Abstract
Tyrosyl-DNA phosphodiesterase 2 (TDP2) is a recently discovered enzyme specifically repairing topoisomerase II (TOP2)-mediated DNA damage. It has been shown that inhibition of TDP2 synergize with TOP2 inhibitors. Herein, we report the discovery of the furoquinolinedione chemotype as a suitable skeleton for the development of selective TDP2 inhibitors. Compound 1 was identified as a TDP2 inhibitor as a result of screening our in-house compound library for compounds selective for TDP2 vs. TDP1. Further SAR studies provide several selective TDP2 inhibitors at low-micromolar range. The most potent compound 74 shows inhibitory activity with IC50 of 1.9 and 2.1 μM against recombinant TDP2 and TDP2 in whole cell extracts (WCE), respectively.
Keywords: tyrosyl-DNA phosphodiesterase, DNA repair, inhibitor, furoquinolinedione, topoisomerase
Graphical abstract
1. Introduction
Tyrosyl-DNA phosphodiesterase 2 (TDP2) is a recently discovered DNA repair enzyme that catalytically hydrolyzes phosphotyrosyl bonds between a DNA 5′-phosphate and a protein tyrosine residue [1, 2]. There are several TDP2 single crystallographic analyses reported [3–6], which show that TDP2 catalytic residues in the enzyme active site indirectly hydrolyze the 5′-phosphotyrosyl bond in one-step with the coordination of divalent metal [6, 7]. In eukaryotic cells, 5′-phosphotyrosyl protein-DNA bonds are mainly derived from abortive topoisomerase II (TOP2)-DNA cleavage complexes (TOP2cc) [8–10], which can be trapped by a broad range of exogenous factors such as TOP2 poisons [8], DNA intercalators [11], cytosine arabinoside [12], ultimately resulting in double-stranded DNA breaks, and subsequently 5′-tyrosyl-DNA adducts. Such adducts are the substrates of TDP2, which functions as a DNA repair enzyme that excises TOP2-DNA covalent complexes resulting from stalled TOP2cc [13, 14]. High expression of TDP2 results in resistance to etoposide [1], a widely used anticancer TOP2 poison [8, 15, 16]. Conversely, TDP2-deletion or knockdown in cell and animal model leads to hypersensitivity to etoposide or elevated levels of TOP2-mediated DNA damage [1, 17–20]. TDP2 has also been identified as the VPg unlinkase enzyme, which is required for the replication of picornaviruses that infect human and livestock worldwide and has been shown to be involved for hepatitis virus replication [21, 22]. Hence, TDP2 inhibitors have the potential to be developed also as novel antiviral drugs.
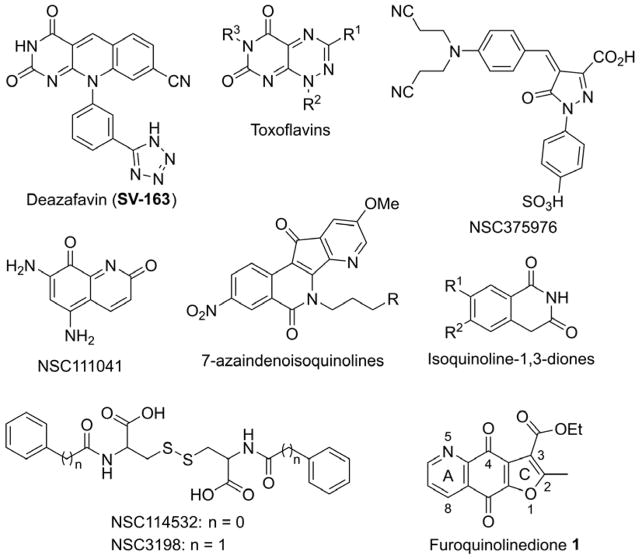
The increasing interest in elucidation the cellular functions of TDP2 and its suitability as drug target [2, 23, 24] prompted the discovery of TDP2 inhibitors. To date, only a few of TDP2 inhibitor chemotypes have been reported, including toxoflavins, deazaflavins, isoquinoline-1,3-diones NSC37596, 5,7-diaminoquinoline-2,8-dione (NSC111041), NSC114532, NSC3198 and 7-azaindenoisoquinolines (Figure 1) [19, 24–28]. It was shown that deazaflavin TDP2 inhibitors synergize with the TOP2 inhibitor etoposide at otherwise non-toxic concentrations [28]. Therefore, there is strong need to discover and develop TDP2 inhibitors as potential anticancer and antiviral drugs, and probes to elucidate the molecular and cellular function of TDP2.
Figure 1.
The reported TDP2 inhibitors and our TDP2 hit 1.
In this study, we report the discovery, synthesis and structure-activity relationship (SAR) of furoquinolinediones as an alternative scaffold of TDP2 inhibitor.
2. Results and Discussion
2.1. Hit Identification
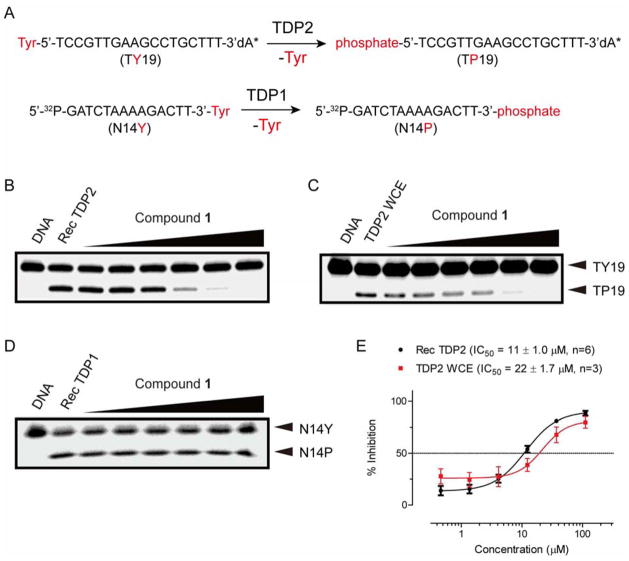
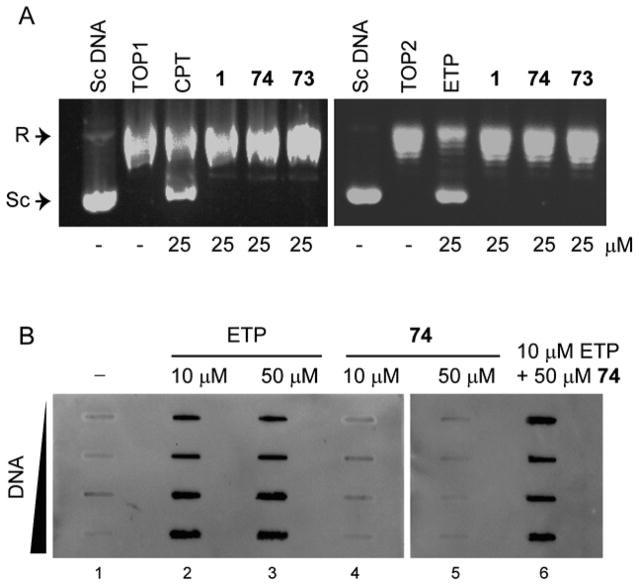
In an effort to discover new chemotypes of TDP2 inhibitors, an in-house chemical library was screened against TDP2 using a single-stranded oligonucleotide TY19 substrate containing 5′-phosphotyrosyl group (Figure 2A) [7]. A furoquinolinedione derivative 1 (Figure 1) was found to inhibit recombinant human TDP2 with an IC50 (the concentration of compound that inhibits 50% of enzyme activity) value of 11 μM (Figure 2B and 2E). Notably, compound 1 did not inhibit TDP1 (a counter enzyme characterized as hydrolyzing a DNA 3′-phosphotyrosyl bond) [2, 10, 29] at concentrations up to 111 μM (Figure 2D), in which assay a single-stranded oligonucleotide N14Y substrate containing 3′-phosphotyrosyl group was used (Figure 2A) [30]. The inhibitory activity of compound 1 against human TDP2 was also tested in whole cell extracts (TDP2 WCE) [28] and showed that 1 remained potent with IC50 value of 22 μM (Figure 2C and 2E). The retention of the TDP2 inhibitory potency in the WCE context shows that 1 is not sequestered by binding to other cellular components, implying that 1 binds specifically to TDP2. To study the selectivity to TDP2, compound 1 was screened against TOP2 and topoisomerase IB (TOP1) by using relaxation assays, which indicated that 1 did not inhibit TOP1 and TOP2 relaxing activity at 25 μM concentration (Figure 4). In addition, TOP1-mediated DNA cleavage assay indicated that compound 1 is unable to induce the formation of the covalent complex of TOP1 and DNA at up to 100 μM (Figure 2S, Supporting Information), not a TOP1 poison [31]. These results indicate that furoquinolinediones represent a suitable chemotype for novel selective TDP2 inhibitors. Therefore, we undertook a systematic exploration of the SAR furoquinolinedione chemotype by probing analogs of 1: 1) by modifying the ethoxy carbonyl group at position 3; and 2) by converting the furan ring into diverse 5-member cycles including pyrrole, isoxazole and pyrazole.
Figure 2.
(A) Schematic representation of TDP2 and TDP1-catalyzed phosphotyrosyl cleavage reaction. Representative inhibition gels of compound 1 against Rec TDP2 (B), TDP2 WCE (C) and TDP1 (D). (E) Dose-response curves of 1 against Rec TDP2 and TDP2 WCE expressed as mean ± SD. Testing concentrations: 0.46, 1.4, 4.1, 12.3, 37 and 111 μM.
Figure 4.
The structures of compounds 77 and AM-8-3.
2.2. Chemistry
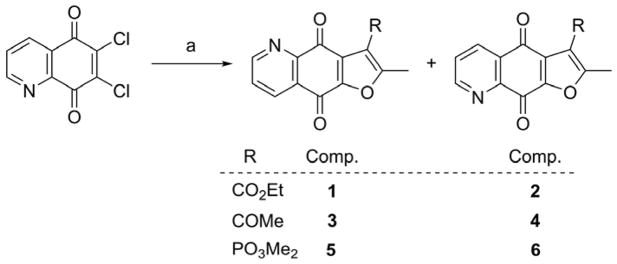
The synthesis of designed furoquinolinedione analogues is outlined in Schemes 1–8. Furoquinolinedione analogues 1–6 could be synthesized in a one-pot reaction as shown in Scheme 1 [32]. The reaction of 6,7-dichloroquinoline-5,8-dione with active methylene agent (AMR), such as ethyl acetoacetate, acetylacetone and dimethyl (2-oxopropyl)phosphonate, gave two furoquinolinedione products, N,O-anti isomer and N,O-syn isomer. The structures of isomers 1 and 2 were characterized with HRMS, 1D and 2D NMR spectroscopy. The structure of 1 was further confirmed with X-ray single crystal analysis (Supporting Information, Figure 1S). The structural modification of the ester at position 3 of compounds 1 and 2 is shown in Schemes 2–6. Compounds 1 and 2 were hydrolyzed with aqueous sodium carbonate in isopropanol to give the corresponding carboxylic acids 7 and 66, respectively. Treatment of acids 7 or 66 with thionyl chloride followed by amidation or esterification afforded the target amides 8–12 (Scheme 2, Table 2), 67 and 68 (Scheme 6), esters 13–55 (Scheme 2, Table 2), and 69 (Scheme 6). The reaction of bromide analogue 50 with pyridine derivatives gave the pyridinium analogues 56 and 57 (Scheme 3). Boc-protecting group of 51–53 was removed by treatment with trifluoroacetic acid leading to oxazole derivative 58 and anilines 59 and 60 (Scheme 4). The alkynes 54 and 55 were introduced into “click” reaction with various alkyl azides to prepare triazoles 61–64 (Scheme 5) [33]. In addition, 3-methyl analogue 77 (Figure 4) was synthesized according to Cherkaoui method (Supporting Information, Scheme S1) [34].
Scheme 1.
Synthesis of compounds 1–6.
Reagents and conditions: (a) CH3COCH2R, MeCN, K2CO3, reflux.
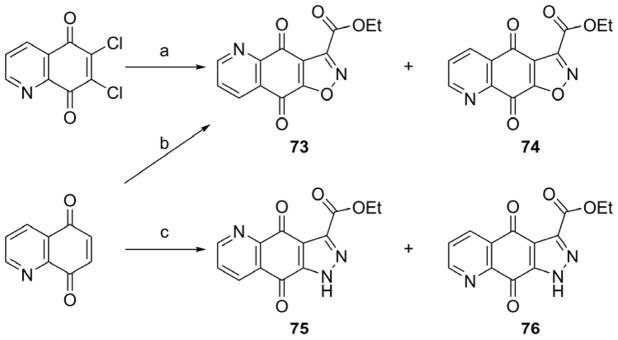
Scheme 8.
Synthesis of compounds 73–76.
Reagents and conditions: (a) MeCN, K2CO3, ethyl nitroacetate, reflux. (b) AcOH, Mn(OAc)3, ethyl nitroacetate, 70 °C. (c) ethyl diazoacetate, toluene, reflux.
Scheme 2.
Synthesis of compounds 8–55.
Reagents and conditions: (a) 2N Na2CO3, i-PrOH, reflux. (b) i) chloroform, SOCl2, reflux; ii) DMAP, TEA, amines for 8–12 or alcohols for 13–55.
Scheme 6.
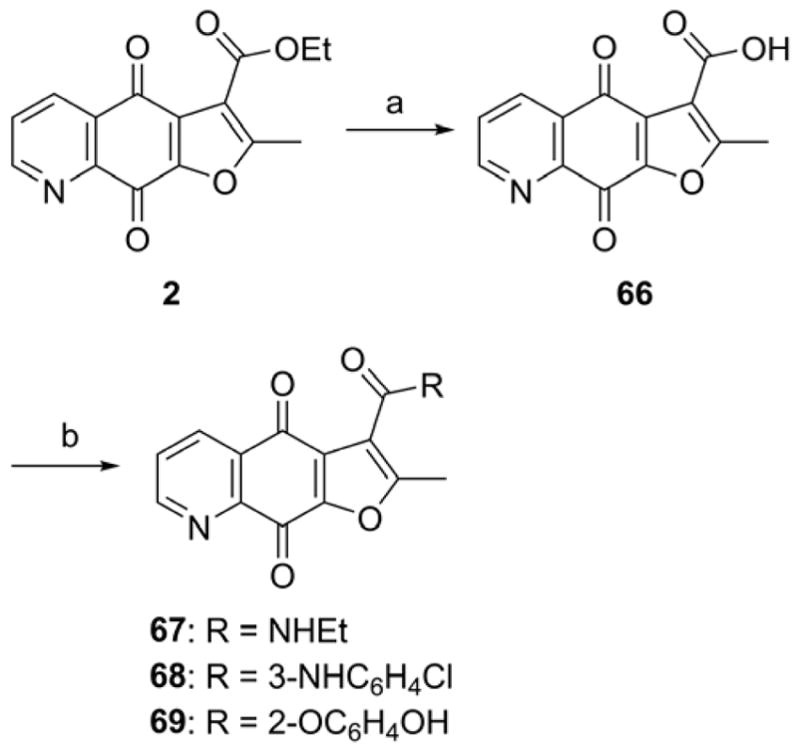
Synthesis of compounds 67–69.
Reagents and conditions: (a) 2N Na2CO3, i-PrOH, reflux. (b) i) chloroform, SOCl2, reflux; ii) DMAP, TEA, amines for 67–68 or catechol for 69.
Table 2.
The structures and inhibitory activity of compounds 8–55 against TDP2.
| |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Cpd. | R | IC50 (μM)a | Cpd. | R | IC50 (μM)a | ||
|
|
|
||||||
| TDP2 | TDP2 WCE |
TDP2 | TDP2 WCE |
||||
| 8 | NHEt | >111 | NDb | 32 | 3-OC6H4Cl | 14 ± 1.8 | 11 ± 1.1 |
| 9 | NHPh | >111 | ND | 33 | 4-OC6H4Cl | 11 ± 1.4 | 7.5 ± 0.56 |
| 10 | 2-NHC6H4OH | >111 | ND | 34 | 2-OC6H4CN | >111 | >111 |
| 11 | 4-NHC6H4OMe | >111 | ND | 35 | 3-OC6H4CN | 12 ± 1.8 | 10 ± 2.7 |
| 12 | 3-NHC6H4Cl | >111 | ND | 36 | 4-OC6H4CN | 15 ± 2.1 | 13 ± 4.5 |
| 13 | OMe | 26 ± 3.3 | 12 ± 1.1 | 37 | 4-OC6H4Br | 35 ± 4.8 | 12 ± 0.92 |
| 14 | OCH2CF3 | 28 ± 11 | 24 ± 6.8 | 38 | 4-OC6H4SO2Me | 14 ± 2.1 | 16 ± 4.2 |
| 15 | OCH2CO2H | 51 ± 20 | 37 ± 6.4 | 39 | 4-OC6H4NO2 | 28 ± 5.7 | ND |
| 16 | O(CH2)2OH | 20 ± 1.3 | 9.8 ± 1.4 | 40 | 2-OC10H7 | >111 | ND |
| 17 | O(CH2)3OH | 17 ± 2.2 | 11 ± 0.62 | 41 | 3-pyridinyloxy | 8.8 ± 1.1 | 7.7 ± 0.61 |
| 18 | O(CH2)4OH | 26 ± 4.2 | 12 ± 0.36 | 42 |
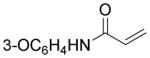
|
71 ± 54 | ND |
| 19 | O(CH2)5OH | 45 ± 17 | 19 ± 3.7 | 43 |
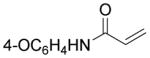
|
68 ± 61 | ND |
| 20 | O(CH2CH2O)2H | 22 ± 3.7 | 13 ± 0.77 | 44 |
|
51 ± 21 | ND |
| 21 |
|
20 ± 1.9 | 11 ± 0.4 | 45 |
|
37 ± 11 | 34 ± 20 |
| 22 |
|
12 ± 0.65 | 14 ± 0.72 | 46 | 4-OC6H4COPh | >111 | ND |
| 23 |
|
50 ± 15 | 15 ± 2 | 47 |
|
23 ± 18 | 17 ± 6.7 |
| 24 | O(CH2)2PO3Me2 | 9.0 ± 0.75 | 17 ± 6.3 | 48 |

|
22 ± 8.5 | 24 ± 6.6 |
| 25 | OCH2CH2CN | 14 ± 4.5 | 13 ± 3.7 | 49 |
|
>111 | >111 |
| 26 | OCH2CH2Ph | 53 ± 3.1 | 24 ± 5.5 | 50 | O(CH2)3Br | ND | ND |
| 27 | OPh | 18 ± 1.1 | 10 ± 1.6 | 51 | 2-OC6H4NHBoc | 36 ± 14 | ND |
| 28 | 4-OC6H4OPO3Me2 | 31 ± 3.5 | ND | 52 | 3-OC6H4NHBoc | 31 ± 6.4 | ND |
| 29 | 2-OC6H4OH | 7.6 ± 2.1 | 6.6 ± 0.38 | 53 | 4-OC6H4NHBoc | 88 ± 33 | ND |
| 30 | 3-OC6H4OH | 29 ± 20 | 8.8 ± 1.2 | 54 | O(CH2)2C≡CH | 22 ± 3.9 | 15 ± 0.47 |
| 31 | 2-OC6H4Cl | 10 ± 1.6 | 14 ± 3 | 55 | 3-OC6H4C≡CH | 43 ± 6.8 | 13 ± 2.4 |
The IC50 was expressed as mean ± SD from at least three independent experiments unless otherwise indicated.
“ND” means “not determined”.
Scheme 3.
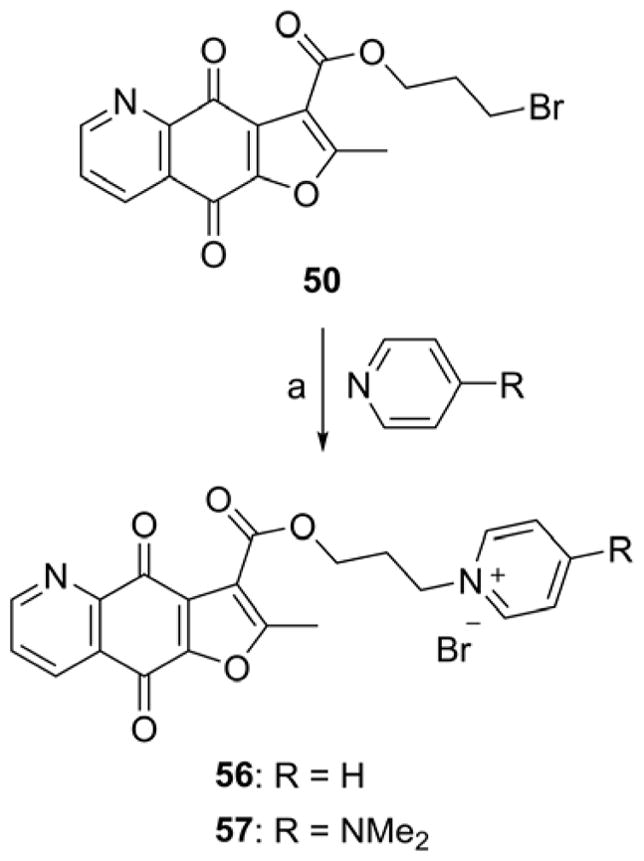
Synthesis of compounds 56 and 57.
Reagents and conditions: (a) THF, reflux.
Scheme 4.
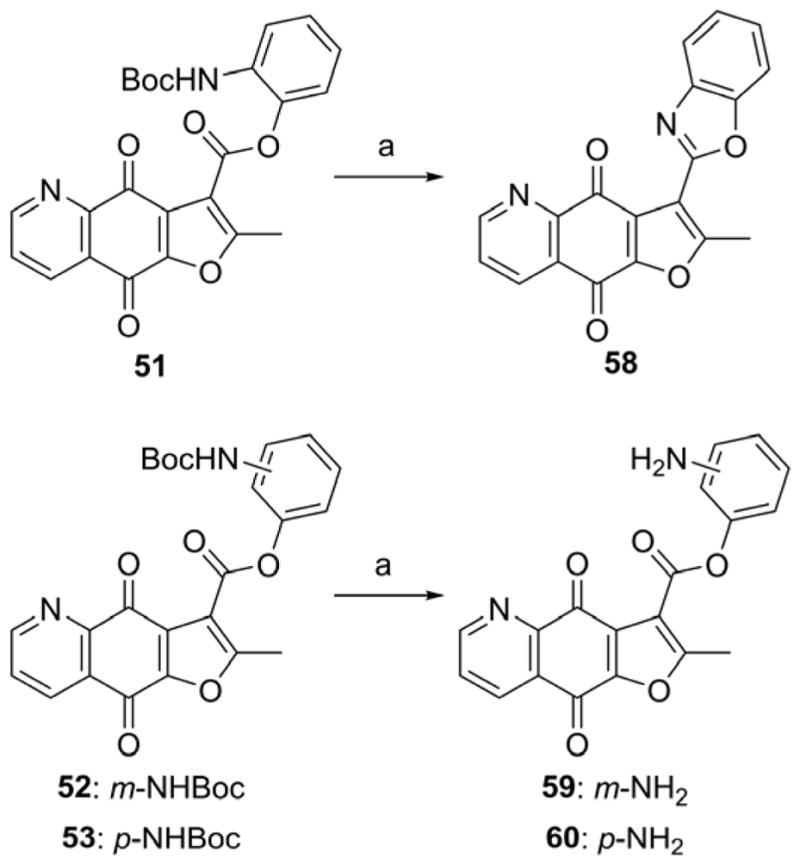
Synthesis of compounds 58–60.
Reagents and conditions: (a) DCM, TFA, rt.
Scheme 5.
Synthesis of compounds 61–65.
Reagents and conditions: (a) i) 3-bromopropanol (for 61, 63) or 3-bromopropanic acid (for 62, 64), MeCN, NaN3, reflux; ii) DMF, H2O, 5 mol% CuSO4/5H2O, 10 mol% L-ascorbic acid sodium, 75 °C. (b) NaOH, EtOH, rt.
To assess the role of furan ring for TDP2 inhibitory potency, analogues 70–76 were designed as shown in Schemes 7 and 8. The N,N-syn pyrroloquinolinedione 70 (Scheme 7) was obtained from the reaction of 7-bromoquinoline-5,8-dione with ethyl 3-aminocrotonate under Mn(OAc)3 catalysis in a low yield (10%). Unfortunately, N,N-anti product 71 was not obtained in sufficient quantities to allow its evaluation as an inhibitor. The reaction of 6,7-dichloroquinoline-5,8-dione with ethyl acetoacetate and followed by treatment with methylamine mainly gave the N,N-syn pyrroloquinolinedione derivative 72. As shown in Scheme 8, the reaction of 6,7-dichloroquinoline-5,8-dione with ethyl nitroacetate gave two isomers 73 and 74 with isoxazole at C ring. Two synthetic pathways (pathway a and b) were investigated, and unfortunately gave the target isomers in low yields (2–11%). The pyrazole analogues 75 and 76 were obtained from the reaction of quinoline-5,8-dione with ethyl diazoacetate (Scheme 8).
Scheme 7.
Synthesis of compounds 70–72.
Reagents and conditions: (a) ethyl 3-aminocrotonate, Mn(OAc)3, MeCN, reflux. (b) i) THF, AcONa, ethyl acetoacetate, reflux; ii) EtOH, MeNH2/H2O, reflux.
2.3. TDP2 and TDP1 inhibition
All prepared compounds were tested at six or eight three-fold dilution concentrations from 111 μM to 0.46 μM or 0.051 μM against recombinant TDP2 and TDP1. For the compounds with high TDP2 inhibitory activity, a further assay against TDP2 whole cell extracts (WCE) was conducted to determine their potency against native TDP2 enzyme in the presence of abundant cellular proteins. The inhibitory results are summarized in Tables 1–3 and expressed as IC50 value. Most compounds were selective against TDP2, as they did not show significant inhibition against TDP1 at the highest concentration tested of 111 μM. Only compounds 39 (TDP1 IC50 48 μM) and 48 (TDP1 IC50 38 μM) showed moderate TDP1 inhibitory potency.
Table 1.
The inhibitory activity of compounds 1–6 against TDP2.
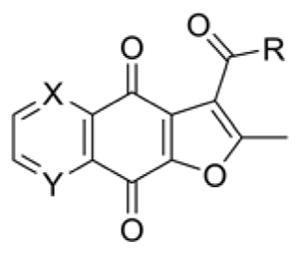
| |||||
|---|---|---|---|---|---|
|
| |||||
| Cpd. | X | Y | R | IC50 (μM)a | |
|
| |||||
| TDP2 | TDP2 WCE | ||||
| 1 | N | CH | CO2Et | 11 ± 1.0 | 22 ± 1.7 |
| 2 | CH | N | CO2Et | 43 ± 5.5 | 54 ± 2.5 |
| 3 | N | CH | COMe | 35 ± 5.1 | 36 ± 2.4 |
| 4 | CH | N | COMe | >111 | NDb |
| 5 | N | CH | PO3Me2 | >111 | ND |
| 6 | CH | N | PO3Me2 | 99 ± 11 | ND |
The IC50 was expressed as mean ± SD from at least three independent experiments unless otherwise indicated.
“ND” means “not determined”.
Table 3.
The inhibitory activity of synthesized compounds against TDP2.
| Cpd. | IC50 (μM)a | Cpd. | IC50 (μM)a | ||
|---|---|---|---|---|---|
|
|
|
||||
| TDP2 | TDP2 WCE | TDP2 | TDP2 WCE | ||
| 56 | 32 ± 1.6 | 24 ± 5.3 | 67 | >111 | ND |
| 57 | 43 ± 17 | 38 ± 8.2 | 68 | >111 | ND |
| 58 | >111 | NDb | 69 | 29 ± 7.4 | 39 ± 4.1 |
| 59 | 16 ± 2.0 | 12 ± 2.1 | 70 | >111 | ND |
| 60 | 19 ± 5.7 | ND | 72 | >111 | ND |
| 61 | 66 ± 11 | 72 ± 18 | 73 | 3.3 ± 0.43 | 3.1 ± 0.1 |
| 62 | 20 ± 1.1 | 16 ± 2.2 | 74 | 1.9 ± 0.28 | 2.1 ± 0.1 |
| 63 | 8.1 ± 0.11 | 8.3 ± 2.0 | 75 | >111 | ND |
| 64 | 22 ± 2.0 | 13 ± 0.03 | 76 | >111 | ND |
| 65 | 50 ± 11 | 28 ± 7.1 | 77 | >111 | ND |
The IC50 was expressed as mean ± SD from at least three independent experiments unless otherwise indicated.
“ND” means “not determined”.
Based on the TDP2 inhibition results, it could be observed that the ester functionality is preferred over amide or methyl groups at position 3 (77, >111 μM), either of which abolished TDP2 inhibitory activity in otherwise similarly substituted derivatives. For example, in ester/amid pairs 1 (IC50 = 11 μM)/8 (inactive, i.e. IC50 >111 μM), 2 (43 μM)/67 (>111 μM), 27 (18 μM)/9 (>111 μM), 29 (7.6 μM)/10 (>111 μM) and 32 (14 μM)/12 (>111 μM), the activity of ester analogues was abolished by changing to amide functionality (Tables 1–3). These findings point out the important role played by the neutral ester functionality in the activity of the furoquinolinedione class as TDP2 inhibitors. Furthermore, the furoquinolinedione core was esterified to a number of different groups both aliphatic and aromatic (Table 2 and 3, 13–65). The phosphorylethyl ester 24 (TDP2 IC50 = 9.0 μM) showed a slightly increased TDP2 inhibition. The phenyl esters 29 (TDP2 IC50 = 7.6 μM and WCE IC50 = 6.6 μM), 33 (11 and 7.5 μM), 63 (8.1 and 8.3 μM) and pyridinyl ester 41 (8.8 and 7.7 μM) showed increased inhibition against both recombinant TDP2 and TDP2 WCE. These results show that steric bulk is tolerated with respect to inhibitory properties. Despite that, a few analogues were found to be adversely affected by the introduction of larger conjugated aromatic groups, 2-naphthyl 40, benzoylphenyl 46, 3,4-methylenedioxyphenyl 49, as these derivatives were determined to be inactive. Various functionality at the end of the conjugated groups electron-donating (e.g. 29, 30) and –withdrawing (e.g. 35, 36, 39) substituents of the aryl analogues as well as amine (44) and acid (64) derivatives are tolerated.
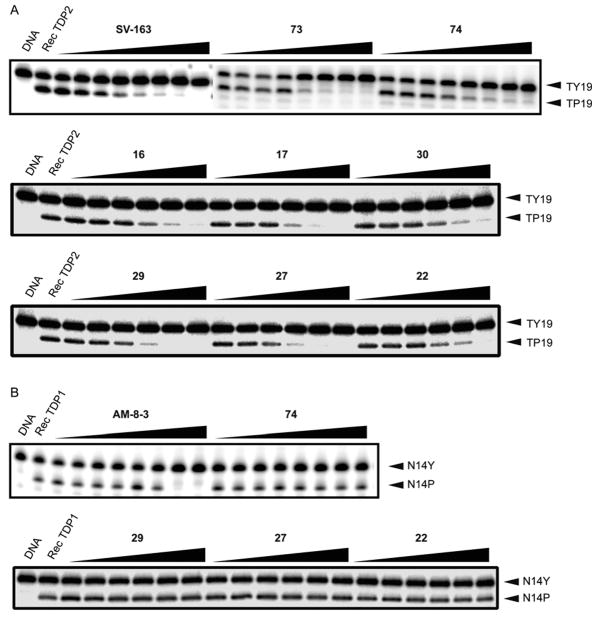
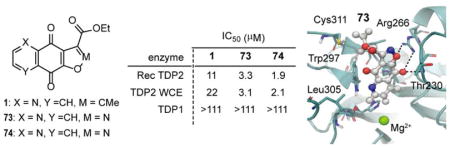
The furan ring modified analogues revealed the importance of the oxygen present in the parent 1 and 2 for the TDP2 inhibitory activity. Conversion of furan to pyrrole 70, N-methyl pyrrole 72 or diazoles 75 and 76 rendered compounds inactive. Isoxazoles 73 (TDP2 3.3 and WCE 3.1 μM) and 74 (1.9 and 2.1 μM) gained an improved inhibitory activity for both recombinant TDP2 and WCE, resulting in the two most potent analogues in the series to date. The representative gels of TDP2 inhibitions is shown in Figure 3A. The deazaflavin SV-163 (Figure 1) was used as the positive control [24]. The tested compounds show full inhibition at the highest concentration (111 μM) and progressive dose-response (Figure 3A). Conversely, they do not inhibit TDP1 up to 111 μM concentration (Figure 3B), in which the indenoisoquinoline AM-8-3 (Figure 4) was used as a positive control [35].
Figure 3.
Representative gels for the testing of compounds against Rec TDP2 (A) and TDP1 (B). The compounds were tested at concentrations increasing from 0.51 to 111 μM (for eight doses) or 0.46 to 111 μM (for six doses). The deazafavin SV-163 was tested as the positive control for TDP2 assay at concentrations increasing from 0.017 to 37 μM. The indenoisoquinoline AM-8-3 was tested as the positive control for TDP1 assay at concentrations increasing from 0.051 to 111 μM.
To study the selectivity of the two most potent compounds 73 and 74, TOP1-mediated cleavage assay and relaxation assay and TOP2-medaited relaxation assay were performed and indicated that compounds 73 and 74 do not have inhibitory potency to TOP1 and TOP2 (Figure 5A and Figure 2S, Supporting Information). Compound 74 was also tested using TOP2-mediated in vivo complex of enzyme (ICE) assay (Figure 5B), which indicated 74 is unable to induce the formation of TOP2cc at concentration up to 50 μM concentration. These results imply they are selective inhibitors of TDP2.
Figure 5.
The inhibition gels compounds against TOP1 and TOP2. (A) TOP1-mediated (left) and TOP2-mediated (right) relaxation assay gels. Lane 1, supercoiled pBR322 DNA alone; lane 2, DNA and enzyme; lanes 3–6, DNA, enzyme and the tested compound at 25 μM concentration, respectively. Camptothecin (CPT) and etoposide (ETP) were used as positive controls for TOP1 and TOP2, respectively. R, relaxed DNA; Sc, supercoiled DNA. (B) Detection of TOP2-DNA covalent cleavage complexes by in vivo complex of enzyme (ICE) assay using CCRF-CEM cells. Lane 1, untreated control; lanes 2 and 3, cells treated with ETP at 10 and 50 μM concentration, respectively; lanes 4 and 5, cells treated with 74 at 10 and 50 μM concentration, respectively; lane 6, cells co-treated with 10 μM ETP and 50 μM 74.
2.4. Molecular Modeling
In order to obtain a molecular view of TDP2 inhibition by furoquinolinediones, we build a hypothetical binding model using in-silico docking. In the absence of the human TDP2 (hTDP2) molecular structure, we have employed homology modeling and constructed a hTDP2 structure using mouse TDP2 (mTDP2) as a template [4]. The assumption of mTDP2 suitability as a homology modeling template is born out of our observation that mTDP2 enzymatic performance and responsiveness to inhibitors can be made to mimic that of hTDP2 with few key mutations in the relative proximity of the catalytic site [28].
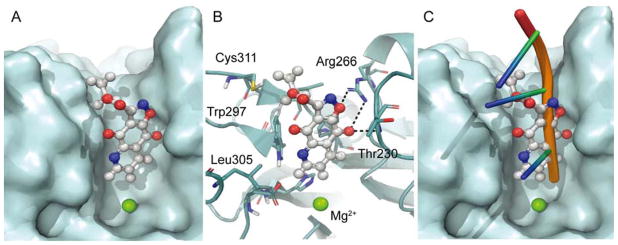
The docking of the furoquinolinediones 1 and 2 as well as their derivatives isoxazoles 73 and 74 into hTDP2 structure gave a theoretical insight into the binding mode of these inhibitors. According to the model of 73-TDP2 (Figure 6), the polycyclic core of 73 is arranged along the DNA binding region and forms polar contacts to the guanidinium group of Arg266 and the backbone amide NH of Thr230 (Figure 6B). The hypothetical binding mode is in general agreement with the empirical SAR observed. The formation of the contact between furan/isoxazole oxygen atom and Arg266 highlights the importance of the hydrogen bond acceptor in that position; the replacement of this oxygen with pyrazole NH of 75 and 76 abolishes TDP2 inhibitory activity. The DNA binding cleft of TDP2 in the proximity of the active site provides tight fit for the polycyclic pharmacophore of furoquinolinediones (Figure 6A). The ester group of 73 points toward solvent-accessible space of the complex, similar to the direction of TDP2-bound DNA in mTDP2 complex, further validating the proposed binding mode (Figure 6C).
Figure 6.
The hypothetical binding mode of isoxazole derivative 73. (A) An inhibitor steric fit in hypothetical binding mode of 73 (gray carbon atoms ball and stick representation) to hTDP2 (cyan surface). (B) Molecular interactions of 73 in complex with hTDP2 (cartoon representation, residues in the proximity of the ligand shown as sticks). (C) Comparison of binding of 73 and DNA (cartoon, from mTDP2 complex, PDB ID 4GZ1). The representations were constructed using PyMOL.
3. Conclusions
The furoquinolinedione scaffold was discovered as a novel chemotype for TDP2 inhibitors. Following the identification of 1 as TDP2 inhibitor, the SAR of furoquinolinediones was explored through a series of analogues designed to probe the potential substitutions at position 3 and alterations of the furan ring motif. The results of this exploration show that the presence of oxygen at position 1 are critical for TDP2 inhibitory potency. Furthermore, the ester functionality at position 3 is preferred, and a bulk substituent at position 3 is tolerated with respect to TDP2 inhibitory potency. The exploration of furoquinolinedione core analogues led to discovery of the isoxazole analogues 73/74 which show the highest TDP2 inhibitory potency of the entire series with IC50 of 3.3/1.9 and 3.1/2.1 μM against TDP2 and TDP2 WCE, respectively, and no activity against TDP1, TOP1 or TOP2, indicating its selectivity. In summary, this report describes a new class of TDP2 inhibitors and provides systematic SAR exploration, providing ground for further development of furoquinolinediones as TDP2 inhibitors as potential anticancer and antiviral agents.
4. Methods and Materials
4.1. General Experiments
The major chemical reagents for synthesis were purchased from Alfa Aesar, Sigma Aldrich Co. or Aladdin Reagent Database Inc (Shanghai), and were used without further purification unless otherwise indicated. The materials, such as quinoline-5,8-diones, 6,7-dichloroquinoline-5,8-dione and 7-bromoquinoline-5,8-dione were prepared in our laboratory according to the reported methods [36, 37]. Chemical reaction courses were monitored by silica gel GF254 thin layer chromatography. Melting points were determined in open capillary tubes on a MPA100 Optimelt Automated Melting Point System without being corrected. Nuclear magnetic resonance spectra were recorded on a Bruker AVANCE III 400 MHz spectrometer using tetramethylsilane as an internal reference. Mass spectra were analyzed on an Agilent 6120 (Quadrupole LC-MS) mass spectrometer. The high-resolution mass spectra were analyzed on an SHIMADZU LCMS-IT-TOF mass spectrometer. All compounds tested for biological activities were analyzed by HPLC and their purities were more than 95%.
4.2. General procedures for the synthesis of furoquinolinediones 1–6
To a solution of 6,7-dichloroquinoline-5,8-dione (0.46 g, 2 mmol) in acetonitrile (30 mL), K2CO3 (1.10 g) and active methylene reagents (2.2 mmol) were added. The yellow reaction solution was stirred and heated under reflux for 3–6 h and cooled to room temperature. The solvent was evaporated under reduced pressure. The residue was purified by silica gel column chromatography to give the N,O-anti and N,O-syn isomers, respectively.
4.2.1. Ethyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (1)
Yellow solid, yield 9%, mp = 173.6–178.0 °C. 1H NMR (CDCl3) δ 9.06 (d, J = 4.0 Hz, 1H), 8.54 (d, J = 7.6 Hz, 1H), 7.70 (dd, J = 7.6, 4.7 Hz, 1H), 4.44 (q, J = 6.7 Hz, 2H), 2.76 (s, 3H), 1.48 (t, J = 7.2 Hz, 3H). 13C NMR (CDCl3) δ 175.4, 171.3, 164.4, 161.0, 153.4, 149.5, 148.1, 133.6, 127.6, 127.3, 126.3, 113.0, 60.7, 13.0. HRMS (ESI) m/z: 284.0551 [M − H]−, calcd for C15H10NO5 284.0564. The structure of compound 1 was confirmed with 2D NMR spectra and X-ray single crystal analysis.
4.2.2. Ethyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[3,2-g]quinoline-3-carboxylate (2)
Yellow solid, yield 11%, mp = 173.5–178.2 °C. 1H NMR (CDCl3) δ 9.04 (d, J = 4.8 Hz, 1H), 8.54 (d, J = 8.0 Hz, 1H), 7.67 (dd, J = 7.6, 4.7 Hz, 1H), 4.46 (q, J = 7.1 Hz, 2H), 2.76 (s, 3H), 1.46 (t, J = 6.7 Hz, 3H). 13C NMR (CDCl3) δ 177.2, 171.5, 165.4, 161.6, 154.2, 151.6, 147.5, 135.4, 130.7, 128.0, 127.6, 113.7, 61.6, 14.3, 14.1. HRMS (ESI) m/z: 284.0574 [M − H]−, calcd for C15H10NO5 284.0564. The structure of compound 2 was confirmed with 2D NMR spectra.
4.2.3. 3-Acetyl-2-methylfuro[2,3-g]quinoline-4,9-dione (3)
Yellow solid, yield 5%, mp = 219.5–220.4 °C. 1H NMR (CDCl3) δ 9.07 (d, J = 4.5 Hz, 1H), 8.55 (dd, J = 7.8, 1.2 Hz, 1H), 7.73 (dd, J = 7.8, 4.7 Hz, 1H), 2.81 (s, 3H), 2.70 (s, 3H). 13C NMR (CDCl3) δ 195.3, 178.3, 172.3, 165.0, 154.4, 150.1, 148.9, 134.7, 128.5, 127.9, 127.6, 121.2, 31.9, 14.4. HRMS (ESI) m/z: 256.0613 [M + H]+, calcd for C14H10NO4 256.0604.
4.2.4. 3-Acetyl-2-methylfuro[3,2-g]quinoline-4,9-dione (4)
Yellow solid, yield 10%, mp = 219.0–220.1 °C. 1H NMR (CDCl3) δ 9.07 (dd, J = 4.7, 1.7 Hz, 1H), 8.54 (dd, J = 7.9, 1.7 Hz, 1H), 7.73 (dd, J = 7.9, 4.7 Hz, 1H), 2.79 (s, 3H), 2.70 (s, 3H). 13C NMR (CDCl3) δ 195.1, 179.0, 171.5, 165.1, 154.6, 151.1, 147.6, 135.4, 130.4, 127.7, 127.1, 120.9, 31.9, 14.5. HRMS (ESI) m/z: 256.0593 [M + H]+, calcd for C14H10NO4 256.0604.
4.2.5. Dimethyl-(2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinolin-3-yl)phosphonate (5)
Yellow solid, yield 9%, mp = 183.4–183.9 °C. 1H NMR (CDCl3) δ 9.06 (d, J = 3.6 Hz, 1H), 8.55 (d, J = 7.8 Hz, 1H), 7.73 (dd, J = 7.7, 4.7 Hz, 1H), 3.95 (s, 3H), 3.92 (s, 3H), 2.83 (s, 3H). 13C NMR (CDCl3) δ 177.0, 172.2, 169.3, 169.0, 154.4, 151.4, 151.3, 148.7, 134.8, 131.2, 131.1, 128.6, 127.5, 108.2, 106.1, 53.6, 53.6, 14.5. HRMS (ESI) m/z: 322.0473 [M + H]+, calcd for C14H13NO6P 322.0475. The structure of compound 5 was confirmed with 2D NMR spectra.
4.2.6. Dimethyl-(2-methyl-4,9-dioxo-4,9-dihydrofuro[3,2-g]quinolin-3-yl)phosphonate (6)
Yellow solid, yield 10%, mp = 193.1–194.1 °C. 1H NMR (CDCl3) δ 9.06 (dd, J = 4.7, 1.7 Hz, 1H), 8.53 (dd, J = 7.9, 1.7 Hz, 1H), 7.73 (dd, J = 7.9, 4.7 Hz, 1H), 3.92 (s, 3H), 3.89 (s, 3H), 2.83 (d, J = 2.0 Hz, 3H). 13C NMR (CDCl3) δ 177.8, 171.3, 169.4, 169.1, 154.4, 152.4, 152.3, 147.7, 135.3, 130.3, 130.2, 130.2, 127.6, 107.5, 105.4, 53.4, 53.3, 14.5. HRMS (ESI) m/z: 322.0485 [M + H]+, calcd for C14H13NO6P 322.0475. The structure of compound 6 was confirmed with 2D NMR spectra.
4.3. Hydrolysis of compounds 1 and 2
To a solution of compound 1 or 2 (4 mmol) in isopropanol (300 mL), a Na2CO3 aqueous solution (2N, 30 mL) was added. The mixture solution was stirred and heated under reflux for 30 min and cooled to room temperature. The solvent was evaporated under reduced pressure. The residue was purified by silica gel column chromatography to give the target compound 7 or 66, respectively.
4.3.1. 2-Methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylic acid (7)
Yellow solid, yield 75%, 1H NMR (CDCl3) δ 9.12 (dd, J = 4.7, 1.7 Hz, 1H), 8.59 (dd, J = 7.9, 1.7 Hz, 1H), 7.82 (dd, J = 7.9, 4.7 Hz, 1H), 2.93 (s, 3H). ESI-MS m/z: 258.1 [M +H]+.
4.3.2. 2-Methyl-4,9-dioxo-4,9-dihydrofuro[3,2-g]quinoline-3-carboxylic acid (66)
Yellow solid, yield 72%, mp = 176.9–177.5 °C. 1H NMR (DMSO) δ 13.33 (s, 1H), 9.02 (d, J = 3.3 Hz, 1H), 8.43 (d, J = 7.6 Hz, 1H), 7.89-7.83 (m, 1H), 2.68 (s, 3H). 13C NMR (DMSO) δ 178.6, 171.6, 164.4, 163.1, 154.3, 152.1, 148.0, 135.2, 130.9, 128.3, 127.4, 114.0, 14.2. HRMS (ESI) m/z: 258.0409 [M + H]+, calcd for C13H8NO5 258.0397.
4.4. General procedures for the esterification or amidation of compounds 7 and 66
To a yellow solution of compound 7 or 66 (1 mmol) in new distilled chloroform (80 mL), a solution of thionyl chloride (1.5 mL) in new distilled chloroform (10 mL) was added dropwise. The mixture solution was stirred and heated under reflux for 5 h and cooled to room temperature. The solvent was evaporated under reduced pressure. The yellow residue was dissolved in new distilled chloroform (40 mL) and was added with a solution of DMAP (0.1 mmol), Et3N (1.2 mmol) and corresponding amines or alcohols (1.2 mmol) in new distilled chloroform (20 mL). The reaction mixture was stirred and heated under reflux for 5 h and cooled to room temperature. The solvent was evaporated under reduced pressure. The residue was purified by silica gel column chromatography to give the amide or ester targets, respectively.
4.4.1. N-Ethyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxamide (8)
Yellow solid, yield 55%, mp = 237.6–238.5 °C. 1H NMR (CDCl3) δ 9.52 (s, 1H), 9.08 (d, J = 3.3 Hz, 1H), 8.56 (d, J = 7.4 Hz, 1H), 7.76 (dd, J = 7.7, 4.7 Hz, 1H), 3.50 (quint, J = 6.4 Hz, 2H), 2.93 (s, 3H), 1.34 (t, J = 7.2 Hz, 3H). 13C NMR (CDCl3) δ 181.0, 171.9, 167.3, 160.7, 154.5, 150.5, 148.6, 134.8, 128.6, 128.1, 126.6, 115.9, 34.5, 15.1, 14.3. HRMS (ESI) m/z: 307.0699 [M + Na]+, calcd for C15H12N2O4Na 307.0689.
4.4.2. N-Phenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxamide (9)
Yellow solid, yield 41%, mp = 295.6–296.2 °C. 1H NMR (DMSO) δ 11.10 (s, 1H), 9.06 (d, J = 3.2 Hz, 1H), 8.51 (dd, J = 7.8, 1.5 Hz, 1H), 7.91 (dd, J = 7.7, 4.7 Hz, 1H), 7.76 (d, J = 7.8 Hz, 2H), 7.42 (t, J = 7.1 Hz, 2H), 7.17 (d, J = 7.4 Hz, 1H), 2.78 (s, 3H). 13C NMR (CDCl3) δ 180.6, 170.9, 167.2, 157.8, 153.6, 149.6, 147.5, 137.2, 133.9, 128.0, 127.6, 127.3, 125.1, 123.5, 119.2, 115.4, 14.4. HRMS (ESI) m/z: 331.0723 [M − H]−, calcd for C19H11N2O4 331.0724.
4.4.3. N-(2-hydroxyphenyl) 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxamide (10)
Orange solid, yield 49%, mp = 290.1–292.3 °C. 1H NMR (DMSO) δ 11.19 (s, 1H), 9.07 (d, J = 3.2 Hz, 1H), 8.51 (dd, J = 7.8, 1.5 Hz, 1H), 7.96 (s, 1H), 7.92 (dd, J = 7.7, 4.7 Hz, 1H), 7.59 (d, J = 8.2 Hz, 1H), 7.45 (t, J = 8.1 Hz, 1H), 7.23 (d, J = 7.8 Hz, 1H), 2.76 (s, 3H). 13C NMR (DMSO) δ 180.2, 172.6, 168.8, 164.7, 159.8, 154.4, 150.5, 148.2, 140.3, 134.8, 133.8, 131.3, 129.1, 128.7, 127.5, 124.3, 119.5, 118.4, 14.4. HRMS (ESI) m/z: 347.0656 [M − H]−, calcd for C19H11N2O5 347.0673.
4.4.4. N-(4-Methoxyphenyl) 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxamide (11)
Red solid, yield 54%, mp = 332.7–333.2 °C. 1H NMR (DMSO) δ 11.00 (s, 1H), 9.07 (d, J = 3.3 Hz, 1H), 8.50 (dd, J = 7.8, 1.6 Hz, 1H), 7.91 (dd, J = 7.7, 4.7 Hz, 1H), 7.67 (d, J = 9.0 Hz, 2H), 6.99 (d, J = 9.0 Hz, 2H), 3.77 (s, 3H), 2.77 (s, 3H). 13C NMR (DMSO) δ 180.0, 172.1, 164.1, 158.5, 155.8, 153.8, 150.0, 148.6, 134.2, 133.4, 131.5, 128.6, 128.2, 126.9, 121.1, 114.2, 55.2, 14.0. HRMS (ESI) m/z: 361.0817 [M − H]−, calcd for C20H13N2O5 361.0830.
4.4.5. N-(3-Chlorophenyl) 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxamide (12)
Orange solid, yield 50%, mp = 311.8–312.5 °C. 1H NMR (DMSO) δ 11.11 (s, 1H), 10.02 (s, 1H), 9.05 (d, J = 3.3 Hz, 1H), 8.49 (d, J = 6.7 Hz, 1H), 8.18 (d, J = 7.8 Hz, 1H), 7.90 (dd, J = 7.7, 4.7 Hz, 1H), 6.99 (t, J = 7.2 Hz, 1H), 6.94 (d, J = 7.0 Hz, 1H), 6.82 (t, J = 7.3 Hz, 1H), 2.83 (s, 3H). 13C NMR (DMSO) δ 180.0, 172.2, 165.7, 159.0, 153.8, 150.1, 148.6, 147.8, 134.1, 128.5, 128.1, 126.2, 124.7, 121.8, 118.9, 115.6, 115.1, 14.7. HRMS (ESI) m/z: 367.0480 [M + H]+, calcd for C19H12N2O4Cl 367.0501.
4.4.6. Methyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (13)
Yellow solid, mp = 176.2–178.4 °C. 1H NMR (CDCl3) δ 9.06 (dd, J = 4.7, 1.7 Hz, 1H), 8.54 (dd, J = 7.9, 1.7 Hz, 1H), 7.71 (dd, J = 7.9, 4.7 Hz, 1H), 3.99 (s, 3H), 2.76 (s, 3H). 13C NMR (CDCl3) δ 176.5, 172.3, 165.5, 162.3, 154.4, 150.6, 149.2, 134.6, 128.7, 128.4, 127.3, 113.6, 52.4, 14.1. HRMS (ESI) m/z: 294.0386 [M + Na]+, calcd for C14H9NO5Na 294.0373.
4.4.7. 2,2,2-Trifluoroethyl 4,9-dihydro-2-methyl-4,9-dioxofuro[2,3-g]quinoline-3-carboxylate (14)
Yellow solid, yield 61%, mp = 166.9–167.4 °C. 1H NMR (CDCl3) δ 9.06 (d, J = 3.5 Hz, 1H), 8.53 (dd, J = 7.8, 1.5 Hz, 1H), 7.71 (dd, J = 7.8, 4.7 Hz, 1H), 4.77 (q, J = 8.3 Hz, 2H), 2.78 (s, 3H). 13C NMR (CDCl3) δ 175.1, 171.3, 165.6, 159.1, 153.6, 149.9, 148.0, 133.7, 127.4, 127.3, 126.4, 121.8 (q, 1JCF = 275.7 Hz), 111.1, 60.0 (q, 2JCF = 36.9 Hz), 13.3. HRMS (ESI) m/z: 362.0266 [M + Na]+, calcd for C15H8NO5F3Na 362.0247.
4.4.8. 2-((2-Methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carbonyl)oxy)acetic acid (15)
Yellow solid, yield 39%, mp = 205.0 °C (decomposing). 1H NMR (CD3OD) δ 8.95 (dd, J = 4.7, 1.6 Hz, 1H), 8.58 (dd, J = 7.9, 1.6 Hz, 1H), 7.84 (dd, J = 7.9, 4.8 Hz, 1H), 4.07 (s, 2H), 2.78 (s, 3H). 13C NMR (CDCl3) δ 176.0, 171.8, 169.0, 166.0, 160.6, 153.7, 150.4, 148.4, 134.5, 128.2, 128.1, 127.3, 112.5, 60.9, 13.6. HRMS (ESI) m/z: 338.0283 [M + Na]+, calcd for C15H9NO7Na 338.0271.
4.4.9. 2-Hydroxyethyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (16)
Yellow solid, yield 24%, mp = 184.7–186.1 °C. 1H NMR (CDCl3) δ 9.08 (d, J = 3.2 Hz, 1H), 8.56 (d, J = 7.6 Hz, 1H), 7.73 (dd, J = 7.4, 4.4 Hz, 1H), 4.49 (t, J = 4.4 Hz, 2H), 4.00 (t, J = 4.4 Hz, 2H), 2.80 (s, 3H). 13C NMR (CDCl3) δ 176.8, 172.3, 166.1, 162.3, 154.4, 150.6, 149.1, 134.7, 128.3, 128.3, 127.4, 113.6, 62.9, 59.5, 31.3, 14.1. HRMS (ESI) m/z: 300.0514 [M − H]−, calcd for C15H10NO6 300.0499.
4.4.10. 3-Hydroxypropyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (17)
Yellow solid, yield 25%, mp = 169.1–169.5 °C. 1H NMR (CDCl3) δ 9.06 (d, J = 3.3 Hz, 1H), 8.54 (d, J = 7.0 Hz, 1H), 7.71 (dd, J = 7.6, 4.7 Hz, 1H), 4.55 (t, J = 5.9 Hz, 2H), 3.91 (t, J = 5.9 Hz, 2H), 2.77 (s, 3H), 2.09 (quint, J = 5.8 Hz, 2H). 13C NMR (CDCl3) δ 176.8, 172.3, 166.1, 162.3, 154.4, 150.6, 149.1, 134.7, 128.3, 127.4, 113.6, 62.9, 59.5, 50.8, 31.3, 14.1. HRMS (ESI) m/z: 314.0653 [M − H]−, calcd for C16H12NO6 314.0670.
4.4.11. 4-Hydroxybutyl 4,9-dihydro-2-methyl-4,9-dioxofuro[2,3-g]quinoline-3-carboxylate (18)
Yellow solid, yield 24%, mp = 128.0–128.5 °C. 1H NMR (CDCl3) δ 9.06 (d, J = 3.5 Hz, 1H), 8.54 (dd, J = 7.8, 1.1 Hz, 1H), 7.71 (dd, J = 7.8, 4.6 Hz, 1H), 4.43 (t, J = 6.4 Hz, 2H), 3.75 (t, J = 6.3 Hz, 2H), 2.76 (s, 3H), 1.95 (quint, J = 6.4 Hz, 2H), 1.81 (quint, J = 6.4 Hz, 2H). 13C NMR (CDCl3) δ 175.5, 171.3, 164.8, 161.1, 153.3, 149.6, 148.0, 133.7, 127.4, 127.3, 126.4, 112.8, 64.6, 61.3, 28.2, 24.0, 13.1. HRMS (ESI) m/z: 352.0810 [M + Na]+, calcd for C17H15NO6Na 352.0792.
4.4.12. 5-Hydroxypentyl 4,9-dihydro-2-methyl-4,9-dioxofuro[2,3-g]quinoline-3-carboxylate (19)
Yellow solid, yield 42%, mp = 107.7–108.8 °C. 1H NMR (CDCl3) δ 9.05 (dd, J = 4.7, 1.7 Hz, 1H), 8.53 (dd, J = 7.9, 1.7 Hz, 1H), 7.70 (dd, J = 7.9, 4.7 Hz, 1H), 4.40 (t, J = 6.5 Hz,2H), 3.71 (t, J = 6.1 Hz, 2H), 2.76 (s,3H), 1.93-1.85 (m,2H), 1.73-1.55 (m, 4H). 13C NMR (CDCl3) δ 176.5, 172.3, 165.6, 162.1, 154.3, 150.6, 149.1, 134.6, 128.6, 128.3, 127.3, 113.8, 65.6, 62.4, 32.2, 28.2, 22.2, 14.0. HRMS (ESI) m/z: 366.0966 [M + Na]+, calcd for C18H17NO6Na 366.0948.
4.4.13. 2-(2-Hydroxyethoxy)ethyl 4,9-dihydro-2-methyl-4,9-dioxofuro[2,3-g]quinoline-3-carboxylate (20)
Brown solid, yield 31%, mp = 134.6–135.4 °C. 1H NMR (CDCl3) δ 9.05 (dd, J = 4.7, 1.7 Hz, 1H), 8.53 (dd, J = 7.9, 1.7 Hz, 1H), 7.71 (dd, J = 7.9, 4.7 Hz, 1H), 4.56-4.53 (m, 2H), 3.94-3.91 (m, 2H), 3.80-3.77 (m, 2H), 3.71-3.68 (m, 2H), 2.76 (s, 3H). 13C NMR (CDCl3) δ 176.7, 172.3, 165.9, 161.9, 154.4, 150.6, 149.1, 134.6, 128.6, 128.3, 127.3, 113.5, 72.5, 68.6, 64.5, 61.6, 14.0. HRMS (ESI) m/z: 368.0752 [M + Na]+, calcd for C17H15NO7Na 368.0741.
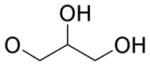
4.4.14. 2,3-Dihydroxypropyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (21)
Yellow solid, yield 26%, mp = 193.7–195.3 °C. 1H NMR (CDCl3) δ 9.08 (dd, J = 4.7, 1.7 Hz, 1H), 8.55 (dd, J = 7.9, 1.7 Hz, 1H), 7.73 (dd, J = 7.9, 4.7 Hz, 1H), 4.47 (d, J = 5.2 Hz, 2H), 4.18 (quint, J = 5.2 Hz, 1H), 3.83-3.74 (m, 2H), 2.80 (s, 3H). 13C NMR (CDCl3) δ 177.4, 172.2, 166.9, 161.9, 154.5, 150.9, 149.0, 134.8, 128.3, 127.8, 127.7, 112.9, 69.8, 66.9, 63.2, 14.1. HRMS (ESI) m/z: 330.0604 [M − H]−, calcd for C16H12NO7 330.0619.
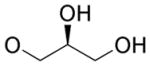
4.4.15. (2’S)-2,3-Dihydroxypropyl 4,9-dihydro-2-methyl-4,9-dioxofuro[2,3-g]quinoline-3-carboxylate (22)
Yellow solid, yield 24%, mp = 173.4–174.8 °C. 1H NMR (CDCl3) δ 9.09 (dd, J = 4.6, 1.6 Hz, 1H), 8.57 (dd, J = 7.8, 1.5 Hz, 1H), 7.75 (dd, J = 7.8, 4.7 Hz, 1H), 4.48 (d, J = 4.7 Hz, 2H), 4.19 (quint, J = 5.3 Hz, 1H), 3.85-3.76 (m, 2H), 2.81 (s, 3H). 13C NMR (CDCl3) δ 177.4, 172.2, 167.0, 161.9, 154.5, 150.9, 149.0, 134.8, 128.3, 127.8, 127.7, 112.9, 69.8, 66.9, 63.2, 14.1. HRMS (ESI) m/z: 332.0750 [M + H]+, calcd for C16H14NO7 332.0765.
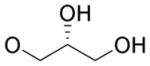
4.4.16. (2’R)-2,3-Dihydroxypropyl 4,9-dihydro-2-methyl-4,9-dioxofuro[2,3-g]quinoline-3-carboxylate (23)
Yellow solid, yield 48%, mp = 159.0–162.7 °C. 1H NMR (CDCl3) δ 9.01 (dd, J = 4.7, 1.7 Hz, 1H), 8.48 (dd, J = 7.9, 1.7 Hz, 1H), 7.66 (dd, J = 7.9, 4.7 Hz, 1H), 4.40 (d, J = 5.2 Hz, 2H), 4.14-4.08 (m, 1H), 3.76-3.67 (m, 2H), 2.73 (s, 3H). 13C NMR (CDCl3) δ 177.4, 172.2, 167.0, 161.9, 154.5, 150.9, 149.0, 134.8, 128.3, 127.8, 127.7, 112.9, 69.8, 66.9, 63.2, 14.1. HRMS (ESI) m/z: 354.0596 [M + Na]+, calcd for C16H13NO7Na 354.0584.
4.4.17. 2-(Dimethoxyphosphoryl)ethyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (24)
Yellow solid, yield 81%, mp = 182.5–183.7 °C. 1H NMR (CDCl3) δ 9.06 (dd, J = 4.6, 1.6 Hz, 1H), 8.54 (dd, J = 7.9, 1.6 Hz, 1H), 7.71 (dd, J = 7.9, 4.7 Hz, 1H), 4.64-4.58 (m, 2H), 3.78 (d, J = 11.2 Hz, 6H), 2.77 (s, 3H), 2.46 (dt, J = 19.2, 7.8 Hz, 2H). 13C NMR (CDCl3) δ 176.5, 172.2, 165.8, 161.5, 154.4, 150.6, 149.0, 134.7, 128.4, 128.3, 127.3, 113.7, 59.6, 52.5 (d, 2JCP = 6.3 Hz), 24.9 (d, 1JCP = 139.5 Hz), 14.0. HRMS (ESI) m/z: 392.0538 [M − H]−, calcd for C17H15NO8P 392.0541.
4.4.18. 2-Cyanoethyl 4,9-dihydro-2-methyl-4,9-dioxofuro[2,3-g]quinoline-3-carboxylate (25)
Yellow solid, yield 49%, mp = 182.5–183.9 °C. 1H NMR (CDCl3) δ 9.06 (dd, J = 4.6, 1.6 Hz, 1H), 8.54 (dd, J = 7.9, 1.7 Hz, 1H), 7.72 (dd, J = 7.9, 4.7 Hz, 1H), 4.60 (t, J = 6.5 Hz, 2H), 2.99 (t, J = 6.5 Hz, 2H), 2.78 (s, 3H). 13C NMR (CDCl3) δ 176.5, 172.2, 166.3, 161.2, 154.5, 150.8, 149.0, 134.7, 128.34, 128.32, 127.4, 116.8, 112.8, 59.7, 17.8, 14.2. HRMS (ESI) m/z: 311.0684 [M + H]+, calcd for C16H11N2O5 311.0662.
4.4.19. Phenethyl 4,9-dihydro-2-methyl-4,9-dioxofuro[2,3-g]quinoline-3-carboxylate (26)
Yellow solid, yield 53%, mp = 141.8–144.1 °C. 1H NMR (CDCl3) δ 9.07 (dd, J = 4.7, 1.7 Hz, 1H), 8.54 (dd, J = 7.9, 1.7 Hz, 1H), 7.70 (dd, J = 7.9, 4.7 Hz, 1H), 7.33-7.28 (m, 4H), 7.25-7.20 (m, 1H), 4.60 (t, J = 7.3 Hz, 2H), 3.19 (t, J = 7.3 Hz, 2H), 2.65 (s, 3H). 13C NMR (CDCl3) δ 176.5, 172.3, 165.5, 161.8, 154.4, 150.6, 149.1, 137.6, 134.6, 129.0, 128.6, 128.5, 128.4, 127.3, 126.6, 113.8, 66.1, 34.8, 14.0. HRMS (ESI) m/z: 384.0864 [M + Na]+, calcd for C21H15NO5Na 384.0842.
4.4.20. Phenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (27)
Yellow solid, yield 36%, mp = 234.8–246.5 °C. 1H NMR (DMSO) δ 9.03 (dd, J = 4.7, 1.7 Hz, 1H), 8.49 (dd, J = 7.9, 1.7 Hz, 1H), 7.87 (dd, J = 7.9, 4.7 Hz, 1H), 7.62-7.46 (m, 2H), 7.44-7.29 (m, 2H), 2.78 (s, 3H). 13C NMR (CDCl3) δ 176.5, 172.3, 166.2, 160.4, 154.5, 150.7, 150.4, 149.1, 134.7, 129.4, 128.4, 127.4, 126.2, 121.6, 113.5, 14.1. HRMS (ESI) m/z: 332.0563 [M − H]−, calcd for C19H10NO5 332.0564.
4.4.21. 4-((Dimethoxyphosphoryl)oxy)phenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (28)
Yellow solid, yield 62%, mp = 118.7–120.3 °C. 1H NMR (CDCl3) δ 9.06 (dd, J = 4.7, 1.7 Hz, 1H), 8.56 (dd, J = 7.9, 1.7 Hz, 1H), 7.73 (dd, J = 7.9, 4.7 Hz, 1H), 7.44 (d, J = 8.9 Hz, 2H), 7.29 (d, J = 8.4 Hz, 2H), 3.89 (d, J = 11.2 Hz, 6H), 2.82 (s, 3H). 13C NMR (CDCl3) δ 176.6, 172.3, 166.4, 160.3, 154.5, 150.7, 149.1, 148.3 (d, 2JCP = 6.8 Hz), 147.3, 134.7, 128.5, 128.4, 127.4, 122.9, 120.8 (d, 3JCP = 4.8 Hz), 113.2, 55.0 (d, 2JCP = 6.2 Hz), 14.1. HRMS (ESI) m/z: 458.0654 [M + H]+, calcd for C21H17NO9P 458.0635.
4.4.22. 2-Hydroxyphenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (29)
Yellow solid, yield 42%, mp = 252.1–253.2 °C. 1H NMR (DMSO) δ 9.81 (s, 1H), 9.03 (d, J = 3.9 Hz, 1H), 8.49 (d, J = 7.7 Hz, 1H), 7.87 (dd, J = 7.6, 4.7 Hz, 1H), 7.23 (d, J = 7.8 Hz, 1H), 7.16 (t, J = 7.6 Hz, 1H), 7.00 (d, J = 7.9 Hz, 1H), 6.90 (t, J = 7.5 Hz, 1H), 2.80 (s, 3H). 13C NMR (DMSO) δ 177.0, 172.8, 165.4, 159.7, 154.3, 151.4, 149.4, 149.3, 138.5, 134.6, 129.0, 128.5, 128.2, 127.6, 123.4, 119.7, 117.4, 116.1, 112.9, 14.5. HRMS (ESI) m/z: 348.0512 [M − H]−, calcd for C19 H10NO6 348.0514.
4.4.23. 3-Hydroxyphenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (30)
Yellow solid, yield 20%, mp = 241.3–243.2 °C. 1H NMR (DMSO) δ 9.81 (s, 1H), 9.03 (d, J = 3.1 Hz, 1H), 8.49 (d, J = 7.2 Hz, 1H), 7.87 (dd, J = 6.6, 4.9 Hz, 1H), 7.27 (d, J = 8.2 Hz, 1H), 6.88-6.68 (m, 4H), 2.76 (s, 3H). 13C NMR (CDCl3) δ 180.5, 176.1, 170.2, 164.2, 162.0, 158.0, 155.1, 154.7, 152.7, 138.9, 133.7, 132.4, 131.7, 117.4, 117.2, 116.4, 112.7, 110.8, 17.8. HRMS (ESI) m/z: 348.0499 [M − H]−, calcd for C19H10NO6 348.0514.
4.4.24. 2-Chlorophenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (31)
Light yellow solid, yield 51%, mp = 212.4–214.0 °C. 1H NMR (CDCl3) δ 9.06 (dd, J = 4.6, 1.6 Hz, 1H), 8.56 (dd, J = 7.9, 1.7 Hz, 1H), 7.72 (dd, J = 7.9, 4.7 Hz, 1H), 7.51 (ddd, J = 9.7, 8.1, 1.4 Hz, 2H), 7.38 (td, J = 7.8, 1.5 Hz, 1H), 7.31-7.23 (m, 1H), 2.85 (s, 3H). 13C NMR (CDCl3) δ 176.4, 172.2, 166.2, 159.2, 154.4, 150.7, 149.0, 146.6, 134.7, 130.3, 128.7, 128.4, 128.0, 127.5, 127.4, 126.8, 123.9, 112.8, 14.2. HRMS (ESI) m/z: 368.0335 [M + H]+, calcd for C19H11NO5Cl 368.0320.
4.4.25. 3-Chlorophenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (32)
Light yellow solid, yield 49%, mp = 212.9–214.2 °C. 1H NMR (CDCl3) δ 9.07 (dd, J = 4.6, 1.6 Hz, 1H), 8.56 (dd, J = 7.9, 1.6 Hz, 1H), 7.72 (dd, J = 7.9, 4.7 Hz, 1H), 7.52 (s, 1H), 7.37 (dd, J = 8.4, 4.7 Hz, 1H), 7.29 (dd, J = 4.4, 1.9 Hz, 1H), 2.83 (s, 1H). 13C NMR (CDCl3) δ 176.5, 172.2, 166.6, 160.0, 154.5, 150.8, 150.7, 149.0, 134.7, 130.1, 128.4, 128.4, 127.4, 126.5, 122.2, 120.0, 112.9, 14.1. HRMS (ESI) m/z: 368.0341 [M + H]+, calcd for C19H11NO5Cl 368.0320.
4.4.26. 4-Chlorophenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (33)
Light yellow solid, yield 71%, mp = 217.0–217.8 °C. 1H NMR (CDCl3) δ 9.07 (dd, J = 4.6, 1.6 Hz, 1H), 8.56 (dd, J = 7.9, 1.6 Hz, 1H), 7.72 (dd, J = 7.9, 4.7 Hz, 1H), 7.41 (br. s, 4H), 2.82 (s, 3H). 13C NMR (CDCl3) δ 176.6, 172.3, 166.5, 160.2, 154.5, 150.8, 149.1, 148.9, 134.7, 131.6, 129.5, 128.4, 128.4, 127.5, 123.0, 113.1, 14.1. HRMS (ESI) m/z: 368.0334 [M + H]+, calcd for C19H11NO5Cl 368.0320.
4.4.27. 2-Cyanophenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (34)
Light yellow solid, yield 53%, mp = 255.7–256.3 °C. 1H NMR (CDCl3) δ 9.06 (dd, J = 4.7, 1.7 Hz, 1H), 8.57 (dd, J = 7.9, 1.7 Hz, 1H), 7.92-7.61 (m, 4H), 7.42 (td, J = 7.5, 1.4 Hz, 1H), 2.87 (s, 3H). 13C NMR (DMSO) δ 176.3, 172.3, 166.1, 159.1, 153.9, 151.9, 151.1, 148.8, 135.3, 134.1, 133.8, 128.5, 127.7, 127.4, 123.4, 119.5, 115.2, 111.0, 106.3, 14.2. HRMS (ESI) m/z: 381.0503 [M + Na]+, calcd for C20H10N2O5Na 381.0482.
4.4.28. 3-Cyanophenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (35)
Light yellow solid, yield 62%, mp = 221.0–221.8 °C. 1H NMR (CDCl3) δ 9.08 (dd, J = 4.7, 1.7 Hz, 1H), 8.57 (dd, J = 7.9, 1.7 Hz, 1H), 7.82 (d, J = 1.5 Hz, 1H), 7.79-7.73 (m, 2H), 7.63–7.53 (m, 2H), 2.84 (s, 3H). 13C NMR (CDCl3) δ 176.6, 172.2, 166.9, 159.8, 154.6, 150.8, 150.5, 149.0, 134.8, 130.4, 129.9, 128.4, 128.3, 127.5, 126.6, 125.4, 117.8, 113.5, 112.6, 14.1. HRMS (ESI) m/z: 359.0679 [M + H]+, calcd for C20H11N2O5 359.0662.
4.4.29. 4-Cyanophenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (36)
Light yellow solid, yield 53%, mp = 244.5–244.7 °C. 1H NMR (CDCl3) δ 9.08 (dd, J = 4.7, 1.7 Hz, 1H), 8.57 (dd, J = 7.9, 1.7 Hz, 1H), 7.76 (d, J = 8.8 Hz, 2H), 7.74 (dd, J = 7.8, 4.7 Hz, 1H), 7.63 (d, J = 8.8 Hz, 2H), 2.84 (s, 3H). 13C NMR (CDCl3) δ 176.6, 172.2, 167.0, 159.6, 154.6, 153.6, 150.9, 149.0, 134.8, 133.7, 128.3, 128.2, 127.6, 122.7, 118.2, 112.6, 110.1, 14.1. HRMS (ESI) m/z: 381.0496 [M + Na]+, calcd for C20H10N2O5Na 381.0482.
4.4.30. 4-Bromophenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (37)
Light yellow solid, yield 69%, mp = 214.3–215.8 °C. 1H NMR (CDCl3) δ 9.07 (dd, J = 4.7, 1.7 Hz, 1H), 8.56 (dd, J = 7.9, 1.7 Hz, 1H), 7.72 (dd, J = 7.9, 4.7 Hz, 1H), 7.56 (d, J = 8.8 Hz, 2H), 7.36 (d, J = 8.8 Hz, 2H), 2.82 (s, 3H). 13C NMR (CDCl3) δ 176.6, 172.3, 166.5, 160.1, 154.5, 150.8, 149.5, 149.1, 134.7, 132.5, 128.4, 127.4, 123.4, 119.3, 113.1, 14.1. HRMS (ESI) m/z: 411.9820 [M + H]+, calcd for C19H11NO5 Br 411.9815.
4.4.31. 4-(Methylsulfonyl)phenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (38)
Light yellow solid, yield 57%, mp = 243.8–44.8 °C. 1H NMR (CDCl3) δ 9.10 (dd, J = 4.7, 1.7 Hz, 1H), 8.59 (dd, J = 7.9, 1.7 Hz, 1H), 8.07-8.03 (m, 2H), 7.76 (dd, J = 7.9, 4.7 Hz, 1H), 7.76-7.63 (m, 2H), 3.11 (s, 3H), 2.86 (s, 3H). 13C NMR (CDCl3) δ 176.7, 172.3, 167.0, 159.8, 154.6, 154.4, 150.9, 149.1, 138.3, 134.8, 129.2, 128.4, 127.6, 122.7, 112.7, 44.7, 14.2. HRMS (ESI) m/z: 434.0334 [M + Na]+, calcd for C20H13NO7SNa 434.0305.
4.4.32. 4-Nitrophenyl 4,9-dihydro-2-methyl-4,9-dioxofuro[2,3-g]quinoline-3-carboxylate (39)
Yellow solid, yield 44%, mp = 191.4–193.3 °C. 1H NMR (CDCl3) δ 9.11 (dd, J = 4.7, 1.7 Hz, 1H), 8.60 (dd, J = 7.9, 1.7 Hz, 1H), 8.39-8.34 (m, 2H), 7.77 (dd, J = 7.9, 4.7 Hz, 1H), 7.74-7.68 (m, 2H), 2.87 (s, 3H). 13C NMR (CDCl3) δ 176.6, 172.2, 167.2, 159.5, 155.0, 154.6, 150.9, 149.0, 145.6, 134.8, 128.4, 128.2, 127.6, 125.2, 122.5, 112.5, 14.2. HRMS (ESI) m/z: 379.0583 [M + H]+, calcd for C19H11N2O7 379.0561.
4.4.33. 2-Naphthalenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (40)
Yellow solid, yield 78%, mp = 235.0–236.8 °C. 1H NMR (CDCl3) δ 9.08 (d, J = 4.4 Hz, 1H), 8.56 (d, J = 8.0 Hz, 1H), 7.95-7.86 (m, 4H), 7.72 (dd, J = 7.8, 4.8 Hz, 1H), 7.59 (dd, J = 8.8, 2.4 Hz, 1H), 7.54-7.47 (m, 2H), 2.86 (s, 3H). 13C NMR (CDCl3) δ 176.6, 172.3, 166.3, 160.5, 154.4, 150.7, 149.1, 148.1, 134.7, 133.7, 131.6, 129.4, 128.5, 128.4, 127.8, 127.8, 127.4, 126.6, 125.8, 121.0, 118.6, 113.4, 14.1. HRMS (ESI) m/z: 382.0706 [M − H]−, calcd for C23H12NO5 382.0721.
4.4.34. Pyridin-3-yl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (41)
Light yellow solid, yield 62%, mp = 218.2–220.1 °C. 1H NMR (CDCl3) δ 9.08 (dd, J = 4.7, 1.7 Hz, 1H), 8.77 (d, J = 2.5 Hz, 1H), 8.58 (dd, J = 3.6, 1.5 Hz, 1H), 8.56 (d, J = 1.7 Hz, 1H), 7.87 (ddd, J = 8.3, 2.7, 1.4 Hz, 1H), 7.73 (dd, J = 7.9, 4.7 Hz, 1H), 7.43 (dd, J = 8.3, 4.8 Hz, 1H), 2.85 (s, 3H). 13C NMR (CDCl3) δ 176.6, 172.3, 166.9, 160.0, 154.6, 150.9, 149.1, 147.4, 147.3, 143.5, 134.8, 129.2, 128.4, 127.5, 124.0, 112.7, 14.2. HRMS (ESI) m/z: 335.0673 [M + H]+, calcd for C18H11N2O5 335.0662.
4.4.35. 3-Acrylamidophenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (42)
Yellow solid, yield 51%, mp = 202.3–205.1 °C. 1H NMR (DMSO) δ 10.40 (s, 1H), 9.03 (dd, J = 4.7, 1.6 Hz, 1H), 8.49 (dd, J = 7.9, 1.6 Hz, 1H), 7.87 (dd, J = 7.9, 4.7 Hz, 1H), 7.85 (t, J = 2.0 Hz, 1H), 7.59-7.54 (m, 1H), 7.46 (t, J = 8.1 Hz, 1H), 7.11-7.07 (m, 1H), 6.46 (dd, J = 17.0, 10.1 Hz, 1H), 6.29 (dd, J = 17.0, 1.9 Hz, 1H), 5.80 (dd, J = 10.1, 1.9 Hz, 1H), 2.77 (s, 3H). 13C NMR (DMSO) δ 177.1, 172.8, 165.7, 163.8, 160.5, 154.3, 151.2, 150.7, 149.4, 140.7, 134.6, 132.1, 130.2, 129.0, 128.4, 128.2, 127.9, 117.4, 117.1, 113.0, 112.6, 14.3. HRMS (ESI) m/z: 403.0924 [M + H]+, calcd for C22H15N2O6 403.0925.
4.4.36. 4-Acrylamidophenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (43)
Yellow solid, yield 44%, mp = 206.5–208.8 °C. 1H NMR (DMSO) δ 10.28 (s, 1H), 9.03 (dd, J = 4.5, 1.3 Hz, 1H), 8.47 (dd, J = 7.8, 1.3 Hz, 1H), 7.87 (dd, J = 7.8, 4.7 Hz, 1H), 7.79 (d, J = 8.8 Hz, 2H), 7.33 (d, J = 8.9 Hz, 2H), 6.46 (dd, J = 17.0, 10.1 Hz, 1H), 6.29 (dd, J = 17.0, 1.7 Hz, 1H), 5.78 (dd, J = 10.1, 1.7 Hz, 1H), 2.76 (s, 3H). 13C NMR (DMSO) δ 176.5, 172.3, 165.1, 163.1, 160.2, 153.8, 150.7, 148.8, 145.6, 137.0, 134.1, 131.7, 128.5, 127.9, 127.7, 127.0, 121.9, 120.3, 112.2, 13.8. HRMS (ESI) m/z: 403.0919 [M + H]+, calcd for C22H15N2O6 403.0925.
4.4.37. 4-(3-Morpholinopropyl)phenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (44)
Yellow solid, yield 35%, mp > 330 °C. 1H NMR (CDCl3) δ 9.06 (dd, J = 4.7, 1.7 Hz, 1H), 8.56 (dd, J = 7.9, 1.7 Hz, 1H), 7.72 (dd, J = 7.9, 4.7 Hz, 1H), 7.38-7.31 (m, 2H), 6.99-6.91 (m, 2H), 4.05 (t, J = 6.3 Hz, 2H), 3.74 (t, J = 4.6 Hz, 4H), 2.82 (s, 3H), 2.54 (t, J = 6.3 Hz, 2H), 2.50 (t, J = 4.6 Hz, 4H), 1.95 (t, J = 6.3 Hz, 2H). 13C NMR (CDCl3) δ 176.6, 172.3, 166.1, 160.8, 157.0, 154.5, 150.7, 149.1, 143.9, 134.7, 128.6, 128.4, 127.4, 122.4, 115.05, 113.5, 67.0, 66.5, 55.5, 53.7, 26.4, 14.1. HRMS (ESI) m/z: 477.1650 [M + H]+, calcd for C26H24N2O7 477.1656.
4.4.38. 4-(4-Acetylpiperazin-1-yl)phenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (45)
Brown solid, yield 61%, mp = 209.1–210.0 °C. 1H NMR (CDCl3) δ 9.06 (d, J = 4.6 Hz, 1H), 8.56 (d, J = 7.8 Hz, 1H), 7.72 (dd, J = 7.8, 4.7 Hz, 1H), 7.35 (d, J = 8.8 Hz, 2H), 6.98 (d, J = 8.8 Hz, 2H), 3.81-3.74 (m, 2H), 3.67-3.62 (m, 2H), 3.21-3.18 (m, 2H), 3.17-3.14 (m, 2H), 2.82 (s, 3H), 2.16 (s, 3H). 13C NMR (CDCl3) δ 176.6, 172.4, 169.0, 166.2, 160.8, 154.5, 150.7, 149.2, 149.1, 144.0, 134.8, 128.6, 128.4, 127.4, 122.2, 117.5, 113.5, 50.1, 49.8, 46.2, 41.3, 21.4, 14.2. HRMS (ESI) m/z: 460.1503 [M + H]+, calcd for C25H22N3O6 460.1503.
4.4.39. 4-Benzoylphenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (46)
Yellow solid, yield 71%, mp = 187.8–190.2 °C. 1H NMR (CDCl3) δ 9.07 (d, J = 1.8 Hz, 1H), 8.56 (d, J = 7.7 Hz, 1H), 7.92 (d, J = 7.8 Hz, 2H), 7.82 (d, J = 7.4 Hz, 2H), 7.76-7.70 (m, 1H), 7.60 (m, 3H), 7.51 (t, J = 7.3 Hz, 2H), 2.85 (s, 3H). 13C NMR (CDCl3) δ 195.6, 176.6, 172.3, 166.8, 160.0, 154.6, 153.5, 150.8, 149.0, 137.4, 135.4, 134.8, 132.6, 131.7, 130.0, 128.39, 128.36, 127.5, 121.6, 113.0, 14.2. HRMS (ESI) m/z: 438.0970 [M + H]+, calcd for C26H16NO6 438.0972.
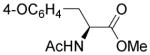
4.4.40. (S)-4-(2-Acetamido-3-methoxy-3-oxopropyl)phenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (47)
Yellow solid, yield 81%, mp = 201.4–202.2 °C. 1H NMR (CDCl3) δ 9.06 (dd, J = 4.7, 1.7 Hz, 1H), 8.56 (dd, J = 7.9, 1.7 Hz, 1H), 7.72 (dd, J = 7.9, 4.7 Hz, 1H), 7.38 (d, J = 8.5 Hz, 2H), 7.17 (d, J = 8.5 Hz, 2H), 5.98 (d, J = 7.5 Hz, 1H), 4.91 (dt, J = 7.7, 5.7 Hz, 1H), 3.75 (s, 3H), 3.17 (dd, J = 5.6, 3.4 Hz, 2H), 2.82 (s, 3H), 2.02 (s, 3H). 13C NMR (CDCl3) δ 176.6, 172.3, 171.9, 169.7, 166.3, 160.3, 154.5, 150.7, 149.6, 149.1, 134.7, 133.9, 130.3, 128.6, 128.4, 127.4, 121.7, 113.3, 53.1, 52.4, 37.3, 23.1, 14.1. HRMS (ESI) m/z: 477.1298 [M + H]+, calcd for C25H21N2 O8 477.1292.
4.4.41. 2-(Methoxycarbonyl)thiophen-3-yl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (48)
Yellow solid, yield 78%, mp = 218.0–221.7 °C. 1H NMR (CDCl3) δ 9.06 (s, 1H), 8.56 (d, J = 6.9 Hz, 1H), 7.71 (s, 1H), 7.55 (d, J = 4.1 Hz, 1H), 7.20 (d, J = 4.7 Hz, 1H), 3.81 (s, 3H), 2.87 (s, 3H). 13C NMR (DMSO) δ 176.1, 172.2, 165.4, 160.2, 158.3, 153.7, 150.9, 149.5, 148.6, 134.0, 132.0, 128.3, 127.7, 127.5, 123.8, 118.3, 111.4, 52.0, 14.0. HRMS (ESI) m/z: 398.0326 [M + H]+, calcd for C19H12NO7S 398.0329.
4.4.42. Benzo[d][1,3]dioxol-5-yl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (49)
Yellow solid, yield 58%, mp = 198.2–199.8 °C. 1H NMR (CDCl3) δ 9.06 (dd, J = 4.6, 1.8 Hz, 1H), 8.54 (dd, J = 7.8, 1.8 Hz, 1H), 7.72 (dd, J = 7.8, 4.6 Hz, 1H), 6.97 (d, J = 2.4 Hz, 1H), 6.89 (dd, J = 8.4, 2.4 Hz, 1H), 6.81 (d, J = 8.4 Hz, 1H), 6.01 (s, 2H), 2.81 (s, 3H). 13C NMR (CDCl3) δ 176.6, 172.3, 166.2, 160.7, 154.5, 150.7, 149.1, 148.0, 145.6, 144.7, 134.7, 128.6, 128.4, 127.4, 114.1, 113.3, 107.9, 103.8, 101.8, 14.1. HRMS (ESI) m/z: 378.0626 [M + H]+, calcd for C20H12NO7 378.0608.
4.4.43. 3-Bromopropyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (50)
Light yellow solid, yield 58%, mp = 177.7–179.1 °C. 1H NMR (CDCl3) δ 9.06 (dd, J = 4.7, 1.7 Hz, 1H), 8.54 (dd, J = 7.9, 1.7 Hz, 1H), 7.70 (dd, J = 7.9, 4.7 Hz, 1H), 4.54 (t, J = 5.8 Hz, 2H), 3.75 (t, J = 5.4 Hz, 2H), 2.78 (s, 3H), 2.45-2.36 (m, 2H). 13C NMR (CDCl3) δ 176.5, 172.3, 166.3, 162.0, 154.4, 150.6, 149.0, 134.6, 128.3, 128.2, 127.4, 113.3, 63.2, 31.4, 30.3, 14.1. HRMS (ESI) m/z: 377.9996 [M + H]+, calcd for C16H13NO5 377.9972.
4.4.44. 2-((Tert-butoxycarbonyl)amino)phenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (51)
Yellow solid, yield 55%, mp = 115.9–117.6 °C. 1H NMR (CDCl3) δ 9.07 (dd, J = 4.7, 1.7 Hz, 1H), 8.68 (s, 1H), 8.56 (dd, J = 7.9, 1.7 Hz, 1H), 8.25 (d, J = 7.2 Hz, 1H), 7.74 (dd, J = 7.9, 4.7 Hz, 1H), 7.26-7.17 (m, 2H), 7.08-7.00 (m, 1H), 2.85 (s, 3H), 1.52 (s, 9H). 13C NMR (CDCl3) δ 177.5, 172.3, 167.8, 160.2, 154.5, 153.1, 150.9, 149.1, 138.8, 134.7, 131.4, 128.4, 128.0, 127.7, 126.9, 122.1, 122.0, 120.4, 112.4, 80.4, 28.3, 14.2. HRMS (ESI) m/z: 471.1177 [M+Na]+, calcd for C24H20N2O7Na 471.1163.
4.4.45. 3-((Tert-butoxycarbonyl)amino)phenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (52)
Yellow solid, yield 75%, mp = 181.6–183.3 °C. 1H NMR (CDCl3) δ 9.06 (dd, J = 4.6, 1.6 Hz, 1H), 8.55 (dd, J = 7.9, 1.6 Hz, 1H), 7.71 (dd, J = 7.9, 4.7 Hz, 1H), 7.54 (s, 1H), 7.34 (t, J = 8.0 Hz, 1H), 7.27 (d, J = 7.6 Hz, 1H), 7.11 (d, J = 8.0 Hz, 1H), 6.65 (s, 1H), 2.81 (s, 3H), 1.52 (s, 9H). 13C NMR (CDCl3) δ 176.5, 172.3, 166.2, 160.2, 154.5, 152.5, 150.8, 150.7, 149.1, 139.6, 134.7, 129.7, 128.6, 128.4, 127.4, 116.1, 116.0, 113.4, 111.9, 80.8, 28.3, 14.1. HRMS (ESI) m/z: 449.1340 [M + H]+, calcd for C24H21N2O7 449.1343.
4.4.46. 4-((Tert-butoxycarbonyl)amino)phenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (53)
Yellow solid, yield 90%, mp = 220.4–221.2 °C. 1H NMR (CDCl3) δ 8.97 (d, J = 3.4 Hz, 1H), 8.47 (d, J = 7.7 Hz, 1H), 7.63 (dd, J = 7.6, 4.8 Hz, 1H), 7.36 (d, J = 8.5 Hz, 2H), 7.28 (d, J = 8.8 Hz, 2H), 6.58 (s, 1H), 2.73 (s, 3H), 1.45 (s, 9H). 13C NMR (CDCl3) δ 176.6, 172.3, 166.2, 160.5, 154.4, 152.7, 150.7, 149.1, 145.7, 136.4, 134.7, 128.6, 128.4, 127.4, 122.0, 119.3, 113.4, 80.6, 28.3, 14.1. HRMS (ESI) m/z: 449.1350 [M + H]+, calcd for C24H21O7N2 449.1343.
4.4.47. But-1-ynyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (54)
Light yellow solid, yield 77%, mp = 193.4–195.2 °C. 1H NMR (CDCl3) δ 9.06 (d, J = 4.5 Hz, 1H), 8.54 (d, J = 7.7 Hz, 1H), 7.70 (dd, J = 7.8, 4.6 Hz, 1H), 4.50 (t, J = 7.0 Hz, 2H), 2.88-2.64 (m, 5H), 2.02 (t, J = 2.6 Hz, 1H). 13C NMR (CDCl3) δ 176.4, 172.3, 165.7, 161.4, 154.4, 150.64, 149.1, 134.6, 128.6, 128.3, 127.3, 113.5, 79.9, 70.1, 63.2, 18.8, 14.2. HRMS (ESI) m/z: 332.0544 [M + Na]+, calcd for C17H11NO5Na 332.0529.
4.4.48. 3-Ethynylphenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (55)
Light yellow solid, yield 69%, mp = 221.3–222.8 °C. 1H NMR (CDCl3) δ 9.07 (dd, J = 4.6, 1.5 Hz, 1H), 8.56 (dd, J = 7.9, 1.7 Hz, 1H), 7.72 (dd, J = 7.9, 4.7 Hz, 1H), 7.60 (s, 1H), 7.50-7.34 (m, 3H), 3.12 (s, 1H), 2.83 (s, 3H). 13C NMR (CDCl3) δ 176.5, 172.3, 166.4, 160.1, 154.5, 150.7, 150.1, 149.1, 134.7, 130.0, 129.4, 128.5, 128.4, 127.4, 125.2, 123.6, 122.4, 113.2, 82.6, 78.2, 14.1. HRMS (ESI) m/z: 380.0543 [M + Na]+, calcd for C21H11NO5Na 380.0529.
4.4.49. N-Ethyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[3,2-g]quinoline-3-carboxamide (67)
Yellow solid, yield 89%, mp = 243.5–244.6 °C. 1H NMR (CDCl3) δ 9.41 (s, 1H), 9.09 (d, J = 4.0 Hz, 1H), 8.56 (d, J = 7.8 Hz, 1H), 7.75 (dd, J = 7.8, 4.7 Hz, 1H), 3.56-3.46 (m, 2H), 2.92 (s, 3H), 1.34 (t, J = 7.2 Hz, 3H). 13C NMR (CDCl3) δ 181.8, 171.2, 167.4, 160.7, 155.0, 151.3, 147.6, 135.5, 130.09, 127.6, 125.9, 115.5, 34.4, 15.1, 14.5. HRMS (ESI) m/z: 285.0882 [M + H]+, calcd for C15H13N2O4 285.0870.
4.4.50. N-3-Chlorophenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[3,2-g]quinoline-3-carboxamide (68)
Yellow solid, yield 83%, mp = 262.0–265.2 °C. 1H NMR (DMSO) δ 11.13 (s, 1H), 9.07 (d, J = 3.9 Hz, 1H), 8.55 (d, J = 7.8 Hz, 1H), 7.93 (s, 1H), 7.90 (dd, J = 7.8, 4.8 Hz, 1H), 7.57 (d, J = 8.0 Hz, 1H), 7.44 (t, J = 8.0 Hz, 1H), 7.22 (d, J = 8.0 Hz, 1H), 2.76 (s, 3H). 13C NMR (DMSO) δ 181.5, 171.4, 164.7, 159.7, 154.8, 151.6, 148.0, 140.2, 135.5, 133.8, 131.3, 130.6, 128.4, 126.4, 124.3, 119.4, 118.4, 116.0, 14.4. HRMS (ESI) m/z: 367.0494 [M + H]+, calcd for C19H12N2O4Cl 367.0480.
4.4.51. 2-Hydroxyphenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[3,2-g]quinoline-3-carboxylate (69)
Yellow solid, yield 48%, mp = 266.7–268.3 °C. 1H NMR (CDCl3) δ 9.10 (dd, J = 4.7, 1.7 Hz, 1H), 8.61 (dd, J = 7.9, 1.7 Hz, 1H), 8.52 (s, 1H), 7.76 (dd, J = 7.9, 4.7 Hz, 1H), 7.38 (dd, J = 8.1, 1.4 Hz, 1H), 7.23-7.15 (m, 1H), 7.12 (dd, J = 8.2, 1.6 Hz, 1H), 6.96-6.85 (m, 1H), 2.87 (s, 3H). 13C NMR (DMSO) δ 178.1, 171.7, 165.5, 159.7, 154.3, 152.6, 149.4, 148.0, 138.5, 135.3, 131.0, 128.4, 127.6, 127.3, 123.4, 119.7, 117.4, 112.5, 14.5. HRMS (ESI) m/z: 350.0657 [M + H]+, calcd for C19H12NO6 350.0659.
4.5. Synthesis of compounds 56 and 57
The reaction solution of compound 50 (40 mg, 0.1 mmol) and pyridines (0.5 mmol) in THF (20 mL) was stirred and heated under reflux for 8 h and cooled to room temperature. The reaction solution was added with ether (40 mL), and yellow precipitate appeared. The resulting precipitate was filtrated and purified by silica gel column chromatography to give target compound.
4.5.1. 1-(3-((2-Methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carbonyl)oxy)propyl)pyridin-1-ium bromide (56)
Yellow solid, yield 57%, mp > 332 °C. 1H NMR (CD3OD) δ 9.20 (dd, J = 5.8, 6.1 Hz, 2H), 9.02 (dd, J = 4.7, 1.6 Hz, 1H), 8.74-8.53 (m, 2H), 8.29-8.08 (m, 2H), 7.89 (dd, J = 7.9, 4.8 Hz, 1H), 5.08 (t, J = 7.4 Hz, 2H), 4.46 (t, J = 5.6 Hz, 2H), 2.74 (s, 3H), 2.64-2.48 (m, 2H). 13C NMR (CD3OD) δ 178.9, 173.3, 167.8, 163.4, 154.9, 152.6, 150.1, 147.1, 146.5, 146.5, 136.2, 130.1, 129.7, 129.7, 129.4, 128.9, 113.7, 62.4, 60.3, 31.5, 14.0. HRMS (ESI) m/z: 377.1149 [M − Br]+, calcd for C21H17N2O5Br 377.1132.
4.5.2. 4-(Dimethylamino)-1-(3-((2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carbonyl)oxy)propyl)pyridin-1-ium bromide (57)
Yellow solid, yield 69%, mp = 136.9–138.7 °C. 1H NMR (CD3OD) δ 9.01 (dd, J = 4.7, 1.5 Hz, 1H), 8.61 (dd, J = 7.9, 1.5 Hz, 1H), 8.35 (d, J = 7.7 Hz, 2H), 7.89 (dd, J = 7.9, 4.8 Hz, 1H), 7.01 (d, J = 7.7 Hz, 2H), 4.57 (t, J = 7.0 Hz, 2H), 4.40 (t, J = 5.6 Hz, 2H), 3.21 (s, 6H), 2.74 (s, 3H), 2.45-2.31 (m, 2H). 13C NMR (CD3OD) δ 178.6, 173.3, 167.7, 163.4, 158.0, 154.9, 152.5, 150.1, 143.5, 136.2, 130.0, 129.4, 128.9, 113.8, 109.1, 62.7, 56.0, 40.3, 30.9, 14.0. HRMS(ESI) m/z: 420.1555 [M − Br]+, calcd for C23H22N3O5Br 420.1554.
4.6. Deprotection of Boc protective group of compounds 51–53
To a solution of Boc-protected compound (90 mg, 0.2 mmol) in dichloromethane (6 mL), trifluoroacetic acid (1.3 mL) was added. The reaction solution was stirred for 1 h at room temperature. The solvent was evaporated under reduced pressure. The resulting solid was washed with ether to give the target product 59 and 60. The compound 58 was obtained through the purification by silica gel column chromatography.
4.6.1. 3-(Benzo[d]oxazol-2-yl)-2-methylfuro[2,3-g]quinoline-4,9-dione (58)
Yellow solid, yield 84%, mp = 266.0 °C (decomposing). 1H NMR (CDCl3) δ 9.10 (dd, J = 4.7, 1.7 Hz, 1H), 8.60 (dd, J = 7.9, 1.7 Hz, 1H), 7.87-7.83 (m, 1H), 7.74 (dd, J = 7.9, 4.7 Hz, 1H), 7.69 (dd, J = 6.2, 2.5 Hz, 1H), 7.48-7.40 (m, 2H), 2.96 (s, 3H). 13C NMR (CDCl3) δ 176.8, 172.2, 163.8, 155.7, 154.4, 151.0, 150.7, 149.1, 141.5, 134.7, 128.6, 127.4, 125.8, 124.8, 120.2, 111.1, 109.9, 14.3. HRMS (ESI) m/z: 331.0710 [M + H]+, calcd for C19H11N2O4 331.0713.
4.6.2. 3-Aminophenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxamide (59)
Brown solid, yield 84%, mp > 300.0 °C. 1H NMR (CD3OD) δ 8.95 (d, J = 3.7 Hz, 1H), 8.57 (d, J = 7.7 Hz, 1H), 7.85 (dd, J = 7.8, 4.8 Hz, 1H), 7.54 (t, J = 8.1 Hz, 1H), 7.36-7.30 (m, 2H), 7.22 (d, J = 7.8 Hz, 1H), 2.76 (s, 3H). 13C NMR (CD3OD) δ 176.7, 171.8, 166.5, 160.1, 153.4, 151.4, 151.2, 148.6, 135.6, 134.7, 130.6, 128.6, 127.9, 127.7, 119.6, 118.8, 114.8, 112.1, 12.7. HRMS (ESI) m/z: 349.0813 [M + H]+, calcd for C19H13N2O5 349.0819.
4.6.3. 4-Aminophenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxamide (60)
Red solid, yield 62%, mp = 170.5 °C (decomposing). 1H NMR (DMSO) δ 9.03 (d, J = 3.7 Hz, 1H), 8.49 (d, J = 7.3 Hz, 1H), 7.88 (dd, J = 7.7, 4.7 Hz, 1H), 7.23 (d, J = 8.6 Hz, 2H), 7.02 (d, J = 8.5 Hz, 2H), 2.76 (s, 3H). 13C NMR (CD3OD) δ 176.8, 171.8, 166.6, 160.2, 153.4, 151.2, 150.0, 148.6, 134.8, 130.0, 128.6, 127.9, 127.7, 123.4, 123.2, 112.1, 12.7. HRMS (ESI) m/z: 349.0807 [M + H]+, calcd for C19H13N2O5 349.0819.
4.7. General procedures for the synthesis of compounds 61–64
According to the reported “click chemistry” method with slight modification [33], A solution of 3-bromopropanol or 3-bromopropanic acid (1 mmol) and sodium azide (195 mg, 2 mmol) in acetonitrile (20 mL) was stirred and heated under reflux for 8 h, and cooled to room temperature. The solvent was evaporated under reduced pressure to give white solid. The resulting white solid was dissolved in water (5 mL) and was added with DMF (5 mL) and acetylene analog (54 or 55, 0.5 mmol). The mixture solution was added with sodium ascorbate (20 mg, 0.1 mmol) and CuSO4/5H2O (12 mg, 0.05 mmol). The reaction solution was stirred and heated at 75 °C for 3 h. The resulting suspension was diluted with water (50 mL). The aqueous solution was extracted with dichloromethane (20 mL × 2). The combined organic layer was washed with water (10 mL × 2) and saturated aqueous saline (10 mL) and dried with anhydride MgSO4. The solvent was evaporated under reduced pressure. The residue was purified by silica gel column chromatography to give the target compound.
4.7.1. 2-(1-(3-Hydroxypropyl)-1H-1,2,3-triazol-4-yl)ethyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (61)
Brown solid, yield 24%, mp = 172.7–173.5 °C. 1H NMR (CDCl3) δ 9.02 (d, J = 4.5 Hz, 1H), 8.57 (dd, J = 7.9, 1.6 Hz, 1H), 7.98 (s, 1H), 7.75 (dd, J = 7.9, 4.7 Hz, 1H), 4.70 (t, J = 5.6 Hz, 2H), 4.58 (t, J = 5.6 Hz, 2H), 3.68 (t, J = 5.8 Hz, 2H), 3.27 (t, J = 5.6 Hz, 2H), 2.76 (s, 3H), 2.15 (quint, J = 5.9 Hz, 2H). 13C NMR (CDCl3) δ 176.5, 172.0, 166.5, 162.3, 154.0, 150.6, 148.7, 144.3, 135.2, 128.4, 128.1, 127.7, 123.4, 113.5, 65.1, 57.8, 46.4, 32.3, 25.6, 14.2. HRMS (ESI) m/z: 433.1142 [M + Na]+, calcd for C20H18N4O6Na 433.1119.
4.7.2. 3-(4-(2-((2-Methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carbonyl)oxy)ethyl)-1H-1,2,3-triazol-1-yl)propanoic acid (62)
Brown solid, yield 19%, mp = 216.6–217.4 °C. 1H NMR (CDCl3) δ 8.99 (dd, J = 4.8, 1.4 Hz, 1H), 8.65 (dd, J = 7.9, 1.5 Hz, 1H), 8.19 (s, 1H), 7.83 (dd, J = 7.9, 4.8 Hz, 1H), 4.78 (t, J = 5.6, 2H), 4.70 (t, J = 5.0, 2H), 3.24 (t, J = 5.0, 2H), 2.92 (t, J = 5.6, 2H), 2.79 (s, 3H). 13C NMR (CD3OD) δ 178.1, 173.3, 166.9, 163.2, 154.8, 152.4, 150.1, 145.3, 136.0, 130.0, 129.3, 129.2, 124.9, 114.2, 65.4, 47.3, 35.9, 26.1, 14.1. HRMS (ESI) m/z: 423.0923 [M − H]−, calcd for C20H15N4O7 423.0946.
4.7.3. 3-(1-(3-Hydroxypropyl)-1H-1,2,3-triazol-4-yl)phenyl 2-methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carboxylate (63)
Light yellow solid, yield 55%, mp = 181.6–182.4 °C. 1H NMR (CDCl3) δ 9.08 (dd, J = 4.7, 1.7 Hz, 1H), 8.58 (dd, J = 7.9, 1.7 Hz, 1H), 7.93 (s, 1H), 7.85 (d, J = 1.8 Hz, 1H), 7.82 (s, 1H), 7.74 (dd, J = 7.9, 4.7 Hz, 1H), 7.52 (t, J = 7.9 Hz, 1H), 7.44-7.37 (m, 1H), 4.61 (t, J = 6.7 Hz, 2H), 3.72 (t, J = 5.8 Hz, 2H), 2.85 (s, 3H), 2.27-2.16 (m, 2H). 13C NMR (CDCl3) δ 175.6, 171.3, 165.3, 159.3, 153.5, 149.7, 149.7, 148.0, 145.8, 133.8, 131.2, 128.9, 127.5, 127.4, 126.5, 122.4, 120.3, 119.7, 117.8, 112.3, 57.7, 45.9, 31.5, 13.2. HRMS (ESI) m/z: 457.1138 [M − H]−, calcd for C24H17N4O6 457.1154.
4.7.4. 3-(4-(3-((2-Methyl-4,9-dioxo-4,9-dihydrofuro[2,3-g]quinoline-3-carbonyl)oxy)phenyl)-1H-1,2,3-triazol-1-yl)propanoic acid (64)
Yellow solid, yield 80%, mp = 218.6–220.6 °C. 1H NMR (DMSO) δ 12.54 (s, 1H), 9.04 (dd, J = 4.6, 1.6 Hz, 1H), 8.68 (s, 1H), 8.50 (dd, J = 7.9, 1.6 Hz, 1H), 7.89 (dd, J = 7.9, 4.7 Hz, 1H), 7.85 (d, J = 1.9 Hz, 1H), 7.82 (d, J = 7.8 Hz, 1H), 7.60 (t, J = 7.9 Hz, 1H), 7.35 (dd, J = 8.1, 1.4 Hz, 1H), 4.63 (t, J = 6.7 Hz, 2H), 2.97 (t, J = 6.7 Hz, 2H), 2.81 (s, 3H). 13C NMR (DMSO) δ 176.6, 172.3, 171.7, 165.3, 160.0, 153.8, 150.8, 150.6, 148.8, 145.2, 134.1, 132.4, 130.3, 128.5, 127.9, 127.7, 122.9, 122.2, 121.0, 118.2, 112.1, 45.6, 33.9, 13.9. HRMS (ESI) m/z: 471.0920 [M − H]−, calcd for C24H15N4O7 471.0946.
4.8. Synthesis of sodium salt 65
To a solution of acid 64 (0.2 mmol) in ethanol (10 mL), a solution of NaOH in ethanol (20%, 0.2 mmol) was added dropwise at room temperature. The reaction solution was stirred at room temperature for 30 min. The resulting precipitate was filtered, washed with ethanol and dried to give the yellow solid 66 (85%). HRMS (ESI) m/z: 495.0938 [M + H]+, calcd for C24H16N4O7Na 495.0911.
4.9. Synthesis of ethyl 2-methyl-4,9-dioxo-4,9-dihydro-1H-pyrrolo[3,2-g]quinoline-3-carboxylate (70)
The reaction solution of 7-bromoquinoline-5,8-dione (0.48 g, 2 mmol), Mn(OAc)3 (0.80 g, 3 mmol) and ethyl 3-aminocrotonate (0.31 g, 2.4 mmol) in acetonitrile (40 mL) was stirred and heated under reflux for 3 h and cooled to room temperature. The solvent was evaporated under reduced pressure. The residue was purified by silica gel column chromatography to give a yellow solid (70, 57 mg), yield 10%, mp = 215.3–217.6 °C. 1H NMR (CDCl3) δ 11.41 (s, 1H), 8.91 (d, J = 3.7 Hz, 1H), 8.51 (d, J = 7.6 Hz, 1H), 7.63 (dd, J = 7.7, 4.7 Hz, 1H), 4.43 (q, J = 7.2 Hz, 2H), 2.64 (s, 3H), 1.44 (t, J = 7.2 Hz, 3H). 13C NMR (CDCl3) δ 177.8, 173.9, 163.8, 153.2, 148.2, 143.7, 135.5, 132.3, 131.6, 127.4, 125.5, 113.8, 61.0, 14.3, 13.6. HRMS (ESI) m/z: 285.0870 [M + H]+, calcd for C15H13N2O4 285.0870. The structure of compound 70 was confirmed with 2D NMR spectra.
4.10. Synthesis of ethyl 1,2-dimethyl-4,9-dioxo-4,9-dihydro-1H-pyrrolo[3,2-g]quinoline-3-carboxylate (72)
The solution of 6,7-dichloroquinoline-5,8-dinoe (230 mg, 1 mmol), sodium acetate (160 mg, 2 mmol) and ethyl acetoacetate (0.13 mL, 1.1 mmol) in THF (10 mL) was stirred and heated under reflux for 3 h and cooled to room temperature. The solvent was evaporated under reduced pressure. The resulting residue was dissolved in dichloromethane (100 mL). The organic solution was washed with water (20 mL × 2), saturated aqueous saline (20 mL) and concentrated under reduced pressure to give yellow oil. The crude yellow oil was dissolved in ethanol (10 mL), and added with methylamine aqueous solution (40%, 0.5 mL, 4 mmol). The reaction solution was stirred and heated under reflux for 5 h and cooled to room temperature. The solvent was evaporated under reduced pressure. The residue was purified by silica gel column chromatography to give a yellow solid (72, 27 mg), yield 9%, mp = 209.3–210.8 °C. 1H NMR (CDCl3) δ 8.97 (dd, J = 4.6, 1.6 Hz, 1H), 8.48 (dd, J = 7.8, 1.6 Hz, 1H), 7.62 (dd, J = 7.8, 4.7 Hz, 1H), 4.44 (q, J = 7.1 Hz, 2H), 4.10 (s, 3H), 2.52 (s, 3H), 1.44 (t, J = 7.1 Hz, 3H). 13C NMR (CDCl3) δ 197.6, 177.9, 174.3, 164.1, 153.5, 148.7, 143.1, 134.8, 130.9, 130.5, 127.0, 113.9, 61.2, 33.2, 14.1, 11.0. HRMS (ESI) m/z: 229.1017 [M + H]+, calcd for C16H15N2O4 229.1026.
4.11. The synthesis of isoxazole analogues 73 and 74
Procedure A
Following “General Procedures for the synthesis of furoquinolinediones”, the reaction of 6,7-dichloroquinoline-5,8-dione with ethyl nitroacetate gave the target products 73 (5%) and 74 (2%), respectively.
Procedure B
The isoxazole analogues were synthesized according Chuang method [38]. Briefly, a solution of quinoline-5,8-dione (1 mmol), ethyl nitroacetate (540 mg, 4 mmol) and manganese (III) acetate (1.61 g, 6 mmol) in acetic acid (15 mL) was stirred and heated at 70 °C overnight, followed by the addition of ethyl nitroacetate (540 mg, 4 mmol) and manganese (III) acetate (1.61 g, 6 mmol). The reaction solution was stirred and heated at 70 °C overnight again. The reaction mixture was diluted with dichloromethane (100 mL) and washed with saturated aqueous sodium bisulfite (50 mL), water (50 mL × 3) and aqueous saturated sodium bicarbonate (50 mL × 3). The solvent was evaporated under reduced pressure. The residue was purified by silica gel column chromatography to give the target compound 73 (11%) and 74 (8%), respectively.
4.11.1. Ethyl 4,9-dioxo-4,9-dihydroisoxazolo[5,4-g]quinoline-3-carboxylate (73)
Yellow solid, mp = 122.7–124.3 °C. 1H NMR (CDCl3) δ 9.14 (s, 1H), 8.62 (d, J = 7.9 Hz, 1H), 7.86-7.81 (m, 1H), 4.59 (q, J = 6.9 Hz, 2H), 1.50 (t, J = 6.9 Hz, 3H). 13C NMR (CDCl3) δ 175.1, 170.6, 165.8, 157.6, 155.2, 153.4, 147.5, 135.9, 130.7, 128.6, 120.0, 63.6, 14.0. HRMS (ESI) m/z: 273.0494 [M + H]+, calcd for C13H9N2O5 273.0506. The structure of compound 73 was confirmed with 2D NMR spectra.
4.11.2. Ethyl 4,9-dioxo-4,9-dihydroisoxazolo[4,5-g]quinoline-3-carboxylate (74)
Yellow solid, yield 8%, mp = 119.5–120.6 °C. 1H NMR (CDCl3) δ 9.17 (dd, J = 4.7, 1.7 Hz, 1H), 8.64 (dd, J = 7.9, 1.7 Hz, 1H), 7.82 (dd, J = 7.9, 4.7 Hz, 1H), 4.59 (q, J = 7.1 Hz, 2H), 1.51 (t, J = 7.1 Hz, 3H). 13C NMR (CDCl3) δ 174.3, 171.6, 165.1, 157.6, 155.8, 153.8, 148.9, 135.6, 128.8, 128.0, 120.4, 63.7, 14.0. HRMS (ESI) m/z: 273.0496 [M + H]+, calcd for C13H9N2O5 273.0506. The structure of compound 74 was confirmed with 2D NMR spectra.
4.12. Synthesis of compounds 75 and 76
A solution of quinoline-5,8-dinoe (318 mg, 2 mmol) and ethyl diazoacetate (0.24 mL, 2.2 mmol) in toluene (60 mL) was stirred and heated under reflux overnight. The precipitate was filtered and purified by silica gel column chromatography to give two isomers 75 and 76.
4.12.1. Ethyl 4,9-dioxo-4,9-dihydro-1H-pyrazolo[3,4-g]quinoline-3-carboxylate (75)
Yellow solid, yield 10%, mp = 208.3–211.2 °C. 1H NMR (CDCl3) δ 9.06 (d, J = 3.7 Hz, 1H), 8.66 (d, J = 7.9 Hz, 1H), 7.77 (dd, J = 7.6, 4.4 Hz, 1H), 4.55 (q, J = 7.1 Hz, 2H), 1.50 (t, J = 7.1 Hz, 3H). 13C NMR (CDCl3) δ 176.3, 174.8, 169.3, 159.7, 154.0, 153.8, 148.2, 136.1, 132.0, 128.3, 120.0, 62.3, 13.9. HRMS (ESI) m/z: 272.0657 [M + H]+, calcd for C13H10N3O4 272.0666. The structure of compound 75 was confirmed with 2D NMR spectra.
4.12.2. Ethyl 4,9-dioxo-4,9-dihydro-1H-pyrazolo[4,3-g]quinoline-3-carboxylate (76)
Yellow solid, yield 11%. mp = 208.6–210.3 °C. 1H NMR (CDCl3) δ 9.07 (d, J = 4.3 Hz, 1H), 8.58 (d, J = 7.9 Hz, 1H), 7.67 (dd, J = 7.8, 4.2 Hz, 1H), 4.51 (q, J = 7.1 Hz, 2H), 1.49 (t, J = 7.1 Hz, 3H). 13C NMR (CDCl3) δ 176.1, 175.4, 167.9, 159.5, 154.6, 153.9, 149.9, 135.3, 129.6, 127.4, 120.2, 62.3, 13.7. HRMS (ESI) m/z: 272.0658 [M + H]+, calcd for C13H10N3O4 272.0666. The structure of compound 76 was confirmed with 2D NMR spectra.
4.13. Recombinant TDP1 assay [30]
A 5′-[32P]-labeled single-stranded DNA oligonucleotide containing a 3′-phosphotyrosine (N14Y) was incubated at 1 nM with 10 pM recombinant TDP1 in the absence or presence of inhibitor for 15 min at room temperature in LMP1 assay buffer containing 50 mM Tris HCl, pH 7.5, 80 mM KCl, 2 mM EDTA, 1 mM DTT, 40 μg/mL BSA, and 0.01% Tween-20. Reactions were terminated by the addition of 1 volume of gel loading buffer [99.5% (v/v) formamide, 5 mM EDTA, 0.01% (w/v) xylene cyanol, and 0.01% (w/v) bromophenol blue]. Samples were subjected to a 16% denaturing PAGE with multiple loadings at 12-min intervals. Gels were dried and exposed to a PhosphorImager screen (GE Healthcare). Gel images were scanned using a Typhoon 8600 (GE Healthcare), and densitometry analyses were performed using the ImageQuant software (GE Healthcare).
4.14. Recombinant TDP2 assay
TDP2 reactions were carried out as described previously with the following modifications [7]. The 18-mer singlestranded oligonucleotide DNA substrate (TY18, 32P-cordycepin-3′-labeled) was incubated at 1 nM with 25 pM recombinant human TDP2 in the absence or presence of inhibitor for 15 min at room temperature in the LMP2 assay buffer containing 50 mM Tris-HCl, pH 7.5, 80 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 40 μg/mL BSA, and 0.01% Tween 20. Reactions were terminated and treated similarly to recombinant TDP1 reactions (see above).
4.15. Whole cell extract TDP2 assay
DT40 knockout cells (1 × 107) for TDP2 (TDP2−/−) complemented with human TDP2 were collected, washed, and centrifuged [28]. Cell pellets were then resuspended in 100 μL of CellLytic M cell lysis reagent (SIGMA-Aldrich C2978). After 15 min on ice, lysates were centrifuged at 12000 g for 10 min, and supernatants were transferred to a new tube. Protein concentrations were determined using a Nanodrop spectrophotometer (Invitrogen), and whole cell extracts were stored at −80 °C. The TY19 DNA substrate was incubated at 1 nM with 5 μg/mL of whole cell extracts in the absence or presence of inhibitor for 15 min at room temperature in the LMP2 assay buffer. Reactions were terminated and treated similarly to recombinant TDP1 reactions (see above).
4.16. Detection of TOP2-DNA covalent cleavage complexes in vivo
In vivo complex of enzyme (ICE) assay was conducted according to the reported method [39]. Briefly, 60 × 105 CCRF-CEM cells were treated for 2 h prior lysis with 1% Sarkosyl. DNA was prepared, and TOP2-DNA complexes were detected by Western slot blot using Ki-S1 mouse anti-human TOP2α antibody from EMD Millipore.
4.17. Molecular modeling
The active furoquinolinedione analogues were docked into the hTDP2. The hTDP2 model was built using SWISS-MODEL [40], using mouse TDP2 struture as a template. The sequence of the mouse TDP2 crystal structure (PDB: 4GZ1) was trimmed by removing one of the monomers, bound DNA, magnesium ions, and water oxygen atoms [4]. The prepared structure and hTDP2 sequence (UniProtKB - O95551) were submitted to SWISS-MODEL for homology modeling. The hTDP2 structure was further geometry-optimized MacroModel [41], using AMBER* force field with 300 iterations of Steepest Descent (SD) minimization. The strutures of ligands were prepared in Maestro [42], and geometry-optimized in MacroModel using MMFFs force field with 2500 iteration of Polak-Ribier Conjugate Gradient (PRCG) minimization. The docking of the active TDP2 inhibitors was performed with Glide [43], using Extra Precision setting [44]. The grid for ligand docking was set to encompass the entire DNA binding area.
4.18. Statistical Analysis
All data are expressed as the mean ± standard deviation. Statistical comparisons were conducted using a one-way analysis of variance (ANOVA) using the Prism statistical software package (GraphPad Software, USA).
Supplementary Material
Research highlight.
A novel TDP2 hit (1) was found through screening from in-house chemical library.
Seventy seven furoquinolinedione analogues were synthesized.
Compound 74 showed the most TDP2 inhibition with IC50 at low micromolar range.
The SAR was analyzed.
Furoquinolinedinone chemotype represents a novel skeleton for novel TDP2 inhibitors.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81373257), Natural Science Foundation of Guangdong Province (No. S2013010015609), and supported by the Intramural Program of the National Cancer Institute (Center for Cancer Research), National Institutes of Health, Bethesda, Maryland, USA (Z01 BC 006150-19). We wish to thank Dr. Jun-Min Quan, Shenzhen Graduate School of Peking University, for his helpful suggestions on compounds’ design.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- 2.Pommier Y, Huang SY, Gao R, Das BB, Murai J, Marchand C. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2) DNA repair. 2014;19:114–129. doi: 10.1016/j.dnarep.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi K, Kurahashi K, Gao R, Tsutakawa SE, Tainer JA, Pommier Y, Aihara H. Structural basis for recognition of 5’-phosphotyrosine adducts by Tdp2. Nat Struct Mol Biol. 2012;19:1372–1377. doi: 10.1038/nsmb.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schellenberg MJ, Appel CD, Adhikari S, Robertson PD, Ramsden DA, Williams RS. Mechanism of repair of 5’-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2. Nat Struct Mol Biol. 2012;19:1363–1371. doi: 10.1038/nsmb.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hornyak P, Askwith T, Walker S, Komulainen E, Paradowski M, Pennicott LE, Bartlett EJ, Brissett NC, Raoof A, Watson M, Jordan AM, Ogilvie DJ, Ward SE, Atack JR, Pearl LH, Caldecott KW, Oliver AW. Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2. Biochem J. 2016;473:1869–1879. doi: 10.1042/BCJ20160180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schellenberg MJ, Perera L, Strom CN, Waters CA, Monian B, Appel CD, Vilas CK, Williams JG, Ramsden DA, Williams RS. Reversal of DNA damage induced topoisomerase 2 DNA-protein crosslinks by Tdp2. Nucleic Acids Res. 2016;44:3829–3844. doi: 10.1093/nar/gkw228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao R, Huang SY, Marchand C, Pommier Y. Biochemical characterization of human tyrosyl-DNA phosphodiesterase 2 (TDP2/TTRAP): a Mg(2+)/Mn(2+)-dependent phosphodiesterase specific for the repair of topoisomerase cleavage complexes. J Biol Chem. 2012;287:30842–30852. doi: 10.1074/jbc.M112.393983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashour ME, Atteya R, El-Khamisy SF. Topoisomerase-mediated chromosomal break repair: an emerging player in many games. Nat Rev Cancer. 2015;15:137–151. doi: 10.1038/nrc3892. [DOI] [PubMed] [Google Scholar]
- 10.Pommier Y, Sun Y, Huang SN, Nitiss JL. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol. 2016;17:703–721. doi: 10.1038/nrm.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan QA, Kohlhagen G, Marshall R, Austin CA, Kalena GP, Kroth H, Sayer JM, Jerina DM, Pommier Y. Position-specific trapping of topoisomerase II by benzo[a]pyrene diol epoxide adducts: implications for interactions with intercalating anticancer agents. Proc Natl Acad Sci U S A. 2003;100:12498–12503. doi: 10.1073/pnas.2032456100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cline SD, Osheroff N. Cytosine arabinoside lesions are position-specific topoisomerase II poisons and stimulate DNA cleavage mediated by the human type II enzymes. J Biol Chem. 1999;274:29740–29743. doi: 10.1074/jbc.274.42.29740. [DOI] [PubMed] [Google Scholar]
- 13.Rao T, Gao R, Takada S, Al Abo M, Chen X, Walters KJ, Pommier Y, Aihara H. Novel TDP2-ubiquitin interactions and their importance for the repair of topoisomerase II-mediated DNA damage. Nucleic Acids Res. 2016;44:10201–10215. doi: 10.1093/nar/gkw719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao R, Schellenberg MJ, Huang SY, Abdelmalak M, Marchand C, Nitiss KC, Nitiss JL, Williams RS, Pommier Y. Proteolytic degradation of topoisomerase II (Top2) enables the processing of Top2.DNA and Top2.RNA covalent complexes by tyrosyl-DNA-phosphodiesterase 2 (TDP2) J Biol Chem. 2014;289:17960–17969. doi: 10.1074/jbc.M114.565374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu CC, Li TK, Farh L, Lin LY, Lin TS, Yu YJ, Yen TJ, Chiang CW, Chan NL. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science. 2011;333:459–462. doi: 10.1126/science.1204117. [DOI] [PubMed] [Google Scholar]
- 16.Do PM, Varanasi L, Fan S, Li C, Kubacka I, Newman V, Chauhan K, Daniels SR, Boccetta M, Garrett MR, Li R, Martinez LA. Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev. 2012;26:830–845. doi: 10.1101/gad.181685.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez-Herreros F, Romero-Granados R, Zeng Z, Alvarez-Quilon A, Quintero C, Ju L, Umans L, Vermeire L, Huylebroeck D, Caldecott KW, Cortes-Ledesma F. TDP2-dependent non-homologous end-joining protects against topoisomerase II-induced DNA breaks and genome instability in cells and in vivo. PLOS Genet. 2013;9:e1003226. doi: 10.1371/journal.pgen.1003226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng Z, Cortes-Ledesma F, El Khamisy SF, Caldecott KW. TDP2/TTRAP is the major 5’-tyrosyl DNA phosphodiesterase activity in vertebrate cells and is critical for cellular resistance to topoisomerase II-induced DNA damage. J Biol Chem. 2011;286:403–409. doi: 10.1074/jbc.M110.181016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kont YS, Dutta A, Mallisetty A, Mathew J, Minas T, Kraus C, Dhopeshwarkar P, Kallakury B, Mitra S, Uren A, Adhikari S. Depletion of tyrosyl DNA phosphodiesterase 2 activity enhances etoposide-mediated double-strand break formation and cell killing. DNA Repair. 2016;43:38–47. doi: 10.1016/j.dnarep.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 20.Maciejewski S, Nguyen JH, Gomez-Herreros F, Cortes-Ledesma F, Caldecott KW, Semler BL. Divergent requirement for a DNA repair enzyme during enterovirus infections. mBio. 2016;7:e01931–01915. doi: 10.1128/mBio.01931-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Virgen-Slane R, Rozovics JM, Fitzgerald KD, Ngo T, Chou W, Noort GJvdHv, Filippov DV, Gershon PD, Semler BL. An RNA virus hijacks an incognito function of a DNA repair enzyme. Proc Natl Acad Sci U S A. 2012;109:14634–14639. doi: 10.1073/pnas.1208096109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caldecott KW. Tyrosyl DNA phosphodiesterase 2, an enzyme fit for purpose. Nat Struct Mol Biol. 2012;19:1212–1213. doi: 10.1038/nsmb.2455. [DOI] [PubMed] [Google Scholar]
- 23.Nitiss JL, Nitiss KC. Tdp2: a means to fixing the ends. PLOS Genet. 2013;9:e1003370. doi: 10.1371/journal.pgen.1003370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raoof A, Depledge P, Hamilton NM, Hamilton NS, Hitchin JR, Hopkins GV, Jordan AM, Maguire LA, McGonagle AE, Mould DP, Rushbrooke M, Small HF, Smith KM, Thomson GJ, Turlais F, Waddell ID, Waszkowycz B, Watson AJ, Ogilvie DJ. Toxoflavins and deazaflavins as the first reported selective small molecule inhibitors of tyrosyl-DNA phosphodiesterase II. J Med Chem. 2013;56:6352–6370. doi: 10.1021/jm400568p. [DOI] [PubMed] [Google Scholar]
- 25.Kankanala J, Marchand C, Abdelmalak M, Aihara H, Pommier Y, Wang Z. Isoquinoline-1,3-diones as selective inhibitors of tyrosyl DNA phosphodiesterase II (TDP2) J Med Chem. 2016;59:2734–2746. doi: 10.1021/acs.jmedchem.5b01973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kossmann BR, Abdelmalak M, Lopez S, Tender G, Yan C, Pommier Y, Marchand C, Ivanov I. Discovery of selective inhibitors of tyrosyl-DNA phosphodiesterase 2 by targeting the enzyme DNA-binding cleft. Bioorg Med Chem Lett. 2016;26:3232–3236. doi: 10.1016/j.bmcl.2016.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang P, Elsayed MSA, Plescia CB, Ravji A, Redon CE, Kiselev E, Marchand C, Zeleznik O, Agama K, Pommier Y, Cushman M. Synthesis and biological evaluation of the first triple inhibitors of human topoisomerase 1, tyrosyl-DNA phosphodiesterase 1 (Tdp1), and tyrosyl-DNA phosphodiesterase 2 (Tdp2) J Med Chem. 2017;60:3275–3288. doi: 10.1021/acs.jmedchem.6b01565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchand C, Abdelmalak M, Kankanala J, Huang SY, Kiselev E, Fesen K, Kurahashi K, Sasanuma H, Takeda S, Aihara H, Wang Z, Pommier Y. Deazaflavin inhibitors of tyrosyl-DNA phosphodiesterase 2 (TDP2) specific for the human enzyme and active against cellular TDP2. ACS Chem Biol. 2016;11:1925–1933. doi: 10.1021/acschembio.5b01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Interthal H, Pouliot JJ, Champoux JJ. The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. Proc Natl Acad Sci U S A. 2001;98:12009–12014. doi: 10.1073/pnas.211429198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dexheimer TS, Gediya LK, Stephen AG, Weidlich I, Antony S, Marchand C, Interthal H, Nicklaus M, Fisher RJ, Njar VC, Pommier Y. 4-Pregnen-21-ol-3,20-dione-21-(4-bromobenzenesulfonate) (NSC 88915) and related novel steroid derivatives as tyrosyl-DNA phosphodiesterase (Tdp1) inhibitors. J Med Chem. 2009;52:7122–7131. doi: 10.1021/jm901061s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 32.Hu HY, Zhu Y, Wang L, Xu JH. A direct one-pot synthesis of naphtho[2,3-b]furan-4,9-dione derivatives via C,O-dialkylation of b-dicarbonyl compounds by 2,3-dichloro-1,4-naphthoquinones. Synthesis. 2005;10:1605–1610. [Google Scholar]
- 33.Wu P, Feldman AK, Nugent AK, Hawker CJ, Scheel A, Voit B, Pyun J, Frechet JM, Sharpless KB, Fokin VV. Efficiency and fidelity in a click-chemistry route to triazole dendrimers by the copper(i)-catalyzed ligation of azides and alkynes. Angew Chem Int Ed. 2004;43:3928–3932. doi: 10.1002/anie.200454078. [DOI] [PubMed] [Google Scholar]
- 34.Cherkaoui O, Nebois P, Fillion H. Regiospecific hetero Diels-Alder synthesis of furo[2,3-g] and furo[3,2-glquinoline-4,9-diones. Tetrahedron. 1996;52:9499–9508. [Google Scholar]
- 35.Nguyen TX, Morrell A, Conda-Sheridan M, Marchand C, Agama K, Bermingham A, Stephen AG, Chergui A, Naumova A, Fisher R, O’Keefe BR, Pommier Y, Cushman M. Synthesis and biological evaluation of the first dual tyrosyl-DNA phosphodiesterase I (Tdp1)-topoisomerase I (Top1) inhibitors. J Med Chem. 2012;55:4457–4478. doi: 10.1021/jm300335n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi HY, Chi DY. Simple preparation of 7-alkylamino-2-methylquinoline-5,8-diones: regiochemistry in nucleophilic substitution reactions of the 6- or 7-bromo-2-methylquinoline-5,8-dione with amines. Tetrahedron. 2004;60:4945–4951. [Google Scholar]
- 37.Cheng Y, An LK, Wu N, Wang XD, Bu XZ, Huang ZS, Gu LQ. Synthesis, cytotoxic activities and structure-activity relationships of topoisomerase I inhibitors: indolizinoquinoline-5,12-dione derivatives. Bioorg Med Chem. 2008;16:4617–4625. doi: 10.1016/j.bmc.2008.02.036. [DOI] [PubMed] [Google Scholar]
- 38.Chuang CP, Wu YL, Jiang MC. Manganese(III) acetate initiated oxidative free radical reaction between 1,4-naphthoquinones and ethyl nitroacetate. Tetrahedron. 1999;55:11229–11236. [Google Scholar]
- 39.Nitiss JL, Soans E, Rogojina A, Seth A, Mishina M. Topoisomerase assays. Current protocols in pharmacology. 2012;Chapter 3(Unit 3):3. doi: 10.1002/0471141755.ph0303s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 41.Schrödinger Release 2017-4: MacroModel. Schrödinger, LLC; New York, NY: 2017. [Google Scholar]
- 42.Schrödinger Release 2017-4: Maestro. Schrödinger, LLC; New York, NY: 2017. [Google Scholar]
- 43.Schrödinger Release 2017-4: Glide. Schrödinger, LLC; New York, NY: 2017. [Google Scholar]
- 44.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.