Abstract
With the advent of modern day high-throughput technologies, the bottleneck in biological discovery has shifted from the cost of doing experiments to that of analyzing results. clubber is our automated cluster-load balancing system developed for optimizing these “big data” analyses. Its plug-and-play framework encourages re-use of existing solutions for bioinformatics problems. clubber’s goals are to reduce computation times and to facilitate use of cluster computing. The first goal is achieved by automating the balance of parallel submissions across available high performance computing (HPC) resources. Notably, the latter can be added on demand, including cloud-based resources, and/or featuring heterogeneous environments. The second goal of making HPCs user-friendly is facilitated by an interactive web interface and a RESTful API, allowing for job monitoring and result retrieval. We used clubber to speed up our pipeline for annotating molecular functionality of metagenomes. Here, we analyzed the Deepwater Horizon oil-spill study data to quantitatively show that the beach sands have not yet entirely recovered. Further, our analysis of the CAMI-challenge data revealed that microbiome taxonomic shifts do not necessarily correlate with functional shifts. These examples (21 metagenomes processed in 172 min) clearly illustrate the importance of clubber in the everyday computational biology environment.
Keywords: cluster job scheduler, high performance computing, job management, load balancing
1. Introduction
Fast-paced growth of high performance computing (HPC), coupled with the recent appearance of new cloud computing solutions, created a new scope of possibilities for applications in today’s science. At the same time, more advanced and less expensive high throughput experimental assays have led to exponential growth of new biological datasets. Having access to sufficient computational resources to deal with the growing “big data” is therefore essential not only for computational, but also for experimental biology research labs, particularly those working in genomics. Less than two decades ago the first human genome took 13 years and $2.7 billion to sequence [1]. Today sequencing a genome takes a day and $1000, with costs projected to go even lower in the near future. Recent projects like the 1000 Genomes Project [2] and others currently under way [3], [4] will provide the field with an unprecedented amount of data, opening up new possibilities to significantly improve current models and tools.
These developments come at a cost, as traditional HPC is quite expensive both in purchase and maintenance. Research labs espouse different models for dealing with this computing need – some have their own computational power, others share machines across an institute or outsource their computing to collaborators. Although usability varies significantly across setups, compute nodes rarely reach the often-targeted utilization rates of 75–85 % consistent workload. Usage usually peaks with a specific high priority project running on the cluster for a limited time or with short-term jobs submitted through a web interface, where timing and responsiveness are essential. Cloud computing offers new alternatives, but is not always an adequate replacement for traditional HPC. The nature of cloud solutions often creates new challenges, such as the transfer of enormous amounts of data to and from the remote cloud storage.
Both from time and performance points of view, there is a clear advantage in making use of all available computational resources when necessary. However, this is a considerable challenge as the, often distributed and setup-disparate, clusters have distinct runtime pre-requisites. Ideally all resources would “speak” the same language, i.e. have a shared common base (OS, executables, job scheduler, etc.) Existing tools [5], [6] for bringing together disjoint computational resources and for distributing jobs among them require significant IT-related knowledge to get up and running. Moreover, none of these were designed explicitly for evaluations and approaches common in computational biology. Their capability is mostly limited to retrieval of job results from compute clusters and does not extend to downstream processing. Thus, post-processing and publishing of results is not automated and has to be dealt with individually.
Here we describe our novel clubber (CLUster-load Balancer for Bioinformatics E-Resources) framework, available at http://services.bromberglab.org/clubber. clubber is designed specifically to facilitate and accelerate common computational biology experimental workflows and used in conjunction with existing methods or scripts to efficiently process large-scale datasets. Using clubber is as simple as downloading and installing Docker [7], a software container platform available for every environment, and using a single command to run the Docker-cloud clubber container [8]. From there, any interaction for basic configuration, job submission, monitoring and displaying results is achieved via the clubber web interface. Note that clubber can also be run from command-line using an interactive console, or from within a Python project by importing the clubber package. Due to our method’s modular design, all of its main components (Manager, Database, Web Interface) can run separately on different environments/machines. Further, clubber can be easily configured to use any of the databases or webservers and thus to directly integrate into existing external services. Results can be accessed directly from the clubber web interface, either as downloadable files or as searchable data tables (given an appropriate output format). A RESTful [9] API provides programmatic access to the jobs managed by clubber, enabling other frameworks to monitor individual job progress and retrieve and display the final results. Very importantly, the clubber API facilitates integration into existing and new web services; i.e. tasks submitted through a web interface can be simply “handed over” to clubber and results queried once available. clubber can be set up on a dedicated server to be accessible by all members of a research group or by a selected few authenticated via a built-in user authentication module.
Existing workflow frameworks like Galaxy [10] and Nextflow [11] allow users to create computational pipelines to process and analyze biological data. Although both environments are highly usable, they have some limitations. Galaxy, for example, requires some time for setup of all components and limits the selection of available tools to those for which corresponding plugins have been written. Nextflow, on the other hand, has limited data filtering and visualization capabilities. Further, both tools can be configured to run jobs on a remote cluster, and Galaxy additionally provides means to make results accessible via a web interface. However, in both cases, jobs are submitted sequentially to only one previously configured cluster. Distributing jobs to multiple resources requires manual interaction and, potentially, adaptation of the necessary submission scripts. This leads to extensive computation times, directly correlated with the amount of processed data.
We designed clubber to deal with the challenges of growing datasets, which are particularly obvious in genome research. The current clubber package includes built-in methods to simplify parallelized job submission, e.g. splitting a single multi-sequence input file to submit parallel jobs, each containing a user-defined number of sequences. All of these features make clubber an essential tool for processing and analyses of vast amounts of biological data in a parallel, efficient, and (very) fast fashion.
2. Methods
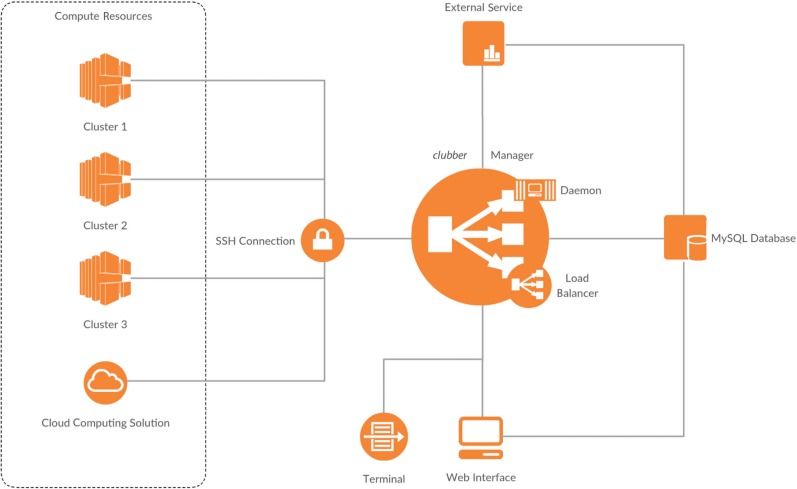
clubber works in all environments and integrates seamlessly with existing workload managers. We made clubber available as a ready-to-launch Docker image. Adding a computing resource (an HPC cluster) requires only a valid username and password combination for a user who is eligible to submit jobs on this specific resource. Note that there is no need for any additional software to be installed on these resources. The standalone clubber python installation has only two requirements: (i) access to a MySQL database (version 5.x) and (ii) availability of python (version 3.x). The optional web interface additionally requires access to a webserver with installed PHP module (version 5.6.x). Detailed installation instructions and sources can be found online [8]. Figure 1 illustrates the clubber workflow. clubber’s three components, Manager, Database, and Web Interface, are independent from each other. The Manager accesses registered clusters via Secure Shell (SSH) and communicates with the Database using MySQL queries. The Web Interface interacts with the Database to register jobs, monitor their progress and retrieve results.
Figure 1:
The clubber pipeline. Jobs can be submitted either through a web interface or via command-line to the clubber manager. These are registered and managed using a relational database. The manager uses an automated balancing approach to distribute jobs among available clusters; the manager daemon runs locally and communicates with available clusters, transferring completed job results and storing them locally or, optionally, in the database.
clubber bundles computational resources, providing an interface for a simple centralized submission. clubber can be used in two different ways: through an interactive web interface or via command-line. First, a clubber project is created, defining basic parameters like project name, selection of clusters to use and the environment variables necessary for job submission. Projects can contain binaries or database files required by the associated jobs. Note that single jobs can be submitted without creating a project; these will automatically be assigned to a default project with no environment variables set. After a project has been created and automatically initiated on the specified clusters, jobs can be submitted using the web interface or from command-line. Additional environment and job specific variables are defined in a simple syntax described in the clubber documentation. The manager uses an auto balancing approach to automatically distribute new jobs between registered clusters. Three factors determine how many jobs are submitted to each cluster during the auto balancing process. These are, in decreasing priority: (i) the cluster workload, (ii) the expected queuing time and (iii) the average job runtime. Cluster workload is calculated as a percentage of total possible workload, with 100 % representing a fully occupied cluster. The expected queuing time and the average job runtime are normalized to a [0,1] range, with one representing the maximum amount of time spent in either queue or run state, respectively, over all jobs of the same project among all active clusters. Both factors are set to one by default and are updated automatically during the progression of a project. In order to obtain the cluster specific load balancing factor (LBF) they are combined with the respective cluster workload (Eq. 1).
| (1) |
clubber communicates with clusters exclusively via encrypted SSH. The rsync [12] utility and Secure Copy (SCP) are used to transfer files to and from the clusters. Since some of the inquiries sent to the many clusters take minutes to process, all communication is threaded to avoid blocking faster transactions. This architecture enables clubber to efficiently distribute and retrieve jobs in a highly parallelized fashion.
To track and update current job states clubber relies on a relational database. This approach results in very robust job exception handling, both regarding errors on remote clusters and exceptions like lost connections on the machines running the clubber manager. The database also allows independent services, which use clubber as a job manager, to monitor current job states and retrieve results. Job success is continuously and extensively validated, ensuring that a project with millions of jobs is completed correctly even after allowing for power failures and compute node breakdowns. Once a job is identified as finished, the validation pipeline ensures that the expected results are present and correctly retrieved from the clusters. In case of errors, jobs are reset and re-sent out for computation. A detailed logging and notification module tracks these processes and notifies the user if specific jobs produce recurring errors.
clubber is designed to be used with existing software tools. Our plug-and-play framework makes it possible to use any existing tools or scripts within the clubber environment. User-defined specific pre- and post-processing actions can also be re-used with clubber projects. This allows for manipulation of input data prior to batch processing (e.g. converting fastq to fasta format) and for automatic processing of job results once they have been retrieved from the clusters. In its initial release, clubber includes two built-in methods for specific pre- and post-processing to simplify parallelized job submission. They allow to automatically split a single multi-sequence input file to submit parallel jobs and merge results once all jobs have been computed. The number of sequences used for each parallel job is user-defined. We expect that with increasing use of clubber (available as Git repository hosted on bitbucket) [8], the community will produce a larger repertoire of common pre- and post-processing tools, e.g. file conversion, filtering, etc., commonly applied in every-day computational biology.
3. Results and Discussion
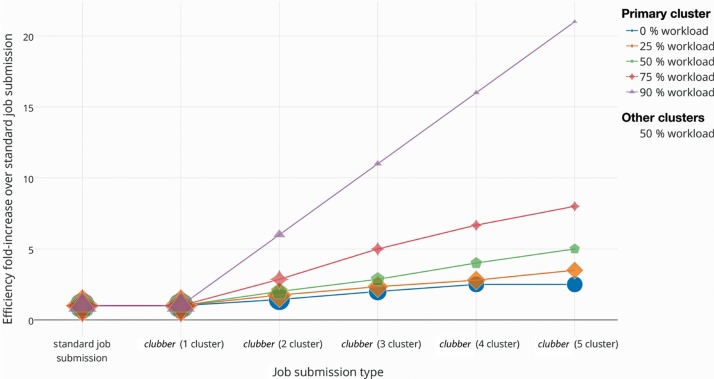
clubber significantly reduces the “real-world” compute time by parallelizing and optimizing the workload distribution across available resources. We evaluated clubber performance by measuring the time required to complete one thousand individual jobs, requiring 1-min CPU time each. Note, that these jobs did not require any data to be transferred to remote clusters. The evaluation was performed in various scenarios. We compared the required time at different cluster workloads when using clubber with one to five separate clusters available vs. a standard job submission (Figure 2). A standard job submission is defined as a manual submission of a single shell script running all thousand jobs on a single local HPC cluster. Note that workloads for remote clusters registered with clubber are conservatively estimated to be consistently at 50 %; the actual gain in computation efficiency could be substantially higher. Also note that 0 % workload is here defined as the ability to run at most 100 jobs in parallel. For the (ideal, but also rare) case of no (0 %) workload on the local cluster, only two additional registered clusters, both exhibiting a workload average, reduce the overall computation time by approximately 50 %. The total gain in computation time is directly correlated to the current workloads on the remote clusters. clubber’s auto-balancing job submission ensures that clusters with a low workload are preferentially selected, optimizing and reducing to a minimum the total required computation time. As expected, the more clusters are registered with clubber the less effect single clusters with a high load have on the final computation time. The advantage of using clubber is particularly obvious in a scenario where only one cluster is available for computation vs. having two clusters – a local and one additional remote cluster. Using clubber speeds up computation by up to 100 %. Note that simply logging into another cluster and submitting job subsets is tedious task, which would not, even in the best case scenario, achieve similar speed up – as one cluster finishes, the other is still only somewhat through its assigned computation.
Figure 2:
Efficiency fold-change of clubber vs. standard job submission: Efficiency fold increase in submitting jobs using clubber as compared to a standard job submission. Primary cluster workload is varied between 0 % and 90 %, where 0 % workload is defined here as the ability to run at most 100 jobs in parallel (100 CPU cluster). Compute time is measured for a submission of 1000 jobs, each requiring 1-min CPU time and no data transfer. Active workloads for remote clusters registered with clubber are conservatively estimated to be consistently at 50 % of possible total. None of these clusters dropped below that threshold in our use experience. They have, however, gone significantly higher. Thus, the actual gain in computation efficiency could be even higher than that displayed.
clubber facilitates fast evaluation of millions of sequences. Our recent work required a total of 19.4 million bacterial sequences to be analyzed for all-to-all pairwise similarity using BLAST [13]. We estimated that our single local cluster of 640 compute cores in its entirety would have taken roughly 4 months to perform the computation. This estimate is based on a 24 day-long 3,797,793 job BLAST run against the 19.4 M sequence database. Using clubber to run on three additional clusters (800, 1536, and 3120 cores, respectively; of varied load, but no more than 50 % of any one cluster available at any given time), speeds up this time to a bit over 2 months (70 days, a factor of 1.8).
Deepwater oil spill metagenome analysis using mi-faser. Our lab’s recently created web service [14], mi-faser [15], uses clubber to rapidly annotate gigabytes of genomic sequence read data for the molecular functionality encoded by the “read-parent genes” without the need for assembly. For every input metagenome, mi-faser computes a function profile – a list of Enzyme Commission (EC) numbers and the associated read abundances. To illustrate clubber functionality, we ran mi-faser on 16 beach sand metagenomes from four phases of the Deepwater Horizon oil spill [16] (BioProject PRJNA260285) study – Pre-oil, two samples of Oil, and Recovery phases (available at http://services.bromberglab.org/mifaser/example). Analysis of this data (73GB sequence reads) using the mi-faser web interface with a clubber back-end was done in only 1 h, with clubber distributing a total of 4.5 k jobs among three compute clusters. Note that running these jobs using only our local cluster (640 cores) with an average workload (unavailability of nodes) of 30 %, took 170 min – 3-fold slower than clubber.
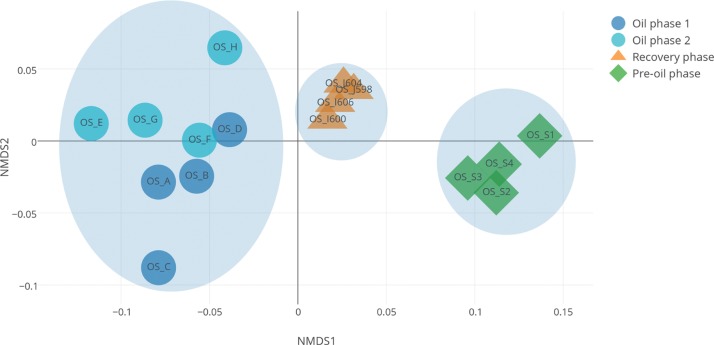
For further analysis, we removed sample-specific functions and normalized the individual entries of the function profile vectors by the total number of annotated reads. We found that microbiome functional profiles of samples from different phases significantly differ from each other (Figure 3, non-metric multidimensional scaling (NMDS) analysis [17]; P < 0.001, permanova test [18]). Interestingly, the samples from the Oil phases show higher variation than the samples from the Pre-oil phase and the Recovery phase, suggesting that “normal” ecosystem microbiomes are functionally more consistent than those in the disturbed ecosystems. The samples from Oil phases are functionally closer to the samples from the Recovery phase than to the Pre-oil phase, indicating that the beach sands have likely not entirely recovered.
Figure 3:
Microbiome functional capabilities of beach sand metagenomes from a study of the Deepwater Horizon oil spill (16) (BioProject PRJNA260285) differ across phases. The samples were collected from four phases, including Pre-oil phase (OS-S1, OS-S2, OS-S3 and OS-S4), Oil phase 1 (OS-A, OS-B, OS-C and OS-D), Oil phase 2 (OS-E, OS-F, OS-G and OS-H) and Recovery phase (OS-I600, OS-I606, OS-J598 and OS-J604). The distances between samples in this non-metric multidimensional scaling (NMDS) graph represent the variation between sample function profiles. Samples from Pre-oil phase, Oil phases and Recovery phase localize separately. Oil phase samples are closer to Recovery phase samples than to Pre-oil phase samples.
Regardless of the significant differences between phases, the fraction of housekeeping functions (compiled from [19]) was highly consistent across samples (22.1±0.5 %); e.g. DNA-directed RNA polymerase (2.7.7.6) is the most abundant function in all samples (about 4∼5 %). As the number of reads encoding a particular functionality is highly correlated to the number of individual cells performing said functionality, these results are not very surprising – all bacterial phyla, no matter how different, carry housekeeping genes. This finding serves as a confirmation of mi-faser’s accuracy, while highlighting its ability to estimate functional diversity in a non-taxon dependent level.
Critical Assessment of Metagenomic Interpretation (CAMI) challenge analysis using mi-faser. We further used mi-faser to evaluate a high complexity data set from the CAMI [20] challenge. The data set contains a time series of five Hiseq samples (15 Gbp each) with small insert sizes sampled from a complex microbial community. With clubber optimizing job submissions, the total computation time for 500 M sequence reads was only 1 h 59 min. Note that the CAMI challenge did not evaluate runtimes for the submitted tools/predictions, but they note that this evaluation is a necessary feature of future method development [20]. Metagenome comparative analysis revealed that the microbiome functional profiles remain highly consistent (Table 1), regardless of a clear community composition shift (Table 2). Interestingly, these results indicate that, over time, microbial species were exchanged, while maintaining the same functional capacity. Thus, the time effect on the microbial community is not as striking as what the taxonomical changes would suggest. This example highlights the fact that inferring microbiome function from its taxonomy composition is misleading. Thus, metagenomic analysis tools such as mi-faser are essential for a deeper understanding of microbiome functional potentials. Note that clubber is uniquely responsible for allowing our lab to make the mi-faser web interface available to the general public for the purposes of extremely fast (and accurate) functional annotation of millions of raw sequence reads.
Table 1:
Spearman correlation between taxonomic profilesa of CAMI metagenomes.
| RH_S001 | RH_S002 | RH_S003 | RH_S004 | RH_S005 | |
|---|---|---|---|---|---|
| RH_S001 | 1 | – | – | – | – |
| RH_S002 | 0.78 | 1 | – | – | – |
| RH_S003 | 0.64 | 0.75 | 1 | – | – |
| RH_S004 | 0.51 | 0.59 | 0.73 | 1 | – |
| RH_S005 | 0.45 | 0.51 | 0.54 | 0.71 | 1 |
aThe taxonomic profiles were obtained from http://cami-challenge.org.
Table 2:
Spearman correlation between functional profilesa of CAMI metagenomes.
| RH_S001 | RH_S002 | RH_S003 | RH_S004 | RH_S005 | |
|---|---|---|---|---|---|
| RH_S001 | 1 | – | – | – | – |
| RH_S002 | 0.99 | 1 | – | – | – |
| RH_S003 | 0.99 | 0.99 | 1 | – | – |
| RH_S004 | 0.99 | 0.99 | 0.99 | 1 | – |
| RH_S005 | 0.99 | 0.99 | 0.99 | 0.99 | 1 |
aThe functional profiles were annotated by mi-faser (15).
Dealing with tool heterogeneity in clubber-accessible resources. Even though clubber is highly successful in facilitating HPC use, there may be still scenarios, which require manual interaction with the individual compute clusters. When creating a clubber project that includes binaries, the user has to validate these binaries on each of the cluster resources. When using pre-installed tools local to each resource, all installs have to be of the same version and produce identical results given identical input. To prevent erroneous results in these scenarios, clubber offers the option to automatically compare cluster environments and submit test jobs before starting a project run on different computing resources. Note that virtualization solutions, e.g. Docker, offer a simple solution to these problems by guaranteeing identical environments on every resource. In this scenario (planned for the next release of our software) clubber distributes a user provided Docker image to the clusters and relays job parameters when starting a Docker container.
Impact of dataset size on clubber performance. clubber was developed to process extremely large datasets using remotely accessed resources. The remoteness of these resources, thus, poses a bottleneck in transferring data between compute clusters. For the larger compute centers, it is safe to assume that an appropriately fast connection is available. For smaller set-ups, data transfer speeds may vary. In testing to evaluate the contribution of transfer times for our collection of clusters, some smaller and some larger ones, we found that times did not vary across remote and local machines and did not affect the relative performance. For all five of our clusters the transfer times varied by as little as 6 %, despite being located in different places of the world (New Brunswick, NJ, USA and Garching, Germany); the speed of transfer of 1Gb of data was 146 ± 8 s. Note that jobs requiring large data transfers would necessarily be slowed down, but roughly in equal measure for local or remote machines. The slow-down is especially visible in cases where the computation time for a single job is fairly short. Increasing the number of jobs processed reduces this initial impact as performance improves by use of additional resources.
Better resource management and faster processing speeds with clubber. Our clubber framework provides a simple way to bundle available, possibly heterogeneous, computational resources and to distribute computations minimizing the required processing time. This approach avoids long computation times associated with an overloaded local cluster when there are in fact additional resources available elsewhere. Simple job submission/monitoring and automated exception handling make clubber easy-to-use and ideal for handling projects with millions of jobs. Its ability to use cloud-computing services like Amazon Web Services (AWS) with clubber on-demand, additionally allows for temporary, large-scale increases in computational resources. With all of these features, web services, the bread-and-butter of the computational biology community, are made extremely responsive with clubber.
With the exponential growth of available data in computational biology waiting to be analyzed, bioinformatics, not experimental analysis, has unexpectedly become the progress bottleneck. By combining the available resources and using them in the most optimal fashion, clubber offers a new approach to tackling this challenge.
Acknowledgements
We thank Max Haggblom, Yannick Mahlich, Alexandra Pushkar, and Yanran Wang (all Rutgers University, New Brunswick, NJ, USA) for many discussions and manuscript review. We thank Bill Abbott and Kevin Abbey (both Rutgers) for technical support. We are grateful to Sonakshi Bhattacharjee (Technical University of Munich, TUM) for advice during the initial development phase. Particular thanks are due to Burkhard Rost (TUM) for his hospitality, valuable discussions, and letting us use the rostlab cluster! We also thank Timothy Karl (TUM), for his help with the rostlab cluster setup within the clubber environment and the rostlab members for quickly adapting clubber after evaluating its functionality. Last but not least, we thank all those who deposit their experimental data in public databases and those who maintain these databases. YB, MM, and CZ were partially supported by the NIH/NIGMS grant U01 GM115486 (to YB). YB and CZ were also supported by the NSF CAREER grant 1553289 (to YB). YB was additionally supported by USDA-NIFA 1015:0228906 grant and the TU München – Institute for advanced study Hans Fischer fellowship, funded by the German Excellence Initiative and the EU Seventh Framework Programme, grant agreement 291763.
Contributor Information
Maximilian Miller, Email: mmiller@bromberglab.org.
Yana Bromberg, Email: yana@bromberglab.org.
Conflict of interest statement:
Authors state no conflict of interest. All authors have read the journal’s publication ethics and publication malpractice statement available at the journal’s website and hereby confirm that they comply with all its parts applicable to the present scientific work.
References
- [1].Gyles C. The DNA revolution. Canadian Vet J. 2008;49:745–6. [PMC free article] [PubMed] [Google Scholar]
- [2].The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].100K Food Pathogen Project: Bart Weimer. 2016 Available from: https://www.ncbi.nlm.nih.gov/bioproject/186441.
- [4].McGrath JA. Rare inherited skin diseases and the Genomics England 100 000 Genome Project. Br J Dermatol. 2016;174:257–8. doi: 10.1111/bjd.14362. [DOI] [PubMed] [Google Scholar]
- [5].Thain D, Tannenbaum T, Livny M. Distributed computing in practice: the Condor experience. Concurr Comp Pract Ex. 2005;17:323–56. [Google Scholar]
- [6].Weitzel D, Sfiligoi I, Bockelman B, Frey J, Wuerthwein F, Fraser D. et al. Accessing opportunistic resources with Bosco. J Phys Conf Ser. 2014;513:032105. [Google Scholar]
- [7]. [Accessed on: April 12th, 2017];Docker, the world’s leading software container platform: the Docker open source project. 2017 Available from: https://www.docker.com/
- [8]. [Accessed on: April 12th, 2017];clubber: Yana Bromberg Lab, Rutgers University. 2017 Available from: https://bitbucket.org/bromberglab/bromberglab-clubber/
- [9]. [Accessed on: April 12th, 2017];Web Services Architecture: World Wide Web Consortium. 2004 Available from: https://www.w3.org/TR/2004/NOTE-ws-arch-20040211/ – relwwwrest.
- [10]. [Accessed on: April 12th, 2017];Galaxy: The Galaxy Project. 2017 Available from: https://galaxyproject.org/
- [11]. [Accessed on: April 12th, 2017];Nextflow: Comparative Bioinformatics group, Barcelona Center for Genomic Regulation (CRG) 2016 Available from: https://www.nextflow.io/
- [12]. [Accessed on: April 12th, 2017];rsync. 2015 Available from: https://rsync.samba.org/
- [13].Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- [14].Zhu C, Miller M, Marpaka S, Vaysberg P, Rühlemann M, Heinsen F-A. Functional sequencing read annotation for high precision microbiome analysis. doi: 10.1093/nar/gkx1209. Submitted, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15]. [Accessed on: April 12th, 2017];mi-faser: Yana Bromberg Lab, Rutgers University. 2017 Available from: http://services.bromberglab.org/mifaser.
- [16].Rodriguez-R LM, Overholt WA, Hagan C, Huettel M, Kostka JE, Konstantinidis KT. Microbial community successional patterns in beach sands impacted by the Deepwater Horizon oil spill. ISME J. 2015;9:1928–40. doi: 10.1038/ismej.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kruskal JB. Nonmetric multidimensional scaling: a numerical method. Psychometrika. 1964;29:115–29. [Google Scholar]
- [18].Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001;26:32–46. [Google Scholar]
- [19].Gil R, Silva FJ, Pereto J, Moya A. Determination of the core of a minimal bacterial gene set. Microbiol Mol Biol Rev. 2004;68:518–37. doi: 10.1128/MMBR.68.3.518-537.2004. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sczyrba A, Hofmann P, Belmann P, Koslicki D, Janssen S, Droege J. et al. Critical Assessment of Metagenome Interpretation − a benchmark of computational metagenomics software. 2017 Jan 9; doi: 10.1101/099127. [DOI] [PMC free article] [PubMed] [Google Scholar]