Abstract
Background and objective:
Millions of people in developing countries lose their lives due to acute respiratory infections, such as Influenza A & B and Adeno viruses. Given the importance of rapid identification of the virus, in this study the researchers attempted to design a method that enables detection of influenza A, B, and adenoviruses, quickly and simultaneously. The Multiplex RT PCR method was the preferred method for the detection of influenza A, B, and adenoviruses in clinical specimens because it is rapid, sensitive, specific, and more cost-effective than alternative methods
Methods:
After collecting samples from patients with respiratory disease, virus genome was extracted, then Monoplex PCR was used on positive samples and Multiplex RT-PCR on clinical specimens. Finally, by comparing the bands of these samples, the type of virus in the clinical samples was determined.
Results:
Performing Multiplex RT-PCR on 50 samples of respiratory tract led to following results; flu A: 12.5%, fluB: 50%, adeno: 27.5%, negative: 7.5%, and 2.5% contamination.
Conclusion:
Reverse transcription-multiplex Polymerase Chain Reaction (PCR) technique, a rapid diagnostic tool, has potential for high-throughput testing. This method has a significant advantage, which provides simultaneous amplification of numerous viruses in a single reaction. This study concentrates on multiplex molecular technologies and their clinical application for the detection and quantification of respiratory pathogens. The improvement in diagnostic testing for viral respiratory pathogens effects patient management, and leads to more cost-effective delivery of care. It limits unnecessary antibiotic use and improves clinical management by use of suitable treatment.
Key Words: Polymerase chain reaction, Influenza A, Influenza B, Adenovirus, Multiplex RT-PCR, Respiratory infection
Introduction
Influenza A and B and Adenoviruses cause most lower respiratory tract viral infections. The diseases reduce immune function and affect heart, lung, and kidney functions and may result in kidney disorders, diabetes and asthma. The probability of becoming infected with influenza A and B increases in pregnant females and elderly as well as immunodeficient individuals, which may lead to death (1).
The influenza virus belongs to the family of Orthomyxoviridae, which is based on antigenic differences in internal structural proteins, Matrix protein (M) and Nucleocapsid (NP), which are subdivided to three types, A, B, and C (2). Due to the mutability and frequency of genetic reassortment of the genome segmented viruses, particularly influenza A, they occasionally cause epidemics worldwide, though in most cases, contamination is sporadic (3).
Sixteen Hemagglutinin (HA) subunits and 9 Neuraminidase (NA) subunits are identified in natural reservoirs (4). Those that are circulating in humans following 3 types of HA (H1, H2, and H3) and two subunits of NA (N1 and N2) are scarce. Wild waterfowl is among the natural hosts of adenoviruses. According to a survey conducted in 2009, (from June 1 to November 11) 2662 swine flu cases were recorded in Iran, of which 58 cases (1.2%) died (5).
Adenoviruses are common causes of cold-like symptoms of upper respiratory tract illness and some others such as ophthalmic, gastrointestinal, and urinary tract diseases in humans (6). Influenza virus is transmitted through respiratory secretions or contact with contaminated hands or objects [7]. Adenoviruses transmissions are mainly fecal and oral, yet they sometimes occur through respiratory droplets or infected objects. This transfer is the cause of infected epithelial cells of the respiratory tract, eyes, digestive system, and urinary tract. The infection is multiplied and results in pneumonia, keratoconjunctivitis, and gastroenteritis (7). The most common cause of annual 51 million visits to physicians is respiratory viral disease. Two-thirds to three-quarters of acute respiratory disease are caused by viruses, belonging to different families [(8, 9). Acute viral respiratory diseases (influenza A, B, and adenovirus) are among the most common human diseases. During the previous century, influenza virus caused millions of deaths around the world [11,12(10, 11). Antigenic shifts in Influenza A and B viruses are sometimes observed in epidemics [(9, 12). Since adenoviruses have the ability to create various diseases in humans, including respiratory illnesses, availability of quick methods for diagnosis of these viruses is very important. There are different methods for the isolation and detection of respiratory viruses, such as influenza and adenovirus. These methods include molecular techniques, such as PCR-based detection, of the viral genome and isolation and identification of the virus in cell culture and serological tests like Enzyme Linked Immunosorbent Assay (ELISA) and hemagglutination inhibition (13, 14)14,15]. The PCR-based method is the preferred one for the detection of influenza and adenovirus in clinical specimens because it is faster, more sensitive, specific, and cost-effective. This study attempted to design a method by which influenza A, B, and adenoviruses are identified simultaneously in clinical samples by multiplex RT-PCR. This strategy saves significant time and costs while it prevents indiscriminate use of antibiotics. Multiplex RT-PCR has also increased diagnostic capacity compared to uniplex PCR (15).
Materials and Methods
Nucleic acid extraction and cDNA synthesis
RNA was extracted from a 100-μL volume of sample (clinical specimens and controls) by a precipitation method (RIBO-prep, ILS). Briefly, the specimen was added to a tube containing 300 μL of lysis buffer and 400 μL of precipitate buffer. It was then mixed and incubated for 10 minutes at room temperature and centrifuged at 13000 rpm for 3 minutes before being washed once with 500 μL of wash buffer, and again with 200 μL of 70% (vol/vol) ethanol (wash buffer). It was dried at 60°C for 10 minutes. The specimen was then re-suspended in 50 μL of RNase-free water and converted to cDNA by RT-PCR. For RT, 5 μL (10μg) of RNA was added to a reaction mixture (15 μL) containing 20 mM of Tris-HCl (pH 8.4), 50 mM KCl, 5.5 mM MgCl2, each dNTP mix at a concentration of 1.5 mM, 20 ng of each random primer (pdN)6, 1.6 U of RNasin, and 200 U of Moloney murine leukemia virus reverse transcriptase (Thermofisher, Germany). The reaction mixture was incubated at room temperature for 10 minutes, at 37°C for 60 minutes, and 95°C for 10 minutes and quenched on ice or stored at 4°C.
Reverse transcription-Uniplex and Multiplex PCR
The primer sets designed and used in this study are shown in Table 1. The properties of the primers were analyzed with Becon designer primer analysis software (PrimerBiosoft Inc.). Each primer pair was used at 10 pmol in the primary amplification and 5 pmol in the Multiplex amplification. For the primary and Multiplex RT-PCR, 5 μL of cDNA (containing 5 μg) was added to 20 μL of a reaction mixture containing 10 mM Tris-HCl (pH 8.8), 2.5 mM KCl, 3.5 mM MgCl2, and 1.5 U of Taq polymerase. Amplification with a DNA engine thermocycler (MWG, Germany) consisted of 1 cycle at 94°C for 15 minutes, followed by 35 cycles at 94°C for 45 seconds, 58°C for 45 seconds, and 72°C for 60 seconds. Products were visualized by Redsafe staining following electrophoresis on 1.5% NuSieve Agarose powder (Thermo fisher) agarose gel.
Table 1.
Primer Sequence for Detection of Influenza A, B and Adeno viruses
| Name | Forward Sequence | Reverse Sequence | Location |
|---|---|---|---|
| FluA | AAG GGC TTT CAC CGA AGA GG | CCC ATT CTC ATT ACT GCT TC | NS* |
| Flu B | ATG GCC ATC GGA TCC TCA AC | TGTCAG CTA TTA TGG AGC TG | NS |
| Adeno | GCC GAG AAG GGC GTG CGC AGG TA | ATG ACT TTT GAG GTG GAT CCC ATG GA | Hexon |
Nonstructural protein
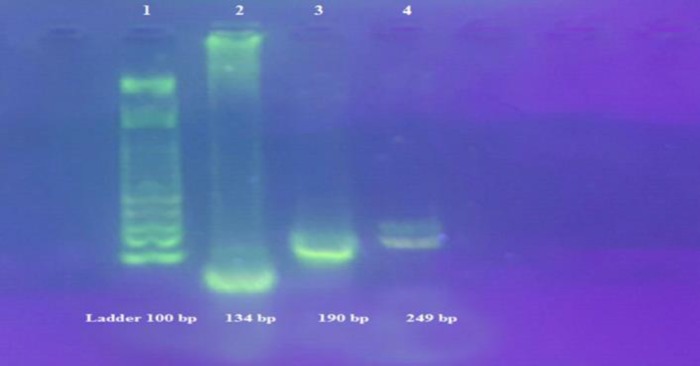
Furthermore, RNA was extracted from Flu A/H1N1/Tehran/p3/70642/1393, Flu A/H3N2/Tehran/p1/72205/1394, Flu B/Tehran/36498/1394, Adeno/P1/M.pod man/87495, and Adeno/P1/J.asmiani/87493 as positive control and water as negative control. Various viruses were distinguished by their unique amplicon sizes for Flu A: 190 bp, Flu B: 249 bp and Adeno: 134 bp, using a 100-bp ladder (Thermo fisher).
Determination of Sensitivity and Specificity of PCR: The sensitivity of PCR was determined with standard strains of viruses. The extracted DNA was serially diluted in 10-fold dilutions and then subjected to PCR. The concentration of extracted DNA was determined spectrophotometrically at 260 nm. The sensitivity of PCR was calculated using different sets of primers, which amplified the target fragment. The specificity of the viral DNAs were amplified using target primers versus human parainfluenza virus types 1 to 3 (n = 3 samples), Respiratory syncytial virus (RSV) (n = 3), herpes simplex virus type 1 (n = 1), untyped enteroviruses (n = 2), and rhinoviruses (n = 4) with the same optimized conditions.
Clinical specimens
Nasopharyngeal aspirates throat swabs were obtained from clinical diagnostic material collected between October 2014 and May 2014 from Mehrad hospital in Tehran. Combined nose and throat acrylic swabs were obtained from patients with acute upper respiratory infections during clinical virological surveillance of influenza in Tehran. Aliquots of material for PCR testing were made immediately on receipt of the specimens and the viral genomes were extracted and stored at −70°C until use. All other clinical material were also stored at −20°C.
Results
Measuring the concentration of the isolated genome
The genome of the positive control and clinical samples were extracted using the precipitation method. After genome extraction from positive control samples, concentration of RNA or DNA was measured (eppendorf) by spectrophotometer. The concentration of the target RNA and DNA viruses were as follows: Influenza A: 13.4 μg/μL, Influenza B: 20.5 μg/μL, and Adeno: 40.3 μg/μL.
Primer design
The oligonucleotide primers were designed to amplify the NP and Matrix regions of the influenza genome, because they are highly conserved and are used for sub-typing of influenza strains into influenza virus A and B types. The G+C contents, melting temperatures and lengths of the primers were chosen and analyzed using Beacon designer primer design software to ensure that they did not only meet the essential criteria for optimal PCR primers yet also could be used together in a multiplex RT- PCR reaction under similar steps (Table1).
Optimizing the best annealing temperature:
To identify the best temperature for multiplex RT-PCR reactions, the gradient temperature of 52°C to 60°C in the PCR device was performed. The best temperature for multiplex RT-PCR reactions was 58°C.
The primers were designed to ensure that the final amplicons could easily be differentiated from each other on the basis of size. All of the primers were 20-26 nucleotide and had G+C contents of less than or equal to 55%. Temperatures of 58°C and 60°C were used for annealing and amplification, respectively, for their ability to produce maximum yield and high specificity. All of the formations of dimmers either within or between pairs were analyzed and no significant theoretical mispriming was identified on any template.
Polymerase Chain Reaction optimization of the amplification conditions for each primer set was performed, and final concentrations of 10 mM Tris-HCl (pH 8.8), 5.5 mM MgCl2, 2.5 mM KCl, and 1.5 U of Taq polymerase (Thermo fisher) were found to be optimal for maximum yield of the specific product. Modifications of the PCR assay protocol included an increase in the concentration of Taq polymerase in the reaction mixture and an increase in the concentration of MgCl2 in mono and multiplex reaction mixtures. An increase in the concentration of Taq in the secondary reaction mixture significantly increased the level of product formation (data not shown). Reduction in pH of the reaction mixture below 8.5 showed a decrease of the sensitivity of detection (data not shown). The final amplification protocol included initial denaturation at 94°C for 15 minutes and then 35 cycles at 94°C for 45 seconds, 58°C for 45 seconds, and 72°C for 60 seconds for the mono and multiplex reaction.
Specificity
The multiplex RT-PCR was tested for its specificity for all of the target viruses. For simultaneous detection of influenza A (H1N1,H3N2), B and adenovirus, multiplex RT-PCR was used. In this reaction, adeno, influenza A, and B primers were added. No mispriming was observed when all of the primer sets were present with either influenza A or B virus or an Adeno virus template. A product of the expected size for each viral template by the multiplex RT-PCR was obtained. The specific products were clearly separated and identified on a 1.5% NuSieve agarose gel. Also, the specific product for target laboratory-adapted virus control materials (tissue culture-grown, adeno and influenza viruses) and for clinical samples containing wild-type strains was shown (Figure 1).
Figure 1.
Positive Sample Bands for Flu A, B and Adenovirus
There was no detectable PCR product following nucleic acid extraction and multiplex RT-PCR amplification from 50 clinical samples (nasopharyngeal aspirates, nose, and throat swabs) containing human parainfluenza virus types 1 to 3, RSV, herpes simplex virus type 1, untyped Enteroviruses, and rhinoviruses. There was no unspecific products or cross reactions to other viruses. No detectable PCR products were observed.
The specificity test comprised of three pooled templates amplified by each pair of primer, individually. Additionally, the measurement of multiplex RT-PCR specificity was determined using 7 respiratory viral RNA preparations: PIV1/2/3, RSV, HSV-1, Rhino virus, and Entroviruses.
Sensitivity
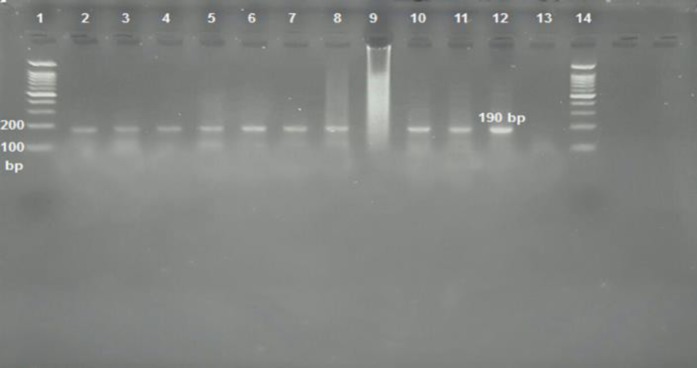
The sensitivity of detection of influenza viruses A and B, and Adeno virus with specific primers sets was determined individually and in a multiplex reaction. The limits of detection (LoD) of the Influenza A, Influenza B, and Adeno virus were determined by analyzing a 10-fold dilution series of RNA extracted from influenza viruses (Flu A/H1N1/Tehran/p3/70642/1393, Flu A/H3N2/Tehran/p1/72205/1394, Flu B/Tehran/36498/1394) and DNA extracted from Adeno virus (Adeno/P1/M.pod man/87495 and Adeno/P1/J.asmiani/87493). A 10-fold dilution series was tested in triplicates for each extracted virus strain. The cDNA synthesis was followed by PCR with the same primer sets in uniplex and multiplex reactions (Figure 4). The limit of detection for Influenza A = > 0.0067 μg/μL, Influenza B = > 0.0102 μg/μL, and for Adeno virus was equal to > 0.0201 μg/μL. A 10-fold serial dilution of extracted genomes was used to compare the sensitivity levels of the multiplex PCR and the single conventional PCR for amplifying each type of influenza and Adeno virus. Next, a pooled solution of all 3 templates was diluted, and detected by multiplex RT-PCR to determine the sensitivity.
Figure 2.
Serial Ten-Fold Dilutions of the Influenza Type A RNA Prepared in Elution Buffer
Clinical Samples
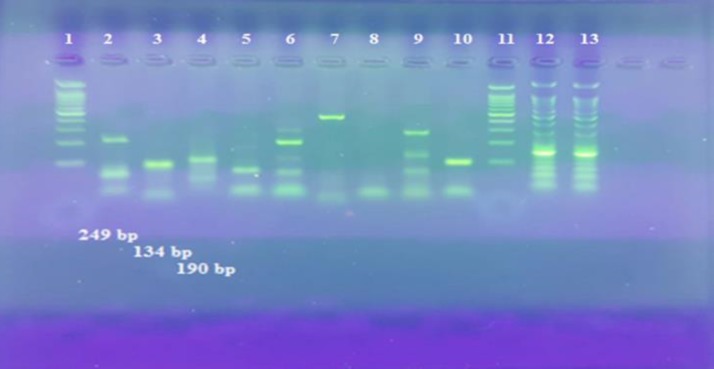
The multiplex RT-PCR was performed on about 50 nasopharyngeal aspirate specimens of the patients referred to Mehrad Hospital of Tehran, Iran. Mean age was 29±13 years old, and there were 15 females (30%) and 35 males (70%). From the tested samples, 27.5% contained Adeno, 12.5% contained Influenza type A, and 50% contained Influenza Type B, and 7.5% of these specimens consisted of negative nasopharyngeal aspirates from which no virus was recovered, and 2.5% was diagnosed as contamination (Figure 3). There was 100% correlation between the type determined by PCR and antigenic type for specimens containing influenza A virus (H1N1 and H3N2), influenza B virus and adeno virus, which were grown in tissue culture (positive controls). Overall, 7.5% of nose and throat swab specimens, which had negative results by PCR, had negative findings by real time PCR method performed in the virology laboratory of Mehrad Hospital. No detectable PCR products were observed.
Figure 3.
Typing and Subtyping of Influenza and Adeno Virus in a Panel of Clinical Specimens by Multiplex Real Time-Polymerase Chain Reaction
Multiple infections
The ability of the multiplex reaction mixture for detection of the presence of more than 1 viral template in the same condition was assessed by the preparation of nose and throat swab specimens with various combinations of viral templates. The multiplex reaction was capable of detecting all three viruses, simultaneously, as well as various combinations of templates both in viral control and in clinical specimens. This indicated that co-infections could be detected by multiplex RT-PCR.
Discussion
In 2000, about 3.5 million and in 2012 about 3.1 million cases of respiratory infections were reported. In Iran, about 112 patients died from swine flu (H1N1) and about 1500 cases of infection were reported. The frequency of acute respiratory diseases is the highest during winter and almost 30 cases per 100 people have been reported [17,18(16, 17). Due to a lack of access to virology tests, most patients with acute respiratory diseases are guided to bacteriological laboratories and are mistakenly diagnosed as being infected by bacteria (18). This occurs despite the fact that in most cases, viruses are the main cause of respiratory infections. Wrong diagnosis leads to medical errors in prescribing antibacterial drugs in the treatment of respiratory viral infections (19). Development of multiplex RT-PCR method could detect viral infections such as influenza A, B, and adeno viruses in the shortest time in patients’ breath samples (20).
However, the use of multiplex RT-PCR for the detection of multiple pathogens with RNA genomes has been much limited, because first the RT step in the RT-PCR or nucleic acid extraction are inefficient, and second, the sample quality is poor [(5, 21). In this study the researchers used a multiplex RT-PCR strategy for detection of the non-structural portion of the NS gene of influenza A and B, and on the Hexon region of the Adeno virus genome. Both of these regions of the respective genomes contain genetic information important in antigenic sub-typing of these viruses, and thus, the PCR products of the reactions could be used for investigation of strain variation, by sequencing or by PCR restriction analysis (PCR-RFLP)(22, 23).
In the extraction method, the researchers used guanidinium thiocyanate and precipitation methods, which are both sensitive for clinical samples containing RNA and DNA viruses and helpful in removing PCR inhibitors, which may be included in the starting material (24). The primer design strategy for the detection of influenza type A or B was slightly different from that for the detection of adeno virus (25).
In this study, the researchers designed a PCR, which was capable of distinguishing influenza type A from influenza type B and adenovirus as well, so it could be used in epidemiological studies of respiratory infections (10, 26, 27).
Furthermore, initial evaluation of the sensitivity and limit of detection of equivalent concentrations of influenza type A and B templates indicated that the level of amplification of the influenza type B template was substantially reduced in the presence of all primer sets compared to that of the Influenza type A template and compared to that in the presence of the Influenza type B primers alone (28). Multiplex RT-PCR optimization of both the primary and the secondary PCR mixtures were alterations of the MgCl2 concentration and the salt and buffer formulations, yet not the buffer pH (29). Increasing the concentration of Taq polymerase in the multiplex reaction mixture produced a substantial increase in sensitivity of Influenza type B, when the multiplex reaction mixture was used, to a sensitivity equivalent to that for Influenza type A detection (data not shown), and these conditions were subsequently used (30).
The researchers compared the RT-PCR endpoints for both influenza virus and adeno virus, which were much more sensitive than the other tests (31). They expected that the sensitivity of detection of a purified influenza RNA template would be approximately equivalent to> 0.0067 μg/μL for influenza type A, > 0.0102 μg/μL for influenza type B, and > 0.0201 μg/μL for adeno virus.
The frequency of co-infection with adeno virus and influenza virus or co-infection with different subtypes of influenza virus is not well documented in the literature and is essentially unknown, however, about 3% to 4% of total respiratory infections occur with other respiratory pathogens respectively (32). It is very interesting that the multiplex RT-PCR developed in the present study was capable of detecting a single pathogen from a clinical sample, and could be amplified more than one pathogen in the co-infections. The multiplex RT-PCR was clearly capable of detecting more than 2 pathogens, simultaneously (33). Successful amplification of all 3 templates in the same reaction mixture with a clinical sample required additional Taq polymerase in the multiplex reaction, because at a limited concentration of Tag, different reaction mixtures containing different copy numbers of various templates and different separate amplicons were not observed (34). Failure to amplify a product also did not appear to depend on the product size. When the concentration of Taq was doubled for the secondary PCR, all three amplicons were clearly visualized. This suggests that the catalytic action of Taq is a rate-limiting factor in achieving simultaneous products of multiple templates in the same reaction mixture (35).
Preparation of the reagent mixture for multiplex RT-PCR containing three primer sets is laborious, and to investigate whether this part of the procedure could be streamlined, the researchers attempted to review the preparation of master mixtures in which reagent compounds containing all the necessary primers, buffers, and dNTP mix could be stored prior to use (36). Performance of the multiplex RT-PCR therefore required only the preparation of nucleic acid template, preparation of a mixture in a tube, and Taq polymerase. Storage of prepared reaction mixtures has not changed the sensitivity of the detection of the virus, even when the mixtures were stored for up to 6 months (37). The authors demonstrated that multiplex RT-PCR could be used for detection and subtyping of adeno and influenza A and B viruses in clinical respiratory samples. The multiplex RT-PCR described here is both highly sensitive and specific for each virus and detection of co-infection in clinical samples. It could be used in studies of viral respiratory infections in both surveillance and laboratory diagnostic tests.
Conclusion
Respiratory diseases are among the most prevalent diseases in humans, and the Influenza A, B, and Adeno take an immense part in such diseases, therefore prompt detection of these viruses is of prime importance. The present research was an effort to design a method (Multiplex RT-PCR) to detect these viruses quickly and simultaneously.
Conflicts of Interest
The authors declared that they had no conflict of interest in this manuscript.
Acknowledgment
This study was performed as an MSc thesis of Mohsen Nakhaie, and was financially supported by the Research Deputy of Tarbiat Modares University, Faculty of Medical Sciences, Tehran, Iran and Department of Medical Microbiology, and Laboratory of Medical Virology in Kerman, Iran. The authors would like to thank Professor Mokhtari Azad for her kind support.
References
- 1.Piewbang C, Rungsipipat A, Poovorawan Y, Techangamsuwan S. Development and application of multiplex PCR assays for detection of virus-induced respiratory disease complex in dogs. J Vet Med Sci. 2016 doi: 10.1292/jvms.16-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long SS. Multiplex PCR testing during respiratory infections: good for the patient, good for infection prevention and antimicrobial stewardship. J Pediatr. 2016;173:1–3. doi: 10.1016/j.jpeds.2016.04.028. [DOI] [PubMed] [Google Scholar]
- 3.Nikonova AA, Uspenskaia ES, Lobodanov SA, Oksanich AS, Gorbalenia AE, Claas EC, et al. [Use of multiplex PCR method with real-time detection for differential diagnosis of respiratory viral infections] Zh Mikrobiol Epidemiol Immunobiol. 2009(1):67–70. [PubMed] [Google Scholar]
- 4.Nickbakhsh S, Thorburn F, B VONW, Mc MJ, Gunson RN, Murcia PR. Extensive multiplex PCR diagnostics reveal new insights into the epidemiology of viral respiratory infections. Epidemiol Infect. 2016;144(10):2064–76. doi: 10.1017/S0950268816000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pabbaraju K, Wong S, Lee B, Tellier R, Fonseca K, Louie M, et al. Comparison of a singleplex real-time RT-PCR assay and multiplex respiratory viral panel assay for detection of influenza "A" in respiratory specimens. Influenza Other Respir Viruses. 2011;5(2):99–103. doi: 10.1111/j.1750-2659.2010.00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker J, Fowler N, Walmsley ML, Schmidt T, Scharrer J, Kowaleski J, et al. Analytical Sensitivity Comparison between Singleplex Real-Time PCR and a Multiplex PCR Platform for Detecting Respiratory Viruses. PLoS One. 2015;10(11):e0143164. doi: 10.1371/journal.pone.0143164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rappo U, Schuetz AN, Jenkins SG, Calfee DP, Walsh TJ, Wells MT, et al. Impact of Early Detection of Respiratory Viruses by Multiplex PCR Assay on Clinical Outcomes in Adult Patients. J Clin Microbiol. 2016;54(8):2096–103. doi: 10.1128/JCM.00549-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck ET, Jurgens LA, Kehl SC, Bose ME, Patitucci T, LaGue E, et al. Development of a rapid automated influenza A, influenza B, and respiratory syncytial virus A/B multiplex real-time RT-PCR assay and its use during the 2009 H1N1 swine-origin influenza virus epidemic in Milwaukee, Wisconsin. J Mol Diagn. 2010;12(1):74–81. doi: 10.2353/jmoldx.2010.090095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iha Y, Kinjo T, Parrott G, Higa F, Mori H, Fujita J. Comparative epidemiology of influenza A and B viral infection in a subtropical region: a 7-year surveillance in Okinawa, Japan. BMC Infect Dis. 2016;16(1) doi: 10.1186/s12879-016-1978-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monavari SH, Mollaie HR, Fazlalipour M. Simultaneous detection of influenza viruses A, B, and swine origin influenza A using multiplex one-step real-time RT-PCR assay. Appl Biochem Biotechnol. 2014;172(2):984–92. doi: 10.1007/s12010-013-0583-6. [DOI] [PubMed] [Google Scholar]
- 11.Hurt AC. The epidemiology and spread of drug resistant human influenza viruses. Curr Opin Virol. 2014;8:22–9. doi: 10.1016/j.coviro.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Kuo RL, Yang SL, Liu YC, Chen LT, Mok CK, Kuo SM, et al. Influenza A/B virus detection and influenza A virus subtyping with emphasis on the novel H7N9 virus by using multiplex real-time RT-PCR. J Virol Methods. 2014;208:41–6. doi: 10.1016/j.jviromet.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 13.Khansarinejad B, Soleimanjahi H, Samiee SM, Hamidieh AA, Paryan M, Sanahmadi Y. Quantitation of human cytomegalovirus DNA in plasma using an affordable in-house qPCR assay. Journal of virological methods. 2012;183(2):170–5. doi: 10.1016/j.jviromet.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Sakthivel SK, Whitaker B, Lu X, Oliveira DB, Stockman LJ, Kamili S, et al. Comparison of fast-track diagnostics respiratory pathogens multiplex real-time RT-PCR assay with in-house singleplex assays for comprehensive detection of human respiratory viruses. Journal of virological methods. 2012;185(2):259–66. doi: 10.1016/j.jviromet.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sultani M, Mokhtari Azad T, Eshragian M, Shadab A, Naseri M, Eilami O, et al. Multiplex SYBR Green Real-Time PCR Assay for Detection of Respiratory Viruses. Jundishapur J Microbiol. 2015;8(8):e19041. doi: 10.5812/jjm.19041v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M, Xie Z, Liu J, Xie L, Deng X, Luo S, et al. Simultaneous detection of four different neuraminidase types of avian influenza A H5 viruses by multiplex reverse transcription PCR using a GeXP analyser. Influenza Other Respir Viruses. 2016;10(2):141–9. doi: 10.1111/irv.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khandaker G, Zurynski Y, Ridley G, Buttery J, Marshall H, Richmond PC, et al. Clinical epidemiology and predictors of outcome in children hospitalised with influenza A(H1N1)pdm09 in 2009: a prospective national study. Influenza Other Respir Viruses. 2014;8(6):636–45. doi: 10.1111/irv.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kidd M. Influenza viruses: update on epidemiology, clinical features, treatment and vaccination. Curr Opin Pulm Med. 2014;20(3):242–6. doi: 10.1097/MCP.0000000000000049. [DOI] [PubMed] [Google Scholar]
- 19.Lazarus R, Lim PL. Avian influenza: recent epidemiology, travel-related risk, and management. Curr Infect Dis Rep. 2015;17(1):456. doi: 10.1007/s11908-014-0456-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez LJ, Perera CL, Coronado L, Rios L, Vega A, Frias MT, et al. Molecular epidemiology study of swine influenza virus revealing a reassorted virus H1N1 in swine farms in Cuba. Prev Vet Med. 2015;119(3-4):172–8. doi: 10.1016/j.prevetmed.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Payungporn S, Phakdeewirot P, Chutinimitkul S, Theamboonlers A, Keawcharoen J, Oraveerakul K, et al. Single-step multiplex reverse transcription-polymerase chain reaction (RT-PCR) for influenza A virus subtype H5N1 detection. Viral Immunol. 2004;17(4):588–93. doi: 10.1089/vim.2004.17.588. [DOI] [PubMed] [Google Scholar]
- 22.Poddar SK, Espina R, Schnurr DP. Evaluation of a single-step multiplex RT-PCR for influenza virus type and subtype detection in respiratory samples. J Clin Lab Anal. 2002;16(3):163–6. doi: 10.1002/jcla.10036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pozzetto B, Grattard F, Pillet S. Multiplex PCR theranostics of severe respiratory infections. Expert Rev Anti Infect Ther. 2010;8(3):251–3. doi: 10.1586/eri.09.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin ZF, Sun J, Lu TK, Zeng SL, Hua QY, Ling QY, et al. Subtyping animal influenza virus with general multiplex RT-PCR and Liquichip high throughput (GMPLex) Virol Sin. 2012;27(2):120–31. doi: 10.1007/s12250-012-3232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanghavi SK, Bullotta A, Husain S, Rinaldo CR. Clinical evaluation of multiplex real-time PCR panels for rapid detection of respiratory viral infections. J Med Virol. 2012;84(1):162–9. doi: 10.1002/jmv.22186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nummi M, Mannonen L, Puolakkainen M. Development of a multiplex real-time PCR assay for detection of Mycoplasma pneumoniae, Chlamydia pneumoniae and mutations associated with macrolide resistance in Mycoplasma pneumoniae from respiratory clinical specimens. Springerplus. 2015;4 doi: 10.1186/s40064-015-1457-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rheem I, Park J, Kim TH, Kim JW. Evaluation of a multiplex real-time PCR assay for the detection of respiratory viruses in clinical specimens. Ann Lab Med. 2012;32(6):399–406. doi: 10.3343/alm.2012.32.6.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Detection of influenza virus subtype A by polymerase chain reaction: WHO external quality assessment programme summary analysis, 2015. Wkly Epidemiol Rec. 2016;91(1):3–11. [PubMed] [Google Scholar]
- 29.Haines AM, Tobe SS, Linacre A. Optimization of Diamond Nucleic Acid Dye for quantitative PCR. Biotechniques. 2016;61(4):183–9. doi: 10.2144/000114458. [DOI] [PubMed] [Google Scholar]
- 30.Cheng YH, Kuo CN, Lai CM. Effective Natural PCR-RFLP Primer Design for SNP Genotyping Using Teaching-Learning-Based Optimization with Elite Strategy. IEEE Trans Nanobioscience. 2016 doi: 10.1109/TNB.2016.2597867. [DOI] [PubMed] [Google Scholar]
- 31.Liu J, Gratz J, Amour C, Nshama R, Walongo T, Maro A, et al. Optimization of Quantitative PCR Methods for Enteropathogen Detection. PLoS One. 2016;11(6):e0158199. doi: 10.1371/journal.pone.0158199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lievens A, Jacchia S, Kagkli D, Savini C, Querci M. Measuring Digital PCR Quality: Performance Parameters and Their Optimization. PLoS One. 2016;11(5):e0153317. doi: 10.1371/journal.pone.0153317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wadle S, Lehnert M, Rubenwolf S, Zengerle R, von Stetten F. Real-time PCR probe optimization using design of experiments approach. Biomol Detect Quantif. 2016;7:1–8. doi: 10.1016/j.bdq.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slavov SN, Otaguiri KK, de Figueiredo GG, Yamamoto AY, Mussi-Pinhata MM, Kashima S, et al. Development and optimization of a sensitive TaqMan(R) real-time PCR with synthetic homologous extrinsic control for quantitation of Human cytomegalovirus viral load. J Med Virol. 2016;88(9):1604–12. doi: 10.1002/jmv.24499. [DOI] [PubMed] [Google Scholar]
- 35.Jacchia S, Nardini E, Savini C, Petrillo M, Angers-Loustau A, Shim JH, et al. Development, optimization, and single laboratory validation of an event-specific real-time PCR method for the detection and quantification of Golden Rice 2 using a novel taxon-specific assay. J Agric Food Chem. 2015;63(6):1711–21. doi: 10.1021/jf505516y. [DOI] [PubMed] [Google Scholar]
- 36.Lizette Del-Toro-Sanchez C, Villasenor-Alvarado S, Zurita-Martinez F, Castellanos-Hernandez OA, Rodriguez-Sahagun A, Isabel Torres-Moran M, et al. Optimization of DNA isolation and PCR protocol for analysis and evaluation of genetic diversity of the medicinal plant, Anemopsis californica using RAPD. Acta Biol Hung. 2013;64(2):184–95. doi: 10.1556/ABiol.64.2013.2.5. [DOI] [PubMed] [Google Scholar]
- 37.Villarreal Camacho JL, Mendoza Torres E, Cadena C, Prieto J, Varela Prieto LL, Villanueva Torregroza DA. The use of factorial design, image analysis, and an efficiency calculation for multiplex PCR optimization. J Clin Lab Anal. 2013;27(3):249–52. doi: 10.1002/jcla.21595. [DOI] [PMC free article] [PubMed] [Google Scholar]