Abstract
Intestinal infection by Cryptosporidium parvum causes significant alterations in the gene expression profile in host epithelial cells. Previous studies demonstrate that a panel of parasite RNA transcripts of low protein-coding potential are delivered into infected host cells and may modulate host gene transcription. Using in vitro models of human intestinal cryptosporidiosis, we report here that trans-suppression of the cadherin 3 (CDH3) and lysyl oxidase like 4 (LOXL4) genes in human intestinal epithelial cells following C. parvum infection involves host delivery of the Cdg7_FLc_1000 RNA, a C. parvum RNA that has been previously demonstrated to be delivered into the nuclei of infected host cells. Downregulation of CDH3 and LOXL4 genes was detected in host epithelial cells following C. parvum infection or in cells expressing the parasite Cdg7_FLc_1000 RNA. Knockdown of Cdg7_FLc_1000 attenuated the trans-suppression of CDH3 and LOXL4 genes in host cells induced by infection. Interestingly, Cdg7_FLc_1000 was detected to be recruited to the promoter regions of both CDH3 and LOXL4 gene loci in host cells following C. parvum infection. Host delivery of Cdg7_FLc_1000 promoted the PH domain zinc finger protein 1 (PRDM1)-mediated H3K9 methylation associated with trans-suppression in the CDH3 gene locus, but not the LOXL4 gene. Therefore, our data suggest that host delivery of Cdg7_FLc_1000 causes CDH3 trans-suppression in human intestinal epithelial cells following C. parvum infection through PRDM1-mediated H3K9 methylation in the CDH3 gene locus, whereas Cdg7_FLc_1000 induces trans-suppression of the host LOXL4 gene through H3K9/H3K27 methylation-independent mechanisms.
Keywords: Cryptosporidium, Intestinal epithelium, CDH3, LOXL4, Gene transcription, Epithelial homeostasis
Graphical Abstract
1. Introduction
Cryptosporidium, a genus of protozoa in the phylum Apicomplexa, represents a group of protozoan parasites that can infect humans and many other species of animals such as mammals, birds and reptiles (O’Donoghue, 1995; Striepen, 2013). The Cryptosporidium parvum and Cryptosporidium hominis spp. cause the majority of Cryptosporidium infections in humans. Whereas C. hominis mostly infects humans, C. parvum has a rather broad host range (Checkley et al., 2015). Clinically, Cryptosporidium remains an important opportunistic pathogen in AIDS patients and is one of the most common pathogens responsible for moderate-to-severe diarrhea in children under 2 years of age in developing regions (Kotloff et al, 2013; Checkley et al., 2015).
The primary infection site of the parasite in humans is the small intestine and one of the pathological hallmarks of intestinal cryptosporidiosis is the inhibition of intestinal epithelial turnover and disturbances in epithelial homeostasis (Savidge et al., 1996; Sasahara et al., 2003). The intestinal mucosa is a monolayer of rapidly self-renewing epithelial cells. New functional epithelial cells are produced from stem cells in the crypt base, differentiated, and migrated from the crypt base to the luminal surface and hence, the entire intestinal epithelium is replaced every 2–3 days in mice (3–5 days in humans) (Creamber et al., 1961; Barker, 2014). Thus, inhibition of epithelial turnover would provide an obvious benefit to parasite replication, particularly for the parasite cell cycle during its intracellular stages (O’Donoghue, 1995). Mechanistic understanding of how C. parvum infection inhibits epithelial turnover and disturbs intestinal epithelial homeostasis would provide new insights into pathogenesis, relevant to the development of novel therapeutic strategies.
Cryptosporidium infection causes significant alterations in the gene expression profile in host epithelial cells (Deng et al., 2004; Yang et al., 2009). These genes that are upregulated in infected intestinal epithelial cells include interleukin 8 (IL8), nitric oxide synthase 2 (NOS2), C-X-C motif chemokine ligand 2, and intercellular adhesion molecule 1, reflecting host defense responses to infection in general (Laurent et al., 1997; Tarver et al., 1998; Alcantara et al., 2003; Deng et al., 2004; Goel et al., 2012). In contrast, many of the downregulated genes code effector proteins important for cell proliferation, differentiation and metabolism, including low density lipoprotein receptor related protein 5 (LRP5), frizzled class receptor 7, polycomb group ring finger 2, and solute carrier family 7 member 8 (SLC7A8) (Deng et al., 2004; Kuhnert et al., 2004; Won et al., 2012). Interestingly, downregulation of this panel of genes appears to be specific to Cryptosporidium infection and may not be a general epithelial cell response to pathogen infection or inflammatory stimulation (Deng et al., 2004; Wang et al., 2017). Nevertheless, how C. parvum infection causes downregulation of specific genes in infected intestinal epithelial cells is still unclear.
The interactions between protozoan parasites and host cells involve exchanges of distinct effector molecules from both sides of the host cell and the parasite at the host-parasite interface (Sibley, 2004). Several C. parvum proteins have been demonstrated to be delivered into host epithelial cells at the host-parasite interface and are involved in parasite intracellular development (Sibley, 2004; O’Connor et al., 2007). In our previous studies (Wang et al., 2016), we demonstrated that several C. parvum RNA transcripts of low protein-coding potential are selectively delivered into intestinal epithelial cells during host-parasite interactions and may modulate gene transcription in infected host cells. Specifically, delivery of parasite Cdg7_FLc_0990 RNA (GenBank accession number: FX115678.1) (Puiu et al., 2004; Yamagishi et al., 2011) into infected intestinal epithelial cells suppresses transcription of the LRP5, SLC7A8 and IL33 genes through histone modification-mediated epigenetic mechanisms (Wang et al., 2016, 2017). Delivery of the parasite Cdg7_FLc_1000 transcript (GenBank accession number: FX115830.1) (Puiu et al., 2004; Yamagishi et al., 2011) causes trans-suppression of the host sphingomyelin phosphodiesterase 3 (SMPD3) gene, resulting in attenuation of intestinal epithelial cell migration (Ming et al., 2017). Downregulation of the cadherin 3 (CDH3) and lysyl oxidase like 4 (LOXL4) genes in intestinal epithelial cells following C. parvum infection has been reported in previous studies (Ming et al., 2017); their protein products have been demonstrated as important regulators of cell migration and adherence (Baek et al., 2010; Paredes et al., 2012; Samuelov et al., 2015; Comptour et al., 2016). Here, we report that host delivery of Cdg7_FLc_1000 promotes the PH domain zinc finger protein 1 (PRDM1)-mediated H3K9 methylation in the CDH3 gene locus, resulting in trans-suppression in infected intestinal epithelial cells; whereas Cdg7_FLc_1000 induces trans-suppression of the LOXL4 gene through H3K9/H3K27 methylation-independent mechanisms.
2. Materials and methods
2.1. Cryptosporidium parvum, cell lines and infection models
Cryptosporidium parvum oocysts of the Iowa strain were purchased from a commercial source (Bunch Grass Farm, Deary, ID, USA). INT cells (FHs 74 Int, CCL-241™) and HCT-8 (CCL-244™) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Models of intestinal cryptosporidiosis using cultured cell lines were employed as previously described; infection was done in serum-free culture medium for 4 h with a 1:1 ratio of C. parvum oocysts and host cells (Ming et al., 2017; Wang et al., 2016, 2017).
2.2. Real-time quantitative PCR (qPCR)
For quantitative analysis of mRNA and C. parvum RNA expression, comparative real-time qPCR was performed as previous reported (Wang et al., 2016, 2017; Ming et al., 2017), using the SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA, USA). Briefly, RNA was extracted using TRI-reagent, treated with a DNA-free™ Kit (Ambion, USA) to remove any remaining DNA. Quantified RNA (500 ng) was reverse-transcribed using T100 thermal cyclers (Bio-Rad, USA). Real-time qPCR was then performed using 25 ng of template cDNA for each RNA gene of interest. Each sample was run in triplicate. The relative abundance of each RNA was calculated using the ΔΔCt method and normalised to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (total mRNA) or the U2 small nuclear RNA (RNU2-1) (a nuclear RNA). The sequences for all the primers are listed in Supplementary Table S1.
2.3. Small interfering RNAs (siRNAs) and plasmids
Custom designed siRNA oligos against Cdg7_FLc_1000 and a scrambled siRNA were synthesized by Integrated DNA Technologies (Coralville, Iowa, USA) and transfected into cells with Lipofectamine RNAimax (Invitrogen, USA). The Cdg7_FLc_1000 expression plasmid was generated by reverse transcription-PCR amplification of Cdg7_FLc_1000 cDNA, using RNA from C. parvum sporozoites (Iowa strain) and cloned into the pcDNA3.1(+) vector in accordance with the manufacturer’s protocol (Invitrogen). Full-Cdg7_FLc_1000 was transfected into cells with lipofectamine 2000 (Invitrogen), a pcDNA3.1(+) empty vector was transfected as a control. The primer sequences for Cdg7_FLc_1000 plasmid generation are listed as following: forward (NheI), 5′-CGGCTAGCAGTTTTTACATTTTGTATCTCAGTT-3′ and reverse (KpnI), 5′-GGGGTACCTGAGCGAAATTAGAGTAGTCTGA-3′. Stable HCT-8-G9a−/− cells were generated through transfection of cells with the G9a-CRISPR/Cas9 KO(h) and G9a-HDR plasmids (Santa Cruz, USA). HCT-8 cells stably expressing the empty vector were selected for controls.
2.4. Whole cell extracts, nuclear extracts and western blots
Whole cell extracts were prepared using the M-PER Mammalian Protein Extraction Reagent (Fisher, USA) supplemented with cocktail protease inhibitors, according to the manufacturer’s instructions. The cell pellet was incubated in the M-PER Mammalian Protein Extraction Reagent on ice for 30 min, centrifuged at 16,100 g for 20 min and the supernatants were saved as the whole cell extracts. Nuclear extracts were obtained using the standard approach, as previously reported (Wang et al., 2016, 2017; Ming et al., 2017). The protein concentration of each fraction or whole cell lysate was determined and subsequently analyzed by western blots. The following antibodies were used for blotting: anti-CDH3 (ThermoFisher, USA), anti-LOXL4 (Santa Cruz), anti-trimethylation of lysine 9 on histone H3 (H3K9me3) (Abcam, USA), anti-trimethylation of lysine 27 on histone H3 (H3K27me3) (Abcam), anti-euchromatic histone lysine methyltransferase 2 (G9a) (Millipore), anti-PRDM1 (Santa Cruz), anti-GAPDH (Santa Cruz) and anti-β-Actin (Sigma).
2.5. Chromatin immunoprecipitation (ChIP) and chromatin isolation by RNA purification (ChIRP)
For ChIP analysis, a commercially available ChIP Assay Kit (Upstate Biotechnologies, USA) was used in accordance with the manufacturer’s instructions. In brief, cells were fixed with 1% formaldehyde for 10 min and the genomic DNA was then sheared to lengths ranging from 200 to 1000 bp by sonication, as previously described (Ma et al., 2016; Wang et al., 2016, 2017; Ming et al., 2017). While one percent of the cell extracts was taken as input, the rest of the extracts were incubated with specific antibodies overnight at 4°C, followed by precipitation with protein G agarose beads. The DNA-protein complex was eluted; after reversal of cross-links with NaCl at 65°C overnight, proteins were digested with proteinase K, and the DNA was detected by real-time qPCR analysis. The following antibodies were used for ChIP analysis: anti-H3K9me3 (Abcam), and anti-H3K27me3 (Abcam), anti-G9a (Millipore), anti-PRDM1 (Santa Cruz).
ChIRP analysis was performed as previously reported (Chu et al., 2011; Wang et al., 2016, 2017; Ming et al., 2017). Briefly, a pool of tiling oligonucleotide probes with affinity specific to the C. parvum Cdg7_FLc_1000 RNA sequences was used and glutaraldehyde cross-linked for chromatin isolation. The DNA sequences of the precipitates from chromatin isolation by RNA purification were confirmed by real-time qPCR using the same primer sets covering the gene promoter regions of interest as for ChIP analysis.
2.6. Statistical analysis
All values are given as mean ± S.E.M. Means of groups were from at least three independent experiments and compared with a Student’s t test (two-tailed unpaired) or the ANOVA test when appropriate. P < 0.05 was considered statistically significant.
3. Results
3.1. Expression of parasite Cdg7_FLc_1000 RNA in cultured human intestinal epithelial cells results in suppression of both CDH3 and LOXL4 genes
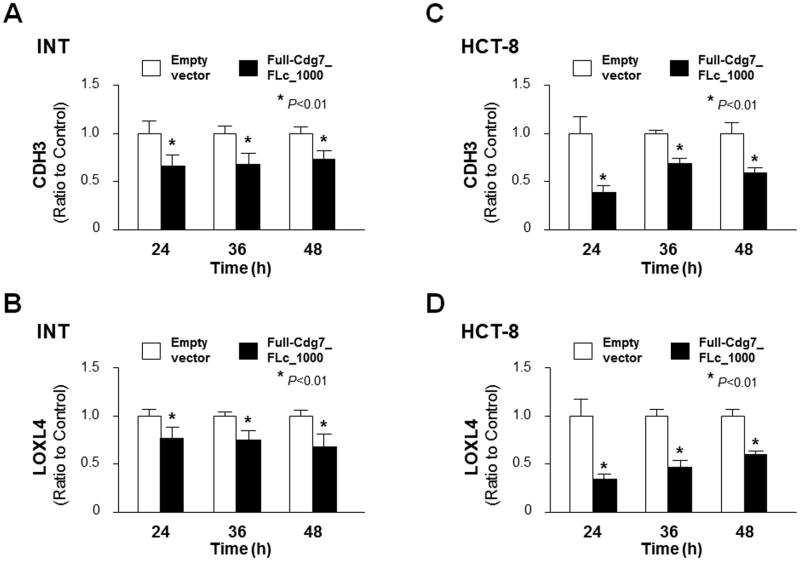
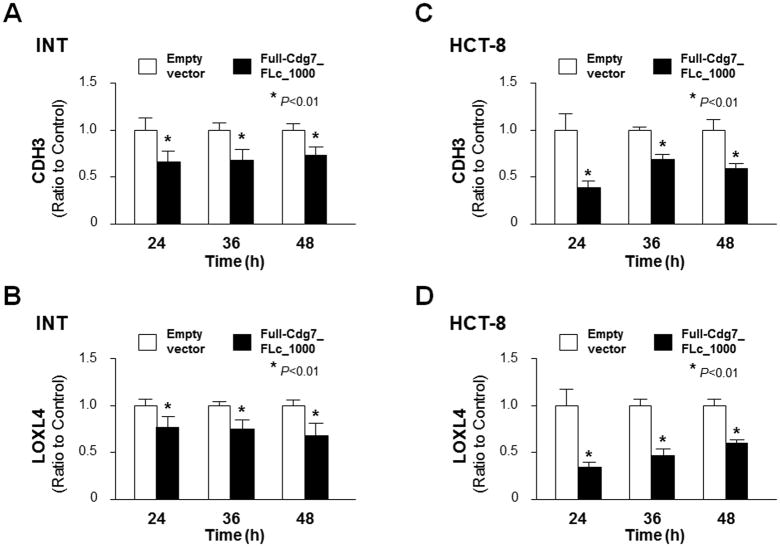
Cryptosporidium parvum infection causes significant dysregulation of intestinal epithelium homeostasis but underlying mechanisms are unclear (Deng et al., 2004; Kuhnert et al., 2004; Won et al., 2012; Wang et al., 2017). In our previous studies (Ming et al., 2017; Wang et al., 2017), we identified that the parasite RNA Cdg7_FLc_1000 is delivered to the nuclei of infected host cells and modulates host gene transcription. Genome-wide array analysis revealed that a panel of host genes are downregulated in INT cells transfected with Full-Cdg7_FLc_1000, including CDH3 and LOXL4; their protein products are important regulators of cell migration and adherence (Baek et al., 2010; Paredes et al., 2012; Samuelov et al., 2015; Comptour et al., 2016). We further confirmed by real-time qPCR the suppression of CDH3 and LOXL4 genes in INT cells transfected with Full-Cdg7_FLc_1000 for various periods of time, compared with cells transfected with the control empty vector (Fig. 1A–B). Suppression of CDH3 and LOXL4 genes was also detected in HCT-8 cells, a different human intestinal epithelial cell line, following transfection with Full-Cdg7_FLc_1000 (Fig. 1C and D).
Fig. 1.
Downregulation of CDH3 and LOXL4 genes in INT and HCT-8 cells expressing Cdg7_FLc_1000. Downregulation of CDH3 and LOXL4 genes associated with Cdg7_FLc_1000 transfection was validated by real-time quantitative PCR in INT and HCT-8 cells. Cells were transfected with the Full-Cdg7_FLc_1000 for 24, 36 and 48 h, followed by real-time quantitative PCR analysis of CDH3 and LOXL4 RNA levels (A–D, respectively). Cells transfected with the empty vector were used as the control. Data represent three independent experiments. *P<0.01, ANOVA compared with empty vector controls.
3.2. Suppression of CDH3 and LOXL4 genes in host cells induced by C. parvum infection is mediated by delivery of Cdg7_FLc_1000
We questioned whether C. parvum infection inhibits expression of CDH3 and LOXL4 genes in infected host cells with the involvement of host delivery of Cdg7_FLc_1000. When INT and HCT-8 cells were exposed to C. parvum infection for 24, 36 and 48 h, the expression levels of both CDH3 and LOXL4 genes were significantly downregulated as measured using real-time qPCR (Fig. 2A–D). Consistent with results from previous studies (Deng et al., 2004; Yang et al., 2009; Ming et al., 2017; Wang et al., 2017), upregulation of several epithelial cell defense genes such as IL8 and NOS2 was detected in INT and HCT-8 cells following C. parvum infection (Supplementary Fig. S1). Downregulation of both CDH3 and LOXL4 genes at the protein level was further confirmed using western blots in HCT-8 cells following C. parvum infection (Fig. 2E and F).
Fig. 2.
Downregulation of the CDH3 and LOXL4 genes in INT and HCT-8 cells following Cryptosporidium parvum infection. (A–D) INT and HCT-8 cells were exposed to C. parvum infection for various periods of time, followed by real-time quantitative PCR analysis of CDH3 and LOXL4 RNA levels. Downregulation of both CDH3 and LOXL4 genes was confirmed in INT and HCT-8 cells following C. parvum infection (A–D, respectively). (E–F) Protein contents of CDH3 and LOXL4 in HCT-8 cells following C. parvum infection for 24, 36 and 48 h, as assessed by western blots. Representative gel images for CDH3 and LOXL4 protein blotting are shown and signal intensities on the blots were quantified by densitometry (E and F, respectively). Data represent three independent experiments. *P<0.01, ANOVA compared with non-infected cells.
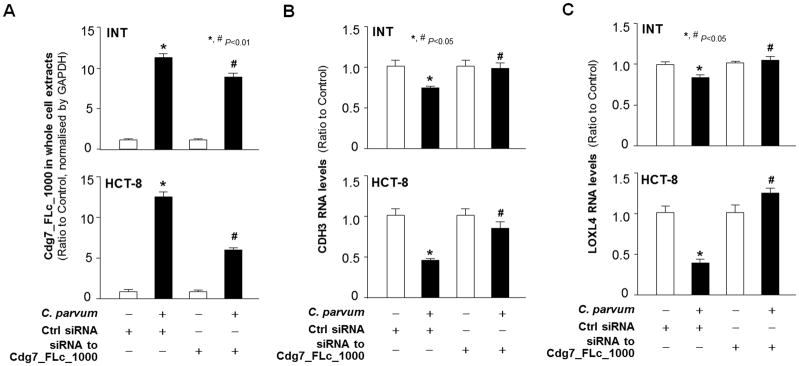
To define whether host delivery of Cdg7_FLc_1000 is involved, we measured the effects of Cdg7_FLc_1000 knockdown on CDH3 and LOXL4 expression in cells following infection. Because conventional genetic tools are very difficult, if not impossible, to use to modify C. parvum genes (Striepen, 2013; Vinayak et al., 2015), we designed an siRNA to Cdg7_FLc_1000 and transfected host cells for 12 h, followed by exposure to C. parvum infection. A non-specific scrambled siRNA was used as a control. The increase in Cdg7_FLc_1000 RNA levels in INT or HCT-8 cells induced by C. parvum infection was partially suppressed by pre-treatment of the siRNA against Cdg7_FLc_1000 (Fig. 3A and Supplementary Fig. S2). Accordingly, suppression of CDH3 and LOXL4 RNA expression induced by C. parvum infection was significantly attenuated through pre-treatment of the siRNA against Cdg7_FLc_1000 in INT and HCT-8 cells (Fig. 3B and C, respectively). These data suggest that suppression of CDH3 and LOXL4 genes in host cells Induced by C. parvum infection may be mediated through host delivery of Cdg7_FLc_1000.
Fig. 3.
Downregulation of the CDH3 and LOXL4 genes in epithelial cells following Cryptosporidium parvum infection is correlated with delivery of Cdg7_FLc_1000 into the infected host cells. (A) Inhibition of delivery of Cdg7_FLc_1000 into infected cells through pre-treatment of host cells with a small interfering RNA (siRNA)to Cdg7_FLc_1000 followed by exposure of cells to C. parvum infection. INT and HCT-8 cells were treated with an siRNA to Cdg7_FLc_1000 for 12 h and then exposed to C. parvum infection for an additional 24 h. Contents of Cdg7_FLc_1000 in the whole infected cells were quantified by real-time quantitative . A non-specific scrambled siRNA was used as the control (Ctrl). (B–C) Inhibition of Cdg7_FLc_1000 in host cells by the siRNA treatment attenuated the downregulation of CDH3 and LOXL4 following C. parvum infection. INT and HCT-8 cells were treated with an siRNA to Cdg7_FLc_1000 for 12 h and then exposed to C. parvum infection for an additional 24 h. Expression levels of CDH3 and LOXL4 in the infected cells were quantified by real-time quantitative PCR. Data represent three independent experiments. *P<0.01, ANOVA compared with non-infected cells treated with the control siRNA; # P<.01, ANOVA compared with infected cells treated with the control siRNA.
3.3. PRDM1/G9a-mediated H3K9 methylation within the regulatory promoter region is correlated with trans-suppression of the CDH3 gene, but not the LOXL4 gene
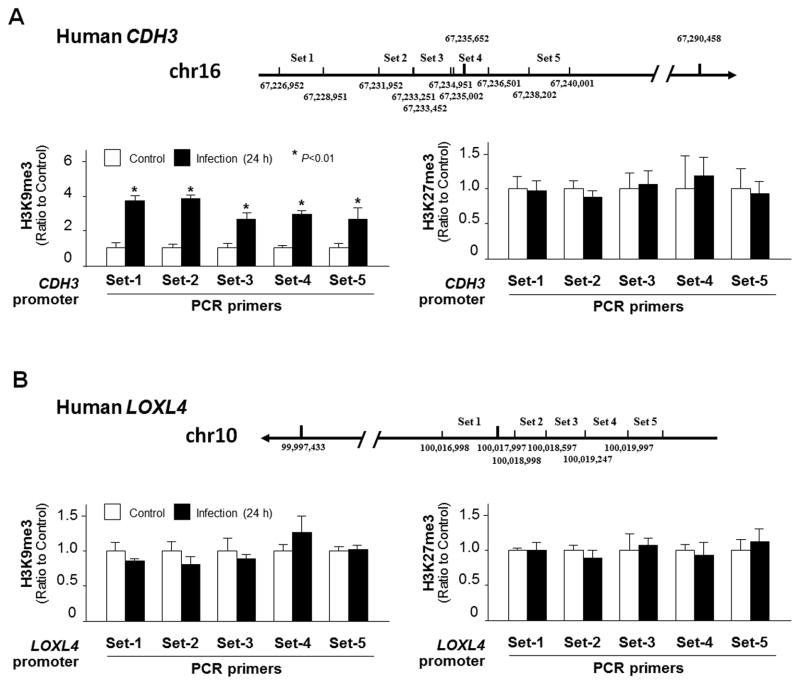
In our previous study (Ming et al., 2017), we demonstrated that nuclear delivery of Cdg7_FLc_1000 is involved in the trans-suppression of the SMPD3 gene in C. parvum-infected intestinal epithelial cells through gene-specific enrichment of H3K9 methylations. We then preformed the ChIP analysis of infected cells using anti-H3K9me3 or anti-H3K27me3 and PCR primer pairs (Set1 – Set5; see more details in Supplementary Table S1) specific to the various promoter regions of the CDH3 and LOXL4 gene loci (Fig. 4A and B, respectively). Increased enrichment of H3K9me3, but not H3K27me3, was detected in the CDH3 gene locus in infected HCT-8 cells (Fig. 4A). Similarly, increased enrichment of H3K9me3, but not H3K27me3, was detected in the CDH3 gene locus in HCT-8 cells after transfection with Full-Cdg7_FLc_1000 (Supplementary Fig. S3). In contrast, no significant enrichment of either H3K9me3 or H3K27me3 was detected in the LOXL4 gene locus in infected HCT-8 cells (Fig. 4B), suggesting that different mechanisms are involved in the trans-suppression of host CDH3 and LOXL4 genes correlated with the delivery of Cdg7_FLc_1000 during C. parvum infection.
Fig. 4.
Enrichment of H3K9me3 within the CDH3 gene locus, but not for the LOXL4 gene locus, in HCT-8 cells following Cryptosporidium parvum infection. (A) Levels of the suppression markers, H3K9me3 and H3K27me3, associated with the CDH3 gene locus in HCT-8 cells following C. parvum infection. Cells were exposed to C. parvum infection for 24 h, followed by Chromatin immunoprecipitation (ChIP) analysis using anti-H3K9me3 or anti-H3K27me3, respectively, and with designed five pairs of PCR primers specific to the various promoter regions of the CDH3 gene (Set1 – Set5; see more details in Supplementary Table S1). The non-infected cells were used as the control (Ctrl). Increased enrichment of H3K9me3, but not H3K27me3, was detected in the CDH3 gene locus in cells following infection. (B) Levels of the suppression markers, H3K9me3 and H3K27me3, associated with the LOXL4 gene locus in HCT-8 cells following infection. Cells were exposed to C. parvum infection for 24 h, followed by ChIP analysis using anti-H3K9me3 or anti-H3K27me3 and with designed PCR primers (Set1–5) covering the regulatory promoter regions of the LOXL4 gene locus. No significant increase in the enrichment of H3K9me3 or H3K27me3 was detected in the LOXL4 gene locus in cells following infection. Data represent means ± S.E.M. from three independent experiments. *P<0.01, ANOVA compared with non-infected cells.
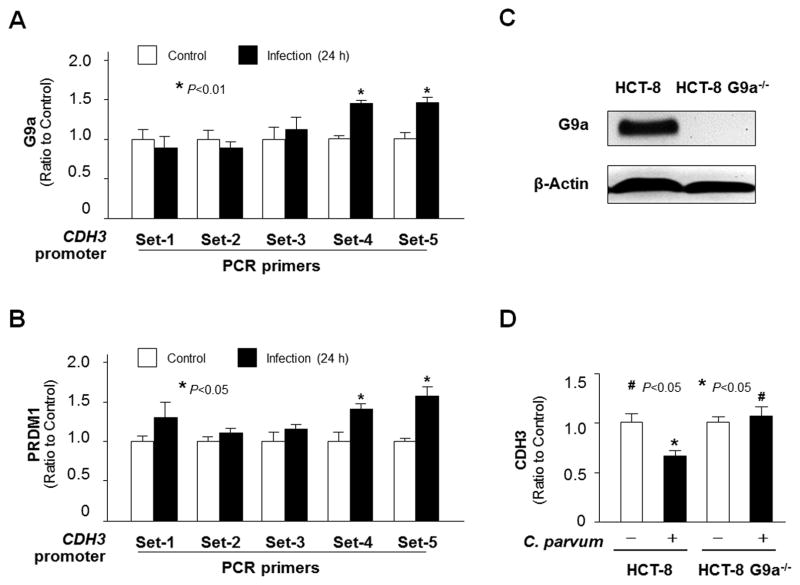
Given the enrichment of H3K9me3 in the promoter region of the CDH3 gene in cells following exposure to C. parvum infection, we then tested the potential involvement of the euchromatic histone lysine methyltransferase 2 (G9a), a histone methyltransferase for H3K9 methylation which mediates gene trans-suppression in many cell types (Shinkai and Tachibana, 2011) and is required for trans-suppression of the SMPD3 gene in C. parvum-infected intestinal epithelial cells (Ming et al., 2017). Increased recruitment of G9a to the CDH3 gene locus was detected in infected HCT-8 cells using anti-G9a and the PCR primer sets as designed for ChIP analysis (Fig. 5A). In addition, PRDM1 (also known as BLIMP-1) is a G9a-interacting protein (Shinkai et al., 2011) and a RNA-binding protein (John and Garrett-Sinha, 2009) that has been implicated in G9a-mediated histone methylation (Gyory et al., 2004). Recruitment of PRDM1 to the CDH3 gene locus was detected in infected HCT-8 cells (Fig. 5B).
Fig. 5.
Enrichment of H3K9me3 within the CDH3 gene locus in HCT-8 cells following Cryptosporidium parvum infection involves the recruitment of G9a and PRDM1. (A–B) Increased recruitment of G9a and PRDM1 to the CDH3 gene locus in HCT-8 cells following C. parvum infection. Cells were exposed to C. parvum infection for 24 h, followed by Chromatin immunoprecipitation (ChIP) analysis using anti-G9a and anti-PRDM1, respectively, and the PCR primer sets as designed. Increased recruitment of Ga9 (A) and PRDM1 (B) was detected in the CDH3 gene locus in cells following infection. (C) Knockdown of G9a in HCT-8 cells. Cells were transfected with the G9a-CRISPR/Cas9 KO(h) and G9a-HDR plasmids; stably transfected cells were cloned and confirmed by western blot analysis. (D) Knockdown of G9a attenuated the downregulation of the CDH3 gene in cells following C. parvum infection. The CDH3 RNA levels were quantified using real-time quantitative PCR in HCT-8 and HCT-8-G9a−/− cells after exposure to C. parvum infection for 24 h. Data represent means ± S.E.M. from three independent experiments. *P<0.05, ANOVA compared with non-infected controls or empty vector controls; #P<0.05, ANOVA compared with infected controls.
To further define the involvement of G9a in Cdg7_FLc_1000-mediated trans-suppression of the host CDH3 gene, we measured the effects of G9a knockdown on CDH3 expression levels in HCT-8 cells following C. parvum infection. We first generated a stable G9a−/− HCT-8 cell line using the G9a-CRISPR/Cas9 KO(h) and G9a-HDR plasmids (Santa Cruz). Deletion of G9a in the stable G9a−/− HCT-8 cell line was confirmed using western blotting (Fig. 5C). Whereas a similar expression level of CDH3 RNA was found between the stable G9a−/− HCT-8 cells and the non-transfected HCT-8 cells, downregulation of the CDH3 gene induced by C. parvum infection was not detected in the G9a−/− HCT-8 cells following infection (Fig. 5D).
3.4. Cdg7_FLc_1000 is recruited to the promoter regions of both CDH3 and LOXL4 gene loci in host cells during C. parvum infection
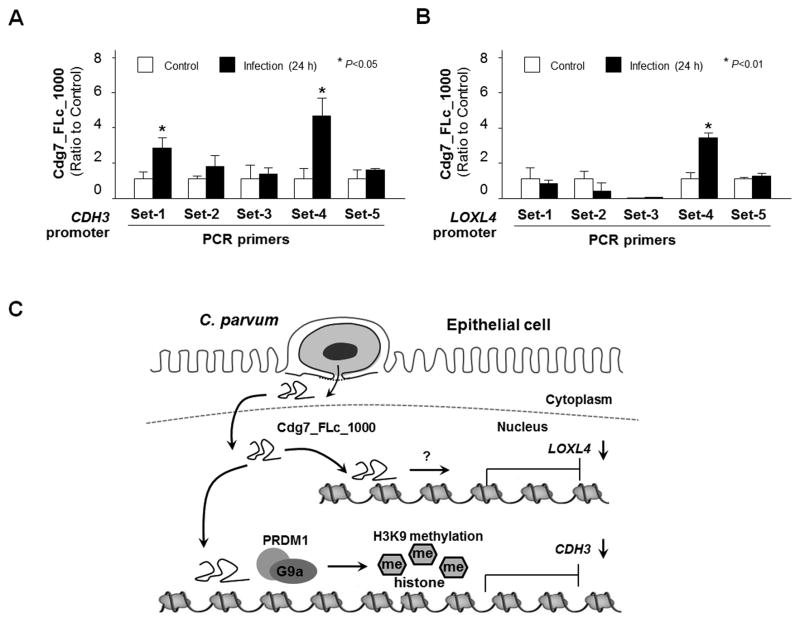
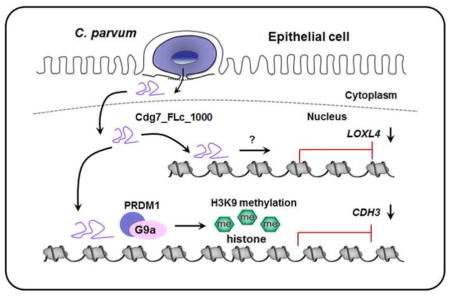
RNA transcripts, particularly long non-coding RNAs, have been demonstrated to be recruited to specific gene loci to modulate gene transcription (Ulitsky and Bartel, 2013). To test whether Cdg7_FLc_1000 RNA is directly recruited to the CDH3 and LOXL4 gene loci, we adapted the ChIRP approach originally developed for the measurement of specific recruitment of long non-coding RNAs to various gene loci (Chu et al., 2011). A pool of biotinylated tiling oligonucleotide probes specific to Cdg7_FLc_1000 were synthesized for the ChIRP analysis (Ming et al., 2017). Recruitment of Cdg7_FLc_1000 was detected within the promoter regions of both CDH3 and LOXL4 gene loci in HCT-8 cells following C. parvum infection (Fig. 6A and B). Sequence alignment analysis of the regions with the recruitment of Cdg7_FLc_1000 within the CDH3 and LOXL4 gene loci revealed no obvious consensus sequences for potential Cdg7_FLc_1000 binding (data not shown). Taken together, our data indicate that host delivery of Cdg7_FLc_1000 promotes PRDM1/G9a-mediated H3K9 methylation in the CDH3 gene locus, resulting in trans-suppression, whereas Cdg7_FLc_1000 induces trans-suppression of the LOXL4 gene through H3K9/H3K27 methylation-independent mechanisms (Fig. 6C).
Fig. 6.
Cdg7_FLc_1000 is recruited to the promoter regions of both CDH3 and LOXL4 gene loci and is associated with their trans-suppression in HCT-8 cells following Cryptosporidium parvum infection through different mechanisms. (A–B) Recruitment of Cdg7_FLc_1000 to the CDH3 and LOXL4 gene loci in HCT-8 cells following C. parvum infection. Cells were exposed to C. parvum infection for 24 h, following by chromatin isolation by RNA purification (ChIRP) analysis using a pool of probes specific to Cdg7_FLc_1000 and the PCR primer sets as designed. Increased recruitment of Cdg7_FLc_1000 was detected in the CDH3 and LOXL4 gene loci in cells following infection or Full-Cdg7_FLc_1000 transfection. Data represent means ± S.E.M. from three independent experiments. *P<0.01, ANOVA compared with non-infected control (Ctrl). (C) Schematic representation of the proposed model for transcriptional suppression of host CDH3 and LOXL4 genes in infected cells through nuclear transfer of C. parvum Cdg7_FLc_1000. Cryptosporidium parvum Cdg7_FLc_1000 transcript is selectively delivered into the nuclei of infected host cells. Host delivery of Cdg7_FLc_1000 promotes PRDM1/G9a-mediated H3K9 methylation in the CDH3 gene locus, resulting in trans-suppression; whereas Cdg7_FLc_1000 induces trans-suppression of the LOXL4 gene through H3K9/H3K27 methylation-independent mechanisms.
4. Discussion
Emerging evidence indicates that histone modifications are key targets for pathogen manipulation of host gene transcription during microbial infection (Hamon and Cossart, 2008; Gómez-Díaz et al., 2012). It is well recognized that the epigenetic modulation of a host’s transcriptional program linked to host defense genes is a relatively common occurrence during pathogenic viral and bacterial infections (Hamon and Cossart, 2008; Paschos and Allday, 2010). A diverse array of bacterial and viral effectors has been identified that either mimic or inhibit the host cellular machinery, thus facilitating the pathogen’s lifecycle. Better understanding of the molecular mechanisms underlying the gene-specific alterations in the host modulated by the pathogens would provide new insights into the pathogenesis. In this regard, our data in this study suggest that trans-suppression of host CDH3 and LOXL4 genes in cultured human intestinal epithelial cells following C. parvum infection involves nuclear delivery of parasite Cdg7_FLc_1000 RNA. Interestingly, although Cdg7_FLc_1000 is recruited to both the CDH3 and LOXL4 gene loci, host delivery of Cdg7_FLc_1000 causes CDH3 trans-suppression through PRDM1/G9a-mediated H3K9 methylation in the CDH3 gene locus, whereas Cdg7_FLc_1000 induces trans-suppression of LOXL4 gene through H3K9/H3K27 methylation-independent mechanisms.
Histone modifications such as H3K9 and H3K27 methylations are generally associated with gene transcriptional suppression (Dong and Weng, 2013). Previous studies demonstrated that nuclear delivery of Cdg7_FLc_1000 RNA suppresses SMPD3 gene transcription in infected host epithelial cells through H3K9 methylation-mediated epigenetic mechanisms (Ming et al., 2017). Similarly, enrichment of H3K9 methylation was detected in the CDH3 gene locus in host cells following infection. Moreover, recruitment of G9a (a key methyltransferase for H3K9) (Shinkai and Tachibana, 2011) and PRDM1 (a G9a-interacting protein) (Gyory et al., 2004; John and Garrett-Sinha, 2009) happened in the promoter region of the host CDH3 gene during C. parvum infection. In addition to its capacity to interact with G9a, PRDM1 has several Zinc-finger C2H2 domains that can interact with DNA and RNA molecules (John and Garrett-Sinha, 2009). Therefore, the interactions between PRDM1 and G9a modulating H3K9 methylation associated with CDH3 suppression in infected cells remain to be further validated. Given the fact that knockdown of G9a can completely attenuate suppression of the CDH3 gene in the infected cells, our data support the notion that host delivery of Cdg7_FLc_1000 causes CDH3 trans-suppression through PRDM1/G9a-mediated H3K9 methylation in the CDH3 gene locus.
How nuclear delivery of Cdg7_FLc_1000 RNA enhances PRDM1/G9a-mediated H3K9 methylation in the CDH3 gene locus is still unclear. Intriguingly, Cdg7_FLc_1000 RNA was physically recruited to the promote region of the CDH3 gene locus. RNA molecules, particularly long non-coding RNAs, can act as scaffold molecules through their interactions with RNA-binding proteins in chromatin remodeling complexes to modulate gene transcription (Ulitsky and Bartel, 2013). In addition, RNAs may interact with DNA molecules to form a triple helical structure, resulting in transcriptional regulation of gene expression (Ulitsky and Bartel, 2013). As such, Cdg7_FLc_1000 may “guide” the initial recruitment of the G9a/PRDM1 complex to the CDH3 gene locus or may function as a scaffold molecule to enhance PRDM1/G9a-mediated gene trans-suppression. Obviously, Cdg7_FLc_1000-mediated trans-suppression of CDH3 through PRDM1/G9a-mediated H3K9 methylation is a gene-specific procedure. Although Cdg7_FLc_1000 is recruited to both CDH3 and LOXL4 gene loci, enrichment of H3K9me3 and increased recruitment of the G9a/PRDM1 complex was detected in the promoter region of the CDH3 gene locus, not the LOXL4 gene. Notably, transfection of host cells with a plasmid expressing Cdg7_FLc_1000 caused trans-suppression of both CDH3 and LOXL4 genes in the transfected cells, suggesting an “initial” role for Cdg7_FLc_1000 in C. parvum-induced trans-suppression of host CDH3 and LOXL4 genes. Furthermore, sequence alignment analysis of the promotor regions of CDH3 and LOXL4 genes with the recruitment of Cdg7_FLc_1000 revealed no obvious consensus sequences for potential Cdg7_FLc_1000 binding. How promoter recruitment of Cdg7_FLc_1000 RNA may suppress LOXL4 transcription in infected cells through H3K9/H3K27 methylation-independent mechanisms is currently under investigation.
Increasing evidence suggests that RNA molecules may play important regulatory roles in diverse biological processes including regulation of gene transcription (Prasanth and Spector, 2007; Ulitsky and Bartel, 2013). Recent genomic research has revealed the expression of novel non-protein coding RNA genes in the protozoan group of parasites such as Plasmodium falciparum and C. parvum (Puiu et al., 2004; Yamagishi et al., 2011; Liao et al., 2014; Vembar et al., 2014). Our recent observations of selective delivery of C. parvum RNA transcripts of low protein-coding potential into infected host epithelial cells (Wang et al., 2016) and consequent trans-suppression of host genes through distinct mechanisms (Ming et al., 2017; Wang et al., 2017) support the notion that Cryptosporidium infection causes transcriptional gene suppression with pathological significance in infected cells through nuclear transfer of specific parasite RNAs. This study on Cdg7_FLc_1000-mediated trans-suppression of CDH3 and LOXL4 genes in infected intestinal epithelial cells provides additional evidence that delivery of parasite RNAs is involved in the parasite-host cell interactions. The pathological significance of CDH3 and LOXL4 trans-suppression in the pathogenesis of intestinal cryptosporidiosis merits future investigation.
Supplementary Material
Upregulation of IL8 and NOS2 genes in HCT-8 and INT cells following Cryptosporidium parvum infection. Cells were exposed to C. parvum oocysts at a 1:1 ratio for 4 h. After extensive washing to remove free parasites, cells were incubated for an additional 20, 32, and 44 h, followed by real-time quantitative PCR analysis of IL8 and NOS2 RNA levels. Upregulation of the IL8 and NOS2 genes was confirmed in INT and HCT-8 cells following C. parvum infection (A and B, respectively). Data represent three independent experiments. *P<0.01, ANOVA compared with non-infected cells.
Inhibition of nuclear delivery of Cdg7_FLc_1000 using the small interfering RNA (siRNA) in INT cells during Cryptosporidium parvum infection. INT cells were treated with an siRNA to Cdg7_FLc_1000 for 12 h and then exposed to C. parvum infection for an additional 24 h. Contents of Cdg7_FLc_1000 in the nuclear extracts of the infected cells were quantified by real-time quantitative PCR. A non-specific scrambled siRNA was used as the control. Data represent three independent experiments. *P<0.01, ANOVA compared with the control siRNA.
Enrichment of H3K9me3, but not H3K27me3, in HCT-8 cells following transfection with Cdg7_FLc_1000. HCT-8 cells were transfected with the Full-Cdg7_FLc_1000 for 24 h, followed by Chromatin immunoprecipitation (ChIP) analysis using anti-H3K9me3 or anti-H3K27me3, respectively, and with designed PCR primers (Set1–5) covering the various regions of the regulatory promoter of the CDH3 gene locus (A). Cells transfected with the empty vector were used as the control. Increased enrichment of H3K9me3 (B), but not H3K27me3 (C), was detected in the CDH3 gene locus in cells transfected with Full-Cdg7_FLc_1000. Data represent means ± S.E.M. from three independent experiments. *P<0.05 ANOVA compared with the empty vector control.
Highlights.
Cryptosporidium parvum infection causes downregulation of CDH3 and LOXL4 genes in host intestinal epithelial cells.
A parasite RNA Cdg7_FLc_1000 is recruited to the promoter regions of both CDH3 and LOXL4 gene loci following infection.
Cdg7_FLc_1000 promotes PRDM1/G9a-mediated H3K9 methylation associated with trans-suppression of CDH3, but not LOXL4.
Acknowledgments
This work was supported by the National Institutes of Health (AI116323) and by the revenue from Nebraska’s excise tax on cigarettes awarded to Creighton University through the Nebraska Department of Health & Human Services (DHHS) (LB595) (to X. M. C.).
We thank Dr. Quanghui Zhao (Northwest A&F University, China) for helpful and stimulating discussions, and Barbara L. Bittner (Creighton University, USA) for her assistance in writing the manuscript. This work was supported by funding from the National Institutes of Health, USA (AI116323 and AI136877) and the Nebraska Stem Cell Research Program, USA (LB606), and by revenue from the State of Nebraska, USA, excise tax on cigarettes awarded to Creighton University through the Nebraska Department of Health & Human Services (DHHS) (LB595). Dr. Zhenping Ming was a visiting scholar supported by the China Scholarship Council and the National Natural Science Foundation of China (NSFC No. 31372194). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the State of Nebraska, DHHS or NSFC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcantara CS, Yang CH, Steiner TS, Barrett LJ, Lima AA, Chappell CL, Okhuysen PC, White AC, Jr, Guerrant RL. Interleukin-8, tumor necrosis factor-alpha, and lactoferrin in immunocompetent hosts with experimental and Brazilian children with acquired cryptosporidiosis. Am J Trop Med Hyg. 2003;68:325–328. [PubMed] [Google Scholar]
- Baek S, Lee YW, Yoon S, Baek SY, Kim BS, Oh SO. CDH3/P-Cadherin regulates migration of HuCCT1 cholangiocarcinoma cells. Anat Cell Biol. 2010;43:110–117. doi: 10.5115/acb.2010.43.2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- Checkley W, White AC, Jaganath D, Arrowood MJ, Chalmers RM, Chen XM, Fayer R, Griffiths JK, Guerrant RL, Hedstrom L, Huston CD, Kotloff KL, Kang G, Mead JR, Miller M, Petri WA, Jr, Priest JW, Roos DS, Striepen B, Thompson RC, Ward HD, Van Voorhis WA, Xiao L, Zhu G, Houpt ER. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect Dis. 2015;15:85–94. doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comptour A, Rouzaire M, Belville C, Bonnin N, Daniel E, Chiambaretta F, Blanchon L, Sapin V. Lysyl oxidase-like 4 involvement in retinoic acid epithelial wound healing. Sci Rep. 2016;6:32688–32705. doi: 10.1038/srep32688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creamber B, Shorter RG, Bamforth J. The turnover and shedding of epithelial cells. I The turnover in the gastro-intestinal tract. Gut. 1961;2:110–118. doi: 10.1136/gut.2.2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M, Lancto CA, Abrahamsen MS. Cryptosporidium parvum regulation of human epithelial cell gene expression. Int J Parasitol. 2004;34:73–82. doi: 10.1016/j.ijpara.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Dong X, Weng Z. The correlation between histone modifications and gene expression. Epigenomics. 2013;5:113–116. doi: 10.2217/epi.13.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel S, Chin EN, Fakhraldeen SA, Berry SM, Beebe DJ, Alexander CM. Both LRP5 and LRP6 receptors are required to respond to physiological Wnt ligands in mammary epithelial cells and fibroblasts. J Biol Chem. 2012;287:16454–16466. doi: 10.1074/jbc.M112.362137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Díaz E, Jordà M, Peinado MA, Rivero A. Epigenetics of host-pathogen interactions: the road ahead and the road behind. PLoS Pathog. 2012;8:e1003007. doi: 10.1371/journal.ppat.1003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyory I, Wu J, Fejér G, Seto E, Wright KL. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol. 2004;5:299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- Hamon MA, Cossart P. Histone modifications and chromatin remodeling during bacterial infections. Cell Host Microbe. 2008;4:100–109. doi: 10.1016/j.chom.2008.07.009. [DOI] [PubMed] [Google Scholar]
- John SA, Garrett-Sinha LA. Blimp1: a conserved transcriptional repressor critical for differentiation of many tissues. Exp Cell Res. 2009;315:1077–1084. doi: 10.1016/j.yexcr.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acácio S, Biswas K, O’Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- Kuhnert F, Davis CR, Wang HT, Chu P, Lee M, Yuan J, Nusse R, Kuo CJ. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci U S A. 2004;101:266–271. doi: 10.1073/pnas.2536800100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent F, Eckmann L, Savidge TC, Morgan G, Theodos C, Naciri M, Kagnoff MF. Cyptosporidium parvum infection of human intestinal epithelial cells induces the polarized secretion of C-X-C chemokines. Infect Immun. 1997;65:5067–5073. doi: 10.1128/iai.65.12.5067-5073.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Q, Shen J, Liu J, Sun X, Zhao G, Chang Y, Xu L, Li X, Zhao Y, Zheng H, Zhao Y, Wu Z. Genome-wide identification and functional annotation of Plasmodium falciparum long noncoding RNAs from RNA-seq data. Parasitol Res. 2014;113:1269–1281. doi: 10.1007/s00436-014-3765-4. [DOI] [PubMed] [Google Scholar]
- Ma S, Ming Z, Gong AY, Wang Y, Chen X, Hu G, Zhou R, Shibata A, Swanson PC, Chen XM. A long noncoding RNA, lincRNA-Tnfaip3, acts as a coregulator of NF-κ B to modulate inflammatory gene transcription in mouse macrophages. FASEB J. 2016;37:1215–1225. doi: 10.1096/fj.201601056R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming ZP, Gong AY, Wang Y, Zhang XT, Li M, Mathy NW, Strauss-Soukup JK, Chen XM. Involvement of Cryptosporidium parvum Cdg7_FLc_1000 RNA in the attenuation of intestinal epithelial cell migration via trans-suppression of host cell SMPD3 gene. J Infect Dis. 2018;217:122–133. doi: 10.1093/infdis/jix392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor RM, Wanyiri JW, Wojczyk BS, Kim K, Ward H. Stable expression of Cryptosporidium parvum glycoprotein gp40/15 in Toxoplasma gondii. Mol Biochem Parasitol. 2007;152:149–158. doi: 10.1016/j.molbiopara.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donoghue PJ. Cryptosporidium and cryptosporidiosis in man and animals. Int J Parasitol. 1995;25:139–195. doi: 10.1016/0020-7519(94)e0059-v. [DOI] [PubMed] [Google Scholar]
- Paredes J, Figueiredo J, Albergaria A, Oliveira P, Carvalho J, Ribeiro AS, Caldeira J, Costa AM, Simões-Correia J, Oliveira MJ, Pinheiro H, Pinho SS, Mateus R, Reis CA, Leite M, Fernandes MS, Schmitt F, Carneiro F, Figueiredo C, Oliveira C, Seruca R. Epithelial E- and P-cadherins: role and clinical significance in cancer. Biochim Biophys Acta. 2012;1826:297–311. doi: 10.1016/j.bbcan.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Paschos K, Allday MJ. Epigenetic reprogramming of host genes in viral and microbial pathogenesis. Trends Microbiol. 2010;18:439–447. doi: 10.1016/j.tim.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- Puiu D, Enomoto S, Buck GA, Abrahamsen MS, Kissinger JC. CryptoDB: the Cryptosporidium genome resource. Nucleic Acids Res. 2004;32:D329–331. doi: 10.1093/nar/gkh050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelov L, Sprecher E, Paus R. The role of P-cadherin in skin biology and skin pathology: lessons from the hair follicle. Cell Tissue Res. 2015;360:761–771. doi: 10.1007/s00441-015-2114-y. [DOI] [PubMed] [Google Scholar]
- Sasahara T, Maruyama H, Aoki M, Kikuno R, Sekiguchi T, Takahashi A, Satoh Y, Kitasato H, Takayama Y, Inoue M. Apoptosis of intestinal crypt epithelium after Cryptosporidium parvum infection. J Infect Chemother. 2003;9:278–281. doi: 10.1007/s10156-003-0259-1. [DOI] [PubMed] [Google Scholar]
- Savidge TC, Shmakov AN, Walker-Smith JA, Phillips AD. Epithelial cell proliferation in childhood enteropathies. Gut. 1996;39:185–193. doi: 10.1136/gut.39.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley LD. Intracellular parasite invasion strategies. Science. 2004;304:248–253. doi: 10.1126/science.1094717. [DOI] [PubMed] [Google Scholar]
- Striepen B. Parasitic infections: Time to tackle cryptosporidiosis. Nature. 2013;503:189–191. doi: 10.1038/503189a. [DOI] [PubMed] [Google Scholar]
- Tarver AP, Clark DP, Diamond G, Russell JP, Erdjument-Bromage H, Tempst P, Cohen KS, Jones DE, Sweeney RW, Wines M, Hwang S, Bevins CL. Enteric beta-defensin: molecular cloning and characterization of a gene with inducible intestinal epithelial cell expression associated with Cryptosporidium parvum infection. Infect Immun. 1998;66:1045–1056. doi: 10.1128/iai.66.3.1045-1056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Scherf A, Siegel TN. Noncoding RNAs as emerging regulators of Plasmodium falciparum virulence gene expression. Curr Opin Microbiol. 2014;20:153–161. doi: 10.1016/j.mib.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayak S, Pawlowic MC, Sateriale A, Brooks CF, Studstill CJ, Bar-Peled Y, Cipriano MJ, Striepen B. Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature. 2015;523:477–480. doi: 10.1038/nature14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won HY, Lee JY, Shin DH, Park JH, Nam JS, Kim HC, Kong G. Loss of Mel-18 enhances breast cancer stem cell activity and tumorigenicity through activating Notch signaling mediated by the Wnt/TCF pathway. FASEB J. 2012;26:5002–5013. doi: 10.1096/fj.12-209247. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gong AY, Ma S, Chen X, Li Y, Su CJ, Norall D, Chen J, Strauss-Soukup JK, Chen XM. Delivery of parasite RNA transcripts into infected epithelial cells during Cryptosporidium infection and its potential impact on host gene transcription. J Infect Dis. 2016;215:636–643. doi: 10.1093/infdis/jiw607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gong AY, Ma S, Chen X, Strauss-Soukup JK, Chen XM. Delivery of parasite Cdg7_FLc_0990 RNA transcript into intestinal epithelial cells during Cryptosporidium parvum infection suppresses host cell gene transcription through epigenetic mechanisms. Cell Microbiol. 2017;19:e12760. doi: 10.1111/cmi.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi J, Wakaguri H, Sugano S, Kawano S, Fujisaki K, Sugimoto C, Watanabe J, Suzuki Y, Kimata I, Xuan X. Construction and analysis of full-length cDNA library of Cryptosporidium parvum. Parasitol Int. 2011;60:199–202. doi: 10.1016/j.parint.2011.03.001. [DOI] [PubMed] [Google Scholar]
- Yang YL, Serrano MG, Sheoran AS, Manque PA, Buck GA, Widmer G. Over-expression and localization of a host protein on the membrane of Cryptosporidium parvum infected epithelial cells. Mol Biochem Parasitol. 2009;168:95–101. doi: 10.1016/j.molbiopara.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Upregulation of IL8 and NOS2 genes in HCT-8 and INT cells following Cryptosporidium parvum infection. Cells were exposed to C. parvum oocysts at a 1:1 ratio for 4 h. After extensive washing to remove free parasites, cells were incubated for an additional 20, 32, and 44 h, followed by real-time quantitative PCR analysis of IL8 and NOS2 RNA levels. Upregulation of the IL8 and NOS2 genes was confirmed in INT and HCT-8 cells following C. parvum infection (A and B, respectively). Data represent three independent experiments. *P<0.01, ANOVA compared with non-infected cells.
Inhibition of nuclear delivery of Cdg7_FLc_1000 using the small interfering RNA (siRNA) in INT cells during Cryptosporidium parvum infection. INT cells were treated with an siRNA to Cdg7_FLc_1000 for 12 h and then exposed to C. parvum infection for an additional 24 h. Contents of Cdg7_FLc_1000 in the nuclear extracts of the infected cells were quantified by real-time quantitative PCR. A non-specific scrambled siRNA was used as the control. Data represent three independent experiments. *P<0.01, ANOVA compared with the control siRNA.
Enrichment of H3K9me3, but not H3K27me3, in HCT-8 cells following transfection with Cdg7_FLc_1000. HCT-8 cells were transfected with the Full-Cdg7_FLc_1000 for 24 h, followed by Chromatin immunoprecipitation (ChIP) analysis using anti-H3K9me3 or anti-H3K27me3, respectively, and with designed PCR primers (Set1–5) covering the various regions of the regulatory promoter of the CDH3 gene locus (A). Cells transfected with the empty vector were used as the control. Increased enrichment of H3K9me3 (B), but not H3K27me3 (C), was detected in the CDH3 gene locus in cells transfected with Full-Cdg7_FLc_1000. Data represent means ± S.E.M. from three independent experiments. *P<0.05 ANOVA compared with the empty vector control.