Abstract
The study of inherited susceptibility to cancer has been one of the most informative areas of research in the past decade. Most of the cancer genetics studies have been focused on the common tumors such as breast and colorectal cancers. As the allelic architecture of these tumors is unraveled, research attention is turning to other rare cancers such as glioma, which are also likely to have a major genetic component as the basis of their development. In this brief review we discuss emerging data on glioma whole genome–association searches to identify risk loci. Two glioma genome-wide association studies have so far been reported. Our group identified 5 risk loci for glioma susceptibility (TERT rs2736100, CCDC26 rs4295627, CDKN2A/CDKN2B rs4977756, RTEL1 rs6010620, and PHLDB1 rs498872). Wrensch and colleagues provided further evidence to 2 risk loci (CDKN2B rs1412829 and RTEL1 rs6010620) for GBM and anaplastic astrocytoma. Although these data provide the strongest evidence to date for the role of common low-risk variants in the etiology of glioma, the single-nucleotide polymorphisms identified alone are unlikely to be candidates for causality. Identifying the causal variant at each specific locus and its biological impact now poses a significant challenge, contingent on a combination of fine mapping and functional analyses. Finally, we hope that a greater understanding of the biological basis of the disease will lead to the development of novel therapeutic interventions.
The study of inherited susceptibility to cancer has been one of the most informative areas of research in the past decade. The major reasons this field of research has been, and will continue to be, of importance are as follows: (1) the identification of susceptibility genes provides a greater understanding of the biological mechanisms of tumors, offering potential targets for therapeutic interventions; and (2) the ability to identify those at increased risk is of immediate clinical relevance in terms of primary and secondary interventions. Furthermore, given the considerable difficulties in unambiguously identifying causative exposures for many cancers, genetic associations may continue to prove extremely valuable via the functional links they reveal, current etiological hypotheses they endorse, or new ones they suggest that merit testing via gene environment–specific hypotheses.
Most of the focus of cancer genetics during the past decade has been on the common tumors such as breast and colorectal cancers. As the allelic architecture of these tumors is unraveled, research attention is turning to other rare cancers such as glioma, which are also likely to have a major genetic component as the basis of their development.
INHERITED SUSCEPTIBILITY TO GLIOMA
Studies have consistently shown that the risk of glioma is elevated 2-fold in first-degree relatives of patients with glioma and other primary brain tumors.1 At present, most cases of glioma cannot be explained by endogenous or exogenous causes. High doses of ionizing radiation and rare genetic syndromes are the only generally accepted well-defined risk factors, and they explain a small percentage of all glioma cases. The genetic basis of inherited susceptibility to glioma outside the context of a few rare mendelian cancer predisposition syndromes (ie, neurofibromatosis, Li-Fraumeni syndrome, and Turcot syndrome) is currently undefined. However, a model of disease susceptibility based solely on high-risk mutations seems unlikely, and, as with other cancers, much of the inherited risk is likely to be a consequence of the coinheritance of multiple low-risk variants, some of which will be common.
RECENT ADVANCES
The common disease/common variant hypothesis implies that conducting association analyses based on scans of single-nucleotide polymorphisms (SNPs) should be a powerful strategy for identifying low-penetrance genes for glioma.
Previous studies aimed at identifying low-penetrance variants for susceptibility to glioma have been based on a candidate gene approach formulated on preconceptions as to the role of specific genes in the development of the disease. Perhaps not surprisingly, most studies to date have evaluated only a restricted number of polymorphisms in a certain pathway, such as those influencing methylation, carcinogen metabolism, DNA repair, the cell cycle, or inflammation. Polymorphisms can affect protein function, promoter activity, messenger RNA stability, and splice variants and therefore can result in a change in the cellular ability to cope with DNA damage, which contributes to altered disease susceptibility. However, without a clear understanding of the biological mechanisms of predisposition, the definition of suitable genes for the disease is inherently problematic, making an unbiased approach to loci selection highly desirable.
Despite much research, no definitive susceptibility alleles have been unequivocally identified through association studies analyzing only a restricted series of candidate genes. As with many other cancers, positive associations have been reported for various polymorphisms in genes such as GST, MGMT, ERCC, and XRCC genes from small studies, but few of the initial positive results have been replicated in subsequent studies. The inherent statistical uncertainty of case-control studies involving just a few hundred cases and controls seriously limits the power of such studies to reliably identify genetic determinants that confer modest but potentially important risks.
After the sequencing of the human genome, large-scale harvests of SNPs were conducted, and more than 10 million were documented (http://www.ncbi.nlm.nih.gov/SNP), along with smaller numbers of insertion/deletion and copy number polymorphisms. Patterns of linkage disequilibrium (LD) between SNPs have been characterized, allowing subsets of SNPs (tagging SNPs) to be selected that, through LD with other variants, capture a large proportion of the common sequence variation in the human genome (http://hapmap.ncbi.nlm.nih.gov). The high-resolution LD maps now available (and hence comprehensive sets of tagging SNPs) coupled with the development of highly efficient analytical platforms allow genome-wide studies for disease associations to be conducted in a cost-effective manner. This approach is agnostic and does not depend on prior knowledge of function or presumptive involvement of any gene in disease causation. Moreover, it avoids the possibility of missing the identification of important variants in hitherto unstudied genes.
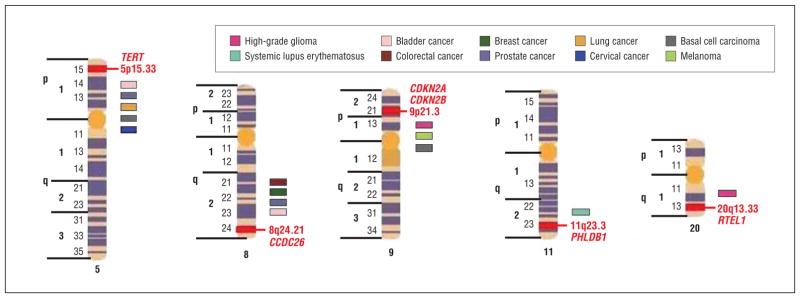
By adopting this approach, we2 and others3 have used the genome-wide association study (GWAS) approach to identify novel susceptibility genes for glioma. In our study, we genotyped 550 000 tagging SNPs in a total of 1878 cases and 3670 controls, with validation in 3 additional independent series totaling 2545 glioma cases and 2953 controls. We have used publicly available controls, and they provide a cost-effective strategy for identifying genetic factors for many diseases. However, such public controls may not be useful for identifying gene environment factors because the environment of interest may not be available for these data sets. We identified 5 risk loci for glioma (shown in the Table and Figure), namely, 5p15.33 (TERT, OMIM 187270), 8q24.21 (CCDC26, OMIM 613040), 9p21.3 (CDKN2A, OMIM 600160, and CDKN2B, OMIM 600431), 20q13.33 (RTEL1, OMIM 608833), and 11q23.3 (PHLDB1, OMIM 612834), and our results were in whole or in part based on the glioma GWAS data of Shete et al.2 The second reported GWAS of glioma, conducted by Wrensch and colleagues,3 was based on an analysis of 275 895 SNPs in 692 adult high-grade glioma cases and 3992 controls with a replication series of 176 high-grade glioma cases and 174 controls. This analysis provided further evidence to implicate the 9p21.3 (CDKN2A-CDKN2B) and 20q13.33 (RTEL1) variants.
Table.
Summary of the 5 Risk Loci in Our Glioma Genome-Wide Association Study
| Chr | Gene | Reported Gene Function | SNP ID | Genic Location | Risk Allele | Ancestral Allele Frequency | OR (95% Cl) | P Value |
|---|---|---|---|---|---|---|---|---|
| 5p15.33 | TERT | Telomerase reverse transcriptase, maintaining telomeres and cell immortalization | rs2736100 | Intron 2 | G | 0.51 | 1.27 (1.19–1.37) | 1.50×10−17 |
| 8q24.21 | CCDC26 | Retinoic acid modulator of differentiation and death | rs4295627 | Intron 3 | G | 0.17 | 1.36 (1.29–1.43) | 2.34×10−18 |
| 9p21.3 | CDKN2A/p14 (ARF)/CDKN2B | Regulates 2 critical cell cycle regulatory pathways, p53 and RB1 | rs4977756 | 5′ UTR | G | 0.40 | 1.24 (1.19–1.30) | 7.24×10−15 |
| 11q23.3 | PHLDB1 | Member of Pleckstrin homology–like domain, family B | rs498872 | 5′ UTR | T | 0.31 | 1.18 (1.13–1.24) | 1.07×10−8 |
| 20q13.33 | RTEL1 | Telomere length regulator, functions in the DNA double-strand break repair pathway by suppressing homologous recombination | rs6010620 | Intron 12 | G | 0.23 | 1.28 (1.21–1.35) | 2.52×10−12 |
Abbreviations: Chr, chromosome; CI, confidence interval; OR, odds ratio; SNP ID, single-nucleotide polymorphism identification number; UTR, untranslated region.
Figure.
Chromosome location of the 5 risk loci identified from the glioma genome-wide association study (GWAS). Each color corresponds to a particular cancer or disease that identified the same region for association from the GWAS.
The 8q24 risk variant localizes to intron 3 of CCDC26, a retinoic acid modulator of differentiation and death. Retinoic acid induces caspase-8 transcription through phosphorylation of cAMP response element binding protein and increases apoptosis to death stimuli in neuroblastoma cells and in glioblastoma cells with downregulation of telomerase activity.4 Variation at 8q24.21 has been implicated in risk of a number of common tumors, including colorectal,5 breast,6 bladder,7 and prostate8 cancer.
The 5p15.33 risk variation is defined by an SNP localizing to intron 2 of TERT, the gene for the reverse transcriptase component of telomerase, essential for telomerase activity in maintaining telomeres and cell immortalization. Although the 5p15.33 copy number change is uncommon in glioblastoma multiforme, TERT expression correlates with glioma grade and prognosis.9 We previously reported that a functional hTERT MNS16A genotype is a potential biomarker for assessment of survival outcome of glioblastoma multiforme.10 Recent data have implicated variation at 5p15.33 (TERT-CLMPTM1L) defined by rs2736098 with risk of multiple tumor types, including lung,11 bladder, prostate, and cervical cancer and basal cell carcinoma.12
The 9p21.3 association implicates genetic variation within the CDKN2A/p14(ARF)/CDKN2B tumor-suppressor genes as a determinant of disease risk. The CDKN2A gene encodes p16(INK4A), a negative regulator of cyclin-dependant kinases, and p14(ARF1), an activator of p53. The CDKN2A/p14(ARF)/CDKN2B gene has an established role in glioma with homozygous deletion in CDKN2A detectable in half of all tumors, and loss of expression has been linked to poor prognosis.13 Regulation of p16/p14ARF has been observed to be important for sensitivity to ionizing radiation, the only environmental factor strongly linked to gliomagenesis.14 Furthermore, germline mutation of the locus, specifically p14ARF, has been reported to cause the melanoma-astrocytoma syndrome.15 Variation at 9p21 in CDKN2A has also been implicated in the risk of melanoma16 and basal cell carcinoma.17
The 20q13.33 association is defined by an SNP within intron 12 of the gene encoding the Rad 3–like helicase RTEL1. Knowledge of the biological characteristics of RTEL1 is limited, but recent work has demonstrated that RTEL1 maintains genomic stability directly by suppressing homologous recombination.18 The RTEL1 gene has been classified as an important member of the DNA repair pathway by the pathway tool PANTHER (Protein Analysis Through Evolutionary Relationships).
The probable basis for the 11q23.3 association is through the effect on the gene encoding PHLDB1. Although there is no direct evidence of a role for PHLDB1 in glioma, 11q23.3 is commonly deleted in neuroblastoma.19 Variation at 11q23 has also been implicated in risk of systemic lupus erythematosus.20
The risk of glioma associated with each of these 5 risk variants is modest (relative risks of 1.2–1.5); however, because they are common, they play a major role in disease predisposition within the population. Moreover, glioma risk increases with increasing numbers of variant alleles for the 5 loci (odds ratio per allele, 1.31; 95% confidence interval, 1.26–1.36; P=1.39×10−74), whereby individuals with 8 or more risk alleles have a greater than 3-fold increase in glioma risk compared with those with a median number of risk alleles.
The risk of glioma increases with increasing numbers of variant alleles for the 5 loci. Although these data provide the strongest evidence to date of the role of common low-risk variants in the etiology of glioma, the SNPs identified are unlikely to be candidates for causality. Identifying the causal variant at each specific locus and its biological impact now poses a significant challenge, contingent on a combination of fine mapping and functional analyses.
CONCLUSIONS AND FUTURE DIRECTIONS
Our knowledge of predisposition to glioma is developing. Recent findings show that common variants influence glioma risk and highlight the importance of variation in genes encoding components of the CDKN2A-CDK4–signaling pathway in glioma. Moreover, this pathway, elucidated through the extended interaction network of CDKN2A, incorporates TERT (through mutual interaction with the HSP90 gene) and other genes (including CCDC26) that we have identified as risk factors.
As with other tumors, the advent of the GWAS enables researchers to identify variants that influence an individual’s susceptibility to develop glioma. Such studies are at the vanguard of the new technologies that will ultimately offer complete interrogation of genetic variation in the human genome. Identifying the sequence changes responsible for causal associations identified should thus provide insight into the biological mechanisms of glioma, and this may lead to the development of etiological hypotheses regarding nongenetic risk factors. Finally, we hope that a greater understanding of the biological basis of the disease will lead to the development of novel therapeutic interventions.
Acknowledgments
Funding/Support: This study was supported by grant C1298/A8362 from the Bobby Moore Fund in England and grants 5R01CA119215 and 5R01CA070917 from the National Institutes of Health in the United States; the American Brain Tumor Association; the National Brain Tumor Society; and the Wellcome Trust (principal funding).
Footnotes
Financial Disclosure: None reported.
Author Contributions: Study concept and design: Shete, Bondy, and Houlston. Acquisition of data: Liu, Robertson, and Bondy. Analysis and interpretation of data: Liu, Shete, and Hosking. Drafting of the manuscript: Liu, Bondy, and Houlston. Critical revision of the manuscript for important intellectual content: Shete, Hosking, Robertson, and Bondy. Statistical analysis: Liu, Hosking, and Bondy. Obtained funding: Bondy. Administrative, technical, and material support: Robertson, Bondy, and Houlston. Study supervision: Shete and Bondy.
References
- 1.Hemminki K, Li X. Familial risks in nervous system tumors. Cancer Epidemiol Biomarkers Prev. 2003;12(11 pt 1):1137–1142. [PubMed] [Google Scholar]
- 2.Shete S, Hosking FJ, Robertson LB, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41(8):899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wrensch M, Jenkins RB, Chang JS, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41(8):905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang M, Zhu K, Grenet J, Lahti JM. Retinoic acid induces caspase-8 transcription via phospho-CREB and increases apoptotic responses to death stimuli in neuroblastoma cells. Biochim Biophys Acta. 2008;1783(6):1055–1067. doi: 10.1016/j.bbamcr.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomlinson I, Webb E, Carvajal-Carmona L, et al. CORGI Consortium. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39(8):984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 6.Easton DF, Pooley KA, Dunning AM, et al. SEARCH Collaborators; kConFab; AOCS Management Group. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiemeney LA, Thorlacius S, Sulem P, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet. 2008;40(11):1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39(5):645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 9.Wager M, Menei P, Guilhot J, et al. Prognostic molecular markers with no impact on decision-making: the paradox of gliomas based on a prospective study. Br J Cancer. 2008;98(11):1830–1838. doi: 10.1038/sj.bjc.6604378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Wei Q, Wang LE, et al. Survival prediction in patients with glioblastoma multiforme by human telomerase genetic variation. J Clin Oncol. 2006;24(10):1627–1632. doi: 10.1200/JCO.2005.04.0402. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Broderick P, Webb E, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40(12):1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rafnar T, Sulem P, Stacey SN, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41(2):221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simon M, Voss D, Park-Simon TW, Mahlberg R, Koster G. Role of p16 and p14ARF in radio- and chemosensitivity of malignant gliomas. Oncol Rep. 2006;16(1):127–132. [PubMed] [Google Scholar]
- 14.Bondy ML, Wang LE, El-Zein R, et al. γ-Radiation sensitivity and risk of glioma. J Natl Cancer Inst. 2001;93(20):1553–1557. doi: 10.1093/jnci/93.20.1553. [DOI] [PubMed] [Google Scholar]
- 15.Randerson-Moor JA, Harland M, Williams S, et al. A germline deletion of p14ARF but not CDKN2A in a melanoma-neural system tumour syndrome family. Hum Mol Genet. 2001;10(1):55–62. doi: 10.1093/hmg/10.1.55. [DOI] [PubMed] [Google Scholar]
- 16.Bishop DT, Demenais F, Iles MM, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet. 2009;41(8):920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stacey SN, Sulem P, Masson G, et al. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet. 2009;41(8):909–914. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barber LJ, Youds JL, Ward JD, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135(2):261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo C, White PS, Weiss MJ, et al. Allelic deletion at 11q23 is common in MYCN single copy neuroblastomas. Oncogene. 1999;18(35):4948–4957. doi: 10.1038/sj.onc.1202887. [DOI] [PubMed] [Google Scholar]
- 20.Han JW, Zheng HF, Cui Y, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41(11):1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]