Abstract
Type IVa pili are filamentous cell surface structures observed in many bacteria. They pull cells forward by extending, adhering to surfaces, and then retracting. We used cryo–electron tomography of intact Myxococcus xanthus cells to visualize type IVa pili and the protein machine that assembles and retracts them (the type IVa pilus machine, or T4PM) in situ, in both the piliated and nonpiliated states, at a resolution of 3 to 4 nanometers. We found that T4PM comprises an outer membrane pore, four interconnected ring structures in the periplasm and cytoplasm, a cytoplasmic disc and dome, and a periplasmic stem. By systematically imaging mutants lacking defined T4PM proteins or with individual proteins fused to tags, we mapped the locations of all 10 T4PM core components and the minor pilins, thereby providing insights into pilus assembly, structure, and function.
Graphical abstract
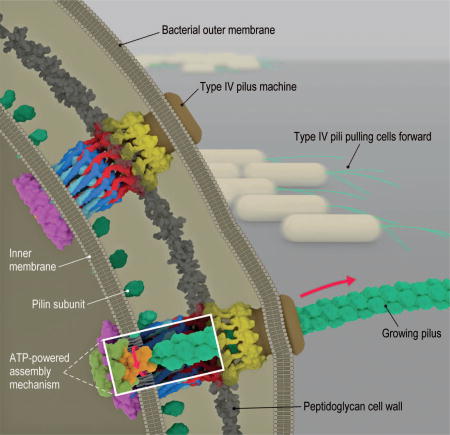
Bacterial type IVa pilus machine (T4PM). Two T4PMs are depicted on the left, spanning the envelope of a Gram-negative bacterial cell. T4PMs extend and retract pili to pull cells forward. The structural data presented here support the hypothesis that ATP hydrolysis in the cytoplasm causes an adapter protein in the inner membrane to rotate, facilitating the transfer of pilin subunits from the inner membrane onto the growing pilus. The process is reversed during retraction.
Type IVa pilus machines (T4PMs) are part of a superfamily of bacterial and archaeal multiprotein assemblies that also include type IVb pilus machines, type II secretion systems (T2SS), DNA uptake systems, and archaeal flagella (1, 2). T4PMs are involved in cell motility (3), host adhesion (4), predation (5), DNA uptake (6), biofilm formation (7, 8), and protein secretion (9). T4PMs consist of an extracellular pilus fiber (T4P) and a cell envelope–spanning complex that we refer to here as the basal body. A key feature of T4Ps is their ability to undergo cycles of extension and retraction (10, 11). Powered by a T4P-assembly adenosine triphosphatase (ATPase), the basal body extracts pilin monomers from the inner membrane (IM) and appends them onto the base of the helical pilus fiber, pushing the fiber outward. The fiber has a diameter of ~6 nm, can extend up to several micrometers in length, and can adhere to specific surfaces (12). Subsequently, the basal body, powered by a T4P-disassembly ATPase, extracts monomers from the pilus base back into the membrane (13). T4PMs retract at rates up to 1 µm/s and generate forces as high as 150 pN to pull the cell forward (14, 15), making T4PMs the strongest molecular motors known. T4Ps have been identified as important virulence factors in several human pathogens, including Neisseria gonorrhoeae, N. meningitidis, andPseudomonas aeruginosa (16).
T4PMs exhibit a multilayered structure spanning the cell envelope of M. xanthus
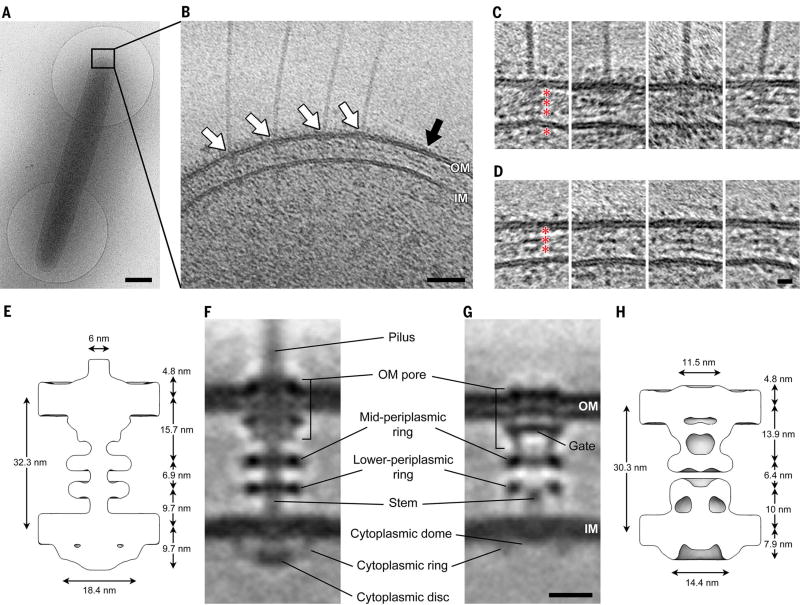
To determine the structure of the T4PM in vivo, we used cryo–electron tomography (also known as electron cryotomography) to image the cell poles of Myxococcus xanthus, a ubiquitous rod-shaped soil-residing predatory bacterium (Fig. 1A). (Examples of a tilt series and a cryotomogram are shown in movies S1 and S2, respectively). T4Ps with diameters of ~6 nm were easily recognized on the cell surface with their basal bodies spanning the cell envelope (Fig. 1B, white arrows). Basal bodies exhibited three layers of densities in the periplasm and a fourth in the cytoplasm (Fig. 1C). In the vicinity of piliated T4PMs, we also observed “empty” nonpiliated basal bodies with similar structures but without long fibers attached (Fig. 1B, black arrow, and Fig. 1D).
Fig. 1. Visualizing the T4PM in intact M. xanthus cells.
(A) A frozen-hydrated M. xanthus cell on an EM grid. (B) Slice through tomogram of the cell pole. White arrows, piliated T4PM basal bodies; black arrow, an empty T4PM basal body. Outer and inner membranes (OM and IM) are indicated. (C and D) Examples of slices through subtomograms containing piliated and empty T4PM basal body structures, respectively. Red asterisks denote periplasmic and cytoplasmic density layers. (E and H) Schematic envelopes of the subtomogram averages of wild-type piliated (E) and ΔpilB empty (H) T4PM basal bodies, showing their molecular dimensions. (F and G) Slices through subtomogram averages of wild-type piliated (F) and ΔpilB empty (G) T4PM basal bodies, with annotations of structural features. Scale bars, 500 nm (A), 50 nm (B), 5 nm [(C) and (D)], 10 nm [(F) and (G)].
To reveal details, we generated subtomogram averages of the piliated T4PM and nonpiliated basal bodies with local resolution between ~2.5 nm and 4.5 nm (Fig. 1F and fig. S1, A and B). We later determined that a ΔpilB mutant (which lacks the PilB T4P-assembly ATPase and cannot assemble T4P) produced better-quality images of the empty basal body (likely due to the basal bodies being stalled in a “pilus preassembly” state and therefore more structurally homogeneous than in the wild type; see below), so we used the average from this strain for subsequent structural interpretation (Fig. 1G).
We found that the structure of the piliated T4PM basal body comprises an outer membrane (OM)–spanning pore including a ring immediately below the OM, two distinct rings in the periplasm, another ring in the cytoplasm surrounding a disc-like structure, and a long stem originating at the IM and passing through the periplasmic rings and OM pore (Fig. 1F). The structure of the empty basal body shows several similar structural features, including an OM pore with a ring immediately below the OM, two periplasmic rings, and a cytoplasmic ring, as well as a gate density in the OM pore, connections between the OM pore and the mid-periplasmic ring, connections between the lower periplasmic ring and the IM, a much shorter stem, and no cytoplasmic disc (Fig. 1G). In the absence of the cytoplasmic disc, a cytoplasmic dome is also apparent. Aligning the averages of piliated and empty basal bodies with the IM reveals clear conformational changes upon piliation (movie S3). Going from the nonpiliated to the piliated state, the OM pore is ~2 nm farther away from the mid-periplasmic ring (13.9 to 15.7 nm). Also, the diameter of the cytoplasmic ring in the piliated state is greater by ~4 nm (18.4 versus 14.4 nm), possibly because of its association with the cytoplasmic disc (Fig. 1, E and H).
Mapping components in the molecular envelope by imaging T4PM mutants
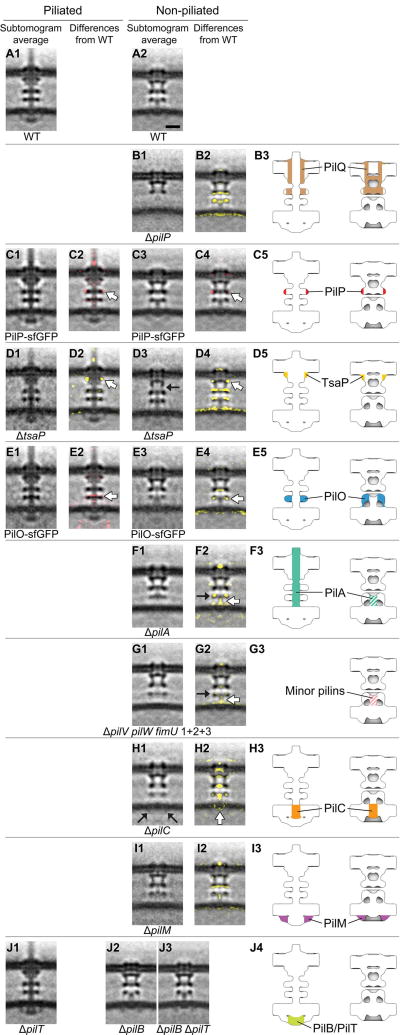
Ten highly conserved proteins are known to constitute the T4PM (16, 17). PilA, the major pilin protein, contains an N-terminal hydrophobic α helix and alternates between being anchored individually in the IM or bundled with other PilA N-terminal α helices to form the central pilus core (18). The remaining nine proteins are divided into three subgroups according to their location and function: the OM pore complex (PilQ and TsaP); the alignment complex (PilM, PilN, PilO, and PilP); and the motor complex (PilB, PilT, and PilC) (19). To systematically localize each component within the basal body, we imaged a series of M. xanthus mutants with individual T4PM proteins either missing or fused to a superfolder green fluorescent protein (sfGFP) (20) tag. Difference maps between the resulting subtomogram averages of the T4PM mutants and the wild-type structures were then calculated (Fig. 2). Combined with information already available about the accumulation, subcellular localization, and incorporation of individual T4PM proteins into the T4PM basal bodies of these mutants (fig. S2) and their connectivities and structures (fig. S3) (17, 18, 21–40), these maps allowed us to pinpoint each component within the T4PM (Figs. 2 and 3A).
Fig. 2. Mapping T4PM components.
First and third columns: Central slices of subtomogram averages of piliated and empty T4PM basal bodies, respectively, from different M. xanthus strains. Second and fourth columns: Differences in the T4PM mutant structures versus the wild type (red and yellow colors respectively denote addition and omission of densities, with opacities of 10%, 20%, 30%, 40%, and 50% corresponding to density differences of 1, 1.5, 2, 2.5, and 3 standard deviations, respectively, overlaid on the wild-type subtomogram averages). White arrows indicate the component locations identified by the difference maps. Fifth column: Schematic representations of piliated (left) and empty (right) T4PM basal bodies showing each identified component location. Scale bar in A2, 10 nm (applies to columns 1 through 4).
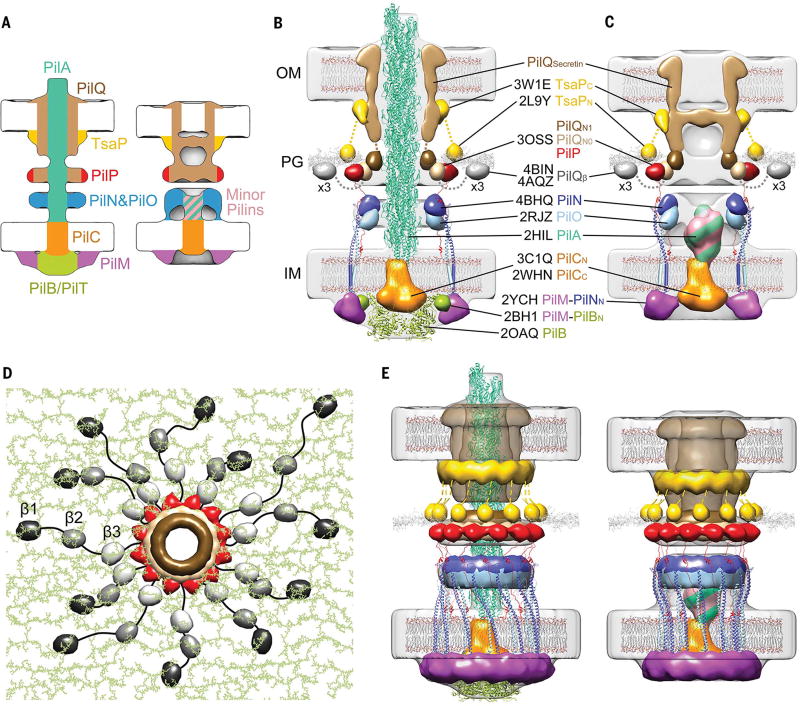
Fig.3. Architectural models of the T4PM.
(A) Summary schematics showing the component locations identified in the piliated and empty T4PM basal body structures. (B and C) Central slices of the architectural models of piliated and empty T4PM basal bodies, respectively, in which atomic models of T4PM components are placed in the in vivo envelopes according to the component maps in (A) and previously reported constraints and filtered to 3-nm resolution. (The process of how each component was placed is detailed in Movie 1 and the supplementary materials.) Models of each component are colored as in (A), with the transmembrane segments of PilN and PilO shown as cylinders; “x3” indicates three AMIN domains per PilQ monomer, only one of which is shown. Note that the empty T4PM basal body is shown with five PilA major pilin subunits in the short stem; however, the short stem likely also contains minor pilins. (D) Top view of the PilP HR domains and the PilQ N0 and N1 domains in the architectural model [colored as in (B) and (C)], with PG model (colored green) as background; 36 AMIN (β) domain models from 12 PilQ proteins are randomly placed on PG and connected by long flexible linkers (black) with lengths within 20 nm between β1 and β2 domains (70 residues), 12 nm between β2 and β3 domains (40 residues), and 12 nm between β3 and N0 domains (40 residues). (E) Overall architectural models of piliated (left) and empty (right) T4PM basal bodies. For clarity, the PilQ AMIN domains displayed in (D) are not shown.
PilQ
A previous study showed that knocking out pilP accelerates degradation of PilM, PilN, and PilO and causes mislocalization of PilC, whereas PilQ and TsaP remain stable and at the cell pole, forming a rudimentary T4PM basal body consisting only of PilQ and TsaP (fig. S2) (21). This suggests that PilP is crucial for stabilizing and linking other components to the OM pore complex. We therefore first imaged the ΔpilP mutant to obtain the structure of the OM pore complex (PilQ and TsaP) alone (Fig. 2, B1). The average showed a cylindrical channel in the OM with a clear gate and a large periplasmic vestibule. The overall structure is reminiscent of single-particle reconstructions of secretin channel complexes in T4PM, T2SS, and type III secretion systems (T3SS) (41–44) (fig. S4). Superposing the highly conserved gate and periplasmic vestibule regions, our in vivo structure was seen to be markedly longer in the transmembrane region than the structures generated from single-particle analyses, probably because the detergent solubilization used in the single-particle reconstructions fails to support this structure. By comparing the ΔpilP mutant structure to that of the wild-type empty basal body (Fig. 2, B2), and noting that the single-particle reconstructions contained no TsaP, we infer that PilQ forms the OM channel as well as part of the mid-periplasmic ring (Fig. 2, B3).
PilP
Because PilP is known to interact directly with PilQ (21, 26) and the ΔpilP structure shows a decrease in density of the mid-periplasmic ring (Fig. 2, B2), we hypothesized that PilP is part of the mid-periplasmic ring. To confirm this, we imaged a mutant in which PilP is fused to sfGFP, giving rise to an active fusion protein (fig. S2). Consistently, in this strain, additional densities appeared at the periphery of the mid-periplasmic ring (Fig. 2, C1 to C5).
TsaP
Single-particle EM reconstructions have shown that TsaP forms a ring-like structure surrounding the PilQ channel, but only top views were obtained (17). To localize TsaP in the OM pore complex in three dimensions, we imaged a ΔtsaP mutant (Fig. 2, D1 and D3). In the ΔtsaP mutant, all other T4PM proteins accumulate and are incorporated into the T4PM basal body as in the wild type (fig. S2) (17). Difference maps pinpointed TsaP’s location to the upper periplasmic region of the OM pore complex just beneath the OM in both the piliated and empty basal bodies [Fig. 2, D2 (white arrow), D4 (white arrow), and D5]. Lack of TsaP caused disengagement of the OM channel from the mid-periplasmic ring in the empty basal body (Fig. 2, D3, black arrow; note that the extensive systematic differences in Fig. 2, D4, reveal a global shift of the periplasmic rings and IM), making the overall length of the complex similar to that of the piliated basal body. In a previous study, lack of TsaP resulted in accumulation of pilus fibers in the periplasm in N. gonorrhoeae and formation of fewer pili in M. xanthus, suggesting a role for TsaP in correct PilQ channel function (17). Consistently, during our imaging, we observed that in the ΔtsaP mutant, about one-tenth as many pili were found on cell poles relative to wild-type cells (table S1).
PilO and PilN
Because knocking out pilO disrupts the entire alignment complex and causes mislocalization of PilC, leaving only PilQ and TsaP in the basal body (fig. S2) (21), we imaged a mutant with an active PilO-sfGFP fusion protein to map the location of PilO (fig. S2). In the piliated structure, we observed additional density in the lower periplasmic ring (Fig. 2, E1 and E2), but in the empty basal body structure, we observed decreased density in this ring (Fig. 2, E3 and E4). Both results suggest that PilO localizes to the lower periplasmic ring (Fig. 2, E5), because the sfGFP tag likely added density to the ring in the piliated form but perturbed the ring in the empty basal body. As with pilO, a ΔpilN mutant also disrupts the entire alignment complex and also causes mislocalization of PilC, leaving only PilQ and TsaP in the basal body (fig. S2) (21). So far, we have been unable to generate a functional PilN protein fused to a tag. Therefore, the same method could not be used to localize PilN in the T4PM basal body. Nonetheless, the two structural homologs PilO and PilN interact directly (21, 30), likely forming heterodimers (30). Therefore, we assume that PilN is also located in the lower periplasmic ring.
PilA and minor pilins
In the wild-type piliated basal body structure, we observed a rod-like stem structure that passes up from the IM through the lower periplasmic ring, mid-periplasmic ring, and OM pore. This stem has the same diameter as the PilA helical polymer (18) (~6 nm) and is directly connected to the pilus outside of the cell. Also, this long stem is missing in the empty basal body structure. These observations suggest that the stem is the part of the pilus fiber located in the periplasm and is associated with the basal body. In the empty basal body, a short stem is also present between the lower periplasmic ring and the IM. To investigate whether this remaining short stem is formed by PilA or by other T4PM components, we first imaged a ΔpilA mutant that lacks the major pilin PilA. As expected (45), no T4P formed in this mutant.
In agreement with the observation that the T4PM assembles in a ΔpilA mutant (fig. S2) (21), we were able to identify empty basal body structures in the cells and generate an average of the ΔpilA mutant, which clearly lacked any stem [Fig. 2, F1 and F2 (white arrow)]. Because the minor pilins in P. aeruginosa were recently suggested to form a complex that primes pilus assembly (46) and the four minor pseudopilins in the T2SS are thought to prime pseudopilus formation by the major pseudopilin (47, 48), we generated mutants lacking as many as nine of the 10 minor pilins encoded in the M. xanthus genome (fig. S5A). Lack of nine of the minor pilins abolished T4P-dependent motility, and T4P did not assemble; however, all 10 core components of the T4PM accumulated at wild-type levels in total cell extracts (fig. S5, B to D). Consistently, when we imaged the mutant deleted for nine of the minor pilin genes, we did not detect T4P (table S1). We did, however, detect empty basal body structures, and they clearly lacked the short stem [Fig. 2, G1 and G2 (white arrow)].
On the basis of these observations, we conclude that the extended stem is made of the major pilin PilA (Fig. 2, F3) and that the short stem is composed of an assembly-priming complex consisting of minor pilins and PilA (Fig. 2, F3 and G3). In the PilA and minor pilin mutants, the structure of the lower periplasmic ring was perturbed and the short stem was absent (Fig. 2, F2 and G2, black arrows). Because it is unlikely for PilA to withdraw its hydrophobic α helix from the IM or pilus fiber to participate in the lower periplasmic ring, the simultaneous changes of the short stem and the lower periplasmic ring suggest a structural/functional linkage between them (see below).
PilC, PilM, PilB, and PilT
The T4PM components PilC, PilM, PilB, and PilT all have folded cytoplasmic domains and are therefore the candidates for the cytoplasmic ring, disc, and dome. Within the set of single-gene knockouts of these four proteins, only empty basal bodies were found on the ΔpilC, ΔpilM, and ΔpilB cells, as expected (Fig. 2, H1, I1, and J2), and only piliated T4PM basal bodies were found on the ΔpilT cells (Fig. 2, J1). In these four mutants, all the remaining T4PM proteins accumulated (fig. S2). Because the cytoplasmic ring was missing in the ΔpilM mutant (Fig. 2, I1) but appeared unperturbed in the ΔpilC, ΔpilB, and ΔpilT mutants, we conclude that the ring is composed of PilM (Fig. 2, I3). Because the ring and dome were retained in the ΔpilB mutant but not the disc, the disc must be PilB (Fig. 2, J2 and J4). The fact that the cytoplasmic disc had a size corresponding to that of a hexameric secretion or traffic ATPase (37–40) also strengthens this conclusion (see below). The cytoplasmic dome and disc were both missing in the ΔpilC mutant (Fig. 2, H2), but the cytoplasmic ring was still present (Fig. 2, H1, black arrows), which suggests that the dome is composed of PilC and that the PilB disc does not localize in its absence (Fig. 2, H3). Notably, the short stem structure was also missing in the ΔpilC structure, revealing that PilC is required to stabilize it.
With the identification of the ring as PilM, the dome as PilC, and the disc as PilB, it follows that (i) the PilM ring assembles in the absence of the PilC dome or PilB disc, (ii) the PilC dome and PilB disc both require the PilM ring, and (iii) the PilC dome assembles without the PilB disc, but the PilB disc requires the PilC dome. These interdependencies match previous observations (fig. S2) except in one regard: Earlier immunofluorescence experiments demonstrated that PilC can be incorporated into the T4PM independently of PilM (21) (fig. S2). Our observation that in the ΔpilM mutant all cytoplasmic densities and the short stem are missing in averages (Fig. 2, I2) clarifies that the PilM cytoplasmic ring is important for consistent incorporation of the other cytoplasmic proteins and formation of the short stem.
After the cytoplasmic ring, dome, and disc had been assigned to PilM, PilC, and PilB, respectively, no additional cytoplasmic density was available to interpret as PilT. The most likely explanation is that PilT occupies the same location as PilB. When generating subtomogram averages, because we could not tell whether any particular pilus was in the extension or retraction state, we averaged all identified particles. The fact that the ΔpilT mutant displayed more T4Ps than wild-type cells in our images (table S1) confirmed that the T4Ps were actively extending and retracting in our wild-type sample. Hence, the wild-type piliated structure was an average of both PilB- and PilT-bound states. The ΔpilT piliated structure (Fig. 2, J1), however, showed no clear addition or loss of cytoplasmic densities relative to the wild type (fig. S1, B and D), which suggests that the differences between PilB- and PilT-bound particles are not visible at this resolution. We conclude that both PilB and PilT form cytoplasmic disc structures (Fig. 2, J4) and bind to the basal body in a mutually exclusive manner.
To test this idea, we generated a ΔpilB ΔpilT double mutant. As expected, this mutant lacked T4P-dependent motility and all other T4PM proteins accumulated as in the wild type (fig. S6). When we imaged the ΔpilB ΔpilT double mutant (Fig. 2, J3), no clear differences were observed on its empty basal body relative to that of ΔpilB and wild-type strains (Fig. 2, A2 and J2), confirming our assignment of both PilB and PilT ATPases to the same cytoplasmic disc. The ATPase density was located ~2 nm away from the IM, preventing any direct interaction between the ATPases and pilin subunits. We therefore conclude that PilC, which forms the cytoplasmic dome structure, lies between the ATPases and the stem and transduces force generated by ATP hydrolysis into pilus extension and retraction. In agreement with this model, the N-terminal cytoplasmic domain of PilC was recently shown to interact directly with PilB, and the C-terminal cytoplasmic domain was suggested to interact with PilT (49).
Placing available component structures into the overall molecular envelope
Because atomic structures are available for homologs of >90% of the domains of the T4PM, we next sought to test the plausibility of our component maps by trying to place these structures into the overall molecular envelope in a way that would satisfy the maps and all known constraints, including domain sizes, structures, and connectivities (see supplementary materials, in particular figs. S7 to S11, and for details, and Movie 1 for a presentation of the process in three dimensions). Remarkably, in this process, the sizes and shapes of all domains analyzed fitted well into the molecular envelopes and resulted in a hypothetical working model of the piliated and nonpiliated basal bodies (fig. S12). Because the 2- to 4-nm resolution of the subtomogram averages was not high enough to reveal the orientation of each component within the EM envelope, we next generated models with most components filtered to 3-nm resolution (Fig. 3, B to E). The process of positioning and connecting the domains in the context of a full hypothetical structural model nonetheless revealed important relationships that rationalize numerous previous observations and suggest insights into T4PM assembly, structure, and function, as described below and shown in Movie 2.
Overall architecture and assembly
All T4PM components except the pilus and PilC appear to form rings. Each of the OM, peptidoglycan (PG), and IM cell envelope layers is negotiated or engaged by a proteinaceous ring, and there is an additional “floating” lower periplasmic ring. Each ring is linked to the rings above and below to create an integrated but flexible structure spanning the entire cell envelope. The subtomogram averages of assembled T4PM subcomplexes in different mutants provide snapshots of the assembly pathway and support the sequence [PilQ, TsaP] → [PilP, PilN, PilO] → [PilM, PilC, PilA, minor pilins]→[PilB, PilT] (fig. S13). The coiled-coil domains of PilN and PilO form a cage-like compartment above and within the IM. Although the exact number of subunit monomers in each ring remains unknown, the 1:1 connectivities between PilQ-PilP, PilP-PilN-PilO, and PilN-PilM suggest that all the rings have the same stoichiometry. We found that 12 copies of the ring components fit best into the EM density, in agreement with observations of secretin channels of T4PM and T2SS in multiple species (41, 42, 50). The entire basal body is therefore robustly anchored to the PG by ~12 TsaP N-terminal LysM domains and ~36 PilQ AMIN domains arranged irregularly (for instance, as in Fig. 3D). PilQ in turn links to PilP, which links to the PilN-PilO ring, which links to PilM, which binds PilB (and PilT). Unless one of these known connectivities is only transitory in vivo, the only components free to rotate are PilC and potentially the pilus.
Pilus assembly and disassembly by a rotating PilC
The structure of the pilus can be thought of as either a 3-start left-handed helix or a 1-start or 4-start right-handed helix. Because no substantial pilus rotation has been observed during pilus retraction or extension, three different assembly structures or mechanisms can be imagined (18): (i) a fixed structure with three active sites adding pilins at each of the three sites needed to extend a 3-start helix; (ii) a fixed structure with four active sites adding pilins at each of the four sites needed to extend a 4-start helix; or (iii) a rotating structure with one or a few active sites adding pilins one at a time as it rotates around the axis of the fiber, extending the fiber as a 1-start helix (3, 51). As described above, the OM pore complex, alignment complex, and PilM-ATPase complexes are all directly or indirectly linked and anchored to the PG. The only T4PM component able to rotate is PilC. We found that the space inside the PilM-PilN-PilO “cage” can accommodate no more than one PilC dimer. It is unlikely that a PilC dimer would have three or four active sites for interacting with different PilA molecules; therefore, our models point to a 1-start assembly mechanism in which PilC rotates as it assembles the helical pilus fiber. It is also known that N. gonorrhoeae T4Ps extend and retract in increments smaller than the length of one helical turn (52), which is most easily explained by a mechanism with more than one step per turn, corresponding to the 1-start assembly mechanism. Another reason to favor models that involve rotation of some component such as PilC is that the homologous archaeal flagellar motor clearly begins to rotate its flagellar filament once this filament has been assembled (53).
In our architectural model of the piliated basal body, the PilC dimer rests on top of (and is presumed to interact with) two opposing PilB subunits in the ATPase hexamer, and the six NTDs of PilB in the ATPase hexamer are clamped in place by interacting with every second PilM subunit in the cytoplasmic ring (fig. S12A). We therefore propose that ATP hydrolysis by the pairs of opposing PilB subunits that contact the PilC dimer causes PilB NTD movements that rotate the asymmetrical PilC dimer. We predict the rotation of PilC to have two consequences: (i) It “scoops” new PilA subunits one at a time out of the membrane and onto the base of the pilus, and (ii) PilC is transferred to the next pair of opposing PilB subunits. The transmembrane segments of PilC likely mate with the tapered tip of the helical pilus fiber in such a way as to extend and anchor the pilus to the basal body, thereby creating a complete binding pocket for the hydrophobic tail of the next PilA subunit to be incorporated. Once a new PilA subunit is incorporated, PilB drives PilC around a fraction of a turn, pushing the pilus up a fraction of the length of a pilin subunit and recreating the binding pocket in the next available position.
One problem with this model is that the hexameric nature of PilB would predict elementary turns of 60° or 120°; neither would result in the exactly 3.6 subunits per turn needed to extend the pilus without any rotation. Perhaps the process does involve some slight rotations or Brownian motions of the proteins that would cause small slips in register. Pilus retraction would be accomplished by a switch from PilB to PilT. Thus, although the exact structure of the PilC dimer and its orientation in the T4PM remain uncertain, the architecture alone implies that both PilB and PilT act as ratchets biasing the assembly or disassembly process: While PilB holds the PilC-tip pocket on the next empty position of the pilus fiber until it is filled, and then quickly rotates to prevent subsequent dissociation, PilT holds the PilC-tip pocket on the last subunit of the tip until it dissociates back into the membrane, after which it quickly rotates to prevent reassociation.
The PilN-PilO and PilM rings likely sense pilus retraction signals and guide ATPase selection
Previous studies have shown that pilus retraction is induced by adding pilin-binding substrates (54) or pulling on the pilus directly (52). Recent studies with antibodies showed that tension induces conformational changes in the pilus itself (55), which could propagate into the basal body. Substrate binding may also induce similar conformational changes. Our results point to a model in which the alignment complex is an IM-crossing transmission module: Pilus retraction signals carried by the pilus itself into the basal body could be sensed in the periplasm by the PilN-PilO ring and then transmitted via the coiled-coil domains through the IM to modulate the conformation of the PilM ring, which in turn governs which ATPase is bound. This model rationalizes the recent report that conformational changes are required in the coiled-coil domains of PilN-PilO during the transition between T4P extension and retraction, which suggests that the alignment complex is not simply a static connector between IM and OM components, but instead plays a critical role in T4P dynamics (56).
The power of combined structural approaches to dissect complicated molecular machines
Solving the structures of large macromolecular machines is challenging. Traditional structural methods such as x-ray crystallography, nuclear magnetic resonance spectroscopy, and single-particle cryo–electron microscopy can deliver near-atomic resolution, but they all rely on purified samples. Many important biological structures such as flagellar motors (57), chemoreceptor arrays (58), and the T4PM studied here, however, may never be purifiable in a native state; we found that even the PilQ (fig. S4), PilC (fig. S10), and PilM-PilN-PilO (fig. S14) subcomplexes lose their native structure when purified. As a result, only structures of isolated subunits and small subcomplexes have been determined. Cryo–electron tomography can reveal at least the architectures of these large machines in situ, as our work has shown. When there is sufficient additional information about the structures and connectivities of the components, working models can be built that provide a path toward structural understanding, where both the structural relationships and the new mechanistic insights they suggest can then be tested.
Supplementary Material
Acknowledgments
We thank C. Oikonomou and D. Ortega for discussions. Supported by NIH grant R01 GM094800B (G.J.J.), the Howard Hughes Medical Institute, the Max Planck Society, and the Deutsche Forschungsgemeinschaft within the framework of the Collaborative Research Center (SFB) 987 “Microbial Diversity in Environmental Signal Response.” The 18 subtomogram averages of T4PMs reported in this study have been deposited in the Electron Microscopy Data Bank with accession numbers EMD-3247 (wild type, piliated); EMD-3248 (wild type, empty); EMD-3249 (ΔpilP, empty); EMD-3250 (PilP-sfGFP, piliated); EMD-3251 (PilP-sfGFP, empty); EMD-3252 (ΔtsaP, piliated); EMD-3253 (ΔtsaP, empty); EMD-3254 (PilO-sfGFP, piliated); EMD-3255 (PilO-sfGFP, empty); EMD-3256 (ΔpilC, empty); EMD-3257 (ΔpilA, empty); EMD-3258 (ΔpilV pilW fimU 1+2+3, empty); EMD-3259 (ΔpilT, piliated); EMD-3260 (ΔpilB, empty); EMD-3261 (ΔpilB ΔpilT, empty); EMD-3262 (ΔpilM, empty); EMD-3263 (ΔpilQβ1–β2, piliated); and EMD-3264 (ΔpilQβ1–β2, empty). The coordinates of the hypothetical T4PM working models have been deposited in the Protein Data Bank with accession numbers 3JC8 (piliated) and 3JC9 (empty), respectively.
Footnotes
Author contributions: Y.-W.C. collected the cryo–electron tomography data, which were analyzed by Y.-W.C. and L.A.R.; Y.-W.C. built the T4PM models and generated the movie describing the modeling process; A.T.-L. and L.S.-A. provided the M. xanthus strains and characterized their motility and T4PM component accumulation and localization; J.I. produced the animation of T4PM dynamics; and Y.-W.C., L.S.-A., and G.J.J. wrote the paper.
References
- 1.Korotkov KV, Sandkvist M, Hol WGJ. The type II secretion system: Biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 2012;10:336–351. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jarrell KF, Albers S-V. The archaellum: An old motility structure with a new name. Trends Microbiol. 2012;20:307–312. doi: 10.1016/j.tim.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Mattick JS. Type IV pili and twitching motility. Annu. Rev. Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 4.Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2004;2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 5.Evans KJ, Lambert C, Sockett RE. Predation by Bdellovibrio bacteriovorus HD100 requires type IV pili. J. Bacteriol. 2007;189:4850–4859. doi: 10.1128/JB.01942-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2004;2:241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 7.Klausen M, Aaes-Jørgensen A, Molin S, Tolker-Nielsen T. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 2003;50:61–68. doi: 10.1046/j.1365-2958.2003.03677.x. [DOI] [PubMed] [Google Scholar]
- 8.O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 9.Hager AJ, et al. Type IV pili-mediated secretion modulates Francisella virulence. Mol. Microbiol. 2006;62:227–237. doi: 10.1111/j.1365-2958.2006.05365.x. [DOI] [PubMed] [Google Scholar]
- 10.Merz AJ, So M, Sheetz MP. Pilus retraction powers bacterial twitching motility. Nature. 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 11.Skerker JM, Berg HC. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. U.S.A. 2001;98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craig L, Li J. Type IV pili: Paradoxes in form and function. Curr. Opin. Struct. Biol. 2008;18:267–277. doi: 10.1016/j.sbi.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morand PC, et al. Type IV pilus retraction in pathogenic Neisseria is regulated by the PilC proteins. EMBO J. 2004;23:2009–2017. doi: 10.1038/sj.emboj.7600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maier B, et al. Single pilus motor forces exceed 100 pN. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16012–16017. doi: 10.1073/pnas.242523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clausen M, Jakovljevic V, Søgaard-Andersen L, Maier B. High-force generation is a conserved property of type IV pilus systems. J. Bacteriol. 2009;191:4633–4638. doi: 10.1128/JB.00396-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pelicic V. Type IV pili: E pluribus unum? Mol. Microbiol. 2008;68:827–837. doi: 10.1111/j.1365-2958.2008.06197.x. [DOI] [PubMed] [Google Scholar]
- 17.Siewering K, et al. Peptidoglycan-binding protein TsaP functions in surface assembly of type IV pili. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E953–E961. doi: 10.1073/pnas.1322889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Craig L, et al. Type IV pilus structure by cryo-electron microscopy and crystallography: Implications for pilus assembly and functions. Mol. Cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Burrows LL. Pseudomonas aeruginosa twitching motility: Type IV pili in action. Annu. Rev. Microbiol. 2012;66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 20.Pédelacq J-D, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 21.Friedrich C, Bulyha I, Søgaard-Andersen L. Outside-in assembly pathway of the type IV pilus system in Myxococcus xanthus. J. Bacteriol. 2014;196:378–390. doi: 10.1128/JB.01094-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bulyha I, et al. Regulation of the type IV pili molecular machine by dynamic localization of two motor proteins. Mol. Microbiol. 2009;74:691–706. doi: 10.1111/j.1365-2958.2009.06891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgiadou M, Castagnini M, Karimova G, Ladant D, Pelicic V. Large-scale study of the interactions between proteins involved in type IV pilus biology in Neisseria meningitidis: Characterization of a subcomplex involved in pilus assembly. Mol. Microbiol. 2012;84:857–873. doi: 10.1111/j.1365-2958.2012.08062.x. [DOI] [PubMed] [Google Scholar]
- 24.Li C, Wallace RA, Black WP, Li YZ, Yang Z. Type IV pilus proteins form an integrated structure extending from the cytoplasm to the outer membrane. PLOS ONE. 2013;8:e70144. doi: 10.1371/journal.pone.0070144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tammam S, et al. PilMNOPQ from the Pseudomonas aeruginosa type IV pilus system form a transenvelope protein interaction network that interacts with PilA. J. Bacteriol. 2013;195:2126–2135. doi: 10.1128/JB.00032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balasingham SV, et al. Interactions between the lipoprotein PilP and the secretin PilQ in Neisseria meningitidis. J. Bacteriol. 2007;189:5716–5727. doi: 10.1128/JB.00060-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayers M, et al. PilM/N/O/P proteins form an inner membrane complex that affects the stability of the Pseudomonas aeruginosa type IV pilus secretin. J. Mol. Biol. 2009;394:128–142. doi: 10.1016/j.jmb.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 28.Tammam S, et al. Characterization of the PilN, PilO and PilP type IVa pilus subcomplex. Mol. Microbiol. 2011;82:1496–1514. doi: 10.1111/j.1365-2958.2011.07903.x. [DOI] [PubMed] [Google Scholar]
- 29.Gu S, et al. Solution structure of homology region (HR) domain of type II secretion system. J. Biol. Chem. 2012;287:9072–9080. doi: 10.1074/jbc.M111.300624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sampaleanu LM, et al. Periplasmic domains of Pseudomonas aeruginosa PilN and PilO form a stable heterodimeric complex. J. Mol. Biol. 2009;394:143–159. doi: 10.1016/j.jmb.2009.09.037. [DOI] [PubMed] [Google Scholar]
- 31.Karuppiah V, Derrick JP. Structure of the PilM-PilN inner membrane type IV pilus biogenesis complex from Thermus thermophilus. J. Biol. Chem. 2011;286:24434–24442. doi: 10.1074/jbc.M111.243535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korotkov KV, et al. Structural and functional studies on the interaction of GspC and GspD in the type II secretion system. PLOS Pathog. 2011;7:e1002228. doi: 10.1371/journal.ppat.1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karuppiah V, Collins RF, Thistlethwaite A, Gao Y, Derrick JP. Structure and assembly of an inner membrane platform for initiation of type IV pilus biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E4638–E4647. doi: 10.1073/pnas.1312313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karuppiah V, Hassan D, Saleem M, Derrick JP. Structure and oligomerization of the PilC type IV pilus biogenesis protein from Thermus thermophilus. Proteins. 2010;78:2049–2057. doi: 10.1002/prot.22720. [DOI] [PubMed] [Google Scholar]
- 35.Abendroth J, et al. The three-dimensional structure of the cytoplasmic domains of EpsF from the type 2 secretion system of Vibrio cholerae. J. Struct. Biol. 2009;166:303–315. doi: 10.1016/j.jsb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abendroth J, Murphy P, Sandkvist M, Bagdasarian M, Hol WGJ. The X-ray structure of the type II secretion system complex formed by the N-terminal domain of EpsE and the cytoplasmic domain of EpsL of Vibrio cholerae. J. Mol. Biol. 2005;348:845–855. doi: 10.1016/j.jmb.2005.02.061. [DOI] [PubMed] [Google Scholar]
- 37.Yamagata A, Tainer JA. Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. EMBO J. 2007;26:878–890. doi: 10.1038/sj.emboj.7601544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Misic AM, Satyshur KA, Forest KT. P. aeruginosa PilT structures with and without nucleotide reveal a dynamic type IV pilus retraction motor. J. Mol. Biol. 2010;400:1011–1021. doi: 10.1016/j.jmb.2010.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satyshur KA, et al. Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility. Structure. 2007;15:363–376. doi: 10.1016/j.str.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu C, et al. Hexamers of the type II secretion ATPase GspE from Vibrio cholerae with increased ATPase activity. Structure. 2013;21:1707–1717. doi: 10.1016/j.str.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berry J-L, et al. Structure and assembly of a transperiplasmic channel for type IV pili in Neisseria meningitidis. PLOS Pathog. 2012;8:e1002923. doi: 10.1371/journal.ppat.1002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reichow SL, Korotkov KV, Hol WGJ, Gonen T. Structure of the cholera toxin secretion channel in its closed state. Nat. Struct. Mol. Biol. 2010;17:1226–1232. doi: 10.1038/nsmb.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kowal J, et al. Structure of the dodecameric Yersinia enterocolitica secretin YscC and its trypsin-resistant core. Structure. 2013;21:2152–2161. doi: 10.1016/j.str.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 44.Tosi T, et al. Structural similarity of secretins from type II and type III secretion systems. Structure. 2014;22:1348–1355. doi: 10.1016/j.str.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 45.Wu SS, Kaiser D. Genetic and functional evidence that Type IV pili are required for social gliding motility in Myxococcus xanthus. Mol. Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- 46.Nguyen Y, et al. Pseudomonas aeruginosa minor pilins prime type IVa pilus assembly and promote surface display of the PilY1 adhesin. J. Biol. Chem. 2015;290:601–611. doi: 10.1074/jbc.M114.616904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sauvonnet N, Vignon G, Pugsley AP, Gounon P. Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J. 2000;19:2221–2228. doi: 10.1093/emboj/19.10.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cisneros DA, Bond PJ, Pugsley AP, Campos M, Francetic O. Minor pseudopilin self-assembly primes type II secretion pseudopilus elongation. EMBO J. 2012;31:1041–1053. doi: 10.1038/emboj.2011.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takhar HK, Kemp K, Kim M, Howell PL, Burrows LL. The platform protein is essential for type IV pilus biogenesis. J. Biol. Chem. 2013;288:9721–9728. doi: 10.1074/jbc.M113.453506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chami M, et al. Structural insights into the secretin PulD and its trypsin-resistant core. J. Biol. Chem. 2005;280:37732–37741. doi: 10.1074/jbc.M504463200. [DOI] [PubMed] [Google Scholar]
- 51.Nunn D. Bacterial type II protein export and pilus biogenesis: More than just homologies? Trends Cell Biol. 1999;9:402–408. doi: 10.1016/S0962-8924(99)01634-7. [DOI] [PubMed] [Google Scholar]
- 52.Clausen M, Koomey M, Maier B. Dynamics of type IV pili is controlled by switching between multiple states. Biophys. J. 2009;96:1169–1177. doi: 10.1016/j.bpj.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Albers S-V, Jarrell KF. The archaellum: How Archaea swim. Front. Microbiol. 2015;6:23. doi: 10.3389/fmicb.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y, et al. Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5443–5448. doi: 10.1073/pnas.0836639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Biais N, Higashi DL, Brujić J, So M, Sheetz MP. Force-dependent polymorphism in type IV pili reveals hidden epitopes. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11358–11363. doi: 10.1073/pnas.0911328107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leighton TL, Dayalani N, Sampaleanu LM, Howell PL, Burrows LL. Novel role for PilNO in type IV pilus retraction revealed by alignment subcomplex mutations. J. Bacteriol. 2015;197:2229–2238. doi: 10.1128/JB.00220-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen S, et al. Structural diversity of bacterial flagellar motors. EMBO J. 2011;30:2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Briegel A, et al. New insights into bacterial chemoreceptor array structure and assembly from electron cryotomography. Biochemistry. 2014;53:1575–1585. doi: 10.1021/bi5000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.