Abstract
Vespertilionid bats (Mammalia: Order Chiroptera) live 3–10 times longer than other mammals of an equivalent body size. At present, nothing is known of how bat fecal metabolic profiles shift with age in any taxa. This study established the feasibility of using a non-invasive, fecal metabolomics approach to examine age-related differences in the fecal metabolome of young and elderly adult big brown bats (Eptesicus fuscus) as an initial investigation into using metabolomics for age determination. Samples were collected from captive, known-aged big brown bats (Eptesicus fuscus) from 1 to over 14 years of age: these two ages represent age groups separated by approximately 75% of the known natural lifespan of this taxon. Results showed 41 metabolites differentiated young (n = 22) and elderly (n = 6) Eptesicus. Significant differences in metabolites between young and elderly bats were associated with tryptophan metabolism and incomplete protein digestion. Results support further exploration of the physiological mechanisms bats employ to achieve exceptional longevity.
Introduction
Studies of wild populations show that bats live longer than terrestrial mammals of comparable body size, and that the vespertilionid family of insectivorous bats displays extreme longevity [1,2] [Fig 1]. Notably, little- and big-brown bats (Myotis and Eptesicus) of the vespertilionid family, have a remarkable lifespan of 19–40+ years, a range that is 3–10 times longer than the expected longevity of size-comparable, non-volant mammals [3–6]. Most investigations into the mechanisms contributing to their extended lifespan are based on adult bats of unknown age as acquisition of known-aged animals requires long-duration banding-recapture studies or the maintenance of captive colonies over decades.
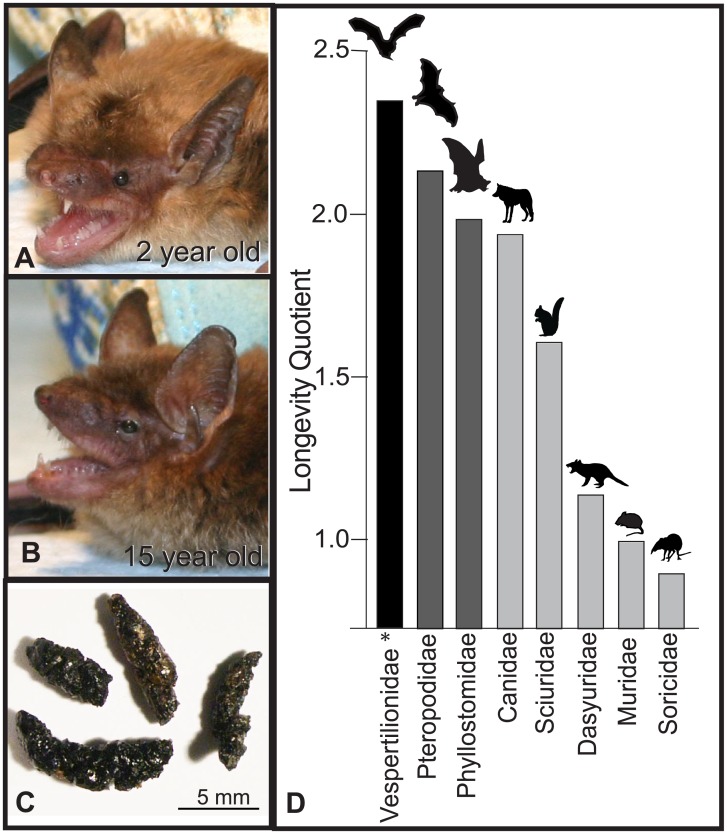
Fig 1. Vespertilionid bats, including big brown bats (Eptesicus fuscus), live 3-10x’s longer than equivalently-sized mammals but lack visible morphological characteristics indicative of age.
Eptesicus may undergo a thinning of pelage hair throughout life as evidenced by a comparison of two-year old (A) and fifteen-year-old (B) adults. Longevity quotients across mammals (D, modified from [1,7]). Longevity quotient (LQ) = Observed/Expected longevity (from NFM regression) *Vespertilionid bar graph in D indicates longevity of Eptesicus (20-year lifespan) and excludes Myotis (over a 40-year lifespan) as it was not available for this study.
Physiological decline that occurs during the natural aging process ultimately leads to an increased prevalence of illness and exposes animals to heightened susceptibility to death. Age-accumulated cellular damage contributes to detrimental alterations that affect a plethora of processes [8] including intercellular communication [9,10], stem cell availability [11] and epigenetic regulation [12]. Cellular aging is partially the result of imbalances in concentrations of pro-oxidants and anti-oxidants [13] and the rate of aging is partially determined by the failure of protein homeostasis [14]. Protein turnover must accommodate challenges associated with oxidation, unfolding, and misfolding. Imbalances can result in the accumulation of insoluble proteins, as is the case in Alzheimer’s and Parkinson’s diseases [15]. Compared to other mammals, bats appear to have increased resistance to cellular damage and cellular aging. The mitochondria of Myotis are more efficient, per unit oxygen consumed, than those of similarly sized terrestrial mammals [16]. Bat liver proteins display increased resistance to oxidative stress and urea-induced unfolding associated with radiation exposure compared to mice [14]. Compared to carnivores and long-lived rodents, the fibroblasts of bats are more resistant to cellular stress when challenged with heavy metals, peroxides, and heat [17–19]. Bats also display increased expression of miRNA-155, a micro-RNA essential for defense against oxidative-induced cellular damage [20], and show positive selection of genes associated with reducing the negative effects of oxidation (e.g., ATM, RAD50, KU80, MDM2 [21]). Furthermore, bats do not demonstrate loss of either protein homeostasis or muscle mass during hibernation [22]. Taken together, bats appear to lack novel modifications to protein repair, degradation and removal [14] and emphasize protein homeostasis.
Accumulation of genetic damage from exogenous and endogenous sources occurs during aging which negatively affects both nuclear and mitochondrial DNA [8,23]. The deleterious impact of this damage can include gene mutations [24], telomere shortening [25] and alteration in gene expression [26]. In bats, novelties in gene sequence and expression patterns probably promote genomic stability, prevent disease, and potentially contribute to their exceptional longevity. Evidence suggests bats promote genomic stability by preventing DNA damage to and pathologies associated with cancer. Specifically, bats display upregulation of genes associated with DNA repair (e.g., UVRAG), inhibition of cell proliferation and tumor formation (miRNA-16 and miRNA-143), and suppression of tumors (e.g., miRNA-101, BRCA1, BRCA2), as well as downregulation of a tumorigenesis promoter, miRNA-221 [20,21]. Furthermore, Myotis, like humans, do not express telomerase in either blood or fibroblasts and enhance DNA repair and prevent telomere shortening via expression of ATM and SETX genes [27]. Together, evidence suggests that bats uniquely promote genomic stability, DNA repair and cell proliferation with age to achieve exceptional longevity.
Research to explicate the mechanisms responsible for extending the health- and lifespan of bats expands our understanding of the natural aging process and the mechanisms used to delay senescence. Unfortunately, most research is comparative and biological innovations, such as unique miRNAs [20], identified in bats but not terrestrial mammals lack functional and phenotypic correlations. To fully understand the systematic and multi-scale approach bats use to combat aging and disease, scientists require access to known-aged animals. This study therefore addresses a critical gap in the field by utilizing a novel metabolomics approach to examine age-related differences in the fecal metabolomes of long-lived big brown bats (Eptesicus fuscus). By establishing the feasibility of utilizing a metabolomics approach to detect age-related differences in bat fecal samples, we lay the foundation for future studies to utilize this non-invasive approach to establish reference ranges in a larger cohort of animals and lay the groundwork for the utility of this method as an eventual means of age determination in bats.
Metabolomics measures low molecular weight compounds (metabolites) present in cells, tissues, or bio-fluids [28–30]. Examinations of excreted metabolites from known-aged bats may provide metabolic ‘snapshots’ of the changes that occur over the lifetime of a bat. When applied to fecal samples from wild-captured or captive-housed bats, metabolomics has the potential to provide insight into age-related differences in physiological processes. Previous metabolomics studies successfully identified age-related biomarkers for C. elegans, rodents, and humans [31,32] as well as biomarkers of human health [33,34]. Within bats, fecal metabolomics allowed for the study of the correlation between cortisol levels and environmental stress exposure in Isabelline Serotine bats (Eptesicus isabellinus; [35]), as well as track testosterone levels during the mating season, in a small, sac-winged bat (Saccopteryx; [36]). By utilizing fecal samples, these studies negated the need for collection of more invasive samples types, such as blood or teeth [36]. It could be that expansion of these methods to address age-related changes in excreted metabolites may eventually generate age-correlated candidate metabolites that offer insights into the mechanisms that uniquely extend the longevity of bats.
To provide insight into the age-related differences in the fecal metabolome of young and elderly adult big brown bats (Eptesicus fuscus) bats, this study utilized liquid chromatrography-quadrapole time-of-flight-mass spectrometry (LC-QTOF-MS) to compare the fecal metabolic profiles of long-lived Eptesicus (Fig 1). Eptesicus have a lifespan of approximately 20 years and our samples, taken from young (1–2 years) and elderly (14+ years) animals, analyzed variations in fecal metabolic profiles of samples separated by approximately 75% of this lifespan. Our work, although limited, significantly furthers the field by applying a metabolomics approach to show that analyses of fecal metabolic profiles can provide rapid, reliable, and non-invasive quantification of chemical biomarkers of aging. This ability to non-invasively compare differences in the fecal metabolome of young and elderly bats in a captive population lays the foundation for future experiments to apply metabolomics in a larger cohort of bats to identify age-specific biomarkers and elucidate specific signatures of senescence and aging mechanisms in bats. Moreover, fostering an understanding of physiological senescence in cardiac, skeletal, and sensory health in bats, may lead to a robust integration of aging and biomedical research.
Material & methods
The Institutional Animal Care and Use Committee (IACUC) of Northeast Ohio Medical University approved the fecal sample collection under Protocol #17-06-142.
Bats
The Cooper laboratory at Northeast Ohio Medical University (NEOMED; Rootstown, Ohio) maintains a known-aged colony of the big brown bat (Eptesicus fuscus; NEOMED IACUC #17-06-142) transferred from an original colony maintained by Dr. Ellen Covey at the University of Washington. While at the University of Washington, bats were banded according to year of capture and/or birth. These bats underwent natural hibernation and were exclusively fed an ab libitum diet of fresh water and meal worms (Tenebrio molitor) of known nutrient content [37]. Mealworm diet consists of non-organic bran supplemented with non-organic apple. These bats were transported to NEOMED and are housed indoors on a 12-hour light/dark artificial light cycle and fed the same ab libitum fresh water and meal worms diet [38].
Longevity quotient estimation
The data utilized to determine LQ of selected mammal species (N = 139) were from a previously published dataset [7]. Predicted longevity was calculated for each species using a linear regression (slope = 0.172; y-intercept = 0.638) fitted to the logged values of body mass (g) and maximum longevity for all non-flying mammals (NFM) following published methods [1]. The longevity quotient was determined following Austad and Fisher [1] as: observed longevity/expected longevity.
Fecal metabolomics
Fecal sample collection
Bats were isolated into individual cages overnight with ab libitum access to fresh water and meal worms. Feces (Fig 1C) were collected three times daily with each sample transferred to individual 1.5mL micro centrifuge tubes and stored immediately at -80°C. Fecal samples were acquired from a total of 28 individual bats ranging from 1 to greater than 15 years of age [1 year (n = 7), 2 years (n = 15), ≥ 14 years (n = 2), ≥ 15 years (n = 4)]. Although future work is aimed at establishing metabolic profiles for middle aged animals, at present, the sample size was too low to be included in this analysis. Samples were then shipped to the Nutritional Metabolomics Research lab at the Canadian Centre for Agri-food Research in Health and Medicine in Winnipeg, Manitoba at -80°C under the specifications of Canadian Food Inspection Agency import permit.
Extraction of metabolites
Frozen fecal samples were extracted, in duplicate, using a modified protocol adapted published protocols [39]. Bat fecal samples were thawed on ice. Aliquots (30mg) of each bat feces sample were mixed with 400 μl ultrapure water (Milli-Q H2O®, EMD Millipore, Billerica, Mass, USA) and homogenized for 1–2 min on ice using a Polytron PT 2600 ET Kinematica benchtop homogenizer (Fisher Scientific, Pittsburgh, Pa, USA). The probe was washed three times with distilled water between samples. After homogenization, samples were kept on ice while 3 μl of sodium azide (400μg/mL concentration; Sigma Aldrich, St. Louis, MO, USA) and 1 mL of ice cold methanol (Chromasolv, HPLC grade 99%; Sigma Aldrich, St. Louis, MO, USA) were added to each fecal sample. Samples were then vortexed for 30 seconds at 3000 rpm and then sonicated in a Branson® 2800 ultrasonic bath (Branson Ultrasonics, Danbury, CT, USA). Samples were then centrifuged at room temperature in an Eppendorf 5415C centrifuge (Eppendorf, Hamburg, Germany) at 12621 rpm for 10 min. The resulting supernatant was removed and transferred to a clean 2 mL Eppendorf tube. One milliliter of ice-cold methanol was added to the remaining fecal sample and the mixture was vortexed for an additional 2 min at 3000 rpm. The sample was then centrifuged again as described above, supernatant removed and added to the first supernatant. The combined supernatants were then dried under a gentle stream of N2 and then stored at -80°C. Prior to LC-QTOF-MS injections, bat fecal sample extracts were reconstituted with 100 μl of acetonitrile: deionized water (4:1) and transferred into glass inserts in brown LC sample vials.
Metabolomics analysis
Analyses were conducted in duplicate using a Rapid Resolution HPLC system (1290 Infinity Agilent Ltd., Santa Clara, CA, USA) equipped with a binary pump, degasser, well-plate auto-sampler (maintained at 6°C throughout the runs) with a thermostat and thermostatic column. This system was coupled to a 6538 UHD Accurate LC-QTOF-MS (Agilent Technologies, Santa Clara, CA, USA) with dual electro-spray ionization (ESI) source. A 2.1 mm x 100 mm Agilent ZORBAX SB-Aq column (Agilent Technologies, Santa Clara, CA, USA) was maintained at 60°C for chromatographic separation of bat fecal samples. Two mobile phases were used, water (A) and acetonitrile (B), and both contained 0.1% formic acid. Run time for the reactions was set at 10 minutes with a gradient of 0–6 min 2% B; 6–8.50 min 60% B; 8.50–8.60 min 2% B and 8.60–10 min 2% B to re-equilibrate the column. A post-run time of 2 min was also instituted prior to the injection of the next sample. Sample carryover during successive injections was minimized by washing the needle in two separate vials of mobile phase (5 washings per sample vial) before each new sample injection. For each individual sample analysis, 3μl of fecal extracts was injected with a flow rate maintained at 0.7 mL/min.
MS data acquisitions were completed in both positive (+) and negative (-) electrospray ionization (ESI) modes. MS parameters included capillary voltage (4000 V), the fragmentor (175V), the skimmer (50V) and the OCT 1 RFVpp (750 V). For drying, nitrogen gas (N2) was utilized at 11 L/min at 300°C with nebulizer settings at 50 psig. MS spectra were collected within the range of 50–1700 m/z and known references masses of 121.0508 and 922.0097 (ESI+) and 112.9860 and 1033.9880 (ESI-) utilized during all runs.
Data processing
The LC-QTOF-MS metabolomics data workflow followed standard protocols (Table 1) and utilized multiple algorithms incorporated in Agilent MassHunter Qualitative (MHQ, 7.01 and Mass Profiler Professional (MPP, version 12.6.1) software programs. Raw chromatographic files, total ion chromatograms (TICs), generated during LC-QTOF-MS runs were acquired and stored as “*.d” files for MHQ data processing. Raw chromatographic files were first processed using both Molecular Feature Extraction (MFE) to detect features with abundances ˃ 4000, and find by formula (fbf) algorithms in MassHunter Qualitative (MHQ) software (Agilent Technologies, Santa Clara, CA, USA) to generate chemical formulae, based on exact masses and annotate detected metabolic entities. The method also provided information regarding [M + H] +, isotope distribution of each candidate molecule and any corresponding sodium adducts. Extracted ions were now considered single features and MHQ algorithms were used to generate potential identifications via Metlin [40] and MHQ/MPP-associated software libraries utilizing chemical formulae. These features, with their potential chemical formulae, associated retention times (RT), exact masses and ion abundances were then converted into compound exchange format (“*.cef”) files. These “.cef” files were exported to MPP once more as part of recursive analyses. Individual “*.cef” files were now binned into the two-designated age groups (1-2-year-old “young” and 14-15-year-old “elderly” individuals). These were then combined, aligned and normalized before application of the ‘Find by ion’ algorithm and subsequent generation of new “*.cef” files. These newly generated “*.cef” files were exported to MHQ for additional data mining. Targeted feature algorithms were also applied to minimize the risk of false positive and false negative features. Another set of individual “*.cef” files were then generated from the original “*.d” files and exported into MPP for statistical analyses. To ensure that potential feature extraction artefacts were not eliminated, features which were detected in a minimum of one condition were excepted using frequency filtration and other MPP filtration methods such as: included charge states set to “all charge states permitted” and number of detected ions was set at “2”. Retention time (RT) compound alignment parameters were set to 0.15 min and the mass tolerance was set at 2.0 mDA. Data were normalized using a percentile shift algorithm (set at 70) and baselined to the median of all samples.
Table 1. Summary of the workflow utilized to investigate the metabolic profile of feces in young and elderly bats.
| Step 1: Metabolomics of Bat Fecal Matters (LC-QTOF-MS and MHQ 7.01) | |
| a) | Non-targeted analysis of bat fecal samples by LC-QTOF-MS in positive and negative ESI modes. |
| b) | Molecular Feature Extraction (MFE) algorithm to extract all detectable compounds. |
| c) | Find-by-Ion algorithm to remove false positive and negative compounds. |
| Step 2: Statistical Analysis (MPP 12.6.1) to Compare Young and Elderly Metabolomes | |
| a) | Mann-Whitney U-test (P < 0.05) followed by robust multiple testing correction of p-values (Benjamini Hochberg FDR multiple testing). |
| Step 3: Biochemical Pathway Analysis—Assignment of Metabolite Function | |
| a) | Identification of biochemical pathways (KEGG, Human Metabolome Database, Metlin) for the metabolites present in fecal matters of young and old bats and their potential correlation with age. |
Metabolic entities were identified using the integrated Metlin database, which contains greater than 200,000 known compounds, within the MHQ and MPP software. Compounds that were not matched with those available in the Metlin database were listed as “endogenous metabolites” and will be the subject of future studies using liquid chromatography-nuclear magnetic resonance (LC-NMR). Because a goal of the study was identification of potential candidates characteristic of an elderly metabolome, candidates were selected that demonstrated significant concentration increases in elderly fecal samples. Common databases such as KEGG [(41)], Human Metabolome Database [41] and published literature were mined to assign potential physiological functions.
Statistical analyses
Statistical analyses (Table 1) included Mann-Whitney U-test (p <, 0.05) with Benjamini Hochberg FDR multiple testing correction. Principle component clustering models (PCA) were constructed in MPP software (version 12.6.1). These statistical analyses of the original identified candidates identified 42 that demonstrated significant (p < 0.05) age-related increases or decreases in concentration between the young and elderly bats (Table 2).
Table 2. Fecal analyses detected 41 metabolites with known biological effects that significantly differentiated metabolome of elderly bats.
Metabolites demonstrated ≥2-fold changes and p < 0.05. Log2 abundance ratios for all metabolites were acquired in ESI+ mode.
| Tryptophan Metabolism and Oxidation | |||||||
|---|---|---|---|---|---|---|---|
| Metabolite | Formula | Non-normalized abundance [young] | Non-normalized abundance [elderly] | Log2 abundance ration ([elderly]/[young]) | p (Corr) | Reference | |
| 1 | 3-Amino-2-naphthoic acid | C11 H9 N O2 | 2.3 | 229712.5 | 16.6 | 0.01010 | [42] |
| 2 | Indole | C8 H7 N | 1.3 | 4025.5 | 11.6 | 0.01010 | [43–46] |
| 3 | 3-Methylindole | C9 H9 N | 0.9 | 704.8 | 9.6 | 0.01010 | [47,48] |
| 4 | 2-aminomuconic acid semialdehyde | C6 H7 N O3 | 0.9 | 457 | 9.0 | 0.01147 | [49] |
| 5 | N, N-Dihydroxy-L-tryptophan | C11 H12 N2 O4 | 2.2 | 477.8 | 7.8 | 0.01010 | [49] |
| Incomplete Protein Digestion/Catabolism | |||||||
| 6 | Threoninyl-Phenylalanine | C13 H18 N2 O4 | 1.9 | 19997.9 | 13.3 | 0.01010 | [50] |
| 7 | Valyl-Isoleucine | C11 H22 N2 O3 | 4.2 | 9355.4 | 11.1 | 0.01010 | [50] |
| 8 | Leu Asp Lys | C16 H30 N4 O6 | 3.1 | 4252.3 | 10.4 | 0.01108 | [50,51] |
| 9 | Prolyl-Lysine | C11 H21 N3 O3 | 1.7 | 1426.9 | 9.7 | 0.01010 | [50] |
| 10 | Ile Gly Arg | C15 H24 N10 | 0.9 | 396.2 | 8.8 | 0.01010 | [50] |
| 11 | Pyrrolidine | C4 H9 N | 4.1 | 1580.7 | 8.6 | 0.01010 | [50] |
| 12 | Pro Val Pro | C15 H28 N4 O4 | 1.3 | 320.2 | 8.0 | 0.01010 | [50] |
| 13 | Leu Lys Ala | C21 H32 N S | 1.3 | 263.7 | 7.6 | 0.01010 | [50] |
| 14 | Leu Pro Lys | C18 H28 N8 | 1.3 | 233.3 | 7.5 | 0.01384 | [50] |
| 15 | Histidinylglycine | C8 H12 N4 O3 | 1.9 | 239.1 | 6.9 | 0.01010 | [50] |
| Protein and Amino Acid Synthesis | |||||||
| 16 | L-Homocitrulline | C7 H15 N3 O3 | 26.2 | 17837.7 | 9.4 | 0.01147 | [52] |
| 17 | L-Lysine 1,6-lactam | C6 H12 N2 O | 2.1 | 316 | 7.3 | 0.01157 | [49] |
| 18 | Ganglioside GD3 (d18:0/12:0) | C64H113N30O29 | 1.4 | 186.8 | 7.1 | 0.01751 | [41] |
| DNA degradation | |||||||
| 19 | N2,N2-Dimethylguanosine | C12H17N5O5 | 0.9 | 243.9 | 8.0 | 0.01157 | [53] |
| Glycation Indicator | |||||||
| 20 | N2-Fructopyranosylarginine | C12 H24 N4 O7 | 2.2 | 700.1 | 8.3 | 0.03993 | [54] |
| Mitochondrial Respiratory chain failure | |||||||
| 21 | Malonylcarnitine | C10 H17 N O6 | 5.3 | 3095.4 | 9.2 | 0.01572 | [55] |
| Fatty Alcohol | |||||||
| 22 | 2,4,6-Octatriyn-1-ol | C8 H6 O | 23.2 | 79422.5 | 11.7 | 0.01157 | [41] |
| Biotin Metabolism | |||||||
| 23 | apo- [3-methylcrotonoyl-CoA: carbon-dioxide ligase (ADP-forming)] | C7 H15 N3 O2 | 5.7 | 5340.6 | 9.9 | 0.02793 | [41] |
| Methane Production | |||||||
| 24 | N-Furfurylformamide | C6 H7 N O2 | 21.5 | 22187.8 | 10.0 | 0.01010 | [56] |
| Cytoketogenesis | |||||||
| 25 | Isopentenyladenine | C10 H13 N5 | 2.8 | 1624.2 | 9.2 | 0.04454 | [57,58] |
| Isothiocyanate | |||||||
| 26 | 6-Isothiocyanato-1-hexene | C7 H11 NS | 1.4 | 990.5 | 9.4 | 0.01108 | [41] |
| Organofloride | |||||||
| 27 | 2-Chloro-1,1,1-trifluoroethane | C2 H2 Cl F3 | 1.3 | 206.1 | 7.3 | 0.01010 | [41] |
| Bacteria Quorum Sensing/ Biofilm inhibition | |||||||
| 28 | N-3-oxo-tetradec-7(Z)-enoyl-L-Homoserine lactone | C18 H29 N O4 | 1.8 | 137.7 | 6.2 | 0.01157 | [59] |
| Fructosylation Product | |||||||
| 29 | N-(1-Deoxy-1 fructosyl) threonine | C10 H19 NO8 | 1.9 | 209 | 6.7 | 0.025300 | [41] |
| Antibiotics/Pharmaceuticals | |||||||
| 30 | Lomustine | C9 H16CI N3 O2 | 2 | 482.3 | 7.9 | 0.01157 | [60] |
| 31 | Methylmercury chloride | C H3 Cl Hg | 2.2 | 465.6 | 7.8 | 0.01010 | [61] |
| 32 | Caldine | C26 H33 NO6 | 1.4 | 286.5 | 7.7 | 0.01010 | [62] |
| 33 | Gaboxadol | C6 H8 N2 O2 | 2.1 | 366.5 | 7.4 | 0.02525 | [63] |
| 34 | 2-Propylglutaric acid | C8 H14 O4 | 0 | 5.7 | 9.6 | 0.01239 | [64] |
| 35 | Istamycin X0 | C14 H30 N4 O4 | 1.4 | 199.6 | 7.2 | 0.02793 | [49] |
| Pesticides/Fungicides | |||||||
| 36 | Bithionol | C12 H6 Cl4 O2 S | 1.7 | 118.4 | 6.1 | 0.01157 | [65] |
| 37 | Zinnimidine | C15 H19 NO3 | 1.9 | 277 | 7.2 | 0.01239 | [66] |
| 38 | Dimethyl phosphate | C2 H7 O4 P | 1.1 | 152.9 | 7.1 | 0.01010 | [67] |
| 39 | Procymidone | C13H11Cl2NO2 | 1.5 | 1499.3 | 9.9 | 0.01010 | [68,69] |
| Endogenous Peptides | |||||||
| 40 | 1-amino-3,3-diethoxypropane | C7 H17 N O2 | 4.1 | 1812.9 | 8.8 | 0.01157 | |
| 41 | α-Thiophenecarboxylic acid | C5 H4 O2 S | 2.8 | 1018.4 | 8.5 | 0.01010 | |
Results
Metabolites
Our metabolomics analysis included 28 Eptesicus, ranging from one to over fourteen years of age. To identify age-associated differences in metabolic profiles, the animals were divided into two different age groups: the “young” group included 22 animals aged 1–2 years, while the “elderly” group contained 6 individuals all 14 years of age and older. The final metabolite dataset contained 2003 entities (unidentified atoms, molecules, compounds and/or compound fragments present in both age groups. Resultant identified metabolites therefore generated a pilot list of candidate metabolites that set the stage for larger age-comparative and physiological studies.
Forty-one compounds within the feces displayed significantly higher concentrations (p < 0.05) in the elderly metabolome (Fig 2, Table 2). Roughly 20% of these compounds are associated with protein and amino acid metabolism (Table 2), where significant increases with age suggest decreases in incomplete protein catabolism and/or absorption. Of these compounds found in the elderly metabolome, the most abundant are dipeptides (i.e., threoninyl-phenylalanine (p = 0.01010), valyl-isoleucine (p = 0.01010), prolyl-lysine (p = 0.01010), and histidinyl-glycine (p = 0.01010)) and tripeptides (leucyl-aspartyl-lysine (Leu Asp Lys; p = 0.01108), isoleucyl-glycyl-arginine (Ile Gly Arg; p = 0.01010), prolyl-valyl-proline (Pro Val Pro; p = 0.01010), leucyl-lysyl-alanine (Leu Lys Ala; p = 0.01010), leucyl-prolyl-lysine (Leu Pro Lys; p = 0.01384)). During normal metabolism in other mammals, these peptides can be either cleaved by proteases, or absorbed by the enterocytes of the intestinal walls via transcytosis or co-transport with ions (e.g., H+, Na+; [47]). Comparisons of digestive efficiency in bats and non-flying mammals showed a similar digestive efficiency for proteins when consuming meal worms [50], which is the diet of our study taxon. Therefore, an increased abundance of di- and tripeptides within the feces of elderly bats may indicate that bats undergo a decline in peptide degradation capacity that may be associated with digestive senescence [70].
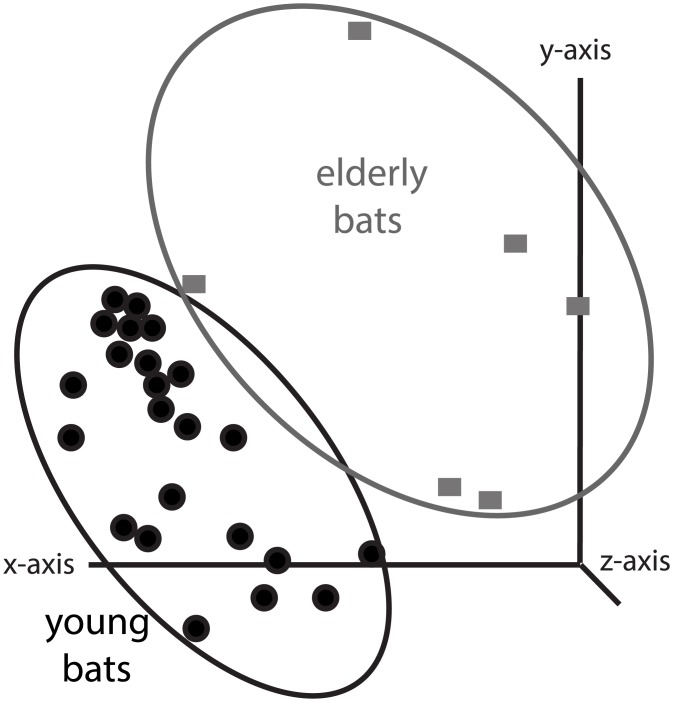
Fig 2. Principle component analysis (PCA) of metabolic profile of fecal samples show separate groupings for age groups.
The young age group is demonstrated by open squares (one-year-old animals) and, filled, black squares (two-year-old animals). The elderly age group is demonstrated by open circles (fourteen-year old animals) and filled, black circles (fifteen-year-old animals). Variation was explained on three axes: x-axis (18.82%), y-axis (7.98%), and z-axis (5.69%).
Of the candidate metabolites, four were functionally associated with metabolism of the amino acid tryptophan (Fig 3; indole (p = 0.01010), 3-Methylindole (skatole; p = 0.01010), 3-amino-2-naphthoic acid (p = 0.01010), and 2-aminomuconate semialdehyde (p = 0.01147)). These compounds displayed some of the highest concentrations of recovered metabolites from the elderly samples, suggesting their concentrations increase with age in Eptesicus. This finding was striking as some of these compounds are known to contribute to longevity and play key roles in immune function including cancer and inflammation [46,71,72].
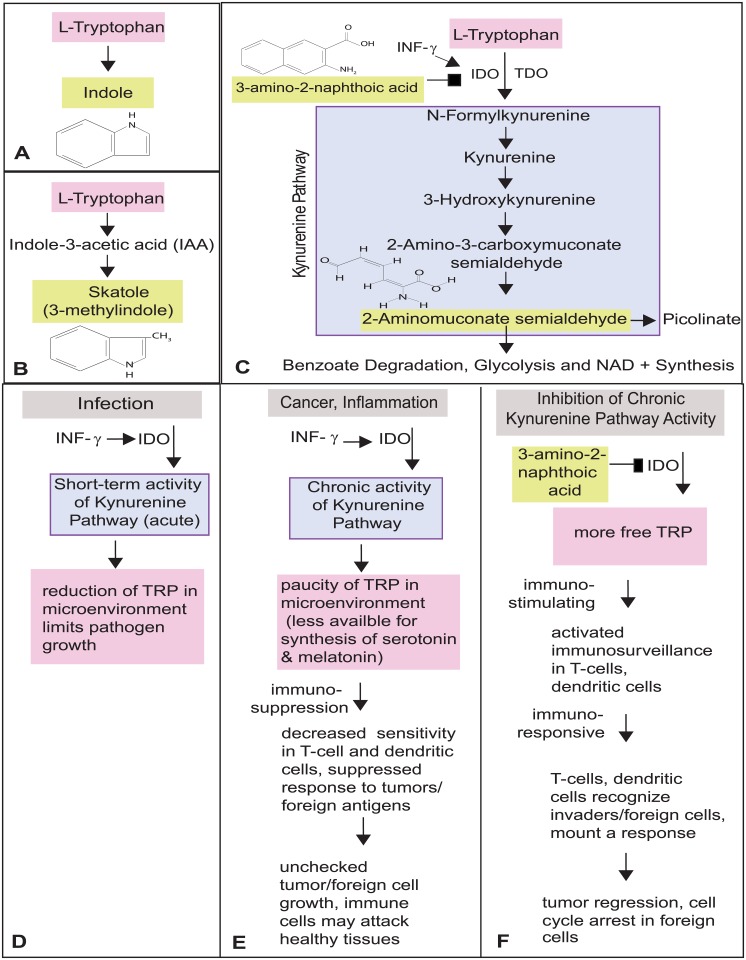
Fig 3. Metabolism of L-Tryptophan can occur as a direct conversion to indole (A), modification to skatole (B), and degraded by the kynurenine pathway (C).
IDO is the most critical enzyme regulating local activity of the Kynurenine Pathway. In response to an infection (via IFN-γ), activation in IDO results a local degradation of tryptophan that limits growth of a pathogen (D). Activation of IDO in cases of chronic inflammation/cancer suppresses the effectiveness of immune cells which creates a sanctuary for pathogen survival and proliferation (E). In contrast, inhibition of the IDO response results in the stimulation of immune cells and catalyzes an immune response that may arrest growth and hinder proliferation of pathogens (F). Metabolites with high concentrations in the feces of elderly Eptesicus are indicated by yellow boxes.
Several synthetic compounds of non-biological origin were also upregulated in elderly bats, but at lower concentrations compared to those functionally associated with tryptophan and protein metabolism. These compounds included an anti-convulsant (2-Propylglutaric acid (p = 0.01239); [64]), an anti-cancer compound (Lomustine (p = 0.01157); [60]), an anti-hypertensive (Caldine, also known as Lacidipine (p = 0.01010); [62]), and an anti-tapeworm and anti-algal metabolite (Bithionol (p = 0.01157); [65]). One potential source of many of these synthetic compounds may be the meal worm food source. The meal worms are raised on a non-organic diet, potentially exposing them to a wide range of chemical contaminants. Future work examining the metabolic profile of the meal worms themselves is planned to test this hypothesis. Evidence of these compounds recovered from feces may indicate impaired detoxication in bats resulting in retention of these compounds.
Beyond skatole and peptides, still other compounds (Table 2) potentially shed light on the physiology of age-related declines in bats. It could be that these compounds offer insight into the long-standing questions of what causes long-lived bats to eventually die. Samples recovered from elderly bats showed increased concentrations of N2-Fructopyranosylarginine (p = 0.03993), a precursor to advanced glycation end products (AGE), which are detrimental accumulations of glycated and oxidized proteins affecting diverse systems like connective tissue integrity, renal function, and aging [54]. The presence and increased concentrations of these compounds in samples from elderly Eptesicus potentially indicate the onset of glycation. Although multiple studies showed modifications associated with mitochondrial function [16,20], results of this study showed increased concentrations of a potential indicator of mitochondrial deficiency in elderly bat samples (Malonylcarnitine; p = 0.01572; [55]). Presence of the nucleoside N2, N2-Dimethylguanosine (p = 0.01157) potentially indicates upregulation of transfer RNA (tRNA) degradation [53]. Presence of these metabolites are suggestive of age-related declines in bat health although further study is required to acquire definitive evidence.
Discussion
The exceptional longevity of many bat species has piqued the interest of researchers aiming to elucidate the physiological adaptations contributing to these extended lifespans [14,19,21]. This study tested the feasibility of a comparative fecal metabolomics approach in young and elderly big brown bats (Eptesicus fuscus). Analyses detected 41 metabolites that demonstrated significant increases with age in Eptesicus. Currently, our analyses lack a larger cohort of elderly animals as well as middle-aged samples which, unfortunately, impedes understanding of how these metabolites do or do not fluctuate across the lifespan in Eptesicus. Future studies will target larger sample sizes of young, middle, and elderly age groups to better assess how fecal metabolites vary with age and better understand the range of individual variation within these animals. Beyond offering the first age-comparative study of fecal metabolomics in bats, our results also provide new insights into how bats maintain physiological health into old age.
Although other tissues and fluids are available for metabolomics analysis (e.g., hair, blood, urine), feces were the focus of this pilot analysis as they offer a reliable and non-invasive means for sample collection that may ultimately be useful for data collection in wild populations. We utilized databases containing identities for over 200,000 metabolites to analyze our fecal samples and 41 metabolites that demonstrated age-associated differences between young and elderly bats. While urine is commonly utilized in conjunction with fecal analyses, the authors found collection of uncontaminated urine to be a major obstacle. Future assays will also address how circulating levels of identified metabolites in blood correlate to levels found in bat fecal samples.
Protein homeostasis and aging
Progressive deterioration of protein homeostasis contributes to age-related functional decline and adversely affects longevity. The cellular mechanisms of translation, folding and clearance cooperatively regulate protein production and stability as well as the refolding or removal of damaged products [73]. With age, incomplete breakdown of proteins becomes more common in mammals [74,75], and it appears to occur in elderly bats as well with increased levels of di- and tripeptides detected in elderly fecal samples. Reactive oxygen species (ROS) are a leading contributor to oxidative protein damage [75], resulting in accumulation of abnormal proteins associated with numerous pathological conditions [76]. Recent evidence suggests that bats possess increased resistance to oxidative stress [14,20], which assists in maintaining protein homeostasis. Heat shock proteins (HSPs) are molecular chaperone proteins induced in response to stress and other stimuli that are critical to the maintaining protein quality [77]. Synthesis of HSPs is impaired in aging in many mammals, including humans [78]. Vespertilionid evening bats (Nycticeius humeralis) and little brown bats (Myotis lucifugus)) have increased HSP levels compared to shorter-lived rodents and marsupials [19]. These elevated chaperone levels may support a healthy stress response [79] and increase longevity in some bat species. Additionally, bats possess lower levels of both protein breakdown and removal and demonstrate an increased resistance to urea-induced protein unfolding compared to mice [14]. Our results suggest elderly bats experience age-related declines in peptide degradation capacity and peptide absorption similar to that seen in other mammals.
Tryptophan metabolism and its influence on longevity
The fecal microbiome is an ecosystem of over 1000 species of bacteria which undergo complex interactions that ultimately influence the physiology and health of their host. Metabolites formed during the bacterial metabolism of the essential amino acid tryptophan (TRP) play key roles in the molecular regulation of host behavior, neuronal activity, immune activity, longevity and extend the healthspan [46,80]. Within our sample of metabolites that demonstrated age-related differences between the young and elderly microbiome, four metabolites play a role in the metabolism of tryptophan (i.e., indole, skatole, 3-amino-2-naphthoic acid, 2-aminomuconate semialdehyde, N, N-dihydroxy-L-tryptophan; Fig 3). Of these, the tryptophan metabolism product indole is the most well-understood. Indole is used as a biomarker indicative of 85 species of gram-negative and gram-positive bacteria [44] and functions to enhance the intestinal barrier [43], support dynamic signaling within microbial communities [81], as well as act as an anti-inflammatory molecule in cell cultures [45] as well as increased healthspan [46]. Because indole is the most prevalent tryptophan degradation product, our results suggest the increased abundance of indole within intestines of elderly bats may function to offset age-related stressor and declines in intestinal health and microbiome-related diseases [46,82] and potentially increase the healthspan of elderly Eptesicus.
The most common naturally occurring analog of indole is skatole (3-methylindole; [48]), also known as the “essence of poop” based on its smell in high concentrations (Fig 3). Although typically found in much lower concentrations than indole, skatole serves as a biomarker for colon cancer [83] and is known to induce kidney damage, pulmonary edema, emphysema, and death in livestock [84]. Skatole also adversely effects biological membranes and promotes the destruction of red blood cells (hemolysis; [47]. Within our sample, elderly bats displayed a marked increase in skatole concentrations. Because skatole is a biomarker for intestinal health [83,85], age-related increases in its concentration in elderly bats suggest some form of physiological decline within the intestines. However, this is not straightforward because elderly bat feces also contain high concentrations of indole, a metabolite that promotes intestinal health.
Tryptophan is most commonly metabolized via the kynurenine pathway (Fig 3), which generates metabolites affecting energy metabolism, neurodegeneration, immune regulation [80], inflammation [86] and bone diseases [87]. Tryptophan degradation along the kynurenine pathway results in increased concentrations of kynurenine and its derivatives (collectively referred to as kynurenines) intracellularly or within the microenvironment [88]. With age, humans increase the rate of tryptophan degradation resulting in higher concentrations of kynurenines relative to tryptophan, a process thought to contribute to aging [89]. Within elderly bats, fecal metabolites showed evidence of the kynurenine pathway of tryptophan degradation. A metabolite nested within the pathway (2-aminomuconate semialdehyde) was increased in elderly bats as were concentrations of 3-amino-2-naphthoic-acid, a molecule that modifies the rate of tryptophan degradation within the kynurenine pathway [42]. Interestingly, the metabolomic profiles of bat feces suggest they may lack this progression toward increased concentrations of kynurenines with age.
The first step of the kynurenine pathway, the breaking of the indole ring of L-tryptophan, is regulated by the enzymatic activities of indoleamine 2, 3-dioxygenase (IDO) and tryptophan 2,3-dioxygenase (TDO; [80]). Normally TDO and IDO act in tandem, but their actions become unsynchronized under pathological conditions [42]. Whereas TPO produces most kynurenine within the blood circulation, IDO is expressed in many cells throughout the body (e.g., mesenchymal stem cells, macrophages) to regulate local tryptophan concentrations. We focus our discussion on IDO because it bears the most relevance to the fecal metabolites of bats. Depending on conditions within the microenvironment, activation of IDO can have beneficial and pathological consequences (Fig 3). During an acute infection, short-term activation of IDO induces a local depression in the concentration of tryptophan, which starves pathogens of this amino acid, thereby reducing the capacity for pathogenic growth and making them more susceptible to cell-cycle arrest and apoptosis [90]. This acute interaction is largely beneficial to the host. However, in response to continuous cytokine production as in chronic inflammatory disease and cancer, the pathogen is not removed from the microenvironment. In this case, long-term or chronic exposure of IDO results in a build-up of kynurenines that modify amino acid and metabolite sensors on antigen-presenting cells (e.g., dendritic cells, T-cells). In response to a lack of sensitivity, these cells fail to recognize tumor cells and therefore tolerate their presence. Tumor/pathogen growth proceeds unchecked by the immune system [80]. In addition, some immune cells instead attack healthy cells of the host and ultimately contribute to age-related and inflammation-related diseases (e.g., sarcopenia, dementia, diabetes; [91]). As such, the kynurenine pathway is a major focus of immunological research.
In contrast, inhibition of IDO promotes immune cell health and activity (Fig 3). Inhibition of IDO results in a greater number of tryptophan molecules within the local microenvironment that are free to be metabolized in other essential pathways influencing behavior and reproduction (serotonin; [92]) and circadian rhythms (melatonin; [93]). Relieved of IDO-dependent suppression, immune system function is improved as T-cells and dendritic cells are sensitized and able to detect the presence of tumor cells/pathogens [94,95]. Activity of these cells ultimately allows for an effective immune response that causes tumor regression via cell-cycle arrest and eventual apoptosis. The feces of elderly bats contain exceptionally high levels of an extremely potent IDO inhibitor, 3-amino-2-naphthoic acid [42]. Presence of this metabolite is rare and typically considered to be of synthetic, rather than biological, origin; although it has been recovered from potatoes and lemon extract [96,97]. While the exact origin of this compound in bat feces is unknown, it could be that 3-amino-2-naphthoic acid is a product of bat metabolism or the bat-specific microbiome. It could be that bats experience inhibition of the kynurenine pathway via inhibition of the enzyme IDO by 3-amino-2-naphthoic acid. If this pathway is in fact suppressed in elderly bats, our results would accordingly suggest that, with age, the activity of immune cells increases with age, making the elderly less susceptible to chronic inflammation and cancer.
Efficacy of evidence obtained by fecal metabolomic assays
The exceptional longevity of bats suggests they might be an ideal model for the study of mammalian senescence. Unfortunately, studies to that effect are hindered in adult animals by the lack of visible age-related physiological changes and the lack of accurate methods of age determination. Here, we employed a non-invasive, fecal metabolomics approach to examine age-related differences in the fecal metabolomes of long-lived big brown bats (Eptesicus fuscus). Results establish the feasibility of this metabolomics approach and lay the foundation for future studies to expand our knowledge of age-related changes in bat fecal metabolic profiles and further develop fecal metabolomics as a means of age determination in bats. Our metabolomics approach, utilizing LC-QTOF-MS, identified at least 40 metabolites that were significantly increased in fecal samples of elderly Eptesicus over 14 years of age. While most of the candidate metabolites identified have known physiological significance in mammals (Figs 2 and 3), fourteen, as-yet unknown compounds, were also detected that demonstrated significant differences between our two age classes. Their exact origins, identity, and/or physiological relevance cannot be confirmed in this study and future work is necessary to identify these compounds.
A LC-QTOF-MS approach is ideal for rapidly and accurately quantifying large numbers of samples as it allows for almost real-time monitoring of concentrations of metabolites within 12–15 minutes. Once acquired, these metabolites can be identified using publicly available databases containing over 200,000 known compounds and, once identified, the physiological significance can be ascertained. Our non-targeted approach generated a list of metabolites that demonstrated significant variation between young and elderly animals. Although this study was limited by the necessity of examining age-related metabolomic changes in captive bats, future work is planned to replicate this study on a larger sampling of wild-caught bats that consume a uniform diet, as well as wild-caught bats that consume a natural diet. Future analyses examining metabolite differences between ad libitum captive diets and wild diets will allow us to address the effects of caloric restriction on age-related differences in fecal metabolic profiles.
If researchers lack access to an LC-QTOF-MS, results of this analysis may still be of interest. Within the metabolome of elderly bats, indole was found in high concentrations compared to younger animals. It could be that examining age-related changes in indole concentration may offer a means of estimating age groups in Eptesicus. A recent study described methods utilizing a spectrophotometer to quantify concentrations of indole within feces [44] and future work will quantify fecal indole concentrations in captive, known-aged and wild-caught unknown-aged Eptesicus. Because specimen preparation is rapid, bats may be briefly held in captivity to allow testing and then unneeded animals can then be released rather than subjected to invasive or fatal tests of age determination. Although quantification of indole alone is not as informative as broad-scale LC-QTOF-MS analyses, it may provide a valuable data point and allow for non-invasive age group estimation in Eptesicus. This study demonstrates non-invasive fecal metabolomics can be utilized to identify and compare differences in the fecal metabolic profiles of young and elderly bats and establishes fecal metabolomics as a promising method to discover unique adaptations to combat age-related declines in exceptionally long-lived mammals.
Supporting information
(XLSX)
Acknowledgments
We thank Dr. Ellen Covey (University of Washington) for her generous donation of a colony of known-aged bats to NEOMED, which provided samples for this study. The Cooper Lab thanks Dr. Alexander Galazyuk for support and challenging us to determine new methods to establish age-classes in Eptesicus. Dr. Takhar Kasumov is thanked by LNC for technical advice and comments on this manuscript. Dr. John McAllister and Aakriti Kharel are thanked by LNC for literature support. LNC and HB also thank members of the Musculoskeletal Biology and Auditory Neurobiology Research Groups within NEOMED’s Department of Anatomy and Neurobiology for ongoing support and comments on this manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the National Science Foundation [grant number NSF-CMMI 1537745 to L.N.C.]. The opinions, findings and conclusions expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
References
- 1.Austad SN, Fischer KE. Mammalian aging, metabolism, and ecology: evidence from the bats and marsupials. J Gerontol [Internet]. 1991;46(2):B47–53. Available from: https://academic.oup.com/geronj/article-lookup/doi/10.1093/geronj/46.2.B47 [DOI] [PubMed] [Google Scholar]
- 2.Tacutu R, Craig T, Budovsky A, Wuttke D, Lehmann G, Taranukha D, et al. Human ageing genomic resources: integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 2013;41(D1):1027–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilkinson GS, South JM. Life history, ecology and longevity in bats. Aging Cell [Internet]. 2002;1(2):124–31. Available from: http://doi.wiley.com/10.1046/j.1474-9728.2002.00020.x [DOI] [PubMed] [Google Scholar]
- 4.Podlutsky AJ, Khritankov AM, Ovodov ND, Austad SN. A new field record for bat longevity. Journals Gerontol—Ser A Biol Sci Med Sci. 2005;60(11):1366–8. [DOI] [PubMed] [Google Scholar]
- 5.Davies KTJ, Tsagkogeorga G, Bennett NC, Dávalos LM, Faulkes CG, Rossiter SJ. Molecular evolution of growth hormone and insulin-like growth factor 1 receptors in long-lived, small-bodied mammals. Gene [Internet]. 2014;549(2):228–36. Available from: 10.1016/j.gene.2014.07.061 [DOI] [PubMed] [Google Scholar]
- 6.Seim I, Fang X, Xiong Z, Lobanov A V., Huang Z, Ma S, et al. Genome analysis reveals insights into physiology and longevity of the Brandt’s bat Myotis brandtii. Nat Commun. 2013;4:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Healy K, Guillerme T, Finlay S, Kane A, Kelly SBA, McClean D, et al. Ecology and mode-of-life explain lifespan variation in birds and mammals. Proc R Soc B Biol Sci [Internet]. 2014;281(1784):20140298–20140298. Available from: http://rspb.royalsocietypublishing.org/cgi/doi/10.1098/rspb.2014.0298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson G, Wordsworth J, Wang C, Jurk D, Lawless C, Martin-Ruiz C, et al. A senescent cell bystander effect: senescence induced senescence. Aging Cell. 2012;11:345–9. 10.1111/j.1474-9726.2012.00795.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang G, Li J, Purkayastha S, Tang Y, Zhang H, Yin Y et al. Hypothalamic programming of systemic ageing involving IKK-b, NF-kB and GnRH. Nature. 2013;497:211–6. 10.1038/nature12143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossi D, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman I. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–9. 10.1038/nature05862 [DOI] [PubMed] [Google Scholar]
- 12.Talens RP, Christensen K, Putter H, Willemsen G, Christiansen L, Kremer D, et al. Epigenetic variation during the adult lifespan: Cross-sectional and longitudinal data on monozygotic twin pairs. Aging Cell. 2012;11(4):694–703. 10.1111/j.1474-9726.2012.00835.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chondrogianni N, Petropoulos I, Grimm S, Georgila K, Catalgol B, Friguet B, et al. Protein damage, repair and proteolysis. Mol Aspects Med [Internet]. 2014;35(1):1–71. Available from: 10.1016/j.mam.2012.09.001 [DOI] [PubMed] [Google Scholar]
- 14.Salmon AB, Leonard S, Masamsetti V, Pierce A, Podlutsky AJ, Podlutskaya N, et al. The long lifespan of two bat species is correlated with resistance to protein oxidation and enhanced protein homeostasis. FASEB J [Internet]. 2009;23(7):2317–26. Available from: http://www.fasebj.org/cgi/doi/10.1096/fj.08-122523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gidalevitz T, Kikis EA, Morimoto RI. A cellular perspective on conformational disease: the role of genetic background and proteostasis networks. Curr Opin Struct Biol [Internet]. 2010;20(1):23–32. Available from: 10.1016/j.sbi.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunet-Rossinni AK. Reduced free-radical production and extreme longevity in the little brown bat (Myotis lucifugus) versus two non-flying mammals. Mech Ageing Dev. 2004;125(1):11–20. [DOI] [PubMed] [Google Scholar]
- 17.Harper JM, Salmon AB, Leiser SF, Galecki AT, Miller RA. Skin-derived fibroblasts from long-lived species are resistant to some, but not all, lethal stresses and to the mitochondrial inhibitor rotenone. Aging Cell. 2007;6(1):1–13. 10.1111/j.1474-9726.2006.00255.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pickering AM, Lehr M, Kohler WJ, Han ML, Miller RA. Fibroblasts from longer-lived species of primates, rodents, bats, carnivores, and birds resist protein damage. Journals Gerontol—Ser A Biol Sci Med Sci. 2014;70(7):791–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pride H, Yu Z, Sunchu B, Mochnick J, Coles A, Zhang Y, et al. Long-lived species have improved proteostasis compared to phylogenetically-related shorter-lived species. Biochem Biophys Res Commun. 2015;457(4):669–75. 10.1016/j.bbrc.2015.01.046 [DOI] [PubMed] [Google Scholar]
- 20.Huang Z, Jebb D, Teeling EC. Blood miRNomes and transcriptomes reveal novel longevity mechanisms in the long-lived bat, Myotis myotis. BMC Genomics [Internet]. 2016;17(1):1–15. Available from: 10.1186/s12864-016-3227-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang G, Cowled C, Shi Z, Huang Z, Bishop-Lilly KA, Fang X, et al. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science (80-). 2013;339(January):456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan YH, Zhang Y, Cui J, Liu Y, McAllan BM, Liao CC, et al. Adaptation of Phenylalanine and Tyrosine Catabolic Pathway to Hibernation in Bats. PLoS One. 2013;8(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park D, Larsson N. Mitochondrial DNA mutations in disease and aging. J Cell Biol. 2011;193:809–18. 10.1083/jcb.201010024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moskalev AA, Shaposhnikov M V., Plyusnina EN, Zhavoronkov A, Budovsky A, Yanai H, et al. The role of DNA damage and repair in aging through the prism of Koch-like criteria. Ageing Res Rev [Internet]. 2013;12(2):661–84. Available from: 10.1016/j.arr.2012.02.001 [DOI] [PubMed] [Google Scholar]
- 25.Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase : the path from maize, Tetrahymena and yeast to human cancer and aging. October. 2006;12(10):1133–8. [DOI] [PubMed] [Google Scholar]
- 26.Harries LW, Hernandez D, Henley W, Wood AR, Holly AC, Bradley-Smith RM, et al. Human aging is characterized by focused changes in gene expression and deregulation of alternative splicing. Aging Cell. 2011;10(5):868–78. 10.1111/j.1474-9726.2011.00726.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foley NM, Hughes GM, Huang Z, Clarke M, Jebb D, Whelan C V., et al. Growing old, yet staying young: The role of telomeres in bats’ exceptional longevity. Sci Adv [Internet]. 2018;4(2):eaao0926 Available from: http://advances.sciencemag.org/lookup/doi/10.1126/sciadv.aao0926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bruce S, Tavazzi I, Parisod V, Rezzi S, Kochhar S, Pa G. Investigation of human blood plasma sample preparation for performing metabolomics using ultrahigh performance liquid chromatography / mass spectrometry. Anal Chem. 2009;81(9):3285–96. 10.1021/ac8024569 [DOI] [PubMed] [Google Scholar]
- 29.Menni C, Kastenmüller G, Petersen AK, Bell JT, Psatha M, Tsai PC, et al. Metabolomic markers reveal novel pathways of ageing and early development in human populations. Int J Epidemiol. 2013;42(4):1111–9. 10.1093/ije/dyt094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seo C, Hwang YH, Kim Y, Joo BS, Yee ST, Kim CM, et al. Metabolomic study of aging in mouse plasma by gas chromatography-mass spectrometry. J Chromatogr B Anal Technol Biomed Life Sci [Internet]. 2016;1025:1–6. Available from: 10.1016/j.jchromb.2016.04.052 [DOI] [PubMed] [Google Scholar]
- 31.Mishur R, Rea S. Applications of mass spectrometry to metabolomics and metabonomics: Detection of biomarkers of aging and of age-related diseases. Mass Spectrom Rev. 2012;31:70–95. 10.1002/mas.20338 [DOI] [PubMed] [Google Scholar]
- 32.Calvani R, Brasili E, Praticò G, Capuani G, Tomassini A, Marini F, et al. Fecal and urinary NMR-based metabolomics unveil an aging signature in mice. Exp Gerontol [Internet]. 2014;49(1):5–11. Available from: 10.1016/j.exger.2013.10.010 [DOI] [PubMed] [Google Scholar]
- 33.Leyva DR, Zahradka P, Ramjiawan B, Guzman R, Aliani M, Pierce GN. The effect of dietary flaxseed on improving symptoms of cardiovascular disease in patients with peripheral artery disease. Rationale and design of the FLAX-PAD randomized controlled trial. Contemp Clin Trials. 2011;32(5):724–30. 10.1016/j.cct.2011.05.005 [DOI] [PubMed] [Google Scholar]
- 34.Ahmed N, Bezabeh T, Ijare OB, Myers R, Alomran R, Aliani M, et al. Metabolic Signatures of Lung Cancer in Sputum and Exhaled Breath Condensate Detected by (1)H Magnetic Resonance Spectroscopy: A Feasibility Study. Magn Reson Insights [Internet]. 2016;9:29–35. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27891048%5Cnhttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC5117486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelm DH, Popa-Lisseanu AG, Dehnhard M, Ibáñez C. Non-invasive monitoring of stress hormones in the bat Eptesicus isabellinus—Do fecal glucocorticoid metabolite concentrations correlate with survival? Gen Comp Endocrinol [Internet]. 2016;226:27–35. Available from: 10.1016/j.ygcen.2015.12.003 [DOI] [PubMed] [Google Scholar]
- 36.Voigt CC, Streich WJ, Dehnhard M. Assessment of fecal testosterone metabolite analysis in free-ranging Saccopteryx bilineata (Chiroptera: Emballonuridae). Acta Chiropterologica [Internet]. 2007;9(2):463–75. Available from: http://www.bioone.org/doi/%0Afull/10.3161/1733-5329%282007%299%5B463%3AAOFTMA%5D2.0.CO%3B2artnerID=tZOtx3y1 [Google Scholar]
- 37.Finke MD. Complete nutrient composition of commercially raised invertebrates used as food for insectivores. Zoo Biol. 2002;21(3):269–85. [Google Scholar]
- 38.Skrinyer AJ, Faure PA, Dannemiller S, Ball HC, Delaney KH, Orman R, et al. Care and Husbandry Flying Mammals. Lab Anim Sci. 2017;(June):24–7. [Google Scholar]
- 39.Sun Y, Liu L, Peng Y, Liu B, Lin D, Li L, et al. Metabolites characterization of timosaponin AIII in vivo and in vitro by using liquid chromatography-mass spectrometry. J Chromatogr B Anal Technol Biomed Life Sci [Internet]. 2015;997:236–43. Available from: 10.1016/j.jchromb.2015.06.015 [DOI] [PubMed] [Google Scholar]
- 40.Smith CA, Maille GO, Want EJ, Qin C, Trauger SA, Brandon TR, et al. Metlin a metabolite mass spectral database. Ther Drug Monit [Internet]. 2005;27(6):747–51. Available from: http://content.wkhealth.com/linkback/openurl?sid=WKPTLP:landingpage&an=00007691-200512000-00016 [DOI] [PubMed] [Google Scholar]
- 41.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–61. 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, et al. HMDB 3.0-The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41(D1):801–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peterson AC, Migawa MT, Martin MJ, Hamaker LK, Czerwinski KM, Zhang W, et al. Evaluation of functionalized tryptophan derivatives and related compounds as competitive inhibitors of indoleamine 2,3-dioxygenase. Med Chem Res. 1994;3:531–44. [Google Scholar]
- 44.Chappell CL, Darkoh C, Shimmin L, Farhana N, Kim D-K, Okhuysen PC, et al. Fecal indole as a biomarher of susceptibility to Cryptosporidium infection. Infect Immun [Internet]. 2016;84(8):IAI.00336-16. Available from: http://iai.asm.org/lookup/doi/10.1128/IAI.00336-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Darkoh C, Chappell C, Gonzales C, Okhuysen P, Dale W. A rapid and specific method for the detection of indole in complex biological samples. 2015;81(23):8093–7. 10.1128/AEM.02787-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin U-H, Lee S-O, Sridharan G, Lee K, Davidson LA, Jayaraman A, et al. Microbiome-Derived Tryptophan Metabolites and Their Aryl Hydrocarbon Receptor-Dependent Agonist and Antagonist Activities. Mol Pharmacol [Internet]. 2014;85(5):777–88. Available from: http://molpharm.aspetjournals.org/cgi/doi/10.1124/mol.113.091165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonowal R, Swimm A, Sahoo A, Luo L, Matsunaga Y, Wu Z, et al. Indoles from commensal bacteria extend healthspan. Proc Natl Acad Sci [Internet]. 2017;201706464. Available from: http://www.pnas.org/lookup/doi/10.1073/pnas.1706464114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bray TM, Carlson JR. The Effects of 3-Methylindole on hemolysis, transport of **Na+, and ATPase activities of bovine erythrocytes (38651). Proceeding Soc Experiemtnal Biol Med. 1975;148:875–9. [DOI] [PubMed] [Google Scholar]
- 49.Jensen MT, Cox RP, Jensen BB, Jensen MT, Cox RP, Jensen BB. 3-Methylindole (skatole) and indole production by mixed populations of pig fecal bacteria. Appl Environ Microbiol. 1995;61(8):3180–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price ER, Brun A, Caviedes-Vidal E, Karasov WH. Digestive adaptations of aerial lifestyles. Physiology [Internet]. 2015;30(1):69–78. Available from: http://physiologyonline.physiology.org/cgi/doi/10.1152/physiol.00020.2014 [DOI] [PubMed] [Google Scholar]
- 51.Stadtman ER, Levine RL. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25(3–4):207–18. 10.1007/s00726-003-0011-2 [DOI] [PubMed] [Google Scholar]
- 52.Piraud M, Vianey-Saban C, Petritis K, Elfakir C, Steghens JP, Morla A, et al. ESI-MS/MS analysis of underivatised amino acids: A new tool for the diagnosis of inherited disorders of amino acid metabolism. Fragmentation study of 79 molecules of biological interest in positive and negative ionisation mode. Rapid Commun Mass Spectrom. 2003;17(12):1297–311. 10.1002/rcm.1054 [DOI] [PubMed] [Google Scholar]
- 53.Topp H, Fusch G, Schöch G, Fusch C. Noninvasive markers of oxidative DNA stress, RNA degradation and protein degradation are differentially correlated with resting metabolic rate and energy intake in children and adolescents. Pediatr Res. 2008;64(3):246–50. 10.1203/PDR.0b013e31817cfca6 [DOI] [PubMed] [Google Scholar]
- 54.Ramasamy R, Vannucci SJ, Du Yan SS, Herold K, Yan SF, Schmidt AM. Advanced glycation end products and RAGE: A common thread in aging, diabetes, neurodegeneration, and inflammation. Glycobiology. 2005;15(7). [DOI] [PubMed] [Google Scholar]
- 55.Santer R, Fingerhut R, Lässker U, Wightman PJ, Fitzpatrick DR, Olgemöller B, et al. Tandem mass spectrometric determination of malonyl-carnitine: Diagnosis and neonatal screening of malonyl-CoA decarboxylase deficiency. Clin Chem. 2003;49(4):660–2. [DOI] [PubMed] [Google Scholar]
- 56.Keltjens J, Vogels G. Conversion of methanol and methylamines to methane and carbon dioxide In: Methanogenesis. Springer; US; 1993. p. 253–303. [Google Scholar]
- 57.Goldstein J, Brown M. Regulation of the mevalonate pathways. Nature. 1990;343:425–30. 10.1038/343425a0 [DOI] [PubMed] [Google Scholar]
- 58.Liao P, Wang H, Hemmerlin A, Nagegowda DA, Bach TJ, Wang M, et al. Past achievements, current status and future perspectives of studies on 3-hydroxy-3-methylglutaryl-CoA synthase (HMGS) in the mevalonate (MVA) pathway. Plant Cell Rep. 2014;33(7):1005–22. 10.1007/s00299-014-1592-9 [DOI] [PubMed] [Google Scholar]
- 59.Wilkinson A, Danino V, Lithgow JK, Downie JA. N -Acyl-Homoserine Lactone Inhibition of Rhizobial Growth Is Mediated by Two Quorum-Sensing Genes That Regulate Plasmid Transfer. 2002;184(16):4510–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee F, Workman P, Roberts J, Bleehen N. Clinical pharmacokinetics of oral CCNU (Lomustine). Cancer Chemother Pharm. 1985;14:125–31. [DOI] [PubMed] [Google Scholar]
- 61.van Vliet E, Morath S, Eskes C, Linge J, Rappsilber J, Honegger P, et al. A novel in vitro metabolomics approach for neurotoxicity testing, proof of principle for methyl mercury chloride and caffeine. Neurotoxicology. 2008;29(1):1–12. 10.1016/j.neuro.2007.09.007 [DOI] [PubMed] [Google Scholar]
- 62.McCormack P, Wagstaff A. Lacidipine. Drugs. 2003;63:2327–56. [DOI] [PubMed] [Google Scholar]
- 63.Krogsgaard-Larsen P, Frølund B, Liljefors T, Ebert B. GABA A agonists and partial agonists: THIP (Gaboxadol) as a non-opioid analgesic and a novel type of hypnotic. Biochem Pharmacol. 2004;68(8):1573–80. 10.1016/j.bcp.2004.06.040 [DOI] [PubMed] [Google Scholar]
- 64.Nau H, Loscher W. Valproic acid and metabolites: pharmacological and toxicological studies. Epilepsia. 1984;25:s1 [DOI] [PubMed] [Google Scholar]
- 65.Bacq Y, Besnier J, Duong T, Pavie G, Metman E, Choutet P. Successful treatment of acute fascioliasis with bithionol. Hepatology. 1991;14:1066–9. [PubMed] [Google Scholar]
- 66.Yang XL, Zhang S, Hu QB, Luo DQ, Zhang Y. Phthalide derivatives with antifungal activities against the plant pathogens isolated from the liquid culture of Pestalotiopsis photiniae. J Antibiot (Tokyo) [Internet]. 2011;64(11):723–7. Available from: 10.1038/ja.2011.82 [DOI] [PubMed] [Google Scholar]
- 67.Sultatos L. Mammalian toxicology of organophosphorus pesticides. J Toxicol Environ Health. 1994;43:271–89. 10.1080/15287399409531921 [DOI] [PubMed] [Google Scholar]
- 68.Georgopoulos S, Sarris M, Ziogas B. Mitotic instability in Aspergillus nidulans caused by the fungicides iprodione, procymidone and vinclozolin. Pest Manag Sci. 1979;10:389–92. [Google Scholar]
- 69.Pappas A, Fisher D. A comparison of the mechanisms of action of vinclozolin, procymidone, iprodione and prochloraz against Botrytis cinerea. Pest Manag Sci. 1979;10:239–46. [Google Scholar]
- 70.Bhutto A, Morley JE. The clinical significance of gastrointestinal changes with aging. Curr Opinio Clin Nutr Metab Care [Internet]. 2008;11:651–60. Available from: http://ovidsp.ovid.com/ovidweb.cgi?T=JS&CSC=Y&NEWS=N&PAGE=fulltext&D=ppvovftj&AN=00075197-200809000-00016 [DOI] [PubMed] [Google Scholar]
- 71.Bansal T, Alaniz RC, Wood TK, Jayaraman A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci [Internet]. 2010;107(1):228–33. Available from: http://www.pnas.org/cgi/doi/10.1073/pnas.0906112107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nachshon-Kedmi M, Yannai S, Haj A, Fares FA. Indole-3-carbinol and 3,3′-diindolylmethane induce apoptosis in human prostate cancer cells. Food Chem Toxicol [Internet]. 2003;41(6):745–52. Available from: http://linkinghub.elsevier.com/retrieve/pii/S0278691503000048 [DOI] [PubMed] [Google Scholar]
- 73.Kikis E, Gidalevitz T, Morimoto RI. Protein homeostasis in models of aging and age-related conformational disease In: Tavernarakis N, editor. Protein Metabolism and Homeostasis in Aging. Springer; US; 2010. p. 138–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kopito RR. Inclusion Bodies and Protein Aggregation. 2000;68(1999):524–30. [DOI] [PubMed] [Google Scholar]
- 75.Koga H, Kaushik S, Cuervo AM. Protein homeostasis and aging: The importance of exquisite quality control. Ageing Res Rev [Internet]. 2011;10(2):205–15. Available from: 10.1016/j.arr.2010.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22(11):1427–38. 10.1101/gad.1657108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–51. 10.1016/j.cell.2006.04.014 [DOI] [PubMed] [Google Scholar]
- 78.Calderwood SK, Murshid A, Prince T. The shock of aging: Molecular chaperones and the heat shock response in longevity and aging—A mini-review. Gerontology. 2009;55(5):550–8. 10.1159/000225957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murshid A, Eguchi T, Calderwood SK. Stress proteins in aging and life span. Int J Hyperth. 2013;29(5):442–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van der Goot AT, Nollen EAA. Tryptophan metabolism: Entering the field of aging and age-related pathologies. Trends Mol Med [Internet]. 2013;19(6):336–44. Available from: 10.1016/j.molmed.2013.02.007 [DOI] [PubMed] [Google Scholar]
- 81.Lee JH, Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev. 2010;34(4):426–44. 10.1111/j.1574-6976.2009.00204.x [DOI] [PubMed] [Google Scholar]
- 82.Shimada Y, Kinoshita M, Harada K, Mizutani M, Masahata K, Kayama H, et al. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS One. 2013;8(11):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zuccato E, Venturi M, Di Leo G, Colombo L, Bertolo C, Doldi SB, et al. Role of bile acids and metabolic activity of colonic bacteria in increased risk of colon cancer after cholecystectomy. Dig Dis Sci [Internet]. 1993;38(3):514–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/8444084 [DOI] [PubMed] [Google Scholar]
- 84.Yokoyama MT, Carlson JR. Microbial metabolites of tryptophan in the intestinal tract with special reference to skatole. Vol. 32, American Journal of Clinical Nutrition. 1979. p. 173–8. [DOI] [PubMed] [Google Scholar]
- 85.Horning E, Dalgliesh C. Association of skatole-forming bacteria in the small intesting with the malabsorption syndrome and certain anaemias. Biochem J. 1958;70:13–4. [Google Scholar]
- 86.Stone TW, Stoy N, Darlington LG. An expanding range of targets for kynurenine metabolites of tryptophan. Trends Pharmacol Sci [Internet]. 2013;34(2):136–43. Available from: 10.1016/j.tips.2012.09.006 [DOI] [PubMed] [Google Scholar]
- 87.Michalowska M, Znorko B, Kaminski T, Oksztulska-Kolanek E, Pawlak D. New insights into tryptophan and its metabolites in the regulation of bone metabolism. J Physiol Pharmacol. 2015;66(6):779–91. [PubMed] [Google Scholar]
- 88.Cervenka I, Agudelo LZ, Ruas JL. Kynurenines: Tryptophan’s metabolites in exercise, inflammation, and mental health. Science (80-). 2017;357(6349). [DOI] [PubMed] [Google Scholar]
- 89.Frick B, Schroecksnadel K, Neurauter G, Leblhuber F, Fuchs D. Increasing production of homocysteine and neopterin and degradation of tryptophan with older age. Clin Biochem. 2004;37(8):684–7. 10.1016/j.clinbiochem.2004.02.007 [DOI] [PubMed] [Google Scholar]
- 90.Grohmann U, Fallarino F, Puccetti P. Tolerance, DCs and tryptophan: Much ado about IDO. Trends Immunol. 2003;24(5):242–8. [DOI] [PubMed] [Google Scholar]
- 91.Refaey M El, McGee-Lawrence ME, Fulzele S, Kennedy EJ, Bollag WB, Elsalanty M, et al. Kynurenine, a Tryptophan Metabolite That Accumulates With Age, Induces Bone Loss. J Bone Miner Res. 2017;32(11):2182–93. 10.1002/jbmr.3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Canli T, Lesch KP. Long story short: The serotonin transporter in emotion regulation and social cognition. Nat Neurosci. 2007;10(9):1103–9. 10.1038/nn1964 [DOI] [PubMed] [Google Scholar]
- 93.Yates C, Herbert J. Differential circadian rhythms in pineal and hypothalamic 5-HT induced by artificial photoperiods or melatonin. Nature. 1976;262:219–20. [DOI] [PubMed] [Google Scholar]
- 94.Hwu P, Du MX, Lapointe R, Do M, Taylor MW, Young HA. Indoleamine 2,3-Dioxygenase Production by Human Dendritic Cells Results in the Inhibition of T Cell Proliferation. J Immunol [Internet]. 2000;164(7):3596–9. Available from: http://www.jimmunol.org/cgi/doi/10.4049/jimmunol.164.7.3596 [DOI] [PubMed] [Google Scholar]
- 95.Uyttenhove C, Pilotte L, Théate I, Stroobant V, Colau D, Parmentier N, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9(10):1269–74. 10.1038/nm934 [DOI] [PubMed] [Google Scholar]
- 96.Ledesma-Escobar CA, Priego-Capote F, Luque De Castro MD. Characterization of lemon (Citrus limon) polar extract by liquid chromatography-tandem mass spectrometry in high resolution mode. J Mass Spectrom. 2015;50(11):1196–205. 10.1002/jms.3637 [DOI] [PubMed] [Google Scholar]
- 97.Tai HH, Worrall K, De Koeyer D, Pelletier Y, Tai GCC, Calhoun L. Colorado Potato Beetle resistance in Solanum oplocense X Solanum tuberosum intercross hybrids and metabolite markers for selection. Am J Potato Res. 2015;92(6):684–96. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.