Abstract
Members of the interferon-inducible PYHIN protein family such as Absent in Melanoma-2 and Interferon Gamma Inducible Protein (IFI) 16 bind double-stranded DNA (dsDNA) and form caspase-1 activating inflammasomes, important in immunity to cytosolic bacteria, DNA viruses or Human Immunodeficiency Virus. IFI16 has also been shown to regulate transcription of type I Interferons during Herpes Simplex Virus infection. The role of other members of the PYHIN protein family in the regulation of immune responses is much less clear. Here, we identified an immune regulatory function for a member of the murine PYHIN protein family, p205 (also called Ifi205). Examination of immune responses induced by dsDNA and other microbial ligands in bone marrow derived macrophages lacking p205 revealed that inflammasome activation by dsDNA as well as ligands that engage the NLRP3 inflammasome was severely compromised in these cells. Further analysis revealed that p205 knockdown cells showed reduced expression of Asc at the protein and RNA level. p205 knockdown resulted in reduced binding of actively transcribing RNA Polymerase II to the endogenous Asc gene resulting in decreased transcription and processing of Asc pre-mRNA. Deletion of p205 in B16 melanoma cells using CRISPR/Cas9 showed similar loss of Asc expression. Ectopic expression of p205 induced expression of an Asc promoter-luciferase reporter gene. Together these findings suggest that p205 controls expression of Asc mRNA to regulate inflammasome responses. These findings expand on our understanding of immune regulatory roles for the PYHIN protein family.
Introduction
Foreign nucleic acids play a critical role in initiating innate and adaptive immune responses. Nucleic acid (NA) sensors expressed in distinct cellular compartments survey the extracellular and intracellular environment for signs of infection and initiate immune defenses against bacterial, viral and eukaryotic pathogens (1). NA sensors include RNA Sensors such as TLR3, 7/8 and RIG-I/MDA5 (2–4) as well as DNA sensors such as TLR9, cGAS, AIM2 and IFI16 (5–8). In addition, recognition of foreign nucleic acids especially dsDNA leads to assembly of an inflammasome, a large caspase-1 activating multiprotein complex that controls the proteolytic processing and release of IL-1β (9). Inflammasome activation also results in a proinflammatory form of cell death called pyroptosis (10). While most inflammasomes are composed of members of the NLR family, the dsDNA-activated inflammasome is formed following dsDNA binding to a PYHIN protein, Absent in Melanoma-2 (AIM2). Work from several labs including our own has defined AIM2 as a cytosolic DNA binding innate immune receptor (7, 11–13). AIM2 binds pathogen-derived dsDNA that accumulates in the cytosol during infection with DNA viruses or cytosolic bacterial pathogens (14, 15). In some instances, AIM2 can also recognize host dsDNA that gains access to the cytosol leading to autoinflammation (16). The related PYHIN protein IFI16 also forms an inflammasome during infection with Kaposi’s Sarcoma Herpes Virus (KSHV) and Human Immunodeficiency Virus 1 (HIV1) (17, 18).
The PYHIN proteins were first characterized as a family of interferon (IFN) inducible proteins that are predominantly nuclear localized (19). PYHINs are constitutively expressed in different hematopoietic cell types, although most members of this family are also inducible by type I IFN in non-hematopoietic cells (19). Phylogenetic analysis of the mammalian PYHIN proteins also called the AIM2-like receptors or ALRs show strong evolutionary and functional diversity (20, 21). The murine PYHIN locus has undergone extensive gene duplication with more than 13 members encoded in the murine genome in contrast to the human gene locus with 5 genes including MNDA, PYHIN1, POP3, IFI16 and AIM2 (19, 22). PYHIN proteins have been implicated in a wide-range of cellular processes including transcription, tumor suppression, cell cycle, cell growth, differentiation and cell death (19). The majority of the PYHIN proteins share the same structural domains. They contain an N-terminal α-helical domain known as the Pyrin domain capable of homotypic protein-protein interactions and one or more HIN200 domains in their C-terminus, except murine p208 and human POP3 that contain only a Pyrin domain and murine p202 that contains only two HIN200 domains (19, 20). Most PYHIN proteins contain a nuclear localization signal in their N-terminus that can be either monopartite, bipartite or both. Some family members also contain a nuclear export sequence that enables them to shuttle between the nucleus and the cytosol. Aim2 is the most conserved family member. Unlike the other PYHINs, AIM2 is localized in the cytosol. Phylogenetic analysis indicates that, with the exception of AIM2, there is a complete lack of orthology among mammalian ALRs (20, 21).
While the role of AIM2 and IFI16 in dsDNA recognition and immune activation has been well established, the role of other members of the PYHIN protein family, especially those in the mouse remains unclear. Recently, genetic studies in mice lacking the entire PYHIN locus and analysis of type I IFN induction following dsDNA treatment in cells from these animals demonstrated no clear link to dsDNA recognition and induction of type I IFN in murine myeloid cells (23). It remains to be defined therefore whether the PYHINs have other immune regulatory functions. In this study we examined the contribution of the murine PYHIN protein p205 (also known as Ifi205) in innate immunity. p205 is primarily expressed in macrophages and granulocytes (24). p205 shares the highest homology with p204/Ifi204 and the human PYHIN protein, MNDA (25). p205 has been implicated as a positive regulator of cell growth, and differentiation during hematopoiesis, adipogenesis and osteogenesis (26–28). To evaluate a possible immune function for p205, we investigated the ability of p205 to control immune gene expression. Using a series of loss of function and gain of function approaches combined with functional studies we demonstrate that p205 controls expression of the inflammasome adapter molecule Asc and as such regulates inflammasome dependent processing of IL-1β. This effect of p205 was not related to the prior work linking AIM2 and IFI16 to sensing of foreign DNA. Rather we find that p205 works in the nucleus to control Asc gene expression. These findings add to our understanding of PYHIN proteins in innate immunity, expanding their functions beyond dsDNA sensing to innate immune gene regulation.
Material and Methods
Reagents and Plasmids
Lipopolysaccharide (LPS) and poly-dAdT (pdAdT) were obtained from Sigma-Aldrich and Immunostimulatory DNA oligonucleotides were synthesized as described (8). Nigericin and ATP were from Invivogen and Sigma respectively. Polyinosinic-polycytidylic acid (poly I:C) was obtained from Invivogen. Sendai virus (Cantrell strain) was purchased from Charles River Laboratories (Wilmington, MA). Lipofectamine 2000® Transfection Reagent was from Invitrogen. GeneJuice was from Novagen (Madison, WI). Universal type I IFN and IFN-γ were from PBL Interferon Source (Piscataway, NJ) and PeproTech (315-05), respectively. S. typhimurium (SL1344 lab strain) was from M. O’Riordan. The plasmids used were p65-pCMV4, c/EBPβ-pcDNA (Addgene), pGL3-enhancer luciferase reporter (Promega). Other plasmids such as Asc in pMSCVneo (Clontech), p205-HA in pRZ-retro, Aim2-FLAG, p204-HA, p205-HA in pEF-BOS or HA-tagged ΔHIN-p205, ΔPYD-p205 and ΔH/ΔP-p205 in pMSCV-PIG (Addgene) were made in the lab.
Cell culture, Stimulations, ELISA and Cell death assays
Primary bone-marrow derived macrophages (BMDM) from C57BL/6J mice, cultured with L929 supernatant as a source for MCSF, were transformed using CreJ2 virus to make immortalized BMDM (iBMDM). The transformation process does not change their ability to induce type I IFNs or other immune genes. The cells were cultured in DMEM with 10% FCS and PenStrep. Antibiotics for selection were used as required. Cells were primed with repurified LPS at 100 ng/ml for 2–3h and stimulated with Salmonella sp., Nigericin or ATP for 1h, pdAdT and Interferon Stimulatory DNA (ISD) for 6h, poly I:C and Sendai virus for overnight. Cells infected with Salmonella typhimurium and media containing gentamicin (100 μg/ml) was added to kill the extracellular bacteria. Supernatants from the stimulated cells were analyzed for the cytokines, IL-1β (eBiosciences) and IFN-β by ELISA. Cell death was measured by quantitating the amount of LDH, which is released into the supernatant upon cell lysis, using CytoTox96 Non Radio Cytotoxicity Assay (Promega) kit. 10% Triton-X was added to the cells as a representation of 100% cell death.
shRNA mediated silencing
The shRNA sequences targeting p205 were cloned into a lentiviral pLKO.1 TRC cloning vector. Two separate shRNA sequences for p205 used either targeted the coding region (KD CDS) (TRCN0000095887) or the 3′ untranslated region (KD 3′UTR) (TRCN0000095884). HEK 293T cells were transfected with 4 μg shRNA along with 3 μg pSPAX (gag/pol) and 1 μg pMD2 (VSV-G) plasmids for production of lentiviral particles. Viral particles were collected at 48 h, filtered and added to immortalized BMDM. As controls, BMDM were transduced with either an empty pLKO.1 vector (EV) or pLKO.1 containing an shRNA sequence targeting GFP (GFP shRNA). The cells were selected for effective transduction by selection with puromycin (5 μg/ml).
qRT-PCR and Nanostring
RNA was extracted using Qiagen RNeasy Kit. cDNA was synthesized from 1 μg total RNA using either iScript cDNA synthesis kit (Bio-Rad) or QuantiTect Reverse Transcription Kit (Qiagen). Quantitative RT-PCR was performed using iQ SYBR Green supermix (Bio-Rad) or QuantiNova SYBR Green PCR Kit (Qiagen). Primers were constructed to respective genes (Supplemental Table 1). Target genes expressions are relative to housekeeping genes expression and were normalized to respective controls. The levels of p204, p205, Mnda, Mndal and Aim2 were measured using nCounter (Nanostring). Briefly, endogenous RNA transcript counting was performed on total RNA hybridized to a custom gene expression CodeSet and analyzed on an nCounter Digital Analyzer (29). The counts were normalized to internal spike-in and endogenous controls per Nanostring Technologies’ specifications. The heatmap was generated using the Morpheus software.
Western Blot and Co-immunoprecipitation
For detecting caspase-1 and IL-1β in the supernatants and lysates, 20% vol RIPA buffer and 30% vol SDS-loading dye were added directly to the wells containing cells and media. The samples were boiled at 100°C for 15′–30′ and were run on 13% polyacrylamide gels. Cytosolic and nuclear fractions for protein detection were prepared by using Active Motif Nuclear Extract Kit. Co-immunoprecipitation assays were carried out with 5×106 cells (treated or untreated) in ice-cold RIPA buffer (without SDS and DOC) using Protein A Dynabeads (Novex/Life Technologies, Cat# 10001D) conjugated with specific antibodies, and immunoblotted for proteins of interest. Antibodies used were against Asc (Santa Cruz, Cat# sc-22514-R or Cell Signaling, Cat# 67824), IL-1β (R&D Systems, Cat# AF-401-NA), caspase-1 (Adipogen, Cat# AG-20B-0042), Aim2 (eBioscience, 14-6008), Nlrp3 (Enzo Life Sciences), Histone 3 (Abcam, Cat# ab1791), c/EBPβ (Santa Cruz, Cat# sc-150), Usf2 (Santa Cruz, Cat# sc-862), Gapdh (Sigma, Cat# G9295), β-actin (Sigma, Cat# A3854) HA-tag (Anti-HA-Peroxidase; Roche, Cat# 12 013 819 001), and FLAG-tag (Sigma, Cat# A8592). An affinity purified polyclonal antibody against p205 was generated using the following peptide: AGLDRLINFCERVPTL-amide) was generated (21st Century Biochemicals).
Confocal Microscopy
HEK 293T cells were transfected with p205 tagged with CFP and 24h post-transfection, the cells were washed with PBS and stained with Acridine Orange and/or Cholera toxin-B (CtxB) (Thermofisher) and were visualized by confocal microscopy (Leica 8000) for localization.
Luciferase assays
The Asc gene promoter from −2000 +10 bp was cloned into pGL3-Enhancer reporter vector upstream of the firefly luciferase gene. The promoter-reporter construct was either transfected alone or co-transfected with plasmids expressing p205, p204, Aim2, p205 deletion mutants, p65/RelA or c/EBPβ or a combination thereof in HEK 293T cells. A plasmid expressing Renilla luciferase gene under thymidine kinase (pGL4-TK Renilla) promoter was included as transfection efficiency control. Data is represented as fold change over Asc reporter construct alone, and relative to transfection efficiency.
Chromatin Immunoprecipitation
Cells were fixed with 1% formaldehyde, lysed and sheared. The DNA was quantified, and 5 μg of total chromatin was immunoprecipitated with specific antibodies and Dynabeads Protein G (Novex/Life Technologies, Cat#10009D). The DNA was then reverse crosslinked, purified and quantitated by qPCR amplification with primers at Asc, Gapdh genes (Supplemental Table 1). Antibodies used were against total RNA Polymerase II (RNAPII; Active Motif Cat# 39097), phospho Serine-2 RNA Polymerase II (Ser2 RNAPII; Abcam Cat# ab5095), phospho Serine-5 RNA Polymerase II (Ser5 RNAPII; Abcam Cat# ab5131) or IgG isotype (Abcam, Cat# AB37415 and Cell Signaling, 5415).
CRISPR/Cas9-mediated gene knockout
B16 mouse melanoma cells were cultured in DMEM containing 10% FBS, 0.5% Ciprofloxacin and 0.0075 % β-mercaptoethanol and transfected with 200 ng of plasmid containing mCherry-Cas9 and a U6 promoter-driven gRNA against p205 (Target Sequence: ATGAAGCCGAAGATGAGACCTGG) using Lipofectamine 2000® according to manufacturer’s protocol (30). Two days after transfection cells were sorted for mCherry expression. Positive cells were plated at limiting dilution to obtain single cell clones. Genotyping of the B16 clones was conducted by deep sequencing (Illumina, MiSeq) as previously described (31) using the following primer sequences: 5′-ACACTCTTTCCCTACACGACGctcttccgatctCGTGAAGAAGATCAAGGCATCTG-3′ and 5′- TGACTGGAGTTCAGACGTGTGctcttccgatctAAATCTCAGGGAGAAGTGGGGGA-3′ (uppercase letters: 1st PCR adapter sequences, lowercase letters: linker sequences, uppercase letters: target site specific primer sequences). Cell clones harboring all-allelic frame shift mutations were then selected as p205 KO cell clones.
Statistics
ELISA and luciferase assays are presented as mean ± SD from three independent biological replicates and are representative of at least three separate experiments. Data was analyzed using two-way ANOVA by Prism 6 Software (GraphPad, San Diego, CA). The p values < 0.05 were considered significant (*p < 0.05, **p < 0.001, ***p < 0.005, ****p < 0.0001) and n.s. = non-significant, unless otherwise indicated.
Results
Lipopolysaccharide (LPS) and Interferon (IFN)-inducible p205 localizes to the nucleus in resting macrophages
The p205 gene is encoded on chromosome 1q, the last of the 13 consecutive genes of the mouse PYHIN locus. p205 is encoded on the reverse strand and flanked by p202b at the 5′-end and several olfactory receptor genes as well as the Spta1 gene at its 3′-end. p205 is expressed in murine primary bone marrow derived macrophages and treatment of these cells with LPS, type-I IFN and IFN-γ treatments further unregulated its expression, similar to other PYHIN genes such as Mnda and p204 (Figure 1A, B and C). The inducible expression of p205 was as robust as the well-characterized immune genes such as Il6 or Cxcl10 that are used as positive controls.
Figure 1. p205 is induced by LPS, Type I and II IFN and localizes to the nucleus.
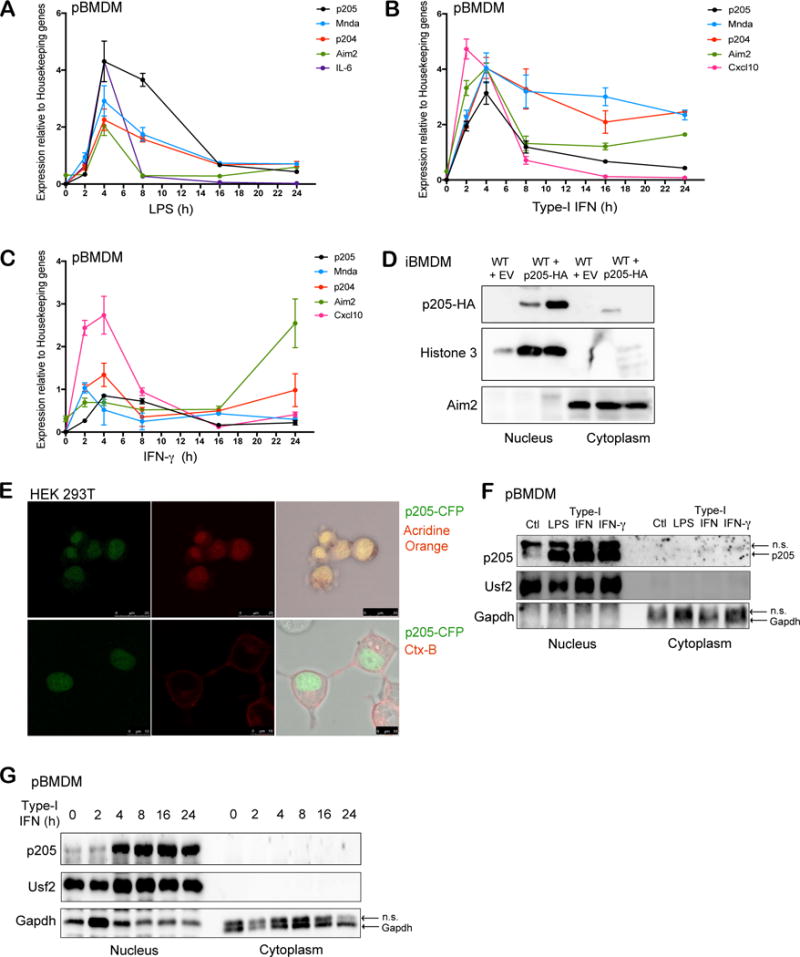
Primary BMDMs stimulated with (A) LPS (200 ng/ml), (B) Type-I IFN (100 U/ml) or (C) IFN-γ (20 ng/ml) at different time points (0, 2, 4, 8, 16, 24 h) were tested for p205 mRNA expression as well as other PYHIN genes, Mnda, p204 and Aim2. Levels of IL-6 or Cxcl10 mRNA were included as positive controls. Gene expression is reported relative to a combination of three housekeeping genes Gapdh, Hprt, β-actin. (D) Immunoblot analysis of the nuclear and cytoplasmic fractions of wild-type BMDM transduced with empty vector (EV) and wild-type BMDM overexpressing HA-tagged p205 using anti-HA antibody. Histone 3 and Aim2 were used as controls for nuclear and cytosolic extracts respectively. (E) Confocal microscopy of CFP-tagged p205 (green) in HEK 293T cells stained for nucleus using Acridine orange in first panel and Cholera Toxin B (CtxB) staining plasma membrane in second panel. Data is representative of two independent experiments. (F) Primary BMDM untreated (Ctl) or treated with LPS for 6h, type-I IFN for 16h or IFN-γ for 16h were separated into nuclear and cytosolic fractions and immunoblotted for endogenous p205 (n.s.- non-specific band). (G) Western blot analysis of endogenous p205 expression in the nuclear and cytosolic extracts of primary macrophages treated with type-I IFN as indicated. Usf2 and Gapdh were used as controls for nuclear and cytosolic fractions respectively (n.s.- non-specific band).
To evaluate the localization of p205 in macrophages, we overexpressed hemagglutinin (HA) tagged p205 into immortalized BMDM and performed subcellular fractionation to purify nuclear and cytosolic extracts. Analysis of these fractions by immunoblotting for HA tagged p205 in two independently transduced BMDM showed that p205 is primarily localized in the nucleus (Figure 1D). We also confirmed the nuclear localization of p205 using confocal microscopy by imaging a CFP-tagged p205 construct expressed in HEK 293T cells (Figure 1E). Treatment of primary BMDM with LPS, type-I IFN and IFN-γ increased p205 protein expression in the nucleus (Figure 1F). Further, we stimulated primary BMDM with type-I IFN at different time points and separated the cell lysates into nuclear and cytosolic fractions, which were immunoblotted for endogenous p205. Upon IFN-treatment, p205 expression was robustly increased over time in the nucleus (Figure 1G).
Knockdown of p205 in bone marrow derived macrophages results in impaired inflammasome activation
To determine the potential immune function of p205, we generated immortalized murine BMDM expressing shRNA-targeting p205. BMDM were transduced with pLKO.1 lentiviral particles either containing short hairpin RNA (shRNA) targeting p205 or GFP as a control. Two shRNA sequences for p205 were used which targeted the 3′ untranslated region (KD 3′UTR) (Figure 2A) and the coding region (KD CDS) (Figure 2B) of the mRNA, respectively. The level of p205 was evaluated by quantitative PCR (qPCR). The shRNA targeting the coding region specifically knocked down p205 mRNA with no apparent effect on the two most closely related murine PYHINs, murine Mnda or p204 mRNA. This effect was also evaluated using Nanostring where p205 mRNA was reduced with little impact on 4 other related PYHIN genes (p204, Mnda, Mndal and Aim2) (Figure 2C).
Figure 2. p205 knockdown using shRNA results in impaired inflammasomes activation.
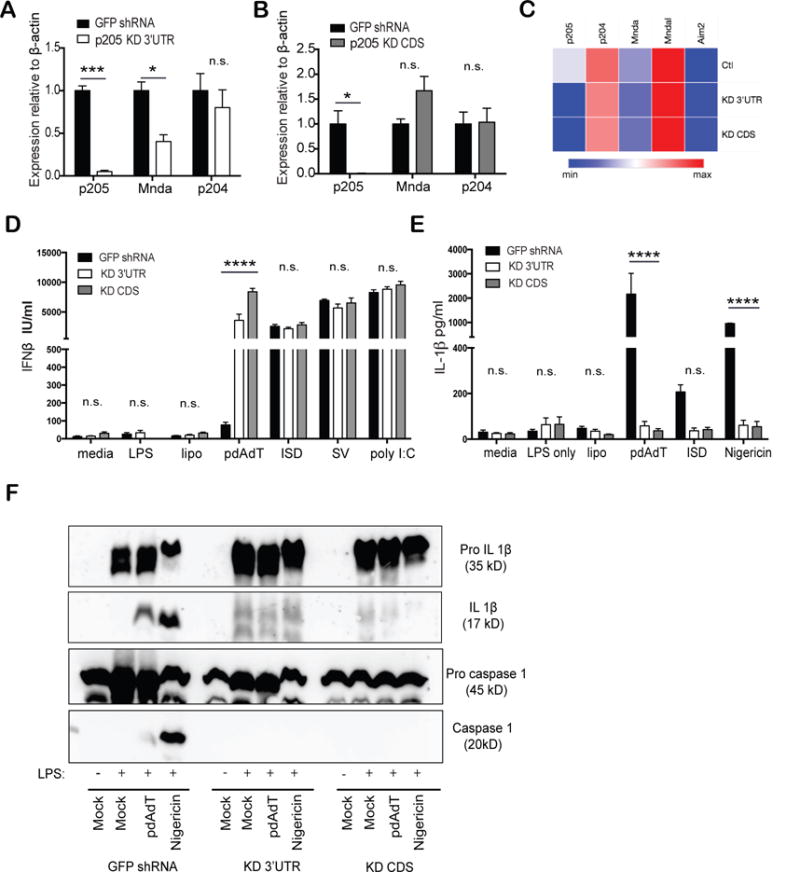
BMDM transduced with shRNA targeting either (A) 3′UTR or (B) CDS of p205 gene were inspected for expression of p205, Mnda and p204 mRNA relative to β-actin mRNA and normalized to expression in GFP shRNA BMDM. (C) Heatmap of PYHIN gene expression in p205 KD 3′UTR and KD CDS BMDM compared to control (Ctl) BMDM. The p205 knockdown BMDMs were primed with LPS (200ng/ml) for 3h and then stimulated with transfected pdAdT (1μg/ml for 6h), transfected ISD (3μM for 6h), Nigericin (10μM for 1h), ATP (5μM) or, stimulated alone with Sendai virus (SV; overnight) or poly I:C (overnight). Secreted (D) IFNβ and (E) IL-1β levels were assessed by ELISA. (F) GFP shRNA CTL, p205 KD 3′UTR and KD CDS were primed with LPS (200ng/ml) for 3h and then stimulated with pdAdT (1 μg/ml for 6h) or Nigericin (10 μM for 1h) and the supernatants and the lysates from the macrophages were immunoblotted for pro-IL1β (35 kD), cleaved form of IL-1β (p17), pro-caspase 1 (45kD) and the active subunit of caspase 1(p20).
Both of these macrophage cell lines with p205 knockdown were then tested for their ability to respond to different immune stimuli and induce cytokine gene expression. We first evaluated the inducible expression of type-I IFN since prior work had identified other PYHIN proteins such as IFI16 and p204 as regulators of type I IFN gene expression. iBMDM expressing shRNA targeting p205 or GFP were stimulated with LPS, pdAdT, Interferon Stimulatory DNA (ISD), Sendai virus (SV) or poly I:C, all of which induced the type I IFN, IFNβ, the levels of which were measured by ELISA. For most of these ligands the inducible expression of IFNβ was largely unaffected by the absence of p205 (Figure 2D). However, stimulation of the p205 knockdown macrophages with pdAdT, a synthetic dsDNA mimetic showed an enhanced IFN-β response compared to the control cells. p205 knockdown cells were also tested for IL-1β production since PYHINs such as AIM2 and IFI16 form inflammasome complexes that regulate IL1β production. The cell lines were treated for two hours with 200 ng/mL of LPS which acts as Signal 1 to prime cells allowing transcriptional induction of pro-IL1β. These cells were then challenged with transfected pdAdT or ISD or Nigericin, which activate the Aim2 and Nlrp3 inflammasomes, respectively. Macrophages lacking p205 had reduced levels of IL-1β in response to all of these stimuli (Figure 2E).
To understand whether the diminished levels of p205 impacted on either the priming (signal 1) or processing of pro-IL-1β (signal 2), the supernatants and lysates from the stimulated macrophages were tested for levels of both the inactive and proteolytically processed forms of both caspase-1 and IL-1β. Cells lacking p205 expressed similar levels of pro-IL-1β following stimulation with LPS, ruling out an effect of p205 on TLR4 signaling and transcription of pro-IL1β (Figure 2F top panel). In contrast the levels of mature processed IL-1β were significantly reduced in the knockdown cells (Figure 2F middle upper panel). Moreover, the activated form of caspase-1 was also undetectable in cells with shRNA knockdown for p205 (Figure 2F, bottom panel), despite comparable amounts of pro-caspase-1 between the control and the knockdown cell lines (Figure 2F middle lower panel). Collectively these observations indicate that knockdown of p205 impacted inflammasome dependent activation of caspase-1 leading to reduced processing of pro-IL1β.
Reduced expression of p205 leads to reduced expression of Asc
To understand the mechanistic basis for the defect in inflammasome activation in cells expressing shRNA-p205, we first measured the expression of Nlrp3 and Aim2 as well as the adapter protein Asc (Apoptosis-associated speck-like molecule containing CARD domain) in these LPS-stimulated macrophages. While there was no difference in the protein levels of either Nlrp3 or Aim2, expression of the Asc protein was completely absent in the p205 knockdown cell lines (Figure 3A). We further confirmed the loss of p205 protein in these cell lines using a newly generated antibody generated against p205, and the LPS-stimulated macrophages lacking p205 showed no Asc protein expression (Figure 3B). The expression of Asc was further investigated by measuring its transcript levels. These studies indicated that cells with shRNA knockdown of p205 had decreased levels of Asc mRNA (Figure 3C). Collectively, these findings indicate that p205 regulates Asc mRNA levels. To discern whether the defective inflammasome responses we observed in the p205-shRNA iBMDM was specifically due to the absence of Asc, we restored Asc expression in these cells using retroviral transduction. The Asc cDNA was cloned into a retroviral vector, pMSCV and viral particles were transduced in p205 knockdown macrophages to renew Asc expression. p205-shRNA cells transduced with the retrovirus had restored Asc levels comparable to that seen in control cell lines (Figure 3D). The reconstituted stable cell lines were then tested for IL-1β release by ELISA. IL-1β release by the macrophages following LPS priming and pdAdT or Nigercin treatment, was fully restored upon re-expression of Asc in these cells (Figure 3E). Salmonella typhimurium also activates the inflammasome. When Salmonella is in its log phase of growth i.e. the bacteria are actively dividing they are recognized by Nlrc4, while when in their non-dividing or stationary growth phase, Salmonella is primarily recognized by Nlrp3. Unlike Nlrp3 or Aim2, Nlrc4 itself contains a CARD domain, and can directly recruit pro-caspase 1 as well as engage Asc to do so. Thus, Nlrc4 can activate the inflammasome in a manner that is only partially dependent on Asc (32). The p205 knockdown cells were challenged with Salmonella in their stationary phase and log phase after LPS stimulation, for one hour. Salmonella in the stationary phase (overnight culture) that primarily engages Nlrp3 to activate the inflammasome showed minimal IL-1β production by ELISA. But when the same cells were infected with the actively dividing Salmonella (4–6hr culture), the cells retained part of their inflammasome activating capability that was again rescued to its full potential upon reconstitution with Asc (Figure 3F). Further, stimulation with LPS and pdAdT in the Asc-reconstituted macrophages showed processing of caspase 1 and IL-1β into their active forms signifying a functional inflammasome (Figure 3G).
Figure 3. Loss of p205 leads to a defect in Asc expression.
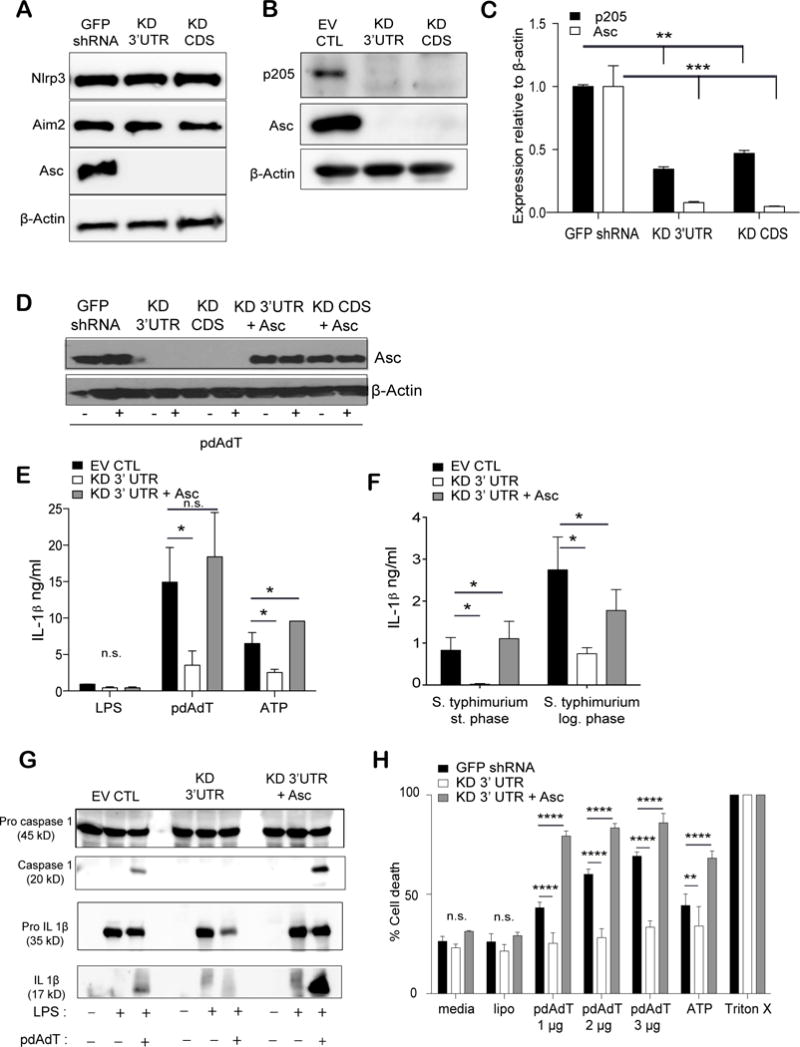
(A) Levels of Nlrp3, Aim2, Asc and β-actin proteins were elucidated by Western blot in LPS stimulated (200 ng/ml) GFP shRNA CTL, p205 KD 3′UTR and KD CDS macrophages. (B) Immunoblot of p205, Asc and β-actin proteins in LPS-stimulated (200 ng/ml for 6h) p205 knockdown macrophages. (C) Levels of p205 and Asc mRNA (relative to β-actin and normalized to GFP shRNA BMDM) were detected by qPCR in the same cell lines. (D) Western blot analysis of Asc overexpression in the p205 knockdown BMDM either left untreated or treated with LPS treated (200 ng/ml for 3h). Asc reconstituted cell lines tested for IL-1β production (E) on stimulation with LPS (200 ng/ml for 3h) and pdAdT (1 μg/ml for 6h) or ATP (5 μM for 1h) (left) and (F) with overnight culture (stationary phase) or log phase culture of Salmonella sp. by ELISA. (G) Levels of caspase 1 and IL-1β processing in the Asc reconstituted cells were detected by Western Blot and (H) cell death was measured by amounts of LDH released with LPS (200 ng/ml for 3h) and pdAdT (1 μg, 2 μg, or 3 μg per ml for 6h) and/or ATP (5 μM for 1h) stimulation.
Inflammasome activation also leads to pyroptotic cell death (10), which can be monitored by examining the release of lactate dehydrogenase enzyme (LDH). Poly dAdT treatment of macrophages leads to Aim2 inflammasome dependent release of LDH. Consistent with the other effects on inflammasome function, the release of LDH was also significantly affected in cells with p205 knockdown. Restoration of Asc in these knockdown cell lines rendered these cells susceptible to cell death similar to the levels seen in the control cell line (Figure 3H). Altogether, these findings attribute the loss of inflammasome responses in p205-knockdown cells directly to p205-dependent effects on Asc expression.
Reconstitution of p205 expression in macrophages restores Asc expression and inflammasome activation
We next restored expression of p205 in the knockdown cells using a HA-tagged retroviral vector. The levels of p205 gene expression were measured by qPCR (Figure 4A). The reconstitution of p205 restored Asc mRNA expression (Figure 4B). Moreover, reconstitution with p205 restored Asc protein expression in these knockdown cell lines (Figure 4C). p205 protein expression was detected using an anti-HA antibody. Cell lysates from GFP shRNA, p205 KD 3′UTR cell lines along with the p205 KD 3′UTR cell line reconstituted with p205-HA were separated into nuclear and cytoplasmic fractions. The reconstituted p205 was found to be primarily nuclear. Levels of Asc in both the nucleus and cytoplasm increased in cells expressing the p205-HA. Levels of Aim2 were largely unchanged upon p205 reconstitution. We also ectopically expressed the HA-tagged p205 in wild-type immortalized BMDM, which resulted in increased Asc mRNA levels compared to that found in control cell lines (Figure 4D). Analysis of IL-1β release and cell death in these cells further demonstrated that reconstitution of p205 in the knockdown cells restored inflammasome dependent responses (Figure 4E and F). Altogether, these findings provide further evidence for a role for p205 as a regulator of Asc expression and inflammasome responses.
Figure 4. Reconstitution of p205 rescues Asc expression and inflammasome activation.
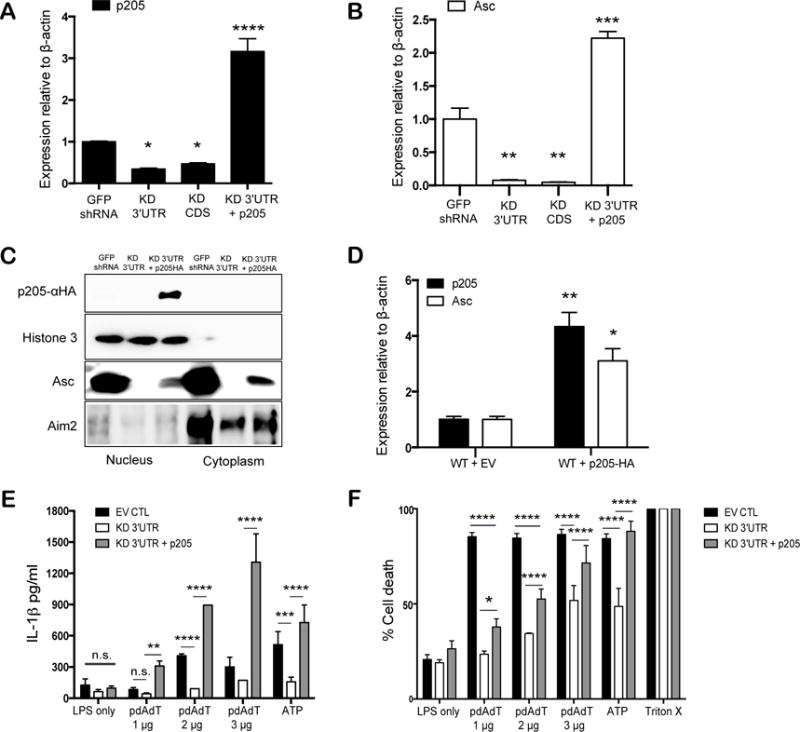
(A) p205 and (B) Asc mRNA expression relative to β-actin and normalized to GFP shRNA BMDM were measured in BMDMs reconstituted with p205. (C) Western blot analysis of nuclear and cytoplasmic fractions of p205 reconstituted cell lines to detect p205-HA, Histone H3, Asc and Aim2. (D) p205 and Asc mRNA levels by qPCR relative to β-actin in p205- overexpressing BMDM. The rescued cells were tested for (E) IL1β production and (F) cell death with LPS (200ng/ml for 3h) and pdAdT (1 μg, 2 μg, or 3μg per ml for 6h) or ATP (5 μM for 1h) by ELISA and by measuring amount of LDH released respectively.
RNA polymerase II occupancy at the Asc locus is reduced in macrophages expressing p205-shRNA
The reduced mRNA levels of Asc in cells lacking p205 could be due to either effects on Asc gene transcription or Asc mRNA stability. To further explore how p205 impacted Asc mRNA levels we performed chromatin immunoprecipitation assays to explore the binding of RNA Polymerase II (RNA Pol II) to the endogenous Asc promoter in both the GFP shRNA and p205 KD BMDM. Figure 5A shows a schematic of the Asc gene locus and the location of the primers used to evaluate RNA Pol II binding. Using antibodies to RNA Pol II and an isotype control IgG, we evaluated RNA Pol II binding to the endogenous Asc promoter. RNA Pol II binding was considerably lower in the p205 KD cell lines using a series of primers as indicated (Figure 5B). We also expanded this analysis using antibodies to RNA Pol II phosphorylated either at position Serine 5 (Serine-5P RNA Pol II) or at Serine 2 (Serine-2P RNA Pol II) in its C-terminal domain. The RNA Pol II carboxy-terminal domain (CTD) contains multiple repeats of a heptapeptide sequence, YSPTSPS that is differentially phosphorylated to control the function of the polymerase (33). When preferentially phosphorylated at Serine 5 alone, there is only abortive transcription where the RNA Pol II can initiate, but not elongate or process functional mRNA (33). The change from an initiating to an actively elongating RNA Pol II occurs with the increase in phosphorylation of the Serine 2 residue on the CTD repeats (33). Consistent with the findings with antibodies to total RNA Pol II, both Serine-5P RNA Pol II (Figure 5C) and Serine-2P RNA Pol II (Figure 5D) distributions on the Asc gene were considerably lower in the p205-shRNA cells compared to the GFP shRNA BMDM. The recruitment of total RNA Pol II, Serine-5P RNA Pol II and Serine-2P RNA Pol II to the housekeeping gene, Gapdh, was equivalent between the GFP shRNA and p205 KD macrophages (Figure 5E).
Figure 5. p205 modulates transcription and splicing from the endogenous Asc gene.
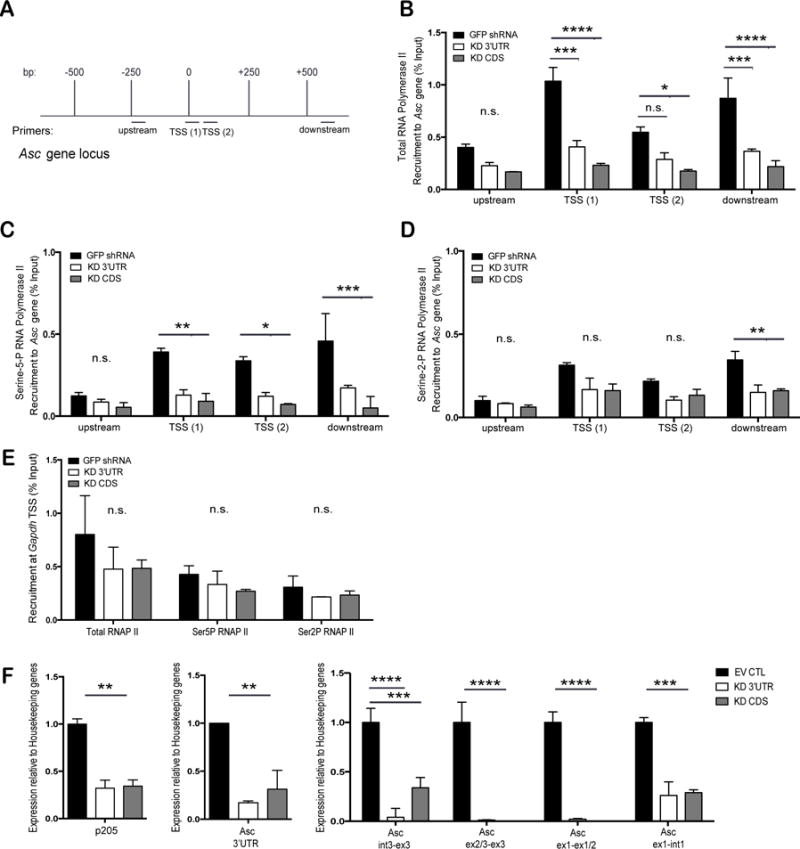
(A) Schematic of the Asc gene locus and Chromatin-IP primers locations on the gene. (B) Recruitment of total RNA Pol II to endogenous Asc gene in GFP shRNA CTL, p205 KD 3′UTR or KD CDS macrophages. Recruitment of (C) Serine-5-P RNA Pol II and (D) Serine-2-P RNA Pol II on endogenous Asc gene. (E) Recruitment of total, Serine-5-P or Serine-2-P RNA Pol II to the Gapdh transcription start site. All values are represented as percent fraction of total input DNA. Data was calculated against the IgG isotype control and is representative of three independent experiments. (D) qRT-PCR analysis of p205, nascent or mature Asc mRNA expression (relative to Hprt and Gapdh, and normalized to EV CTL BMDM) in p205 knockdown macrophages.
Although p205 KD BMDM showed reduced levels of RNA Pol II recruited at the endogenous loci, the levels of Asc mRNA appeared to be affected more severely than the ChIP data indicated. Recent studies have shown that RNA Pol II is also a major player in successful mRNA processing and splicing. Transcription and splicing are dependent on each other- a phenomenon termed co-transcriptional splicing (34). Hence, to understand if p205 was regulating both transcription as well as mRNA processing of the Asc transcript, we measured the levels of Asc pre-mRNA as well as that of the processed and mature mRNA in cells with and without p205. By designing primers that specifically measure pre-mRNA and mature mRNA (Supplemental Table 1), we found that p205 KD BMDM had very low levels of the Asc pre mRNA and little mature Asc mRNA being made (Figure 5F). We conclude from this data that p205 controls Asc expression both at the level of gene transcription as well as processing of mRNA.
p205 drives expression from an Asc promoter-reporter construct in a dose-dependent manner
To gain further insight into the mechanisms involved in the transcription of Asc, we cloned the Asc promoter (−2000 to +10 Asc gene) as a reporter gene upstream of a firefly luciferase gene. This promoter-reporter construct was transfected into HEK 293T cells together with increasing concentrations of p205. Ectopic expression of p205 led to a dose dependent increase in the Asc-luciferase reporter gene. This effect was specific to p205, as transfection of two related PYHIN proteins p204 and Aim2 had no effect (Figure 6A). We also determined the role of specific domains of p205 on reporter gene activity by generating and testing deletion mutants of p205 that either lacked the pyrin domain (ΔPYD), the HIN domain (ΔHIN) or both (ΔH/ΔP). p205 mutants lacking either the HIN or PYD domain failed to induce expression from the Asc promoter (Figure 6B) indicating that both HIN and PYD domains were important.
Figure 6. p205 drives expression from an Asc promoter-luciferase reporter construct.
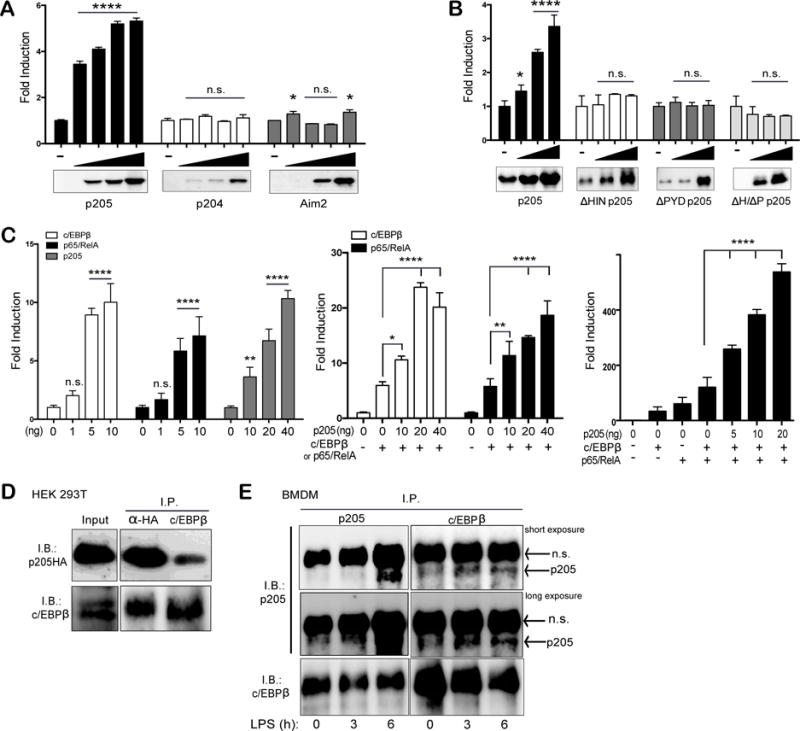
HEK 293T cells were transfected with a mixture containing 10 ng of TK-Renilla luciferase with 1 ng of Asc promoter firefly luciferase reporter and increasing amounts of (A) p205-HA, p204-HA or Aim2-FLAG, or (B) with full length p205 or p205 deletion mutants as indicated. (C) Transfection of increasing concentrations of c/EBPβ, p65/RelA and p205 alone or, co-transfection of increasing concentrations of p205-HA with either c/EBPβ or p65/RelA with Asc promoter-reporter or, transfection of the Asc promoter-reporter with increasing concentrations of p205 with both c/EBPβ and p65/RelA, as indicated. All luciferase values were measured and normalized to Renilla values. Values are displayed as fold change over the Asc reporter construct alone. Data is representative of three independent experiments. (D) Co-immunoprecipitation and immunoblot of overexpressed HA-tagged p205 and c/EBPβ in HEK 293T cells using anti-HA and anti-c/EBPβ antibodies. (E) Co-immunoprecipitation and Western blot of endogenous p205 and c/EBPβ in LPS-stimulated BMDM using antibodies against p205 and c/EBPβ. Data is representative of two independent experiments (n.s.- non-specific band).
Analysis of the Asc promoter for binding sites for transcription factors revealed putative binding sites for c/EBPβ and NF-κB amongst others. Previously, Parsons et al. had shown that TNFα treatment could enhance Asc expression through p65/RelA in MCF7 cell lines (35). Furthermore, p205 has been shown to co-localize and bind directly with the CCAAT/enhancer binding protein-β (c/EBPβ) in mouse adipose-derived stem cells (27). Furthermore, c/EBPβ has also been implicated in the induction of several proinflammatory genes in macrophages (36). Therefore we tested the effect of ectopic expression of c/EBPβ or p65/RelA on Asc reporter gene expression. In both cases these transcription factors increased Asc luciferase reporter gene expression in a dose dependent manner (Figure 6C, left panel). In addition, co-expressing p205 together with c/EBPβ or p65/RelA, further enhanced Asc reporter gene expression (Figure 6C, middle panel). Transfection of c/EBPβ and p65/RelA together with increasing concentrations of p205 further synergized to drive more robust expression from the Asc gene reporter (Figure 6C, right panel). Furthermore, transiently transfected HA-tagged p205 was found to interact with overexpressed c/EBPβ in HEK 293T cells (Figure 6D) as well as endogenously in both resting and LPS-stimulated (200ng/ml for 3h or 6h) wild-type macrophages (Figure 6E) by co-immunoprecipitation assays. Together these observations indicate that p205 can increase Asc reporter gene activation and can collaborate with c/EBPβ or p65/RelA to do so.
Knockout of p205 using CRIPSR/Cas9 impairs Asc expression
Having defined p205 as a regulator of Asc gene expression using shRNA–based knockdown approaches, we also wanted to evaluate the impact of p205 on Asc gene expression using an independent strategy and in a different cell-type that expresses p205. We used B16 melanoma cell lines to generate CRISPR/Cas9 mediated knockout of p205. Plasmids expressing Cas9 and the sgRNA targeting specific regions of p205 were transfected into B16 melanoma cells to generate p205 knockout (p205 KO) cells. A clone of these cells was produced and evaluated for p205 deficiency using sequencing, qPCR and Western blot. Levels of p205 mRNA following overnight IFN-γ stimulation were undetectable in these cells (Figure 7A). In addition, there was no discernible p205 protein expression in p205 knockout cells stimulated with Sendai virus (Figure 7B). Similar to what we had observed in our macrophage system, we found that cells lacking p205 had a loss of mature Asc mRNA and protein (Figure 7C and D). These observations further support our shRNA-based studies and underscore the importance of p205 as a regulator of Asc expression.
Figure 7. CRISPR KO of p205 leads to reduced Asc expression in B16 melanoma.
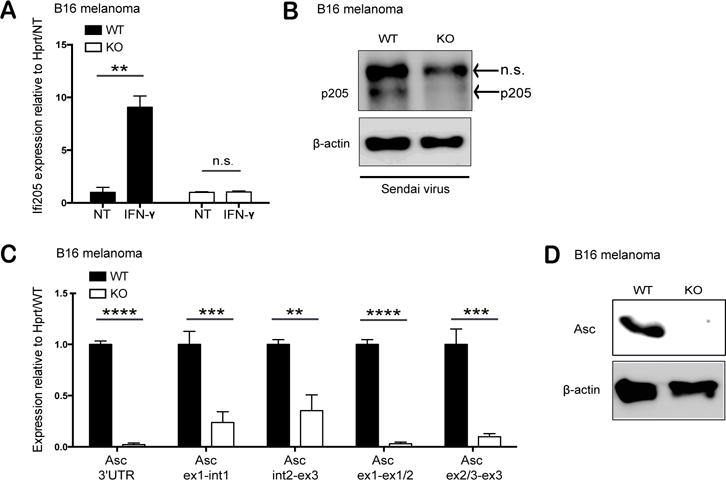
(A) Expression of p205 mRNA in IFN-stimulated wild type (WT) and p205 knockout (KO) B16 melanoma cell lines relative to Hprt and normalized to non-treated (NT). (B) p205 and β-actin protein expression in WT and p205 KO cell lines stimulated with Sendai virus (n.s.- non-specific band) (C) Mature mRNA and pre-mRNA profile of Asc expression (relative to Hprt and normalized to WT) and (D) Western blot analysis of Asc and β-actin in WT and p205 KO B16 melanoma cell lines.
Discussion
Studies over the last decade have defined the importance of AIM2, a cytosolic DNA binding protein as a regulator of caspase-1 activity and proteolytic processing of IL-1β and IL18 (37). The role of the PYHINs in sensing dsDNA was further highlighted by the identification of IFI16 as a regulator of type I IFN gene transcription following HSV-1 infection (8).
Early studies had also linked members of the murine PYHIN family to type I IFN gene regulation. Knockdown of mouse Ifi203 and mouse Ifi204 have been shown to dampen the IFN response to infection with multiple pathogens, including HSV-1, human immunodeficiency virus (HIV), murine leukemia virus (MLV), Francisella tularensis, and Mycobacterium tuberculosis (8, 38, 39). Despite these studies, a recent genetic study from Gray et al. using mice lacking the entire PYHIN locus, lacking all 13 PYHIN genes, found that there was no change in the IFN signature when challenged with various immunostimulatory DNA ligands, DNA virus infection and lentivirus infection (23). This body of work indicates that the murine PYHIN proteins play limited roles in DNA ligand recognition at least in the myeloid cells and fibroblasts tested in these studies, raising the possibility that the PYHINs have alternative functions in these cells.
Prior to the discovery of AIM2 and IFI16 as sensors of microbial DNA, PYHIN proteins were shown to regulate cell growth, differentiation, tumor suppression and DNA damage responses (25). IFI16, IFIX, p202, and p204 regulate cell cycle transcription factors such as p53, p21, pRb, and E2F resulting in cell cycle arrest (40). p202 acts as a transcriptional repressor targeting NF-κB (41), AP-1 (42, 43), MYOD1 (44), and myogenin (19, 43). p204 also regulates gene expression during monocyte/macrophage differentiation and osteoblast differentiation (45–47).
Here we expand upon these studies and define p205, a murine PYHIN protein as an additional regulator of gene expression in innate immunity. Using a series of loss of function approaches (both shRNA and CRISPR/Cas9 mediated gene editing), we report that p205 regulates expression of the inflammasome adapter protein Asc and in doing so controls inflammasome activation pathways broadly. Cells lacking p205 failed to activate caspase-1 and control inflammasome dependent processing of IL1β in response to multiple ligands that engage the Aim2 inflammasome as well as the Nlrp3 inflammasome. The abrogated inflammasome activation in the p205 KD macrophages upon pdAdT stimulation also likely explained the enhanced IFNβ levels detected in these cells, since prior work from our lab and others have shown that activation of the Aim2 inflammasome by intracellular DNA antagonizes the type I IFN pathway (14, 48). In cells with defects in inflammasome responses, there is a more robust dsDNA driven induction of type I IFNs.
By carefully measuring expression levels of key components of the Aim2 and Nlrp3 pathways, we found that cells lacking p205 had reduced expression of Asc. This effect was observed at both the protein and mRNA levels. The compromised inflammasome dependent responses observed in these cells could be fully rescued by ectopically expressing either p205 or Asc itself. Cells lacking p205 had reduced RNA Polymerase II binding to the Asc gene indicating that p205 functioned in part to control Asc gene transcription. In addition, by comparing the levels of the Asc pre-mRNA to those of the mature transcript, we could also observe additional effects on Asc mRNA processing. It is broadly accepted that the CTD of RNA Pol II is involved in efficient transcription as well as mRNA processing (34), and the Serine-2 phosphorylated polymerase determines the rate of mRNA elongation, spliceosome assembly and splicing efficiency (49). Thus we deduced from our ChIP experiments and qPCR analysis of Asc pre-mRNA and mature transcripts, that the absence of p205 not only affects Asc gene transcription but also affected the processing of the immature Asc mRNA.
Further, we show that in regulating gene expression, p205 cooperates with both c/EBPβ and p65/RelA to drive Asc expression. The transcription factor c/EBPβ is important in controlling macrophage differentiation. c/EBPβ remains fully active in resting macrophages and stays positioned on target genes, ready to stimulate transcription with other inducible transcription factors (36). p205 has previously been shown to interact with c/EBPβ in adipocytes. p205 interacts with c/EBPβ in unstimulated macrophages as well, and maintains basal expression of Asc, while under stimulated conditions, LPS-inducible p205 may interact with both c/EBPβ and activated NF-κB transcription factors to further enhance expression.
Previous studies have shown that ASC, originally identified as TMS1 (Target of methylation induced silencing-1) is influenced by DNA methylation. The ASC gene contains a 600-bp long CpG island located near the transcription start site and the methylation status of this CpG island correlates with the expression level of ASC/TMS1. The methylation status of the CpG island, as well as promoter proximal pausing of RNA Polymerase II in multiple cancer cell lines and tissues result in reduced expression of ASC (50–58). Our studies indicate that Asc levels are influenced by p205 in murine macrophages adding additional understanding to the regulation of this important immune gene. Our findings expand on our understanding of transcriptional regulatory roles for the PYHIN proteins. p205 has previously been shown to control p21CIP/WAF gene expression via p53 in Saos2 cell lines and to impact gene expression in adipogenesis and osteogenesis via transcriptional mechanisms. p205 acts as a transactivator synergizing with p53 to induce expression of the cell cycle inhibitor p21 to inhibit cell growth. In adipocytes, p205 interacts with c/EBPβ and c/EBPα to further activate the transcriptional activity of c/EBPα and PPARγ. All of these studies help establish evidence for PYHIN proteins as regulators of gene expression. The discovery of p205 as a regulator of inflammasomes via transcriptional regulation of Asc further supports a role for the PYHIN proteins as regulators of gene expression and in addition emphasizes the importance of PYHIN family members in innate immunity.
Supplementary Material
Acknowledgments
We sincerely thank Zhaozhao Jiang, Jennifer Bernier and the Fitzgerald lab members, Anubhab Nandy and Neal Silverman (University of Massachusetts Medical School), Vijay Rathinam and Sivapriya K. Vanaja (University of Connecticut), and Shruti Sharma (Tufts Medical School) for all their help and insightful comments.
This work was supported by grants from the NIH (AI093752, AI083713 and AI067497).
References
- 1.Sharma S, Fitzgerald KA. Innate immune sensing of DNA. PLoS pathogens. 2011;7:e1001310. doi: 10.1371/journal.ppat.1001310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nature immunology. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 3.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 4.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 5.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 6.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nature immunology. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 10.Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nature reviews Microbiology. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nature immunology. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 12.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, Hume DA, Stacey KJ. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 14.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, Hornung V, Vogel SN, Szomolanyi-Tsuda E, Fitzgerald KA. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nature immunology. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, Eisenlohr L, Landel CP, Alnemri ES. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nature immunology. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baum R, Sharma S, Carpenter S, Li QZ, Busto P, Fitzgerald KA, Marshak-Rothstein A, Gravallese EM. Cutting edge: AIM2 and endosomal TLRs differentially regulate arthritis and autoantibody production in DNase II-deficient mice. Journal of immunology (Baltimore, Md : 1950) 2015;194:873–877. doi: 10.4049/jimmunol.1402573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monroe KM, Yang Z, Johnson JR, Geng X, Doitsh G, Krogan NJ, Greene WC. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schattgen SA, Fitzgerald KA. The PYHIN protein family as mediators of host defenses. Immunological reviews. 2011;243:109–118. doi: 10.1111/j.1600-065X.2011.01053.x. [DOI] [PubMed] [Google Scholar]
- 20.Cridland JA, Curley EZ, Wykes MN, Schroder K, Sweet MJ, Roberts TL, Ragan MA, Kassahn KS, Stacey KJ. The mammalian PYHIN gene family: phylogeny, evolution and expression. BMC evolutionary biology. 2012;12:140. doi: 10.1186/1471-2148-12-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunette RL, Young JM, Whitley DG, Brodsky IE, Malik HS, Stetson DB. Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J Exp Med. 2012;209:1969–1983. doi: 10.1084/jem.20121960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Connolly DJ, Bowie AG. The emerging role of human PYHIN proteins in innate immunity: implications for health and disease. Biochemical pharmacology. 2014;92:405–414. doi: 10.1016/j.bcp.2014.08.031. [DOI] [PubMed] [Google Scholar]
- 23.Gray EE, Winship D, Snyder JM, Child SJ, Geballe AP, Stetson DB. The AIM2-like Receptors Are Dispensable for the Interferon Response to Intracellular DNA. Immunity. 2016;45:255–266. doi: 10.1016/j.immuni.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiler SR, Gooya JM, Ortiz M, Tsai S, Collins SJ, Keller JR. D3: a gene induced during myeloid cell differentiation of Linlo c-Kit+ Sca-1(+) progenitor cells. Blood. 1999;93:527–536. [PubMed] [Google Scholar]
- 25.Asefa B, Klarmann KD, Copeland NG, Gilbert DJ, Jenkins NA, Keller JR. The interferon-inducible p200 family of proteins: a perspective on their roles in cell cycle regulation and differentiation. Blood cells, molecules & diseases. 2004;32:155–167. doi: 10.1016/j.bcmd.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Li H, Jiao Y, Zhang L, Wang C, Zhang X, Guo H, Xu H. The interferon-inducible protein p205 acts as an activator in osteoblast differentiation of mouse BMSCs. Differentiation. 2016;92:318–325. doi: 10.1016/j.diff.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Liu F, Jiao Y, Zhu Z, Sun C, Li H. Interferon-inducible protein 205 (p205) plays a role in adipogenic differentiation of mouse adipose-derived stem cells. Mol Cell Endocrinol. 2014;392:80–89. doi: 10.1016/j.mce.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 28.Dermott JM, Gooya JM, Asefa B, Weiler SR, Smith M, Keller JR. Inhibition of growth by p205: a nuclear protein and putative tumor suppressor expressed during myeloid cell differentiation. Stem cells (Dayton, Ohio) 2004;22:832–848. doi: 10.1634/stemcells.22-5-832. [DOI] [PubMed] [Google Scholar]
- 29.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt T, Schmid-Burgk JL, Hornung V. Synthesis of an arrayed sgRNA library targeting the human genome. Scientific reports. 2015;5:14987. doi: 10.1038/srep14987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmid-Burgk JL, Schmidt T, Gaidt MM, Pelka K, Latz E, Ebert TS, Hornung V. OutKnocker: a web tool for rapid and simple genotyping of designer nuclease edited cell lines. Genome research. 2014;24:1719–1723. doi: 10.1101/gr.176701.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broz P, Newton K, Lamkanfi M, Mariathasan S, Dixit VM, Monack DM. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J Exp Med. 2010;207:1745–1755. doi: 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 34.Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parsons MJ, Vertino PM. Dual role of TMS1/ASC in death receptor signaling. Oncogene. 2006;25:6948–6958. doi: 10.1038/sj.onc.1209684. [DOI] [PubMed] [Google Scholar]
- 36.Bradley MN, Zhou L, Smale ST. C/EBPbeta regulation in lipopolysaccharide-stimulated macrophages. Molecular and cellular biology. 2003;23:4841–4858. doi: 10.1128/MCB.23.14.4841-4858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rathinam VA, Vanaja SK, Fitzgerald KA. Regulation of inflammasome signaling. Nature immunology. 2012;13:333–342. doi: 10.1038/ni.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conrady CD, Zheng M, Fitzgerald KA, Liu C, Carr DJ. Resistance to HSV-1 infection in the epithelium resides with the novel innate sensor, IFI-16. Mucosal Immunol. 2012;5:173–183. doi: 10.1038/mi.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Storek KM, Gertsvolf NA, Ohlson MB, Monack DM. cGAS and Ifi204 cooperate to produce type I IFNs in response to Francisella infection. Journal of immunology (Baltimore, Md : 1950) 2015;194:3236–3245. doi: 10.4049/jimmunol.1402764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ludlow LE, Johnstone RW, Clarke CJ. The HIN-200 family: more than interferon-inducible genes? Experimental cell research. 2005;308:1–17. doi: 10.1016/j.yexcr.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 41.Ma XY, Wang H, Ding B, Zhong H, Ghosh S, Lengyel P. The interferon-inducible p202a protein modulates NF-kappaB activity by inhibiting the binding to DNA of p50/p65 heterodimers and p65 homodimers while enhancing the binding of p50 homodimers. The Journal of biological chemistry. 2003;278:23008–23019. doi: 10.1074/jbc.M302105200. [DOI] [PubMed] [Google Scholar]
- 42.Datta B, Li B, Choubey D, Nallur G, Lengyel P. p202, an interferon-inducible modulator of transcription, inhibits transcriptional activation by the p53 tumor suppressor protein, and a segment from the p53-binding protein 1 that binds to p202 overcomes this inhibition. The Journal of biological chemistry. 1996;271:27544–27555. doi: 10.1074/jbc.271.44.27544. [DOI] [PubMed] [Google Scholar]
- 43.Min W, Ghosh S, Lengyel P. The interferon-inducible p202 protein as a modulator of transcription: inhibition of NF-kappa B, c-Fos, and c-Jun activities. Molecular and cellular biology. 1996;16:359–368. doi: 10.1128/mcb.16.1.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Datta B, Min W, Burma S, Lengyel P. Increase in p202 expression during skeletal muscle differentiation: inhibition of MyoD protein expression and activity by p202. Molecular and cellular biology. 1998;18:1074–1083. doi: 10.1128/mcb.18.2.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lembo M, Sacchi C, Zappador C, Bellomo G, Gaboli M, Pandolfi PP, Gariglio M, Landolfo S. Inhibition of cell proliferation by the interferon-inducible 204 gene, a member of the Ifi 200 cluster. Oncogene. 1998;16:1543–1551. doi: 10.1038/sj.onc.1201677. [DOI] [PubMed] [Google Scholar]
- 46.Liu CJ, Chang E, Yu J, Carlson CS, Prazak L, Yu XP, Ding B, Lengyel P, Di Cesare PE. The interferon-inducible p204 protein acts as a transcriptional coactivator of Cbfa1 and enhances osteoblast differentiation. The Journal of biological chemistry. 2005;280:2788–2796. doi: 10.1074/jbc.M412604200. [DOI] [PubMed] [Google Scholar]
- 47.Liu C, Wang H, Zhao Z, Yu S, Lu YB, Meyer J, Chatterjee G, Deschamps S, Roe BA, Lengyel P. MyoD-dependent induction during myoblast differentiation of p204, a protein also inducible by interferon. Molecular and cellular biology. 2000;20:7024–7036. doi: 10.1128/mcb.20.18.7024-7036.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corrales L, Woo SR, Williams JB, McWhirter SM, Dubensky TW, Jr, Gajewski TF. Antagonism of the STING Pathway via Activation of the AIM2 Inflammasome by Intracellular DNA. Journal of immunology (Baltimore, Md : 1950) 2016;196:3191–3198. doi: 10.4049/jimmunol.1502538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirose Y, Tacke R, Manley JL. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 1999;13:1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Collard RL, Harya NS, Monzon FA, Maier CE, O’Keefe DS. Methylation of the ASC gene promoter is associated with aggressive prostate cancer. Prostate. 2006;66:687–695. doi: 10.1002/pros.20371. [DOI] [PubMed] [Google Scholar]
- 51.Das PM, Ramachandran K, Vanwert J, Ferdinand L, Gopisetty G, Reis IM, Singal R. Methylation mediated silencing of TMS1/ASC gene in prostate cancer. Molecular cancer. 2006;5:28. doi: 10.1186/1476-4598-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Machida EO, Brock MV, Hooker CM, Nakayama J, Ishida A, Amano J, Picchi MA, Belinsky SA, Herman JG, Taniguchi S, Baylin SB. Hypermethylation of ASC/TMS1 is a sputum marker for late-stage lung cancer. Cancer research. 2006;66:6210–6218. doi: 10.1158/0008-5472.CAN-05-4447. [DOI] [PubMed] [Google Scholar]
- 53.Ohtsuka T, Liu XF, Koga Y, Kitajima Y, Nakafusa Y, Ha CW, Lee SW, Miyazaki K. Methylation-induced silencing of ASC and the effect of expressed ASC on p53-mediated chemosensitivity in colorectal cancer. Oncogene. 2006;25:1807–1811. doi: 10.1038/sj.onc.1209204. [DOI] [PubMed] [Google Scholar]
- 54.Riojas MA, Guo M, Glockner SC, Machida EO, Baylin SB, Ahuja N. Methylation-induced silencing of ASC/TMS1, a pro-apoptotic gene, is a late-stage event in colorectal cancer. Cancer biology & therapy. 2007;6:1710–1716. doi: 10.4161/cbt.6.11.4829. [DOI] [PubMed] [Google Scholar]
- 55.Wu L, Zhang C, Wang X, Ding X, Deng J, Liang H. Methylation of ASC/TMS1 promoter is associated with poor prognosis of patients with gastric cancer. Clinical & translational oncology : official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2016;18:296–303. doi: 10.1007/s12094-015-1367-y. [DOI] [PubMed] [Google Scholar]
- 56.Levine JJ, Stimson-Crider KM, Vertino PM. Effects of methylation on expression of TMS1/ASC in human breast cancer cells. Oncogene. 2003;22:3475–3488. doi: 10.1038/sj.onc.1206430. [DOI] [PubMed] [Google Scholar]
- 57.Moriai R, Tsuji N, Kobayashi D, Yagihashi A, Namiki Y, Takahashi H, Watanabe N. A proapoptotic caspase recruitment domain protein gene, TMS1, is hypermethylated in human breast and gastric cancers. Anticancer research. 2002;22:4163–4168. [PubMed] [Google Scholar]
- 58.Virmani A, Rathi A, Sugio K, Sathyanarayana UG, Toyooka S, Kischel FC, Tonk V, Padar A, Takahashi T, Roth JA, Euhus DM, Minna JD, Gazdar AF. Aberrant methylation of TMS1 in small cell, non small cell lung cancer and breast cancer. International journal of cancer. 2003;106:198–204. doi: 10.1002/ijc.11206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


