilis was only collected from M. regulus (Table 2).
Table 2.
Species of chiggers collected from small mammals captured in the southwest area of Korea, from September 2014 to December 2016
| Number of chiggers collected (%) | Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Host species | Leptotrombidium scutellare | Leptotrombidium pallidum | Leptotrombidium orientale | Leptotrombidium palpale | Leptotrombidium zetum | Neotrombicular gardellai | Neotrombicular japonica | Euchoengastia koreaensis | Walchia fragilis | |
| Rodents | ||||||||||
| Apodemus agrarius | 1,866 | 474 | 585 | 237 | 30 | 50 | 2 | 268 | – | 3,512 (92.0) |
| Myodes regulus | 38 | – | 219 | 19 | 2 | – | – | 5 | 18 | 301 (7.9) |
| Shrews | ||||||||||
| Crocidura lasiura | 3 | – | – | – | – | – | – | – | – | 3 (> 0.1) |
| Total | 1,907 (49.9) | 474 (12.4) | 804 (21.1) | 256 (6.7) | 32 (0.8) | 50 (1.3) | 2 (> 0.1) | 273 (7.2) | 18 (0.5) | 3,816 (100) |
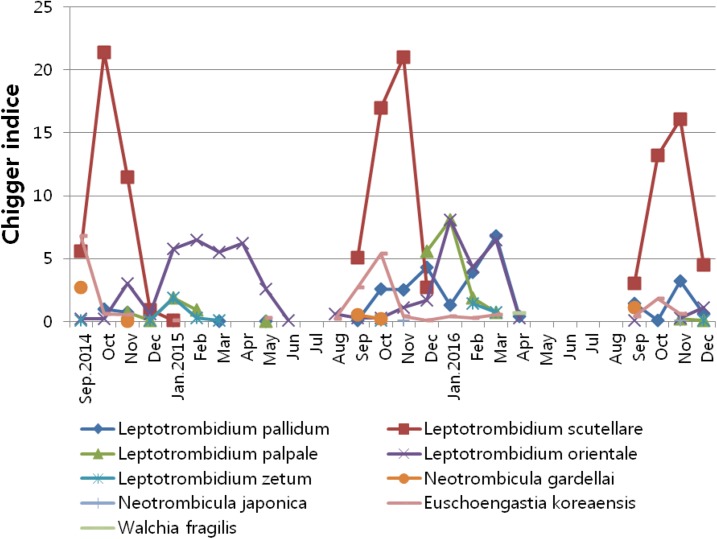
Monthly fluctuation of chiggers by host is presented in Table 3 and Figure 2. We observed seasonal differences in chigger species. During the autumn season from September to December, the predominant species was L. scutellare whereas, during the spring season between March and May, the predominant species was L. orientale.
Table 3.
Monthly distribution of chigger species (positive chigger for Orientia tsutsugamushi) collected from small mammals captured in the southwest area of Korea
| Month | Leptotrombidium pallidum | Leptotrombidium scutellare | Leptotrombidium palpale | Leptotrombidium orientale | Leptotrombidium zetum | Neotrombicular gardellai | Neotrombicular japonica | Euchoengastia koreaensis | Walchia fragilis | Total |
|---|---|---|---|---|---|---|---|---|---|---|
| September 2014 | – | 56 | – | 2 | 1 | 27 | – | 68 | – | 154 |
| October | 12 | 257 (2) | – | 2 | – | – | – | 7 | – | 278 (2) |
| November | 17 | 265 | 15 | 70 | – | 1 | – | 12 | – | 380 |
| December | – | 6 | 1 | 12 | 1 | – | – | – | – | 20 |
| January 2015 | 1 | 1 | 35 | 105 | 3 | – | – | 1 | – | 146 |
| February | – | – | 7 | 52 | 2 | – | – | – | – | 61 |
| March | 5 | – | – | 60 | 1 | – | – | – | – | 66 |
| April | – | – | – | 62 (1) | – | – | – | – | – | 62 |
| May | 1 | – | 1 | 68 | – | – | – | 9 | – | 79 |
| June | – | – | – | 2 | – | – | – | – | 2 | |
| July | – | – | – | – | – | – | – | – | ||
| August | – | – | – | 8 | – | – | – | 2 | – | 10 |
| September | 1 | 56 | – | 3 | – | 5 | – | 30 | – | 95 |
| October | 41 | 272 (2) | – | 4 | 1 | 2 | – | 87 | – | 407 |
| November | 45 | 378 | – | 19 | – | – | 1 | 4 | – | 447 |
| December | 65 | 40 | 84 | 25 | – | – | – | 2 | – | 216 |
| January 2016 | 9 | – | 57 | 57 | – | – | – | 3 | – | 126 |
| February | 31 | – | 15 | 34 | 11 | – | – | 2 | – | 93 |
| March | 102 | – | 10 | 98 | 10 | – | – | 8 | – | 228 |
| April | 44 | – | 24 | 74 | – | – | – | 5 | – | 147 |
| May | 10 | – | – | 7 | – | – | – | – | 18 | 35 |
| June | – | – | – | – | – | – | – | – | – | – |
| July | – | – | – | – | – | – | – | – | – | – |
| August | – | – | – | – | – | – | – | – | – | – |
| September | 19 | 42 | – | 2 | – | 15 | – | 6 | – | 84 |
| October | 1 | 132 (6) | – | – | – | – | – | 18 | – | 151 |
| November | 51 | 258 (1) | 3 | 4 | – | – | 1 | 9 | – | 326 |
| December | 19 | 144 | 4 | 34 | 2 | – | – | – | – | 203 |
| Total | 474 | 1,907 (11) | 256 | 804 (1) | 32 | 50 | 2 | 273 | 18 | 3,816 (12) |
Figure 2.
Monthly chigger indices by species collected from small mammals captured in the southwest area of Korea, from September 2014 to December 2016. This figure appears in color at www.ajtmh.org.
Prevalence of O. tsutsugamushi in samples.
Individual nested PCRs were used to screen 3,816 chiggers for O. tsutsugamushi. A total of 12 chiggers (0.3%; 12/3,816) were positive for O. tsutsugamushi, and 11 samples of L. scutellare and one L. orientale were confirmed. Furthermore, one A. agrarius specimen from the 413 small mammals tested was confirmed to have O. tsutsugamushi (0.2%; 1/413). Finally, 125/402) of the patients with suspected scrub typhus tested positive (Table 4).
Table 4.
Distribution of Orientia tsutsugamushi strains in chigger, small mammals, and humans in the southwest area of Korea
| Host | Test | Positive (%) | Cluster | ||||
|---|---|---|---|---|---|---|---|
| Boryong | Kawasaki | Saitama | Karp | Etc. | |||
| 114 | 4 | 2 | 17 | 1 | |||
| Chiggers | 3,816 | 12 (0.3) | 11 | – | – | – | 1 |
| Leptotrombidium scutellare | 1,907 | 11 (0.6) | 10 | – | – | – | 1 |
| Leptotrombidium pallidum | 474 | – | – | – | – | – | – |
| Leptotrombidium orientale | 256 | 1 (0.4) | 1 | – | – | – | – |
| Leptotrombidium palpale | 804 | – | – | – | – | – | – |
| Leptotrombidium zetum | 32 | – | – | – | – | – | – |
| Neotrombicular gardellai | 50 | – | – | – | – | – | – |
| Neotrombicular japonica | 2 | – | – | – | – | – | – |
| Euchoengastia koreaensis | 273 | – | – | – | – | – | – |
| Walchia fragilis | 18 | – | – | – | – | – | – |
| Small mammals | 413 | 1 (0.2) | – | – | 1 | – | – |
| Apodemus agrarius | 370 | 1 (0.3) | – | – | 1 | – | – |
| Myodes regulus | 15 | – | – | – | – | – | – |
| Crocidura lasiura | 28 | – | – | – | – | – | – |
| Human | 402 | 125 (50.5) | 103 | 4 | 1 | 17 | – |
Phylogenetic analysis for the 56-kDa TSA gene of O. tsutsugamushi.
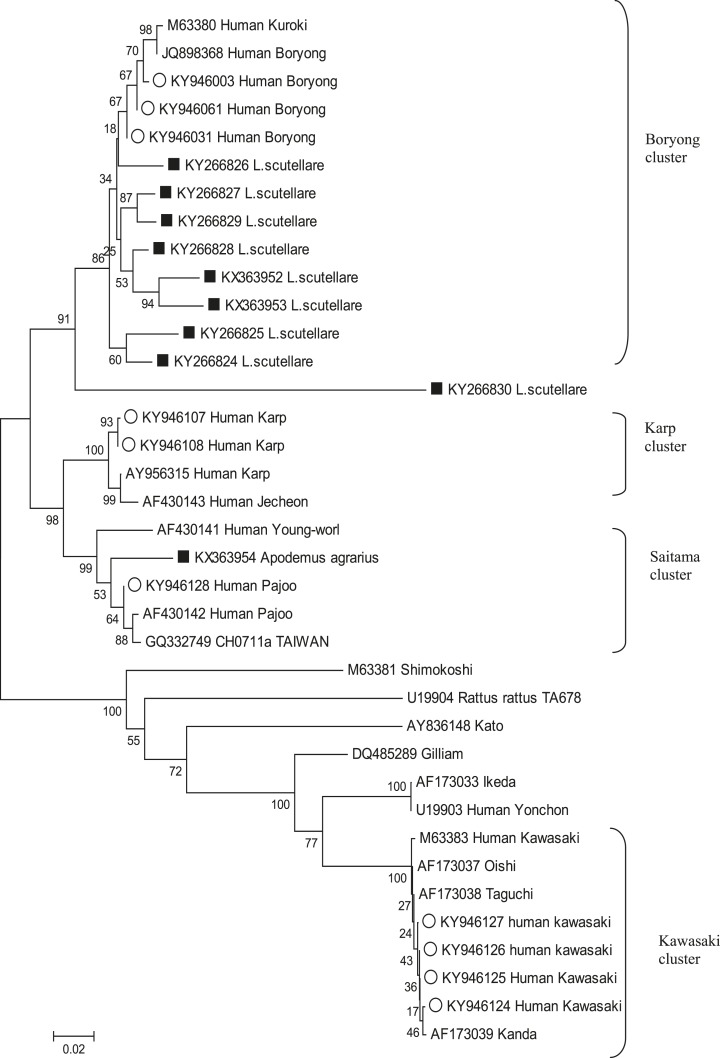
The 56 kDa TSA gene sequence identified in 11 chiggers, one rodent, and 125 patients corresponded to four different strains of O. tsutsugamushi. The Boryong strain was the most common in chiggers (11/12; 95.8%) and patients (103/125; 82.4%). Interestingly, the sequence (KX363954) identified from the rodents was 84.3% homologous to the CH0711a strain (GQ342749), which is related to the Saitama strain cluster (Table 5). Peculiarly, there was a difference between the sequence (KY266830) identified from the chigger and Boryong cluster (Similarity percent: 74.0–79.8%) (Table 5). In patient samples, the 17 sequences (KY946107-KY946123) identified (17/125; 13.6%) are related to the Karp cluster. In addition, the four sequences (KY946124-KY946127) that were identified (4/125; 3.2%) are related to the Kawasaki cluster, one sequence (KY946128) that were identified (1/125; 0.8%) are related to the Saitama cluster (Figure 3; Table 6). For composition of phylogenetic tree, sequences obtained from chiggers (KX363952, KX363953, KY266824-KY266830), rodent (KX363954), human (KY946003, KY946031, KY946061, KY946107, KY946108, and KY946128, KY946124-KY946127) were used (Table 6).
Table 5.
Identity matrix of Orientia tsutsugamushi strains in chigger, small mammals, and humans in the southwest area of Korea
| Percent identity | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Divergence | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | |||
| 1 | – | 92.1 | 92.2 | 76.7 | 67.1 | 67.0 | 67.0 | 67.0 | 85.6 | 87.4 | 88.3 | 93.7 | 73.8 | 70.9 | 72.4 | 70.0 | 82.7 | 66.1 | 71.8 | 81.2 | 95.5 | 87.4 | 65.7 | 67.9 | 68.8 | 68.4 | 87.7 | 87.2 | 87.2 | 90.3 | 89.9 | 90.1 | 89.4 | 72.4 | 84.1 | 83.2 | 82.7 | 1 | KY946003_Human_Boryong | |
| 2 | 0.0 | – | 99.8 | 76.7 | 73.8 | 74.5 | 73.8 | 74.9 | 83.4 | 85.2 | 85.6 | 86.6 | 66.4 | 64.1 | 65.2 | 62.8 | 80.5 | 58.8 | 75.8 | 85.4 | 88.4 | 80.0 | 64.1 | 67.3 | 66.8 | 66.4 | 80.3 | 80.0 | 91.0 | 94.4 | 94.4 | 94.0 | 93.9 | 76.9 | 88.4 | 87.5 | 87.0 | 2 | KY946031_Human_Boryong | |
| 3 | 0.0 | 0.0 | – | 76.5 | 74.0 | 74.7 | 74.0 | 74.7 | 83.2 | 85.0 | 85.7 | 86.8 | 66.6 | 64.3 | 65.3 | 63.0 | 80.3 | 59.0 | 76.0 | 85.6 | 88.6 | 80.1 | 63.9 | 67.5 | 66.6 | 66.2 | 80.5 | 80.1 | 91.2 | 94.6 | 94.6 | 94.2 | 94.0 | 76.9 | 88.6 | 87.7 | 87.2 | 3 | KY946061_Human_Boryong | |
| 4 | 8.4 | 8.9 | 8.9 | – | 59.6 | 59.4 | 58.7 | 59.7 | 82.3 | 80.9 | 81.6 | 71.1 | 57.2 | 57.0 | 57.9 | 55.1 | 84.1 | 51.8 | 60.5 | 76.4 | 72.9 | 77.4 | 71.5 | 55.4 | 61.7 | 61.4 | 75.5 | 78.9 | 67.9 | 71.1 | 71.1 | 70.8 | 70.6 | 57.9 | 65.3 | 64.4 | 63.9 | 4 | KY946128_Human_Pajoo | |
| 5 | 34.7 | 34.7 | 34.6 | 37.5 | – | 99.3 | 98.4 | 98.9 | 64.6 | 66.2 | 67.7 | 61.7 | 81.6 | 82.9 | 91.2 | 68.4 | 62.6 | 77.6 | 70.6 | 67.9 | 63.5 | 62.8 | 64.3 | 93.3 | 92.1 | 91.7 | 61.9 | 62.3 | 68.2 | 69.3 | 69.3 | 69.5 | 69.1 | 58.5 | 63.7 | 63.5 | 63.0 | 5 | KY946124_Human_Kawasaki | |
| 6 | 34.2 | 34.3 | 34.2 | 37.0 | 0.2 | – | 99.1 | 99.6 | 64.4 | 66.1 | 67.5 | 61.6 | 81.4 | 82.3 | 90.6 | 67.7 | 62.5 | 77.1 | 71.3 | 68.6 | 63.4 | 62.6 | 64.1 | 92.8 | 91.9 | 91.5 | 61.7 | 62.1 | 69.0 | 70.0 | 70.0 | 70.2 | 69.9 | 59.2 | 64.4 | 64.3 | 63.7 | 6 | KY946125_Human_Kawasaki | |
| 7 | 33.9 | 34.3 | 34.1 | 36.9 | 0.2 | 0.0 | – | 98.9 | 63.7 | 65.3 | 67.5 | 61.6 | 81.4 | 82.3 | 90.4 | 67.5 | 61.7 | 77.1 | 71.3 | 69.1 | 63.4 | 62.6 | 63.4 | 92.6 | 91.0 | 90.6 | 61.7 | 62.1 | 69.9 | 70.8 | 70.4 | 71.1 | 70.4 | 59.9 | 65.2 | 64.8 | 64.3 | 7 | KY946126_human_kawasaki | |
| 8 | 33.9 | 33.9 | 33.9 | 36.5 | 0.5 | 0.2 | 0.0 | – | 64.8 | 66.4 | 67.5 | 61.6 | 81.4 | 82.3 | 90.3 | 67.3 | 62.8 | 77.1 | 71.3 | 68.4 | 63.4 | 62.6 | 64.4 | 92.4 | 91.9 | 91.5 | 61.7 | 62.1 | 68.8 | 69.9 | 69.9 | 70.0 | 69.7 | 59.2 | 64.3 | 64.1 | 63.5 | 8 | KY946127_human_kawasaki | |
| 9 | 8.6 | 9.2 | 9.2 | 5.5 | 36.5 | 36.1 | 36.0 | 35.7 | – | 98.2 | 84.8 | 82.9 | 68.8 | 64.1 | 67.3 | 64.4 | 96.0 | 62.1 | 64.4 | 79.4 | 84.7 | 86.8 | 68.8 | 60.5 | 71.7 | 71.5 | 91.7 | 87.2 | 74.5 | 77.8 | 77.8 | 77.4 | 77.3 | 64.1 | 71.8 | 70.9 | 70.4 | 9 | KY946107_Human_Karp | |
| 10 | 8.4 | 9.0 | 9.0 | 5.5 | 35.9 | 35.4 | 35.4 | 35.1 | 0.0 | – | 86.5 | 84.5 | 70.6 | 65.7 | 69.0 | 66.1 | 94.4 | 63.5 | 66.1 | 81.0 | 86.3 | 88.3 | 67.3 | 62.1 | 71.8 | 71.5 | 93.3 | 88.6 | 76.4 | 79.6 | 79.6 | 79.2 | 79.1 | 65.5 | 73.6 | 72.7 | 72.2 | 10 | KY946108_Human_Karp | |
| 11 | 9.5 | 9.5 | 9.4 | 0.3 | 34.8 | 34.3 | 34.1 | 34.1 | 6.1 | 6.2 | – | 84.8 | 71.3 | 69.7 | 68.6 | 67.3 | 82.5 | 64.4 | 72.0 | 89.9 | 86.8 | 91.9 | 61.4 | 70.4 | 64.8 | 64.4 | 88.4 | 91.7 | 82.3 | 83.8 | 83.6 | 84.7 | 83.0 | 69.9 | 79.1 | 77.8 | 77.6 | 11 | AF430142_Human_Pajoo | |
| 12 | 1.0 | 0.0 | 0.0 | 8.7 | 34.7 | 34.2 | 33.9 | 33.9 | 10.1 | 9.7 | 9.2 | – | 75.3 | 71.1 | 69.5 | 73.6 | 80.9 | 70.9 | 66.6 | 75.5 | 96.6 | 87.5 | 61.9 | 67.5 | 66.6 | 66.6 | 86.8 | 85.6 | 83.2 | 84.7 | 84.7 | 85.6 | 84.3 | 68.1 | 83.4 | 81.9 | 81.8 | 12 | M63380_Human_Kuroki | |
| 13 | 25.7 | 28.0 | 27.9 | 31.5 | 11.1 | 10.6 | 10.4 | 10.4 | 27.5 | 26.8 | 26.7 | 24.5 | – | 91.2 | 89.7 | 78.2 | 66.4 | 89.2 | 65.0 | 62.6 | 75.1 | 73.8 | 63.9 | 86.3 | 85.9 | 85.6 | 73.3 | 71.1 | 65.3 | 65.3 | 65.0 | 66.4 | 65.0 | 54.9 | 61.6 | 61.0 | 61.4 | 13 | DQ485289_Gilliam | |
| 14 | 31.3 | 33.9 | 33.8 | 33.2 | 10.0 | 10.1 | 9.8 | 9.8 | 32.1 | 31.6 | 30.6 | 30.9 | 7.7 | – | 88.1 | 77.4 | 61.7 | 94.8 | 65.5 | 61.2 | 69.3 | 69.1 | 61.2 | 89.2 | 83.8 | 83.4 | 67.5 | 66.2 | 63.0 | 63.0 | 62.6 | 64.1 | 62.6 | 53.1 | 60.5 | 59.7 | 59.9 | 14 | AF173033_lkeda | |
| 15 | 31.9 | 34.7 | 34.6 | 34.6 | 0.2 | 0.2 | 0.2 | 0.5 | 34.2 | 33.7 | 32.5 | 31.9 | 10.6 | 9.3 | – | 76.5 | 65.9 | 85.9 | 65.7 | 62.5 | 71.3 | 71.1 | 66.1 | 92.4 | 95.5 | 95.1 | 70.2 | 70.2 | 63.5 | 63.9 | 63.5 | 64.6 | 63.4 | 53.2 | 58.5 | 58.3 | 57.8 | 15 | M63383_Human_Kawasaki | |
| 16 | 30.2 | 32.6 | 32.5 | 33.5 | 25.9 | 26.5 | 26.6 | 27.0 | 34.1 | 33.6 | 31.6 | 29.4 | 23.5 | 23.9 | 23.7 | – | 63.0 | 76.2 | 66.6 | 59.4 | 71.3 | 70.8 | 62.5 | 74.5 | 72.4 | 72.0 | 68.2 | 67.3 | 61.6 | 61.7 | 61.4 | 62.5 | 61.0 | 52.7 | 59.4 | 58.1 | 58.3 | 16 | AY836148_Kato | |
| 17 | 8.9 | 9.5 | 9.5 | 5.5 | 36.0 | 35.5 | 35.4 | 35.1 | 1.3 | 1.1 | 5.5 | 9.1 | 27.4 | 32.2 | 32.5 | 32.4 | – | 61.7 | 62.3 | 77.1 | 83.8 | 86.1 | 70.8 | 58.5 | 70.4 | 70.8 | 90.8 | 86.8 | 71.7 | 74.9 | 74.9 | 74.5 | 74.4 | 61.2 | 69.0 | 68.1 | 67.5 | 17 | AY956315_Human_Karp | |
| 18 | 31.2 | 33.9 | 33.8 | 33.2 | 10.0 | 10.1 | 9.8 | 9.8 | 33.1 | 32.3 | 30.6 | 29.8 | 7.5 | 0.0 | 9.4 | 23.3 | 31.1 | – | 60.3 | 56.0 | 69.9 | 69.1 | 58.1 | 83.9 | 82.3 | 82.7 | 68.1 | 66.8 | 57.8 | 57.8 | 57.4 | 58.8 | 57.4 | 47.8 | 57.9 | 57.4 | 57.4 | 18 | U19903_Human_Yonchon | |
| 19 | 31.2 | 31.5 | 31.4 | 33.8 | 35.6 | 35.1 | 34.9 | 34.9 | 35.8 | 35.2 | 32.0 | 30.8 | 32.0 | 32.1 | 34.8 | 28.2 | 35.6 | 32.1 | – | 71.8 | 68.4 | 67.1 | 59.6 | 67.9 | 63.2 | 62.8 | 65.5 | 66.4 | 71.7 | 73.3 | 72.9 | 73.3 | 72.9 | 62.5 | 67.3 | 66.6 | 66.1 | 19 | U19904_Rattus._rattus_TA6.7.8 | |
| 20 | 14.7 | 12.2 | 12.1 | 2.8 | 37.4 | 37.0 | 37.5 | 37.1 | 9.1 | 9.2 | 5.1 | 15.2 | 35.6 | 40.0 | 40.5 | 39.8 | 8.6 | 40.0 | 37.0 | – | 77.3 | 83.2 | 56.9 | 64.6 | 60.5 | 60.1 | 80.0 | 84.3 | 84.7 | 87.9 | 86.8 | 86.8 | 87.2 | 74.0 | 81.4 | 81.0 | 80.7 | 20 | KX363954_Apod.e.mus_agrarius | |
| 21 | 1.0 | 0.0 | 0.0 | 8.7 | 34.7 | 34.2 | 33.9 | 33.9 | 10.1 | 9.7 | 9.0 | 0.2 | 24.6 | 30.6 | 31.9 | 29.4 | 9.1 | 29.3 | 30.8 | 15.2 | – | 90.6 | 63.7 | 66.1 | 68.4 | 68.8 | 90.3 | 88.8 | 85.0 | 86.5 | 86.3 | 87.5 | 86.1 | 69.9 | 81.8 | 80.5 | 80.3 | 21 | JQ898368_Human_Boryong | |
| 22 | 10.2 | 10.5 | 10.5 | 3.4 | 34.8 | 34.3 | 34.1 | 34.1 | 8.6 | 8.4 | 3.4 | 10.1 | 25.5 | 29.7 | 31.0 | 29.2 | 7.3 | 28.8 | 32.0 | 8.0 | 9.8 | – | 62.8 | 65.5 | 68.1 | 68.4 | 92.8 | 94.6 | 76.9 | 78.7 | 78.3 | 79.6 | 78.0 | 64.8 | 73.5 | 72.4 | 72.6 | 22 | AF430141_Human_Young-worl | |
| 23 | 27.0 | 28.3 | 28.3 | 29.7 | 30.3 | 29.8 | 29.7 | 29.3 | 28.2 | 28.2 | 28.6 | 27.0 | 28.6 | 31.5 | 28.2 | 25.1 | 27.8 | 30.9 | 31.4 | 33.3 | 27.0 | 27.4 | – | 59.9 | 69.9 | 69.5 | 62.1 | 62.1 | 57.8 | 59.4 | 59.0 | 59.0 | 58.8 | 51.3 | 53.1 | 52.3 | 52.2 | 23 | M63381_Shimokoshi | |
| 24 | 33.3 | 34.7 | 34.6 | 36.2 | 0.2 | 0.2 | 0.2 | 0.5 | 35.5 | 34.9 | 32.6 | 32.6 | 10.7 | 9.4 | 0.0 | 24.1 | 34.9 | 9.4 | 34.8 | 40.5 | 32.9 | 32.6 | 29.7 | – | 87.9 | 87.5 | 63.5 | 62.8 | 67.0 | 66.2 | 66.1 | 67.7 | 65.9 | 56.5 | 64.1 | 63.5 | 63.4 | 24 | AF173039_Kanda | |
| 25 | 32.5 | 35.0 | 35.0 | 34.1 | 0.5 | 0.0 | 0.0 | 0.2 | 34.0 | 33.9 | 33.5 | 32.3 | 10.4 | 9.8 | 0.2 | 25.3 | 31.9 | 9.8 | 35.9 | 37.8 | 32.3 | 31.7 | 27.8 | 0.2 | – | 99.6 | 67.1 | 67.5 | 60.8 | 61.9 | 61.9 | 62.1 | 61.7 | 51.4 | 56.3 | 56.1 | 55.6 | 25 | AF173038_Taguchi | |
| 26 | 32.5 | 35.0 | 35.0 | 34.1 | 0.5 | 0.0 | 0.0 | 0.2 | 34.2 | 33.9 | 33.5 | 32.2 | 10.4 | 9.8 | 0.2 | 25.3 | 31.7 | 9.8 | 35.9 | 37.8 | 32.1 | 31.5 | 27.8 | 0.2 | 0.0 | – | 67.5 | 67.9 | 60.5 | 61.6 | 61.6 | 61.7 | 61.4 | 51.1 | 56.0 | 55.8 | 55.2 | 26 | AF.173037_Oishi | |
| 27 | 9.3 | 9.5 | 9.4 | 5.8 | 35.9 | 35.5 | 35.2 | 35.2 | 1.9 | 1.7 | 5.9 | 9.2 | 24.2 | 30.2 | 32.0 | 31.4 | 0.9 | 29.5 | 34.5 | 12.1 | 8.9 | 6.7 | 28.2 | 34.0 | 32.7 | 32.6 | – | 93.5 | 76.4 | 78.5 | 78.3 | 79.2 | 77.8 | 64.4 | 73.1 | 72.0 | 71.7 | 27 | AF430143_Human_Jecheon | |
| 28 | 9.5 | 10.0 | 10.0 | 0.8 | 35.2 | 34.8 | 34.5 | 34.5 | 7.6 | 7.5 | 0.6 | 9.5 | 26.3 | 30.8 | 31.5 | 31.2 | 5.9 | 30.1 | 32.7 | 5.6 | 9.4 | 3.3 | 28.2 | 33.6 | 32.1 | 31.9 | 6.2 | – | 75.3 | 77.8 | 77.4 | 78.2 | 76.9 | 63.4 | 72.2 | 71.5 | 70.9 | 28 | GQ332749_CH0711a_TAIWAN | |
| 29 | 5.6 | 4.1 | 4.1 | 14.5 | 35.9 | 35.5 | 35.2 | 35.6 | 14.0 | 13.7 | 15.9 | 6.0 | 31.9 | 38.0 | 36.6 | 37.0 | 14.5 | 38.0 | 36.2 | 17.1 | 6.0 | 16.8 | 32.7 | 37.2 | 36.3 | 36.3 | 15.5 | 15.6 | – | 92.4 | 93.0 | 93.9 | 92.6 | 79.8 | 90.4 | 88.8 | 89.0 | 29 | KY266825_L.scutellare | |
| 30 | 4.1 | 2.1 | 2.1 | 12.0 | 27.2 | 36.8 | 36.9 | 36.9 | 11.8 | 11.5 | 14.4 | 4.8 | 32.8 | 38.9 | 39.5 | 37.5 | 12.2 | 38.9 | 36.0 | 13.3 | 4.8 | 14.7 | 30.6 | 39.7 | 37.6 | 37.6 | 14.4 | 14.5 | 7.3 | – | 96.0 | 95.8 | 96.4 | 79.2 | 90.1 | 88.6 | 88.6 | 30 | KY266826_L.scutellare | |
| 31 | 39 | 1.9 | 1.9 | 11.6 | 36.9 | 36.5 | 36.7 | 36.6 | 11.5 | 11.3 | 14.5 | 4.6 | 33.2 | 39.4 | 39.3 | 38.0 | 11.9 | 39.4 | 35.9 | 14.7 | 4.8 | 15.0 | 31.1 | 39.8 | 37.3 | 37.3 | 14.5 | 14.3 | 6.8 | 4.4 | – | 96.2 | 97.7 | 79.4 | 91.0 | 90.3 | 89.4 | 31 | KY266827_L.scutellare | |
| 32 | 3.0 | 1.6 | 1.6 | 11.4 | 35.5 | 35.1 | 34.9 | 35.2 | 11.3 | 11.1 | 12.6 | 3.2 | 30.1 | 36.0 | 36.0 | 35.5 | 11.7 | 36.0 | 34.3 | 14.0 | 3.0 | 12.9 | 30.0 | 36.1 | 35.9 | 35.9 | 12.4 | 12.5 | 5.4 | 3.9 | 3.7 | – | 96.0 | 79.1 | 91.5 | 91.0 | 90.8 | 32 | KY266828_L.scutellare | |
| 33 | 4.8 | 1.9 | 1.9 | 11.7 | 36.3 | 35.9 | 36.4 | 36.0 | 11.6 | 11.3 | 15.0 | 4.8 | 32.9 | 39.1 | 40.0 | 38.4 | 12.0 | 39.1 | 36.1 | 14.9 | 4.8 | 15.3 | 30.5 | 39.9 | 36.7 | 36.7 | 15.0 | 15.4 | 7.1 | 3.7 | 1.8 | 3.2 | – | 79.4 | 90.1 | 89.4 | 89.4 | 33 | KY266829_L.scutellare | |
| 34 | 25.9 | 22.8 | 23.1 | 33.8 | 57.0 | 56.4 | 56.6 | 56.1 | 30.4 | 29.7 | 35.4 | 26.9 | 53.4 | 60.0 | 59.6 | 55.1 | 31.6 | 60.0 | 54.7 | 32.3 | 26.9 | 35.8 | 49.6 | 60.6 | 56.8 | 56.8 | 33.9 | 34.2 | 24.6 | 24.7 | 24.6 | 25.0 | 24.4 | – | 75.5 | 73.6 | 74.0 | 34 | KY266830_L.scutellare | |
| 35 | 3.9 | 2.1 | 2.1 | 11.8 | 36.8 | 36.4 | 36.6 | 36.5 | 11.9 | 11.7 | 14.2 | 6.5 | 34.5 | 41.9 | 38.1 | 40.8 | 12.3 | 43.9 | 35.9 | 14.3 | 4.6 | 15.3 | 31.9 | 40.6 | 37.2 | 37.2 | 13.8 | 13.6 | 3.9 | 4.6 | 3.7 | 3.2 | 4.2 | 24.0 | – | 92.1 | 92.4 | 35 | KY266824_L.scutellare | |
| 36 | 6.0 | 4.3 | 4.2 | 14.6 | 38.4 | 37.9 | 38.5 | 38.1 | 14.3 | 14.0 | 16.6 | 8.9 | 36.0 | 44.1 | 39.6 | 44.0 | 14.8 | 45.7 | 38.4 | 15.3 | 6.7 | 17.4 | 35.0 | 42.3 | 38.8 | 38.8 | 16.4 | 15.7 | 6.1 | 7.5 | 5.1 | 3.9 | 5.6 | 26.4 | 8.5 | – | 96.0 | 36 | KX363952_L.scutellare | |
| 37 | 6.3 | 4.5 | 4.5 | 15.0 | 38.9 | 38.4 | 39.0 | 38.6 | 14.7 | 14.3 | 16.9 | 9.1 | 35.3 | 43.6 | 40.0 | 43.5 | 15.2 | 45.7 | 38.9 | 15.9 | 6.9 | 17.1 | 34.7 | 42.7 | 39.3 | 39.3 | 16.4 | 16.0 | 6.3 | 7.0 | 6.3 | 4.6 | 5.6 | 26.4 | 8.4 | 4.3 | – | 37 | KX363953_L.scutellare | |
| – | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | – | – | |
Figure 3.
Phylogenetic tree of Orientia tsutsugamushi based on the 56-kDa type specific gene sequence detected in chiggers; closed rectangle ■ (KX363952, KX363953, KY266824-KY266830), rodent (KX363954), human; open circle ○ (KY946003, KY946031, KY946061, KY946107, KY946108, KY946128, KY946124-KY946127). Phylogenetic tree was constructed by neighbor-joining method with the Kimura’s two-parameter model (bootstrap 1,000) using MEGA 6.0. Genbank accession numbers of O. tsutsugamushi are indicated for each sequence.
Table 6.
Strain of Orientia tsutsugamushi obtained in rodent, chigger, and humans from September 2014 to December 2016 in the southwest area of Korea
| Source | Genbank accession number | Collection date | Cluster | Reference |
|---|---|---|---|---|
| Chigger (Leptotrombidium scutellare) | KX363949 | October 2014 | Boryong | 11 |
| Chigger (L. scutellare) | KX363950 | October 2014 | Boryong | 11 |
| Chigger (Leptotrombidium orientale) | KX363951 | May 2015 | Boryong | 11 |
| Chigger (L. scutellare) | KX363952 | October 2015 | Boryong | – |
| Chigger (L. scutellare) | KX363953 | October 2015 | Boryong | – |
| Rodent (Apodemus agrarius) | KX363954 | December 2015 | Saitama | – |
| Chigger (L. scutellare) | KY266824 | October 2016 | Boryong | – |
| Chigger (L. scutellare) | KY266825 | October 2016 | Boryong | – |
| Chigger (L. scutellare) | KY266826 | October 2016 | Boryong | – |
| Chigger (L. scutellare) | KY266827 | October 2016 | Boryong | – |
| Chigger (L. scutellare) | KY266828 | October 2016 | Boryong | – |
| Chigger (L. scutellare) | KY266829 | October 2016 | Boryong | – |
| Chigger (L. scutellare)* | KY266830 | November 2016 | – | – |
| Human | KY946003 | 2014 | Boryong | – |
| Human | KY946031 | 2015 | Boryong | – |
| Human | KY946061 | 2015 | Boryong | – |
| Human | KY946107 | 2014 | Karp | – |
| Human | KY946108 | 2014 | Karp | – |
| Human | KY946128 | 2014 | Saitama | – |
| Human | KY946124 | 2015 | Kawasaki | – |
| Human | KY946125 | 2015 | Kawasaki | – |
| Human | KY946126 | 2015 | Kawasaki | – |
| Human | KY946127 | 2015 | Kawasaki | – |
Different strain from other chiggers.
DISCUSSION
Scrub typhus is a common endemic in the southwest region of Korea. The purpose of this study was to confirm the strains of O. tsutsugamushi for the 56-kDa TSA gene in chiggers, small mammals, and humans with chigger surveillance.
Seasonal chigger infestation rate for small mammals can be a criterion for the prevalence of scrub typhus. Furthermore, the abundance of chiggers influences the number of cases of scrub typhus. The degree of chigger infestation in small mammals varies from species to species. We found a high infestation rate in A. agrarius, which is the main host for scrub typhus in the southwest region of Korea,9 and which is the most abundant rodent species captured in this study.
Leptotrombidium scutellare, the predominant chigger species in the southern area of Korea, begins to appear in September and reaches a peak in autumn from October to November.31 In this study, L. scutellare was also the predominant chigger species identified during this period and was the primary contributor to the increased chigger infestation rate. However, during spring (March to May), L. orientale was the predominant species. Furthermore, the W. fragilis strain was identified from M. regulus. However, W. fragilis has never been reported in chigger species in the southwest region of Korea, but has been previously reported in M. regulus in the central region of Korea.32
Here, we analyzed for the 56-kDa TSA gene sequence of O. tsutsugamushi, and identified the Boryong strain to predominate in chiggers and patients. The Boryong strain is primarily distributed in the southern region of Korea where L. scutellare is found.15 In the present study, the results suggest that the Boryong strain can be transmitted through L. scutellare in the southwest region, and that L. orientale can act as a vector for scrub typhus in the spring. However, strains of O. tsutsugamushi carried by L. scutellare can differ between East Asian nations. For instance, in China, L. scutellare is considered a vector of the Karp strain,24 whereas in Japan, L. scutellare is considered the main vector of the Kawasaki strain.14
Unlike the Boryong strain, the Karp- and Kawasaki-related strains had different genotypes in patients than what was predicted. In the central and southwest regions of Korea, the Karp and Kawasaki strain had already been identified in patients.33,34 The Karp strain was classified into a higher virulence group, and the Boryong and Kawasaki strains were classified into a lower virulence group.16
Furthermore, a Saitama-related strain was found in a single A. agrarius, but was not found in chiggers. The Saitama strain is closely related to the Karp strain based on the 56-kDa TSA gene sequence. This strain was first reported in Japan in 1993 by Tamura et al.35 by identifying it in wild rodents, which tend to carry various strains in East Asia. In addition, Wu et al. (2015) confirmed the Kato, Gilliam, Karp, and TA763 strain in the Guangdong region of China.36,37 These strains were also reported in Taiwan.38 Finally, the CH0711a strain was reported in a patient in 2010 in Taiwan.39
Our study had several strengths. First, this study was conducted on a monthly basis. Secondly, this is the first study to compare the O. tsutsugamushi strains between chiggers, rodents, and patients in the southwest region. Despite a low infection rate, the data presented here provide a foundation for evidence of other strains in chiggers and small mammals. Unlike the strains identified in chiggers and patients, a Saitama-related strain was demonstrated in rodents.
In conclusion, the 56-kDa TSA gene showed variations in O. tsutsugamushi strains from different hosts. We confirmed that the Boryong strain is the most common strain in chiggers and patients, whereas wild rodents harbored a Saitama-related strain. In patients, Kawasaki-related strains prevalent in Japan were observed. To identify strains that are circulating in a host at any given point in time, a survey should be conducted continuously. The results provided here will be helpful for minimizing future incidences of scrub typhus by understanding disease transmission vectors.
Acknowledgments:
This study was financially supported by the Health and Environment Research Institute (HERI) of Gwang-ju, Korea. This was a joint research project between HERI of Gwang-Ju and Chosun University.
REFERENCES
- 1.Kelly DJ, Fuerst PA, Ching WM, Richards AL, 2009. Scrub typhus: the geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin Infect Dis 48 (Suppl 3): S203–S230. [DOI] [PubMed] [Google Scholar]
- 2.Ge Y, Rikihisa Y, 2011. Subversion of host cell signaling by Orientia tsutsugamushi. Microbes Infect 13: 638–648. [DOI] [PubMed] [Google Scholar]
- 3.Coleman RE, et al. 2003. Occurrence of Orientia tsutsugamushi in small mammals from Thailand. Am J Trop Med Hyg 69: 519–524. [PubMed] [Google Scholar]
- 4.Kim DM, et al. 2011. Differences in clinical features according to Boryoung and Karp genotypes of Orientia tsutsugamushi. PLoS One 6: e22731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee HI, Shim SK, Song BG, Choi EN, Hwang KJ, Park MY, Park C, Shin EH, 2011. Detection of Orientia tsutsugamushi, the causative agent of scrub typhus, in a novel mite species, Eushoengastia koreaensis, in Korea. Vector Borne Zoonotic Dis. 11: 209–214. [DOI] [PubMed] [Google Scholar]
- 6.Takhampunya R, Tippayachai B, Promsathaporn S, Leepitakrat S, Monkanna T, Schuster AL, Melendrez MC, Paris DH, Richards AL, Richardson JH, 2014. Characterization based on the 56-Kda type-specific antigen gene fo Orientia tsutsugamushi genotypes isolated from Leptotrombidium mites and th rodent host post infection. Am J Trop Med Hyg 90: 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ree HI, Chang WH, Kee SH, Lee IY, Jeon SH, 1997. Detection of Orientia tsutsugamushi DNA in individual trombiculids using polymerase chain reaction in Korea. Med Ent Zool. 48: 197–209. [Google Scholar]
- 8.Lee IY, Kim HC, Lee YS, Seo JH, Lim JW, Yong WS, Klein TA, Lee WJ, 2009. Geographical distribution and relative abundance of vectors of scrub typhus in the republic of korea. Korean J Parasitol 47: 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song HJ, et al. 1998. Molecular and serologic survey of Orientia tsutsugamushi infection among field rodents in southern Cholla province, Korea. Am J Trop Med Hyg 58: 513–518. [DOI] [PubMed] [Google Scholar]
- 10.Kim HC, Lee IY, Chong ST, Richards AL, Gu SH, Song JW, Lee JS, Klein TA, 2010. Serosurveillance of scrub typhus in small mammals collected from military training sites near the DMZ, northern Gyeonggi-do, Korea, and analysis of the relative abundance of chiggers from mammals examined. Korean J Parasitol 48: 237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JW, et al. 2016. Seroepidemiological survey of zoonotic diseases in small mammals with PCR detection of Orientia tsutsugamushi in chiggers, Gwangju, Korea. Korean J Parasitol 54: 307–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enatsu T, Urakami H, Tamura A, 1999. Phylogenetic analysis of Orientia tsutsugamushi strains based on the sequence homologues of 56-kDa type-specific antigen genes. Fems Micro Let 18: 163–169. [DOI] [PubMed] [Google Scholar]
- 13.Lu HY, Tsai KH, Yu SK, Cheng CH, Yang JS, Su CL, Hu HC, Wang HC, Huang JH, Shu PY, 2010. Phylogenetic analysis of 56-kDa type-specific antigen gene of Orientia tsutsugamushi isolates in Taiwan. Am J Trop Med Hyg 83: 658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohashi N, Nashimoto H, Ikeda H, Tamura A, 1992. Diversity of Immunodominant 56-kDa type-specific antigen(TSA) of Rickettsia tsutsugamushi. J Biochem 18: 12728–12735. [PubMed] [Google Scholar]
- 15.Stover Ck, Marana DP, Carter JM, Roe BA, Mardis E, Oaks EV, 1990. The 56-kilodalton major protein antigen of Rickettsia tsutsugamushi: molecular cloning and sequence analysis of the sta56 gene and precise identification of a strain-specific epitope. Infect Immun 58: 2076–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakayama K, Kurokawa K, Fukuhara M, Urakami H, Yamamoto S, Yamazaki K, Ogura Y, Ooka T, Hayashi T, 2010. Genome comparison and phylogenetic analysis of Orientia tsutsugamushi strains. DNA Res 17: 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuo CC, Lee PL, Chen CH, Wang HC, 2015. Surveillance of potential hosts and vectors of scrub typhus in Taiwan. Parasit Vectors 8: 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shim SK, et al. 2009. Characterization of Orientia tsutsugamushi genotypes from wild rodents and chigger mites in Korea. J Clin Microbiol 15: 311–312. [DOI] [PubMed] [Google Scholar]
- 19.Zhang M, Zhao ZT, Wang ZJ, Li Z, Ding L, Ding SJ, Yang HL, 2013. Genetic variants of Orientia tsutsugamushi in domestic rodents, northern China. Emerg Infect Dis 19: 1135–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roddkvamtook W, Ruangareerate T, Gaywee J, Richards AL, Jeamwattanalert P, Bodhidatta D, Sangjun N, Prasartvir A, Jatisatienr A, Jarisatienr C, 2011. Isolation and characterization of Orientia tsutsugamushi from rodents captured following a scrub typhus outbreak at a military training base, Bothong district, Chonburi province, central Thailand. Am J Trop Med Hyg 84: 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varghese GM, Janardhanan J, Mahajan SK, Tariang D, Trowbridge P, Prakash JAJ, David T, Sathendra S, Abraham OC, 2015. Molecular epidemiology and genetic diversity of Orientia tsutsugamushi from patients with scrub typhus in 3 regions of India. Emerg Infect Dis 21: 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cosson JF, et al. 2015. Detection of Orientia sp. DNA in rodents from Asia, west Africa and Europe. Parasit Vectors 8: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin PR, Tsai HP, Tsut PY, Weng MH, Kuo MD, Lin HC, Chen KC, Jo DD, Chu DM, Liu WT, 2011. Genetic typing, based on the 56-kilodlaton type-specific antigen gene, of Orientia tsutsugamushi strains isolated from chiggers collected from wild-caught rodents in Taiwan. Appl Environ Microbiol 77: 3398–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu YX, Jia N, Xing YB, Suo JJ, Du MM, Jia N, Gao Y, Xie LJ, Liu BW, Ren SW, 2013. Consistency of the key genotypes of Orientia tsutsugamushi in scrub typhus patients, rodents, and chiggers from a new endemic focus of northern China. Cell Biochem Biophys 67: 1461–1466. [DOI] [PubMed] [Google Scholar]
- 25.Korea Centers for Disease Control and Prevention (K-CDC) , 2017. Diseases Web Statistics System. Accessed at: http://is.cdc.go.kr/dstat/index.jsp. Accessed January 5, 2017.
- 26.Kweon SS, Choi JS, Lim HS, Kim JR, Kim KY, Ryu SY, Lee SD, Im HK, Kwon JW, 2009. A community-based case-control study of behavioral factors associated with scrub typhus during the autumn epidemic season in South Korea. Am J Trop Med Hyg 80: 442–446. [PubMed] [Google Scholar]
- 27.Lee HW, Cho PY, Moon SU, Na BK, Kang YJ, Sohn YG, Youn SK, Hong YS, Kim TS, 2015. Current situation of scrub typhus in South Korea from 2001–2013. Parasit Vectors 8: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeong MA, Youn SK, Kim YK, Lee HM, Kim SJ, Sohn A, 2013. Trends in the incidence of scrub typhus: the fastest growing vector-borne disease in Korea. Osong Public Health Res Perspect 4: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park SW, Ha NY, Ryu B, Bang JH, Song HY, Kim Y, Kim GH, Oh MD, Cho NH, Lee JK, 2015. Urbanization of scrub typuhs disease in South Korea. PLoS Negl Trop Dis 9: e0003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ree HI, Chang WH, Kee S, Lee IY, Jeon SH, 1997. Detection of Orientia tsutsugamushi DNA in individual trombiculids using polymerase chain reaction in Korea. Med Entomol Zool 48: 197–209. [Google Scholar]
- 31.Ree HI, 1990. Fauna and key to the chigger mites of Korea (Acarina: Trombiculidae and Leeuwenhoekiidae). Korean J Syst Zool 6: 57–70. [Google Scholar]
- 32.Lee IY, Kim HC, Lee YS, Seo JH, Lim JW, Yong WS, Klein TA, Lee WJ, 2009. Geographical distribution and relative abundance of vectors of scrub typhus in the Republic of Korea. Korean J Parasitol 47: 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee YM, Kim DQ, Lee SH, Jang MS, Neupane GP, 2011. Phylogenetic analysis of the 56kDa protein genes of Orientia tsutsugamushi in southwest area of Korea. Am J Trop Med Hyg 84: 250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeong HW, Choi YK, Baek YH, Seing MH, 2012. Phylognetic analysis of the 56-kDa type-specific protein genes of Orientia tsutsugamushi in central Korea. J Korean Med Sci 27: 1315–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura A, Yamamoto N, Koyama S, Makisaka Y, Takahashi M, Urabe KI, Takaoka M, Nakazawa K, Urakami H, Fukuhara M, 2001. Epidemiological survey of Orientia tsutsugamushi distribution in field rodents in Saitama prefecture, Japan, and discovery of a new type. Microbiol Immunol 45: 439–446. [DOI] [PubMed] [Google Scholar]
- 36.De W, Jing K, Huan Z, Qiong ZH, Monagin C, Min ZJ, Ping H, Wen KC, Yan LJ, 2015. Scrub typhus, a disease with increasing threat in Guangdong, China. PLoS One 10: e0113968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang M, et al. 2013. Molecular epidemiology of Orientia tsutsugamushi in chiggers and ticks from domestic rodents in Shangong, northern China. Parasit Vectors 6: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pey RL, Tsai HP, Weng MH, Lin HC, Chen KC, Kuo MD, Tsui PY, Hung YW, Hsu HL, Liu WT, 2014. Field assessment of Orientia tsutsugamushi infection in small mammals and its association with the occurence of human scrub typhus in Taiwan. Acta Trop 131: 117–123. [DOI] [PubMed] [Google Scholar]
- 39.Lu HY, Tsai KH, Yu SK, Cheng CH, Yang JS, Su CL, Hu HC, Wang HC, Huang JH, Shu PT, 2010. Phylogenetic analysis of 56-kDa type-specific antigen gene of Orientia tsutsugamushi isolates in Taiwan. Am J Trop Med Hyg 83: 658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]