Abstract
We recently showed that maize (Zea mays L.) leaves contain appreciable amounts of phosphoenolpyruvate carboxykinase (PEPCK; R.P. Walker, R.M. Acheson, L.I. Técsi, R.C. Leegood [1997] Aust J Plant Physiol 24: 459–468). In the present study, we investigated the role of PEPCK in C4 photosynthesis in maize. PEPCK activity and protein were enriched in extracts from bundle-sheath (BS) strands compared with whole-leaf extracts. Decarboxylation of [4-14C]aspartate (Asp) by BS strands was dependent on the presence of 2-oxoglutarate and Mn2+, was stimulated by ATP, was inhibited by the PEPCK-specific inhibitor 3-mercaptopicolinic acid, and was independent of illumination. The principal product of Asp metabolism was phosphoenolpyruvate, whereas pyruvate was a minor product. Decarboxylation of [4-14C]malate was stimulated severalfold by Asp and 3-phosphoglycerate, was only slightly reduced in the absence of Mn2+ or in the presence of 3-mercaptopicolinic acid, and was light dependent. Our data show that decarboxylation of Asp and malate in BS cells of maize occurs via two different pathways: Whereas malate is mainly decarboxylated by NADP-malic enzyme, decarboxylation of Asp is dependent on the activity of PEPCK.
C4 plants have been classified as NADP-ME, NAD-ME, and PEPCK types, according to the major decarboxylase involved in the decarboxylation of C4 acids in the BS cells (Gutierrez et al., 1974; Hatch et al., 1975). Maize (Zea mays) belongs to the NADP-ME subgroup of C4 plants and possesses negligible activity of PEPCK, although Gutierrez et al. (1974) suggested that some other NADP-ME species utilize PEPCK as an auxiliary decarboxylase. However, we showed previously that maize leaves contain large amounts of PEPCK (Walker et al., 1997), and Furumoto et al. (1999) identified a PEPCK gene from maize that is specifically expressed in BS cells. Other NADP-ME species such as Echinochloa colona, Echinochloa crus-galli, Digitaria sanguinalis, and Paspalum notatum also contain PEPCK, whereas PEPCK protein was not detectable in Sorghum bicolor, Saccharum officinarum, or Flaveria bidentis (Walker et al., 1997). The reason that the occurrence of PEPCK in these plants has been overlooked may be due to difficulties in measuring PEPCK activity (Walker et al., 1997) or to the lack of an antibody.
PEPCK-type C4 species mainly use Asp as the CO2 donor and decarboxylate the oxaloacetate formed via the following reaction:
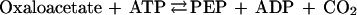 |
In NADP-ME species such as maize, 14CO2 is initially incorporated into the C-4 position of malate and Asp (about 75% and 25%, respectively), and the C-4 of both is subsequently incorporated into other metabolites (Hatch, 1971; Morot-Gaudry and Farineau, 1978). Using isolated BS strands, Chapman and Hatch (1981) showed that BS cells of maize have a significant capacity to decarboxylate Asp, and they assumed that NADP-ME was responsible. In view of the occurrence of PEPCK in maize, the involvement of PEPCK in the decarboxylation of Asp appears more likely, because PEPCK can directly catalyze the decarboxylation of oxaloacetate formed from Asp without previous reduction of oxaloacetate to malate.
To study the role of PEPCK in photosynthesis in maize, we measured the decarboxylation of Asp and malate by BS strands under conditions supporting the activities of either PEPCK or NADP-ME. In addition, we determined the protein contents or activities of other enzymes potentially involved in Asp metabolism.
MATERIALS AND METHODS
Plant Material
Maize (Zea mays L. cv H511) plants were grown in high-nutrient compost (M3, Fisons, Ipswich, UK) in a growth cabinet at a PPFD of 250 μmol m−2 s−1 and a photoperiod of 12 h d−1. The temperature was 28°C during the day and 24°C at night. For all experiments, the second and third leaves of 10- to 15-d-old plants were used.
Isolation of BS Strands
BS strands were isolated by slicing deribbed leaves into 1- to 2-mm sections and blending them five to six times for 10 s with 150 mL of blending buffer (100 mm Bicine-KOH, pH 8.0, 0.3 m sorbitol, 0.1% [w/v] PVP-40, 0.02% [w/v] BSA, 5 mm DTT, 2 mm KH2PO4, and 5 mm K2SO4) in a Waring blender. The strands were collected by filtration through Miracloth (Calbiochem) and washed with 350 mL of blending buffer. Where indicated, a digestion treatment (Hatch and Kagawa, 1974) was included in the procedure. After the sample was blended three times for 10 s each time, the BS strands were washed with blending medium, incubated for 25 min in 20 mm Mes-KOH (pH 5.5), 0.5 m sorbitol, 2 mm MgCl2, 0.2% (w/v) BSA, 1% (w/v) cellulase (Onozuka R-10, Yakult, Honsha, Japan), and 0.1% (w/v) pectinase (Macerozyme R-10, Yakult), washed with blending medium, and blended again (twice for 10 s each time).
Determination of Enzyme Activities
PEPCK was extracted in 180 mm Bicine and 20 mm 3-(cyclohexylamino)-1-propanesulfonic acid (pH 10.0) containing 50 mm DTT, and the carboxylation reaction was measured according to the method of Walker et al. (1995). Cytosolic pyruvate kinase was extracted and assayed as described by Zervoudakis et al. (1997); ADP was omitted for the assay of PEP phosphatase.
Western Analysis
Proteins were extracted in 200 mm Bicine-KOH (pH 9.5), 25 mm DTT, and 1% (w/v) SDS. Extracts were boiled for 90 s with equal volumes of solubilization buffer (62.5 mm Tris, 20% [v/v] glycerol, 2.5% [w/v] SDS, and 5% [v/v] 2-mercaptoethanol, pH 6.8). After SDS-PAGE, the proteins were transferred onto a PVDF membrane (Immobilon-P, Millipore) and probed with antisera raised against PEPCK from cucumber (Walker et al., 1995), PEPC from Amaranthus edulis, Rubisco from Brassica napus, Ala aminotransferase from barley (Muench and Good, 1994), Asp aminotransferase from Panicum maximum (Numuzawa et al., 1989), and NAD-ME (Murata et al., 1989) and NADP-ME from maize (Langdale et al., 1988). A peroxidase-conjugated secondary antibody was used, and immunoreactive bands were visualized with an enhanced chemiluminescence kit (Amersham).
Determination of Asp and Malate Decarboxylation
[4-14C]Aspartate and [4-14C]malate were synthesized enzymatically (Hatch, 1972). The reaction medium for the synthesis of [4-14C]Asp contained 50 mm Tris-HCl (pH 8.1), 1 mm PEP, 1 mm glutamate, 1 mm DTT, 10 mm MgCl2, 1 mm EDTA, 1 unit of PEPC (Boehringer Mannheim), 20 units of Glu:oxaloacetate aminotransferase (Boehringer Mannheim), and 10 MBq of NaH14CO3 (ICN) in 1 mL. The reaction was stopped by adding 10 μL of 5 m HCl. The [4-14C]Asp formed was purified by loading the reaction mixture onto Dowex-50 columns, washing with 5 mL of water, and eluting with 3 mL of 5 m NH4OH. The eluted fractions were evaporated to dryness in a freeze drier (Speed-Vac, Savant Instruments, Holbrook, NY), acidified with HCl, evaporated again, and redissolved in water. [4-14C]Malate was synthesized in a medium containing 50 mm Tris-HCl (pH 8.1), 3 mm PEP, 5 mm NADH, 1 mm DTT, 10 mm MgCl2, 1 mm EDTA, 1 unit of PEPC, 20 units of malate dehydrogenase (Boehringer Mannheim), and 37 MBq NaH14CO3 in a total volume of 1 mL. The [4-14C]malate formed was purified by loading the reaction mixture onto a Dowex-1 column, washing with 2 mL of 20 mm HCl, and eluting with 3 mL of 5 m HCl. The eluted fractions were evaporated, redissolved in water, and boiled for 20 min to destroy any formed [4-14C]oxaloacetate. The identity and purity of the labeled compounds was confirmed by TLC.
For the determination of decarboxylation rates, BS strands isolated without the digestion step were resuspended in reaction medium consisting of blending buffer containing 25 mm d,l-glyceraldehyde to inhibit the reassimilation of CO2 (Stokes and Walker, 1972) and other compounds as indicated in the figure legends. The suspension (0.8 mL) was then incubated with 0.2 mL of [4-14C]Asp or [4-14C]malate solution (reaction medium containing 62.5 mm Asp or malate with a specific radioactivity of 0.2 MBq mmol−1) in sealed vials. After 20 min at 25°C, the reaction was stopped by injecting 0.3 mL of 1 m HCl, and the released 14CO2 was trapped on filter paper moistened with 0.1 mL of 20% (w/v) KOH. Phaeophytin was extracted in 80% acetone and quantified according to the method of Vernon (1960).
Determination of PEP and Pyruvate Formation
For the determination of PEP and pyruvate formation, BS strands were resuspended in reaction medium (blending buffer containing 10 mm 2-oxoglutarate, 1 mm MnCl2, 1 mm MgCl2, and different concentrations of ATP and ADP), and 0.4 mL of the suspension was incubated with 0.1 mL of 62.5 mm Asp (dissolved in reaction medium) at 25°C. The reaction was stopped by adding 0.5 mL of 10% (v/v) HClO4. BS strands were extracted in a glass-in-glass homogenizer, and metabolites were measured according to the method of Lowry and Passonneau (1972).
RESULTS
PEPCK in BS Cells of Maize
Maize leaves contained substantial amounts (Fig. 1) and activities (Table I) of PEPCK. The activity of PEPCK recorded in maize was between one-fifth and one-half of that measured in PEPCK C4 plants (Chapman and Hatch, 1983; Smith and Woolhouse, 1983; Walker et al., 1997).
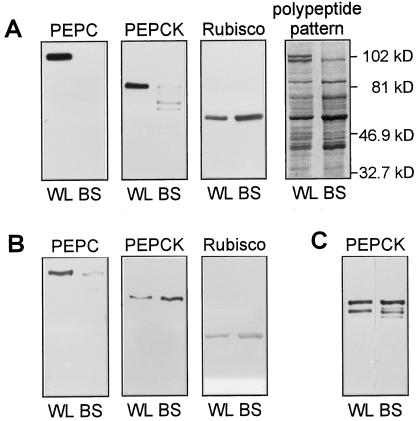
Figure 1.
A and B, Western blots for PEPC, PEPCK, and Rubisco, and the polypeptide pattern in extracts from whole leaves (WL) and BS strands from maize. The BS strands were isolated including (A) or omitting (B) treatment with cellulase and pectinase. C, Western blot for PEPCK in extracts from whole leaves (WL) and BS strands from U. panicoides isolated after treatment with cellulase and pectinase. Three micrograms of total protein was loaded per lane.
Table I.
In vitro activities of PEPCK, pyruvate kinase (PK), and PEP phosphatase (PP) in extracts of whole leaves and of BS strands, and in vivo formation of pyruvate (Pyr) from PEP by BS strands in the presence of 5 mm PEP, 10 mm ADP, 1 mm Mn2+, 1 mm Mg2+, and 40 mm KCl
| Extract | PEPCK | PK | PP | Pyr | |
|---|---|---|---|---|---|
| μmol min−1 g−1 fresh wt | μmol min−1 mg−1 phaeophytin | μmol min−1 mg−1 phaeophytin | |||
| Whole leaves | 4.34 ± 0.22 | 2.26 ± 0.18 | 0.41 ± 0.04 | 0.74 ± 0.12 | n.d. |
| BS strands | n.d. | 3.58 ± 0.39 | 0.51 ± 0.08 | 0.61 ± 0.09 | 0.39 ± 0.02 |
Data are means ± se of three plants. n.d., Not determined.
Because PEPCK from plants is subject to rapid proteolytic cleavage, we first optimized the conditions for the isolation of BS strands, checking the intactness of PEPCK protein. When the isolation procedure included treatment with cellulase and pectinase, strands free of mesophyll contamination, as judged by the absence of PEPC, were obtained (Fig. 1A). Even though this treatment did not lead to general proteolysis, it resulted in the cleavage of PEPCK. PEPCK in plant extracts is rapidly cleaved at low pH or, in the absence of SDS, to a 62-kD form with no loss of activity (Walker et al., 1995, 1997). However, during the isolation of BS strands from maize, PEPCK was degraded further and sometimes no PEPCK protein was detectable on western blots. This indicates that protease activity within intact BS cells was stimulated by low pH. On the other hand, PEPCK in BS strands isolated from the PEPCK-type species Urochloa panicoides was largely intact after treatment of the BS strands with cellulase and pectinase (Fig. 1C).
Maize BS strands isolated without the cell wall-digestion step were contaminated with approximately 6% of the mesophyll protein, as estimated from the distribution of PEPC and Rubisco on western blots (Fig. 1B). PEPCK extracted from these strands was intact and enriched compared with whole-leaf extracts, confirming the location of PEPCK in the BS cells in maize (Walker et al., 1997). The activity of PEPCK was also enriched in BS strands (Table I).
Decarboxylation of [4-14C]Asp
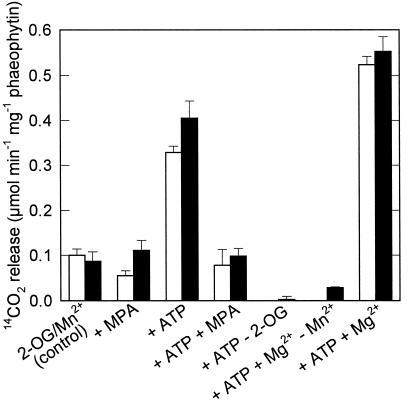
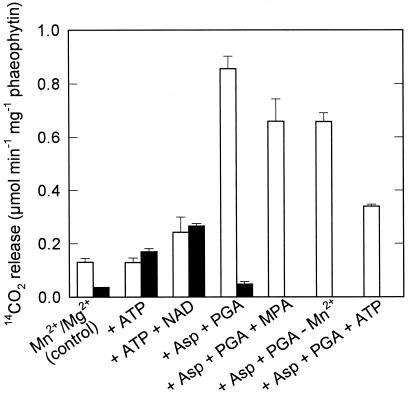
Because of the persistence of open plasmodesmata, BS cells are permeable to metabolites and can be used to study the metabolism of externally added substrates such as Asp. Rates of [4-14C]Asp decarboxylation by BS strands were independent of illumination (Fig. 2); the reaction was completely dependent on the presence of 2-oxoglutarate. ATP, which is a substrate of the PEPCK reaction, stimulated Asp decarboxylation. An inhibitor of PEPCK activity, MPA (Jomain-Baum et al., 1976), inhibited the ATP-stimulated activity. PEPCK has an absolute requirement for Mn2+ and, accordingly, Mg2+ did not substitute for Mn2+ in the decarboxylation of Asp. Mg2+ did, however, stimulate Asp decarboxylation in the presence of Mn2+. The requirements for Mn2+ and ATP and the inhibition by MPA are evidence for an involvement of PEPCK. In contrast, Asp decarboxylation by BS strands from S. bicolor, an NADP-ME species without detectable amounts of PEPCK protein, was low (0.06 μmol min−1 mg−1 phaeophytin), was not stimulated by the addition of ATP, and was not inhibited by MPA (data not shown).
Figure 2.
Decarboxylation of [4-14C]Asp (12.5 mm) by BS strands at a PPFD of 200 μmol m−2 s−1 (white bars) or in the dark (black bars). The incubation medium for the control treatment contained 10 mm 2-oxoglutarate (2-OG) and 1 mm Mn2+. For the other treatments, 400 μm MPA, 10 mm ATP, or 1 mm Mg2+ was added and 2-oxoglutarate or Mn2+ was omitted. Data are means ± se of three assays.
Metabolism of Asp
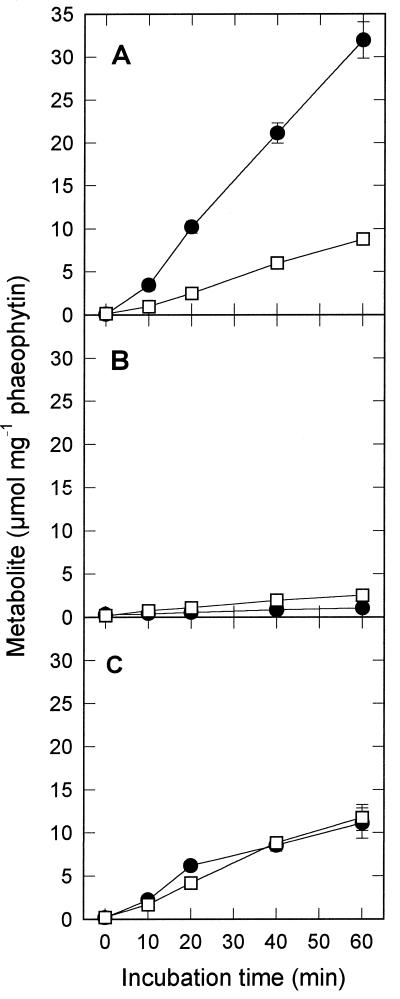
Under the conditions supporting the highest rates of Asp decarboxylation, PEP was the main product of Asp metabolism (Fig. 3A). The rates of PEP formation (0.55 μmol min−1 mg−1 phaeophytin) were comparable to the rates of Asp decarboxylation (Fig. 2). Pyruvate was also formed but at a much lower rate. In the absence of ATP, the rates of both PEP and pyruvate formation were negligible (Fig. 3B). The predominance of PEP formation in the presence of ATP supports the view that PEPCK, and not NADP-ME, was mainly responsible for the decarboxylation of Asp.
Figure 3.
Formation of PEP (•) and pyruvate (□) from Asp in a medium containing 12.5 mm Asp, 10 mm 2-oxoglutarate, 1 mm Mn2+, and 1 mm Mg2+, with (A) or without (B) the addition of 10 mm ATP or with the addition of 1 mm ATP, 1 mm ADP, and 40 mm KCl (C). Data are means ± se of three assays.
To determine whether PEP could be converted into pyruvate by pyruvate kinase, ADP (1 mm) was supplied in addition to ATP. Because high concentrations of ATP inhibit pyruvate kinase (Baysdorfer and Bassham, 1984), the ATP concentration was decreased to 1 mm. Under these conditions, equal amounts of PEP and pyruvate were formed and the sum of PEP and pyruvate formation was reduced to 0.38 μmol min−1 mg−1 phaeophytin (Fig. 3C). This lower rate was probably due to the effect of a changed ATP:ADP ratio on PEPCK (Walker et al., 1997). The accumulation of PEP, even in the presence of ADP, shows that the activity of pyruvate kinase (plus PEP phosphatase) in BS cells is not sufficient to convert all PEP formed by PEPCK into pyruvate. The rate of conversion of PEP into pyruvate by BS strands (Table I) was also lower than the rate of Asp decarboxylation. In addition, the in vitro activities of pyruvate kinase and PEP phosphatase in BS cells were much lower than the activity of PEPCK (Table I).
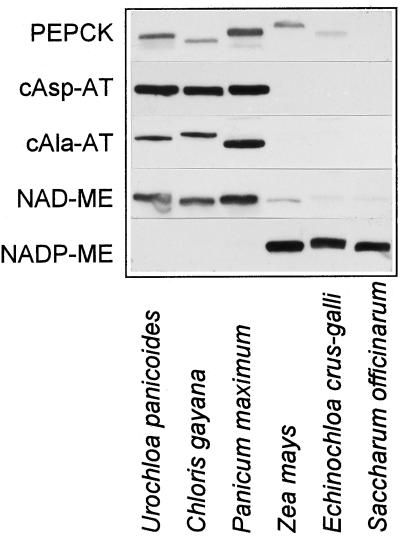
Since PEPCK is a cytosolic enzyme, we determined whether cytosolic aminotransferases are present in maize. Although all of the PEPCK species we tested (U. panicoides, C. gayana, and P. maximum) contained large amounts of cytosolic Asp and Ala aminotransferases, these aminotransferases were not detectable in maize or other NADP-ME species (E. crus-galli and S. officinarum; Fig. 4). In PEPCK species, NAD-ME in combination with the respiratory chain provides ATP for the PEPCK reaction. Although maize contained some NAD-ME protein, the amount was only comparable to that found in C3 species and was lower than in PEPCK species.
Figure 4.
Western blots for PEPCK, cytosolic Asp aminotransferase (cAsp-AT), cytosolic Ala aminotransferase (cAla-AT), NAD-ME, and NADP-ME in leaves of U. panicoides, C. gayana, P. maximum, Z. mays, E. crus-galli, and S. officinarum. Equal amounts of fresh weight were loaded per lane.
Decarboxylation of [4-14C]Malate
In contrast to rates of Asp decarboxylation, those of [4-14C]malate were generally low in the dark (Fig. 5). The stimulation of malate decarboxylation by ATP and NAD, which is needed for the oxidation of malate to oxaloacetate, indicates some contribution of PEPCK to malate decarboxylation in the dark. Under illumination, however, the effect of ATP and NAD was smaller and malate decarboxylation was strongly stimulated by Asp and 3-phosphoglycerate. Stimulation by Asp and 3-phosphoglycerate is characteristic of malate decarboxylation catalyzed by NADP-ME (Hatch and Kagawa, 1976; Chapman and Hatch, 1979; Boag and Jenkins, 1986). The light dependence of malate decarboxylation also suggests the involvement of NADP-ME (Hatch and Kagawa, 1976). In the presence of MPA or in the absence of Mn2+, malate decarboxylation was slightly reduced, but the inhibition was minor compared with the effect on Asp decarboxylation. In the presence of Mn2+, Mg2+, Asp, and 3-phosphoglycerate, ATP inhibited the decarboxylation of malate, possibly by binding the Mg2+ required for NADP-ME.
Figure 5.
Decarboxylation of 12.5 mm [4-14C]malate by BS strands at a PPFD of 200 μmol m−2 s−1 (white bars) or in the dark (black bars). The incubation medium for the control treatment contained 1 mm Mn2+ and 1 mm Mg2+. For the other treatments, 10 mm ATP, 10 mm NAD, 10 mm Asp, 5 mm 3-phosphoglycerate (PGA), or 400 μm MPA was added or, in one treatment, Mn2+ was omitted. Data are means ± se of three assays.
DISCUSSION
The Function of PEPCK in Maize
One reason that the presence of PEPCK in maize has been overlooked may be the instability of this enzyme during the isolation of BS strands (Fig. 1). When the procedure for maize included digestion of the mesophyll cells with cellulase and pectinase, PEPCK protein was almost completely degraded. In contrast, PEPCK from plants that have been classified as PEPCK species appears to be more stable. Intactness of the PEPCK from U. panicoides was not affected by the digestion step, and PEPCK from P. maximum was still fully active in BS protoplasts isolated by treatment with cellulase and pectinase (Chapman and Hatch, 1983).
The formation of PEP from Asp (Fig. 3), the dependence of Asp decarboxylation on Mn2+ and ATP, and the inhibition by MPA (Fig. 2) strongly suggest that PEPCK is involved in the metabolism of Asp in BS cells of maize. The conditions supporting malate decarboxylation (Fig. 5) were different from those supporting Asp decarboxylation. ATP enhanced malate decarboxylation only in the dark, whereas Asp and 3-phosphoglycerate stimulated malate decarboxylation in the light. Since Asp facilitates the transport of malate into the chloroplasts (Chapman and Hatch, 1979; Boag and Jenkins, 1986) and 3-phosphoglycerate is important for the reoxidation of NADPH formed in the NADP-ME reaction (Hatch and Kagawa, 1976), the stimulation by Asp and 3-phosphoglycerate confirms that malate is mainly decarboxylated by NADP-ME and not by PEPCK.
The lack of an effect of illumination on Asp decarboxylation indicates that the activity of PEPCK is not subject to rapid modulation by light. In some C4 plants PEPCK is phosphorylated in the dark and dephosphorylated under illumination (Walker and Leegood, 1996). However, PEPCK from maize is not phosphorylated (Walker et al., 1997), and the putative phosphorylation motif (Walker and Leegood, 1995) is modified (Furumoto et al., 1999). Thus, light-dependent activation of PEPCK by dephosphorylation probably does not occur in maize. Expression of the PEPCK gene, on the other hand, is much higher in the daytime than at night (Furumoto et al., 1999).
Decarboxylation of Asp not only was dependent on Mn2+ but was stimulated when Mg2+ was present in addition to Mn2+. Although previous studies have shown that in vitro activities of PEPCK from P. maximum and U. panicoides are inhibited by Mg2+ at physiological concentrations (Burnell, 1986; Walker et al., 1997), recent work in this laboratory has shown that Mg2+ is an activator at physiological concentrations of Mn2+.
Photosynthetic Metabolism in Maize
The importance of PEPCK in photosynthetic metabolism will depend on the relative fluxes through Asp and malate during C4 photosynthesis. In maize, substantial amounts of Asp are labeled (Hatch, 1971; Créach et al., 1974; Morot-Gaudry and Farineau, 1978; Chapman and Hatch, 1981), more so in N-sufficient than in N-deficient maize (Khamis et al., 1992), but measurements of labeling and/or pools do not give any indication of relative fluxes. The relative capacities for Asp and malate decarboxylation would have major energetic consequences in the BS. Whereas malate decarboxylation by NADP-ME provides one-half of the NADPH required for the reduction of 3-phosphoglycerate, no NADPH is formed during Asp decarboxylation by PEPCK. Typically, BS cells of NADP-ME-type species show little or no capacity for PSII-dependent photoreduction of NADP. The occurrence of PEPCK in NADP-ME-type species may be correlated with PSII activity. Species that contain PEPCK (maize, D. sanguinalis, and Setaria lutescens; Gutierrez et al., 1974; Walker et al., 1997) show some PSII activity in the BS cells (Ku et al., 1974; Chapman et al., 1980), whereas BS cells of S. bicolor and S. officinarum lack PEPCK and have the lowest PSII activity of the C4 species tested (Ku et al., 1974).
Decarboxylation of Asp in the presence of 2-oxoglutarate shows that Asp aminotransferase activity is present in the BS strands. In contrast to PEPCK species, the transamination of Asp to form oxaloacetate as the substrate for the PEPCK reaction is probably not catalyzed by cytosolic Asp aminotransferase (Fig. 4). Chapman and Hatch (1981) showed that the activity of Asp aminotransferase in BS cells of maize is located mainly in the mitochondria. In addition to oxaloacetate, the PEPCK reaction requires ATP. In PEPCK-type species, this ATP is provided by the activity of mitochondrial NAD-ME in combination with the respiratory chain (Hatch, 1987). In maize, some NAD-ME protein was present, but the amount was much lower than in PEPCK-type species (Fig. 4), which is consistent with measurements of its activity (Hatch et al., 1975). An additional source of ATP could be the conversion of PEP to pyruvate by pyruvate kinase. The activity of pyruvate kinase was, however, lower than the activity of PEPCK (Table I), and in vivo rates of pyruvate formation from PEP were not sufficient for the complete conversion of PEP into pyruvate (Fig. 3). Valle and Heldt (1991) measured similar activities of pyruvate kinase in BS strands of maize and concluded that these activities were not high enough to fully account for the formation of Ala from PEP. In addition, activities of pyruvate kinase in PEPCK-type species either are lower than rates of photosynthesis (Smith and Woolhouse, 1983) or are not detectable (Rathnam and Edwards, 1977). Thus, PEP and not pyruvate or Ala is probably the C3 compound mainly transported from the BS to the mesophyll, where it can serve directly as a substrate for PEPC.
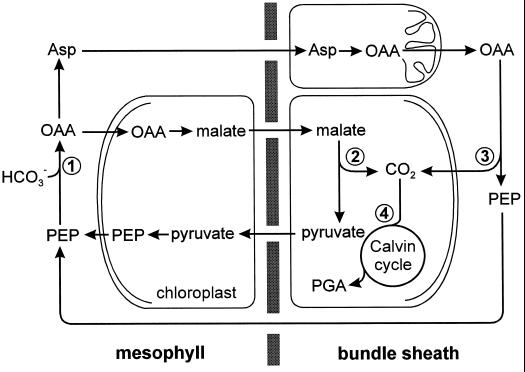
In conclusion, our results show that two distinct pathways for the decarboxylation of malate and Asp occur in maize and probably in other species previously classified as NADP-ME types (Fig. 6): Whereas malate is decarboxylated by NADP-ME in the chloroplasts, resulting in the formation of pyruvate, Asp is decarboxylated by PEPCK in the cytosol, resulting in the formation of PEP. It would appear that NADP-ME species can be subdivided into those with and without PEPCK, extending the biochemical classification of C4 plants. Further studies will show to what extent the PEPCK-dependent pathway contributes to C4 photosynthesis under different environmental conditions and how the activity of PEPCK is regulated.
Figure 6.
Photosynthetic metabolism in maize leaves. The oxaloacetate (OAA) formed by PEPC (1) in the mesophyll is either reduced to malate or transaminated to Asp. Whereas malate is decarboxylated to pyruvate by NADP-ME (2) in the chloroplasts of the BS cells, oxaloacetate formed from Asp in the mitochondria is decarboxylated to PEP by PEPCK (3) in the cytosol of the BS cells. The CO2 produced in the decarboxylation reactions is subsequently fixed by Rubisco (4). PGA, Phosphoglycerate.
ACKNOWLEDGMENTS
We thank A.G. Good (University of Alberta, Edmonton, Alberta, Canada), T. Nelson (Yale University, Hartford, CT), R. Ohsugi (National Institute of Agrobiological Resources, Ibaraki, Japan), and M. Taniguchi (Nagoya University, Japan) for providing antisera.
Abbreviations:
- Asp
aspartate
- BS
bundle sheath
- ME
malic enzyme
- MPA
3-mercaptopicolinic acid
- PEPC
PEP carboxylase
- PEPCK
PEP carboxykinase
Footnotes
This research was supported by the Biotechnology and Biological Sciences Research Council of the United Kingdom (grant no. CO5229).
LITERATURE CITED
- Baysdorfer C, Bassham JA. Spinach pyruvate kinase isoforms. Partial purification and regulatory properties. Plant Physiol. 1984;74:374–379. doi: 10.1104/pp.74.2.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boag S, Jenkins CLD. The involvement of Asp and glutamate in the decarboxylation of malate by isolated bundle sheath chloroplasts from Zea mays. Plant Physiol. 1986;80:115–119. doi: 10.1104/pp.81.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnell JN. Purification and properties of phosphoenolpyruvate carboxykinase from C4 plants. Aust J Plant Physiol. 1986;13:577–587. [Google Scholar]
- Chapman KSR, Berry JA, Hatch MD. Photosynthetic metabolism in bundle sheath cells of the C4 species Zea mays: sources of ATP and NADPH and the contribution of photosystem II. Arch Biochem Biophys. 1980;202:330–341. doi: 10.1016/0003-9861(80)90435-x. [DOI] [PubMed] [Google Scholar]
- Chapman KSR, Hatch MD. Aspartate stimulation of malate decarboxylation in Zea mays bundle sheath cells: possible role in regulation of C4 photosynthesis. Biochem Biophys Res Commun. 1979;86:1274–1280. doi: 10.1016/0006-291x(79)90254-7. [DOI] [PubMed] [Google Scholar]
- Chapman KSR, Hatch MD. Aspartate decarboxylation in bundle sheath cells of Zea mays and its possible contribution to C4 photosynthesis. Aust J Plant Physiol. 1981;8:237–248. [Google Scholar]
- Chapman KSR, Hatch MD. Intracellular location of phosphoenolpyruvate carboxykinase and other C4 photosynthetic enzymes in mesophyll and bundle sheath protoplasts of Panicum maximum. Plant Sci Lett. 1983;29:145–154. [Google Scholar]
- Créach E, Michel JP, Thibault P. Aspartic acid as an internal CO2 reservoir in Zea mays: effect of oxygen concentration and of far-red illumination. Planta. 1974;118:91–100. doi: 10.1007/BF00388386. [DOI] [PubMed] [Google Scholar]
- Furumoto T, Hata S, Izui K. Cloning of cell-type specific genes in maize leaves and characterization of PEP carboxykinase in bundle-sheath-strands. In: Garab G, editor. Photosynthesis: Mechanisms and Effects. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. (in press) [Google Scholar]
- Gutierrez M, Gracen VE, Edwards GE. Biochemical and cytological relationships in C4 plants. Planta. 1974;119:279–300. doi: 10.1007/BF00388331. [DOI] [PubMed] [Google Scholar]
- Hatch MD. The C4-pathway of photosynthesis: evidence for an intermediate pool of carbon dioxide of the donor C4-dicarboxylic acid. Biochem J. 1971;125:425–432. doi: 10.1042/bj1250425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch MD. Synthesis of l-malate-4-14C and determination of label in the C-4 carboxyl of l-malate. Anal Biochem. 1972;47:174–183. doi: 10.1016/0003-2697(72)90290-4. [DOI] [PubMed] [Google Scholar]
- Hatch MD. C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochim Biophys Acta. 1987;895:81–106. [Google Scholar]
- Hatch MD, Kagawa T. Activity, location and role of NAD malic enzyme in leaves with C4-pathway photosynthesis. Aust J Plant Physiol. 1974;1:357–369. [Google Scholar]
- Hatch MD, Kagawa T. Photosynthetic activities of isolated bundle sheath cells in relation to differing mechanisms of C4 pathway photosynthesis. Arch Biochem Biophys. 1976;175:39–53. doi: 10.1016/0003-9861(76)90483-5. [DOI] [PubMed] [Google Scholar]
- Hatch MD, Kagawa T, Craig S. Subdivision of C4-pathway species based on differing C4 acid decarboxylating systems and ultrastructural features. Aust J Plant Physiol. 1975;2:111–128. [Google Scholar]
- Jomain-Baum M, Schramm VL, Hanson RW. Mechanism of 3-mercaptopicolinic acid inhibition of hepatic phosphoenolpyruvate carboxykinase (GTP) J Biol Chem. 1976;251:37–44. [PubMed] [Google Scholar]
- Khamis S, Lamaze T, Farineau J. Effect of nitrate limitation on the photosynthetically active pools of Asp and malate in maize, a NADP malic enzyme C4 plant. Physiol Plant. 1992;85:223–229. [Google Scholar]
- Ku SB, Gutierrez M, Kanai R, Edwards GE. Photosynthesis in mesophyll protoplasts and bundle sheath cells of various types of C4 plants. II. Chlorophyll and Hill reaction studies. Z Pflanzenphysiol. 1974;72:320–337. [Google Scholar]
- Langdale JA, Rothermel BA, Nelson T. Cellular patterns of photosynthetic gene expression in developing maize leaves. Genes Dev. 1988;2:106–115. doi: 10.1101/gad.2.1.106. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Passonneau JV (1972) A Flexible System of Enzymatic Analysis. Academic Press, New York
- Morot-Gaudry J-F, Farineau J. Etude comparée des réactions de carboxylations photosynthétiques chez un Maïs normal (W 64 A) et chez un Maïs mutant opaque 2 (W 64 A o2): mise en évidence de déviations métaboliques chez le mutant. Physiol Veg. 1978;16:451–467. [Google Scholar]
- Muench DG, Good AG. Hypoxically inducible barley alanine aminotransferase: cDNA cloning and expression analysis. Plant Mol Biol. 1994;24:417–427. doi: 10.1007/BF00024110. [DOI] [PubMed] [Google Scholar]
- Murata T, Ikeda J, Takano M, Ohsugi R. Comparative studies of NAD-malic enzyme from leaves of various C4 plants. Plant Cell Physiol. 1989;30:429–437. [Google Scholar]
- Numuzawa T, Yamada S, Hase T, Sugiyama T. Aspartate aminotransferase from Panicum maximum Jacq. var. trichoglume Eyles, a C4 plant: purification, molecular properties, and preparation of antibody. Arch Biochem Biophys. 1989;270:313–319. doi: 10.1016/0003-9861(89)90033-7. [DOI] [PubMed] [Google Scholar]
- Rathnam CKM, Edwards GE. C4-dicarboxylic acid metabolism in bundle-sheath chloroplasts, mitochondria and strands of Eriochloa borumensis Hack., a phosphoenolpyruvate-carboxykinase type C4 species. Planta. 1977;133:135–144. doi: 10.1007/BF00391911. [DOI] [PubMed] [Google Scholar]
- Smith AM, Woolhouse HW. Metabolism of phosphoenolpyruvate in the C4 cycle during photosynthesis in the phosphoenolpyruvate-carboxykinase C4 grass Spartina anglica Hubb. Planta. 1983;159:570–578. doi: 10.1007/BF00409147. [DOI] [PubMed] [Google Scholar]
- Stokes DM, Walker DA. Photosynthesis by isolated chloroplasts: inhibition by dl-glyceraldehyde of carbon dioxide assimilation. Biochem J. 1972;128:1147–1157. doi: 10.1042/bj1281147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle EM, Heldt HW. Alanine synthesis by bundle sheath cells of maize. Plant Physiol. 1991;95:839–845. doi: 10.1104/pp.95.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon LP. Spectrophotometric determination of chlorophylls and phaeophytins in plant extracts. Anal Chem. 1960;32:1144–1150. [Google Scholar]
- Walker RP, Acheson RM, Técsi LI, Leegood RC. Phosphoenolpyruvate carboxykinase in C4 plants: its role and regulation. Aust J Plant Physiol. 1997;24:459–468. [Google Scholar]
- Walker RP, Leegood RC. Purification, and phosphorylation in vivo and in vitro, of phosphoenolpyruvate carboxykinase from cucumber cotyledons. FEBS Lett. 1995;362:70–74. doi: 10.1016/0014-5793(95)00212-r. [DOI] [PubMed] [Google Scholar]
- Walker RP, Leegood RC. Phosphorylation of phosphoenolpyruvate carboxykinase in plants: studies in plants with C4 photosynthesis and Crassulacean acid metabolism and in germinating seeds. Biochem J. 1996;317:653–658. doi: 10.1042/bj3170653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RP, Trevanion SJ, Leegood RC. Phosphoenolpyruvate carboxykinase from higher plants: purification from cucumber and evidence of rapid proteolytic cleavage in extracts from a range of plant tissues. Planta. 1995;196:58–63. [Google Scholar]
- Zervoudakis G, Georgiou CD, Mavroidis M, Kokolakis G, Angelopoulos K. Characterization of purified leaf cytosolic pyruvate kinase from the C4 plant Cynodon dactylon. Physiol Plant. 1997;101:563–569. [Google Scholar]