Abstract
Microglia and macrophages are the main non-neuronal subsets of myeloid origin in the brain, and are critical regulators in neurodegenerative disorders, where inflammation is a key factor. Since HIV infection results in neurological perturbations that are similar to those in aging, we examined microglial and infiltrating myeloid subsets in the search for changes that might resemble the ones in aging. For that, we used the SIV infection in rhesus macaques to model neuroAIDS. We found that Sirt-1, a molecule that impacts survival and health in many models, was decreased in cell preparations containing a majority of microglia and myeloid cells from the brain of infected macaques. The role of Sirt-1 in neuroAIDS is unknown. We hypothesized that Sirt-1 silencing functions are affected by SIV. Mapping of Sirt-1 binding patterns to chromatin revealed that the number of Sirt-1-bound genes was 29.6% increased in myeloid cells from infected animals with mild or no detectable neuropathology, but 51% was decreased in severe neuropathology, compared to controls. Importantly, Sirt-1-bound genes in controls largely participate in neuroinflammation. Promoters of type I IFN pathway genes IRF7, IRF1, IFIT1, and AIF1, showed Sirt-1 binding in controls, which was consistently lost after infection, together with higher transcription. Loss of Sirt-1 binding was also found in brains from old uninfected animals, suggesting a common regulation. The role of Sirt-1 in regulating these inflammatory markers was confirmed in two different in vitro models, where Sirt-1 blockage modulated IRF7, IRF1 and AIF1 levels both in human macrophage cell lines and in human blood-derived monocytes from various normal donors, stimulated with a TLR9 agonist. Our data suggests that Sirt-1-inflammatory gene silencing is disturbed by SIV infection, resembling aging in brains. These findings may impact our knowledge on the contribution of myeloid subsets to the neurological consequences of HIV infection, aggravated and overlapping with the aging process.
Introduction
Microglia along with infiltrating macrophages are the main innate immune cells present in the brain, and are critical regulators of brain health. Microglia cells, which derive from erytho-myeloid precursors in the yolk sac, and macrophages, which are originated later from myeloid precursors in the bone marrow(Ginhoux et al. 2010; Greter et al. 2015; Greter and Merad 2013), differ in localization, longevity and functions(London et al. 2013), but share surface markers and the ability to respond to insults through the development of pro-inflammatory phenotypes(Kierdorf et al. 2013; Kierdorf and Prinz 2013; Prinz 2014; Prinz et al. 2014). The inflammatory response in the brain plays a key role in neurotoxicity, but also in immune-regulation and regeneration, which may be detrimental to neuronal health in aging, and in response to chronic stresses such as misfolded proteins (e.g., Alzheimer's Disease) and HIV. Using Simian Immunodeficiency Virus (SIV) in rhesus macaques as a model of neuroAIDS, we examined the possibility that cognitive dysfunction and inflammation in neuroAIDS are associated to the development of characteristics that resemble aging in microglia and macrophage phenotype. While seeking the identification of regulatory pathways that are triggered by SIV infection in cells of the brain innate immune system isolated from the central nervous system (CNS), we found a profound decrease in the transcription levels of Sirt-1 (silent information regulator 1), which is a molecule involved in lifespan, were triggered by SIV infection (Chaudhuri et al. 2013).
Sirt-1(silent information regulator 1) is an intriguing molecule, as it may be linked to mechanisms associated to health and aging phenotypes in several experimental models(Herranz and Serrano 2010b). Pathways correlated to aging in humans include histone acetylation(Kumar et al. 2013). In various organs, aging is also associated with inflammation and oxidative stress, and a decrease in the ability to perform tissue repair, all conditions that are also part of the pathogenesis in SIV and HIV infection. Sirt-1 belongs to the family of sirtuins, which are class III deacetylases, whose activity is nutrient-responsive and modulated by NAD+/NADH ratios that change with respiratory activity. Thus, it has emerged as a key regulator of health and life span in several organisms(Frankel and Rogina 2006; Haigis and Guarente 2006; Kaeberlein et al. 1999; Rogina and Helfand 2004; Tissenbaum and Guarente 2001). In mammalian cells, DNA damage triggers Sirt-1 redistribution to DNA breaks for repair, resulting in transcriptional changes that are similar to those in the aging mouse brain(Oberdoerffer et al. 2008). It also modulates non-histone substrates involved in transcriptional regulation (Zhang and Kraus 2010). In addition, increased Sirt-1 expression promotes survival in a mouse model of genomic instability and suppresses age-dependent transcriptional changes. Thus, redistribution of Sirt-1 and other chromatin-modifying proteins due to DNA damage may be a conserved mechanism of aging in eukaryotes(Oberdoerffer et al. 2008). Sirt1 regulates transcription factors that function in cell metabolism, growth, differentiation, survival, apoptosis, or stress response, and, in inflammation. For these reasons, it has a protective role on age-associated pathologies, such as neurodegenerative diseases (Herranz and Serrano 2010a; Herranz and Serrano 2010b; Palacios et al. 2010). It is also a positive regulator of telomere length and DNA repair (Palacios et al. 2010). Sirt1 is induced by Egr1, which we also have found to be decreased in association with SIV encephalitis (SIVE) (Gersten et al. 2009). The role of Sirt1 in neuroAIDS has not been explored.
Chromatin consists of DNA wrapped around histones, and acetylation may promote chromatin expansion. Thus, DNA expression is in part regulated by histone acetylation and deacetylation. Given the role of Sirt-1 in this process described above, we asked what is the relationship between Sirt-1 and pathogenesis. We predicted that SIV induced changes in the Sirt-1 mediated epigenetic regulation, under the hypothesis that Sirt-1 could negatively influence the expression of several molecules, and be responsible for repressing, among other targets, pro-inflammatory genes as well as genes that maintain the CNS physiological balance, and as a result, preventing pathogenesis. On the other hand, we predicted that changes in the Sirt-1 mediated gene regulation could be triggered in the CNS by the infection, de-repressing critical inflammatory genes in a process that may have commonalities with the aging brain. Thus we investigated the implications of changes of Sirt-1 transcriptional levels and activity in predominantly innate immune cells isolated from the infected brain, especially regarding the resulting pro-inflammatory phenotype that may result from the un-repressing of inflammatory promoters.
These questions are important because in mouse studies the lack or decrease of Sirt-1 has a profound impact on survival. In addition, the Sirt1−/− mice that survive to adulthood exhibit abnormal metabolic rates, elevated rates of spontaneous activity, decreased fertility, elevated autoimmunity, osteoarthritis, and compromised cognition(Boily et al. 2009; Boily et al. 2008; Bosch-Presegue and Vaquero 2011; Bosch-Presegue and Vaquero 2013; Gabay et al. 2013; Gagarina et al. 2010; Herranz and Serrano 2010b; Kolthur-Seetharam et al. 2009; Lemieux et al. 2005). These are characteristics of aging that are also found in HIV-infected individuals at various degrees. Given the role of Sirt1 at maintaining gene silencing through histone deacetylation, identifying which genes are affected by the presence and activity of Sirt-1 seemed critical. Therefore, using ChIP and NextGen sequencing, we have identified Sirt-1 genome-wide targets in microglia and myeloid cells isolated from controls, SIV-infected with mild or without inflammatory CNS pathology (SIV), defined by the absence or low frequency of mononuclear infiltrate, and SIV-infected with CNS pathology (SIV encephalitis, SIVE) brains, defined by the abundance of perivascular and parenchymal inflammatory infiltrate, as well as the presence of multinucleated giant cells. A list of molecular targets with a described role in inflammation was generated, and the increased expression of these Sirt-1 target molecules was confirmed in the infected brain tissue. Thus, there were dynamic changes of Sirt-1 levels and binding to chromatin after SIV infection, which may be critical in the control of inflammation, through epigenetic mechanisms. Due to the role of Sirt-1 in the aging process, we examined the potential for the same Sirt-1 target molecules to be involved in the “inflammaging”. The results may have implications to the current overlapping and synergism of HIV infection with the normal aging process, ultimately aggravating CNS disorders in the aging HIV+ population.
Results
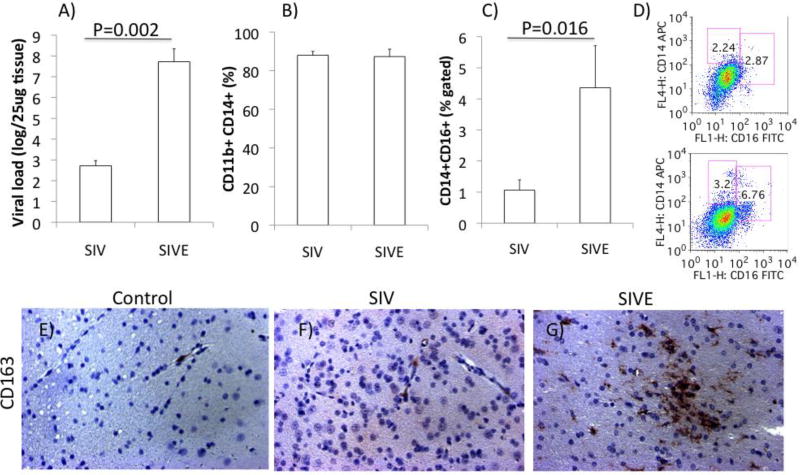
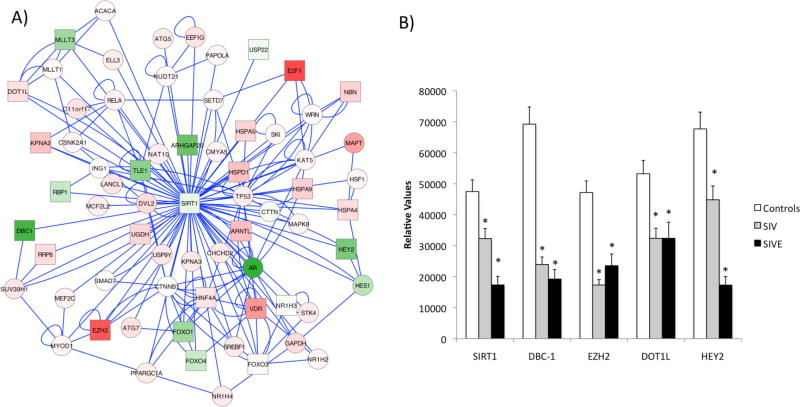
Using qRT-PCR, we found that Sirt-1 is strongly downregulated by SIV infection in brain cell suspensions composed of a majority of microglia and innate immune myeloid subsets (Figure 1), even in animals without CNS disease, and more in cases of spontaneous development of encephalitis (SIVE) (Figure 2). The transcriptional decrease in Sirt-1 levels was not detected in whole brain samples (not shown), but only in the mononuclear cells isolated from the brain, which were highly enriched for innate immune CD45LCAlow CD11b+ microglial cells (~85%), suggesting that this phenomenon is highly associated to the innate immune compartment of the CNS in the context of infection. Infiltrating macrophages (CD11b+ CD14+ CD16+), present both in controls and in SIV animals, were enriched from 2 – 6 fold in SIVE animals compared to SIV (Figure 1C), along with brain viral load (Figure 1A). In gene arrays performed in the brain cell isolates, we identified a down regulation of several of the Sirt-1 molecular gene partners and co-regulators, such as HEY2, DBC-1 and others, suggesting potential changes in Sirt-1 activity (Figure 2A). The decrease of the important Sirt-1 molecular partners, DBC-1, EZH2, DOT1L and HEY2, was confirmed using qRT-PCR on isolated microglia cells from controls, SIV and SIVE animals (Figure 2B).
Figure 1. Characterization of the cellular isolate and definition of SIVE.
A) Viral load in the brain tissue was measured and compared between animals with mild or severe signs of pathology, separated in SIV and SIVE groups. The quality of the cell suspensions isolated from the brain was examined using antibodies against subset-defining surface markers, by flow cytometry. B) Percentage of CD11b+ CD14+ myeloid cells in the total cell suspension. C) Percentage of CD14+CD16+ macrophages in CD11b-gated cells. D) Representative scatter plots of cell suspensions from SIV (upper) and SIVE (lower panel), showing CD14 and CD16 distribution in gated CD11b+ cells. Further characterization of the CNS pathology was performed by IHC using anti-CD163 antibodies, which detect inflammatory infiltrate, and allow the distinction of severe encephalitis (SIVE). The figures show the staining and distribution of CD163+ cells in representative frontal cortex sections in E) uninfected controls, F) SIV and G) SIVE animals.
Figure 2. Decrease in transcription of Sirt-1 and of molecules associated to its activity was identified using a systems biology approach applied to gene expression profile data from microglia cells that were isolated from SIV-infected compared to uninfected macaques.
Biogrid was utilized to identify high score modules using a cut-off of 3 significantly changed neighboring genes, as previously described(Gersten et al. 2009). A) Sirt-1-centered node generated J Modules pathway analysis based on Biogrid, upon comparison between Control and SIV microglia. Green color shades represents down regulated genes and red color shades represent upregulated genes. Squares represent genes that are significantly changed (p<0.01). B) The changes in mRNA transcription of Sirt-1 molecular partners were confirmed using SyBr Green qRT-PCR. Values were normalized against the expression of GAPDH. *p<0.05 compared to controls.
Given the demonstrated role of Sirt-1 in lifespan and aging, controlling gene expression by histone deacetylation at the chromatin level, we investigated the hypothesis that in SIV infection a Sirt-1-regulated gene network may be involved in producing phenotypic changes in innate immune cells of the CNS, which are associated to the development of an inflammatory environment, and that may resemble the phenotype found in the aging population. To understand that, we used Chromatin Immunoprecipitation (ChIP) and NextGen ChIP-sequence(Oberdoerffer et al. 2008) to investigate genome-wide Sirt-1 binding targets, in the cell preps from control and SIV-infected macaques, and the changes produced by infection were compared in animals that exhibited or not severe encephalitis, as defined by a detectable increase in infiltrating inflammatory cell foci both perivascularly and in the parenchyma at histopathology, and expressing innate immune and myeloid markers such as CD163, which correlate with CNS inflammation (Figures 1E, F and G). After the identification of relevant Sirt1 targets in chromatin, we further investigated the protein expression and distribution of selected candidate molecules in the brain of uninfected but aging monkeys (>19 years old).
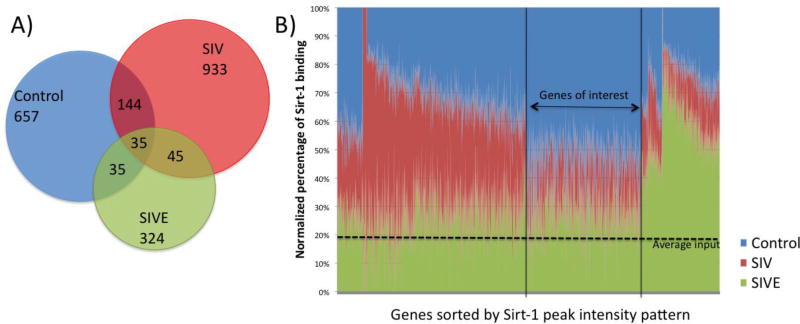
The differences of Sirt-1/promoter binding between Controls, SIV and SIVE were analyzed in depth for the identification of molecular candidates in correlation with the inflammatory etiology of neuroAIDS. ChIP-Seq revealed that the binding of Sirt-1 to gene targets suffers a drastic shuffle after infection (SIV), and that the gene targets largely differ in animals with encephalitis (SIVE) compared to animals with mild or no signs of inflammation (SIV). Figure 2A shows the number of gene targets in uninfected Controls, SIV and SIVE animals, illustrating the strong dynamic of Sirt-1 binding to gene targets. With a focus on in-promoter, upstream gene sequences where Sirt-1 enrichment was noted, we have identified changes of abundance and of chromatin-binding localization. Interestingly, the number of Sirt-1-bound genes increased in brain cell isolates from SIV infected macaques. On the other hand, in encephalitis there was a significantly decrease of genes with Sirt-1–binding activity, which occurred in correlation with a lower Sirt-1 transcription. Consistently, gene promoters from which Sirt-1 binding was observed in controls but lost with infection were annotated to inflammation, cell cycle and tumorigenesis. Intriguingly, in controls, Sirt-1 showed binding to the exons of NR3C2 and Lamin A (LMNA), both markers of aging in human brains(Kumar et al. 2013), and this binding was lost in infected microglia, suggesting that Sirt-1 is a potential silencer of these and probably other aging markers.
Compared to controls, SIV-derived cells had a 29.6% increase in the number of genes found to have Sirt-1 binding, while SIVE-derived cells showed a 51% decrease, making up to a 75.3% decrease in genes showing Sirt1 binding compared to SIV (Figure 3A). In addition, Sirt-1 binding was highly redistributed, with only 10.6% of conserved binding sites across groups (Figure 3). The shuffling, together with the changes in Sirt-1 activity produced by the infection, generated different binding patterns (Figure 3B), where genes with a potential role in early stages of inflammation, were located in the fraction with high Sirt1 binding in controls, and lost after infection, both in SIV and in SIVE (Figure 3B). Cluster analysis pointed to a number of processes associated to regulation of inflammation. The genes that presented Sirt-1 binding motifs in controls were involved in several pathways that are also potentially disturbed in aging (Table 1), including cancer, innate and adaptative immune responses, energy balance and circadian rythmicity, suggesting a role for Sirt-1 as a silencer and regulator of a number of pathogenic processes.
Figure 3. Dynamics of Sirt-1 binding sites on the chromatin in Controls, SIV and SIVE macaque microglia cells.
A) Number of Sirt-1 targets in the groups. B) Normalized percentage of Sirt-1 binding peak intensity demonstrating the distinctive pattern and dynamics in the groups. Blue is control, Red is SIV and Green is SIVE. Sirt-1 binding peak intensity shows the distinctive binding patterns and dynamics. We have focused on genes that showed Sirt-1 binding in controls, and decreased binding in SIV and SIVE.
Table 1.
Pathway analysis of Sirt-1 target genes in control microglia. Genes with Sirt-1 binding peaks identified in control microglia were subjected to DAVID Bioinformatics resource v6.7 for pathway value identification, as described (Huang da et al. 2009; Huang et al. 2007).
| Category | Term | Count | P-Value | Benjamini |
|---|---|---|---|---|
| KEGG_PATHWAY | Pathways in cancer | 43 | 3.7E-2 | 3.9E-1 |
| KEGG_PATHWAY | Huntington's disease | 30 | 8.6E-2 | 5.6E-1 |
| KEGG_PATHWAY | Endocytosis | 28 | 2.6E-2 | 3.3E-1 |
| KEGG_PATHWAY | Spliceosome | 27 | 5.1E-4 | 8.4E-2 |
| KEGG_PATHWAY | Wnt signaling pathway | 23 | 1.6E-2 | 2.9E-1 |
| KEGG_PATHWAY | Ubiquitin mediated proteolysis | 22 | 1.3E-2 | 2.7E-1 |
| KEGG_PATHWAY | Cell cycle | 18 | 5.8E-2 | 4.9E-1 |
| KEGG_PATHWAY | Chronic myeloid leukemia | 17 | 2.0E-3 | 1.6E-1 |
| KEGG_PATHWAY | Prostate cancer | 16 | 1.1E-2 | 2.8E-1 |
| KEGG_PATHWAY | Melanogenesis | 16 | 6.0E-2 | 4.7E-1 |
| KEGG_PATHWAY | T cell receptor signaling pathway | 16 | 9.9E-2 | 5.6E-1 |
| KEGG_PATHWAY | B cell receptor signaling pathway | 15 | 6.6E-3 | 3.2E-1 |
| KEGG_PATHWAY | ErbB signaling pathway | 14 | 4.7E-2 | 4.5E-1 |
| KEGG_PATHWAY | Colorectal cancer | 14 | 9.2E-2 | 5.5E-1 |
| KEGG_PATHWAY | Acute myeloid leukemia | 12 | 1.1E-2 | 3.2E-1 |
| KEGG_PATHWAY | Phosphatidylinositol signaling system | 12 | 6.2E-2 | 4.6E-1 |
| KEGG_PATHWAY | Pancreatic cancer | 12 | 8.9E-2 | 5.5E-1 |
| KEGG_PATHWAY | Hedgehog signaling pathway | 11 | 2.2E-2 | 3.2E-1 |
| SP_PIR_KEYWORDS | phosphoprotein | 10 | 6.2E-3 | 1.2E-1 |
| KEGG_PATHWAY | Nucleotide excision repair | 10 | 1.8E-2 | 2.9E-1 |
| KEGG_PATHWAY | Basal cell carcinoma | 10 | 6.0E-2 | 4.8E-1 |
| GOTERM_MF_FAT | nucleotide binding | 7 | 1.3E-3 | 7.3E-2 |
| SP_PIR_KEYWORDS | nucleus | 7 | 1.4E-2 | 1.7E-1 |
| KEGG_PATHWAY | Homologous recombination | 7 | 3.4E-2 | 3.9E-1 |
| GOTERM_MF_FAT | ATP binding and activity | 6 | 2.1E-3 | 6.0E-2 |
| GOTERM_MF_FAT | adenyl ribonucleotide binding | 6 | 2.1E-3 | 6.0E-2 |
| GOTERM_MF_FAT | purine nucleoside binding | 6 | 3.0E-3 | 5.8E-2 |
| GOTERM_MF_FAT | nucleoside binding | 6 | 3.0E-3 | 5.8E-2 |
| GOTERM_MF_FAT | adenyl nucleotide binding | 6 | 3.0E-3 | 5.8E-2 |
| GOTERM_MF_FAT | purine ribonucleotide binding | 6 | 4.2E-3 | 6.1E-2 |
| GOTERM_MF_FAT | ribonucleotide binding | 6 | 4.2E-3 | 6.1E-2 |
| GOTERM_MF_FAT | purine nucleotide binding | 6 | 5.7E-3 | 5.5E-2 |
| COG_ONTOLOGY | Cell division and chromosome partitioning / Cytoskeleton | 6 | 6.7E-2 | 9.3E-1 |
| SP_PIR_KEYWORDS | atp-binding | 5 | 5.4E-3 | 1.6E-1 |
| KEGG_PATHWAY | Circadian rhythm | 5 | 9.9E-3 | 3.5E-1 |
| GOTERM_MF_FAT | ATP-dependent helicase activity | 3 | 4.8E-3 | 5.6E-2 |
| GOTERM_MF_FAT | purine NTP-dependent helicase activity | 3 | 4.8E-3 | 5.6E-2 |
| SP_PIR_KEYWORDS | helicase | 3 | 5.3E-3 | 2.8E-1 |
| GOTERM_MF_FAT | helicase activity | 3 | 9.4E-3 | 7.8E-2 |
For validation of the involvement of Sirt-1 regulated gene network in SIV-induced CNS pathogenesis, we have prioritized genes where Sirt-1 was bound to in-promoter regions in control microglia, but this binding was decreased or lost in SIV, and in SIVE-derived microglia. In addition, we have focused on genes that have a demonstrated role in inflammation in other models (Table 2). Importantly, we have examined the expression of the genes putatively regulated by Sirt-1 in gene arrays, and they were mostly confirmed to be upregulated by SIV infection and even more in SIVE, in correlation with the loss of Sirt-1 binding (Table 2).
Table 2. Genes with a role in inflammation that had in-promoter Sirt1-binding in Controls, which was decreased with SIV, and with SIVE, and fold change transcript values from gene array data in SIVE compared to SIV.
These genes may be kept under control by Sirt-1 and are expected to become unrepressed in association with infection-driven CNS pathology.
| Gene Symbol |
Gene Name | Chromosome | Active Region length (bp) |
ChIP Peak Value Control |
ChIP Peak Value SIV |
ChIP Peak Value SIVE |
Positive Interval (from TSS) |
Transcriptional fold change SIV/Ctr |
P value |
Transcriptional fold change SIVE/SIV |
P value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| IRF7 | Interferon regulatory factor 7 | 14 | 1098 | 6.3 | 3.8 | 3.4 | Upstream | 2.63 | 0.004 | 4.99 | 0.001 |
| IFIT1 | Interferon-induced protein with tetratricopeptide repeats 1 | 9 | 659 | 5.6 | 0 | 1.6 | Upstream | 1.84 | 0.024 | 3.31 | 0.004 |
| AIF1 | Allograft inflammatory factor 1 (Iba-1) | 4 | 1122 | 6.5 | 3.2 | 4.0 | Downstream | 3.64 | 0.049 | 2.34 | 0.054 |
| IRF1 | Interferon regulatory factor 1 | 6 | 801 | 6.9 | 4.8 | 3.9 | Upstream | 1.25 | 0.26 | 1.88 | 0.050 |
Of the genes in Table 2, we have prioritized the examination of AIF1, IRF7 and IFIT1, due to the involvement of these molecules in inflammation, and also a potential involvement in aging.
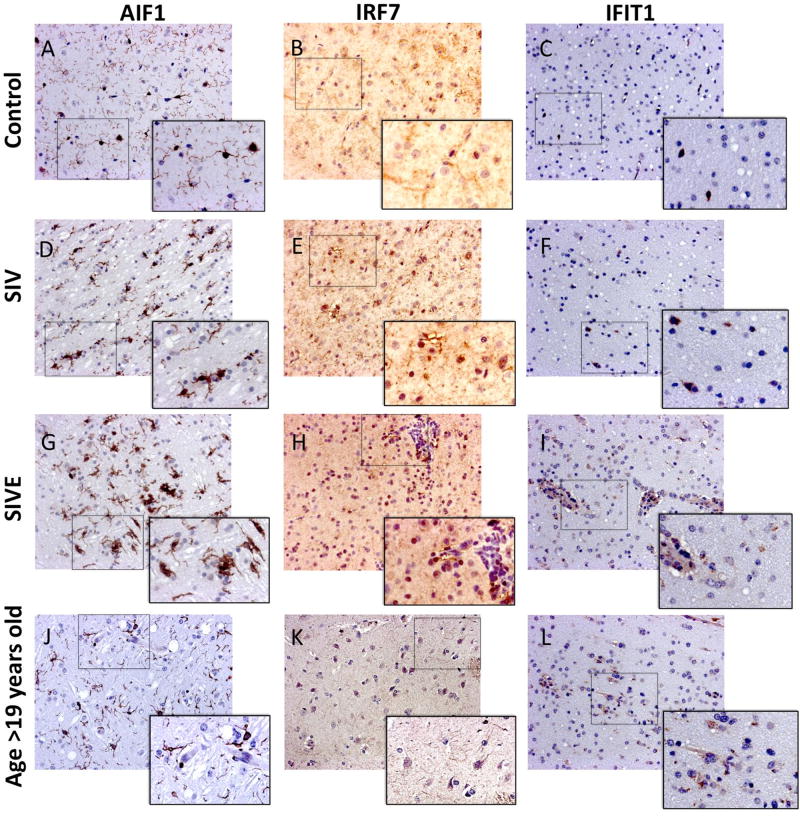
One gene promoter where Sirt-1 binding was found in controls, but significantly lost in SIV and SIVE, is AIF1 (Iba-1)(Figure 4A). AIF1 is a molecule that defines functionally activated microglia(Schwab et al. 2001), validating our hypothesis of upregulation of Sirt-1 targets on microglia cells after infection. The increase of AIF1 (Iba1) expression has been extensively documented by us and by others (Burudi et al. 2002; Roberts et al. 2004a; Roberts et al. 2004b; Roberts et al. 2003) (see Table 2). By IHC, cells with a strong AIF1 staining were detected in correlation with inflammation, after SIV infection (Figure 5A, D and G). AIF1-positive microglia cells were also identified in uninfected monkeys at late age (>19 years old) (Figure 5J).
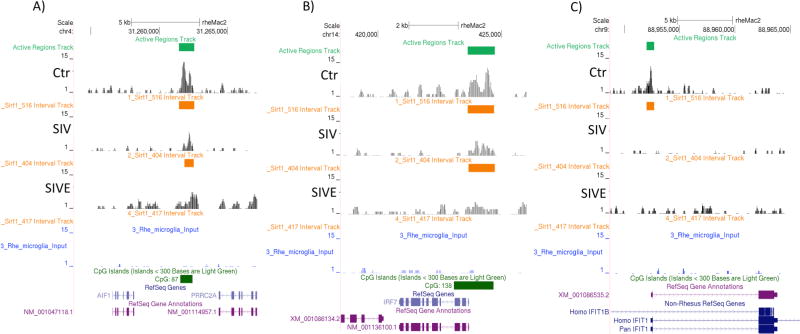
Figure 4. Sirt-1 regulation of gene promoters and expression of Sirt-1 regulated genes in the brain.
(A, B, C) A ChIP reaction was performed on microglia chromatin, with anti-Sirt-1 Ab (Millipore). ChIP DNA was processed into an Illumina ChIP-Seq library and sequenced to generate >2 million reads, aligned to the M.mulatta annotation (MacaM/ Oct 2014). We obtained 6.2 million unique aligns (removed duplicates). A signal map of fragment densities along the genome was visualized in Integrated Genome Browser (IGB). MACS peak finding was used to identify significant peaks. Control data was derived from 5.1 million (positive control) and 5.8 million (negative control) alignments. With default settings, 2141 Sirt-1 meaningful genome-wide peaks were found. (A, B, C) Peaks show binding of Sirt-1 onto annotated inflammation-relevant gene sequences. Sirt-1 binding onto (A) AIF1, (B) IRF7, and (C) IFIT1, in controls, SIV and SIVE pooled cases (n=6/group).
Figure 5. Detection and distribution of inflammatory molecules coded by genes that lost Sirt-1 binding in the brain of SIV-infected and aged macaques.
IHC was used for confirming the expression of (A, D, G, J) AIF1, (B, E, H, K) IRF7 and (C, F, I, L) IFIT1, in representative frontal lobe sections from (A, B, C) Controls, (D, E, F) SIV, (G, H, I) SIVE and from (J, K, L) a representative old macaque’s brain.
The IRF7 sequence (NM_001136100.1) in chromosome 14, was characterized by strong Sirt-1 binding peaks to the promoter region in control animals (Figure 4B), but these were decreased by 50% after infection both in animals with and without encephalitis, suggesting this as a putative gene silenced by Sirt-1 in healthy animals and un-repressed following infection. Indeed, the expression of IRF 7 was increased after infection both in SIV and SIVE (Table 2). IRF7 is an important marker of inflammation in the brain, as it has been involved in the transcriptional activation of several virus-inducible genes, especially IFN alpha(Honda et al. 2005b). In the context of HIV, IRF7 is at the basis of the modulation of Interferon-induced genes(Kim et al. 2013). In addition, overexpression of IRF7 has been shown to be associated to higher HIV replication(Sirois et al. 2011). In the macaque model of neuroAIDS we have confirmed changes of IRF7 levels in the brain tissue of SIV-infected macaques both by qPCR, and by IHC. Positive IRF7 staining was associated to the surface and processes of cells with morphology of microglia, but also of neurons. After infection, IRF7 enrichment was detectable in these cells, and also in association with the presence of inflammatory infiltrate (Figure 5B, E, and H). IRF7-positive cells were also detected in uninfected old macaques’ brains, at a higher frequency than in young controls (Figure 5K), but in cells with morphology of neurons.
The next gene promoter with a strong Sirt-1 binding motif in controls was IFIT1 (M_001068535.2) (Figure 4C). This molecule is an important pattern recognition receptor that can sense viral genomic 5’-triphosphated RNA, and is a non-specific anti-viral factor triggered by IFNs(Yan and Chen 2012). In the SIV/macaque model, we found IFIT upregulation in microglia of SIVE (Table 2), which was confirmed by IHC (Figure 5). IFIT1-positive cells were more frequent after infection (Figures 5C and 5F), but highly upregulated in SIV, in cells localized in the perivascular domain (Figure 5I). This finding is contrasting with other findings in the literature, showing that IFIT1 is upregulated in HIV+ elite controllers(Krishnan et al. 2014), or in cancer patients, where high levels of IFIT1 correlate with improvement following treatment(Danish et al. 2013). On the other hand, it has been shown that a high expression of IFIT1 in areas of the brain is a marker of aberrant IFN response in aging(Baruch et al. 2014). In fact, cells that were positive to IFIT1 were identified in old, uninfected macaques (Figure 5L), both perivascularly, and in the parenchyma. Importantly, IFIT1 has been identified as a key enhancer of innate immune function(McDermott et al. 2012). IRF1-positive staining was very pale, and we did not detect changes in the staining pattern in SIV or SIVE, compared to Controls. The transcriptional upregulation of molecules putatively regulated by Sirt-1 was confirmed by qRT-PCR (Figure 6).
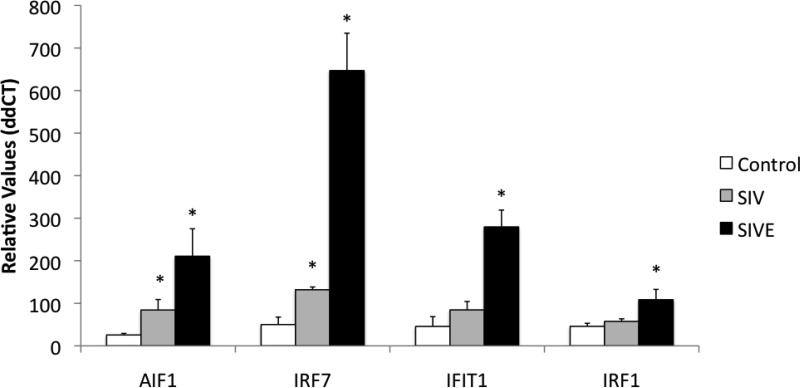
Figure 6. Transcription of putative Sirt-1 regulated genes in microglia from SIV-infected macaques.
Isolated microglia cells from controls and SIV-infected macaques were used to examine the transcriptional expression of AIF1, IRF7, IFIT1 and IRF1, using SyBr Green qRT-PCR. Values were normalized againt the expression of GAPDH. *p<0.05 compared to controls.
The Sirt1-mediated control of those inflammatory targets was examined in non-infectious models, using both THP1 myeloid cell lines stimulated with CpG Oligo-deoxynucleotides (CpG ODN), a TLR9 agonist with pro-inflammatory effects, and primary human monocytes stimulated with the HIV Tat peptide, which has a reported effect on inflammatory gene transcription(Carvallo et al. 2017). These in vitro models verified the consistency of the ability of Sirt-1 to modulate the transcriptional expression of inflammatory markers prioritized in this study.
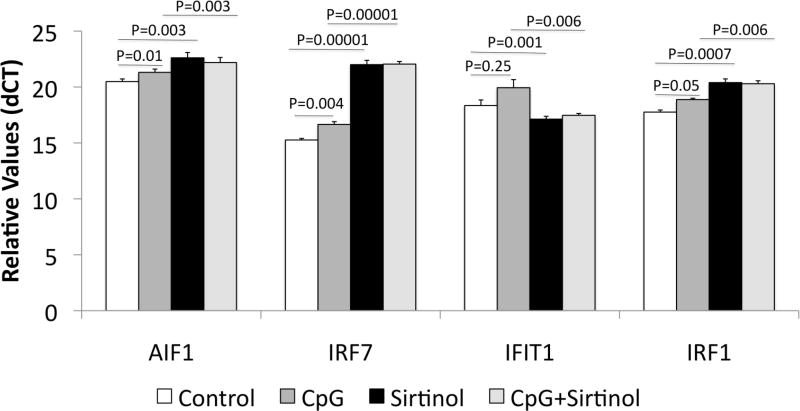
Human THP1 cells are an innate immune myeloid cell line that can be induced to express AIF1, IRF7, and IFIT1, as well as other genes that had Sirt1 binding, such as IRF1, with a 24-hr incubation with the TLR9 agonist CpG ODN. The incubation of THP1 cells with a Sirt-1 blocker, Sirtinol, significantly enhanced the transcription of AIF1, IRF7 and IRF1 (Figure 7), but a decrease in the expression of IFIT1. The increased transcription of AIF1, IRF7 and IRF1 by Sirtinol were above the induction caused by CpG positive control. Moreover, Sirtinol further increased the CpG-induced stimulation of these genes. These findings inform of a role of Sirt-1 as a silencer and modulator of innate immune inflammatory genes, and a potential role for TLR signaling in the induction of changes in Sirt-1 activity.
Figure 7. Sirt-1 modulates transcription of inflammatory genes in macrophages.
Cells of the THP1 macrophage cell line were stimulated with 6ug/ml of CpG ODN, and/or the 100uM of the Sirt-1 inhibitor Sirtinol for 24 hrs. RNA was extracted from the cell pellets for determination of AIF1, IRF7, IFIT1 and IRF1 using SyBr Green qRT-PCR. Values of 3 independent experiments were normalized against the expression of GAPDH. P values represent results from Bonferroni’s posthoc test, which followed ANOVA.
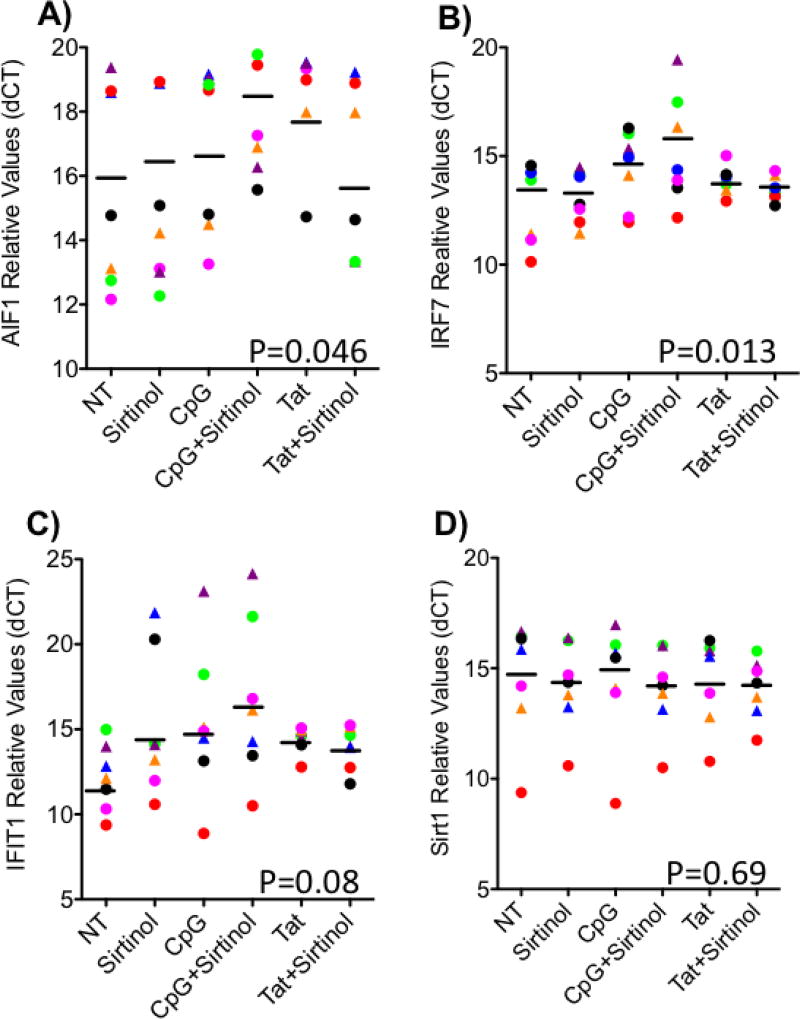
A similar trend was identified in a normal donor primary human monocyte system used to further examine the capacity of Sirt-1 to silence these markers. For that assay, CD14+ monocytes were magnetically isolated from normal donors blood (4 males and 3 females). The cells were stimulated for 24 hrs with CpG ODN, or HIV-1 Tat peptide, with or without Sirtinol, and compared to unstimulated controls, in triplicates (Figure 8). Sirtinol further increased the CpG-induced expression of AIF1 in 4 of the 7 subjects, but decreased in one of the 7 individuals (p=0.046, repeated measures ANOVA; pairing effect p=0.0113, R2=0.301, F=2.586)(Figure 8A). IRF7 (Figure 8B) and IFIT1 (Figure 8C) were both further increased by Sirtinol in CpG-stimulated cells from 4 of the 7 subjects (IRF7 p=0.0131, repeated measures ANOVA; pairing effect p=0.0053, R2=0.36, F=3.495) (IFIT1 p=0.08, repeated measures ANOVA; pairing effect p= 0.0136, R2=0.25, F=2.07). IRF1 was not detectable in human peripheral cells. Tat had no significant effect in the expression of most genes, in the majority of the tested individuals, regardless of Sirtinol. The treatments did not affect the transcription of Sirt-1 (Figure 8D). However, the data suggests that TLR signaling may be important to modify Sirt1 activity in the active infection.
Figure 8. Sirt-1 affects transcription of induced inflammatory genes in human peripheral blood monocytes.
Blood monocytes isolated from normal peripheral blood were stimulated with 6ug/ml of CpG ODN, or HIV-1 Tat peptide (10ng/ml), and/or the 100uM of the Sirt-1 inhibitor Sirtinol for 24 hrs. RNA was extracted from the cell pellets for determination of (A) AIF1, (B) IRF7, (C) IFIT1 and (D) Sirt1 using SyBr Green qRT-PCR. Values of 6 independent experiments, each one performed in triplicate, were normalized against the expression of GAPDH. Individual subject sample behaviors can be observed, as different donors have been displayed with different colors. Circles are males and triangles are females. P values represent results from repeated measures ANOVA.
Discussion
We identified dynamic changes of Sirt-1 binding to in-promoter chromatin regions that correspond to genes involved in inflammation after HIV/SIV infection. It is interesting that the number of Sirt-1- bound genes was increased with SIV infection in preparations where the large majority of the cells (>85%) corresponds to microglia, in animals with mild or no signs of inflammation, in spite of the transcriptional decrease of Sirt-1. This suggests that a boost on Sirt-1 activity may not depend on transcription or on neo-production of that molecule, but perhaps on the levels of other molecules that partner with Sirt-1 to aid on its activity, such as methyltransferases. This possibility remains to be examined. It is also interesting that there was a drastic shuffling on the Sirt-1 binding targets caused by the infection, and that the collection of Sirt-1 targets is largely different in encephalitis. In vitro, the HIV Tat peptide, which is known for its ability to stimulate transcription, was not able to induce a similar effect on innate immune cell systems, while and agent that stimulates TLR9 signaling did. It is known that one of the factors in the recruitment of Sirt-1 is DNA damage(Wang et al. 2006), which is certainly a factor in non-neuronal cells of the brain upon SIV and HIV infection(Wiley et al. 2000). However, this adds a random element to the loss of Sirt-1 silencer activity to favor its repositioning aiming at repair, with the production of a range of outcomes, beneficial, but also potentially detrimental(Song and Surh 2012).
Following the identification of genome-wide targets of Sirt-1, we have generated a list of Sirt-1-controlled molecules with the potential of affecting microglia and myeloid cells towards inflammation in the context of SIV infection, and in a way that resembles the aging process(Franceschi 2007; Franceschi et al. 2007). The Sirt-1 regulated candidate genes were prioritized based on their relevance in the development of an inflammatory process, and may not be simply part of an adaptive microglial response to SIV, but because they become chronically upregulated, there may result in adverse consequences for neurons and/or synapses. These genes were AIF-1, IRF7, IFIT1, and IRF1. We confirmed the increase of AIF1, IRF7 and IFIT1 in the tissue of SIV-infected macaques, and also in the brain of old, uninfected macaques, suggesting a common ground. Interestingly, we did not observe Sirt-1 binding to any of these genes in pre-frontal cortical samples from the aging macaques (data not shown).
AIF1 (also known as Iba-1) is a highly conserved EF-handed, putative calcium binding peptides, which is a sensitive marker of microglial activation, and helps define functionally activated microglia(Postler et al. 2000; Schwab et al. 2001). AIF1-upregulation is part of the early microglial response leading to activation and proliferation, essential for the acute as well as chronic response to CNS injury(Schwab et al. 2001). Sirt-1 binding was decreased by SIV, in correlation with transcription and with the augmentation of the protein on microglial cell surface.
Similarly, IRF7 is involved in the transcriptional activation of various virus-inducible genes, especially IFN alpha(Honda et al. 2005b). IRF7 becomes activated by innate receptor signaling, such as TLR3, 7, 9 and RIG-I(Ning et al. 2011), resulting in translocation to the nucleus and induction of type I IFN(Seth et al. 2006), which leads to further induction of IRF7, so creating a feed-forward loop to amplify production of type I IFN. Mice lacking IRF7 are deficient in type I IFN responses and consequently lack innate responsiveness to viruses(Honda et al. 2005a; Honda et al. 2005b; Honda et al. 2005c). Sirt-1 is bound to the IRF7 promoter in microglia and the binding is highly decreased in infected macaques. This correlates with the upregulation of IRF7 in infected mice, both transcriptionally and at the protein level in the brain, associated to cells with macrophages and activated microglia morphology. In SIV-infected animals, the IRF7-expressing cells are both diffuse and in perivascular clusters, which are more frequent in animals with encephalitis. There is the possibility that infiltrating cells that do not have Sirt-1 binding to the IRF7 promoter contribute to the observed reduction associated to the infection. However, the decrease in Sirt-1 binding to the IRF7 promoter due to SIV was around 40%, which is significantly higher than the contribution of infiltrating cells to the brain-derived cell isolate, in conditions of mild inflammation. This suggests that there is loss of Sirt-1 binding from the IRF7 promoter. A comparison of the Sirt-1 binding pattern between infiltrating, perivascular and parenchyma populations could be instrumental to determine whether Sirt-1 regulation is more inherent to resident cells. In the brain, the expression of IRF7 in injury has been predominantly associated to innate immune and glial cells(Khorooshi and Owens 2010), but in aged animals, the IRF7-staining was also associated to cells with neuronal morphology, suggesting that there are cell specific differences in the expression of this gene, regarding the Sirt-1-mediated control of inflammatory gene expression or other cell-specific regulatory factors that were not explored here.
Another gene that raised our interest was IFIT1, which is a type I IFN-responsive recognition, highly conserved pattern receptor, with an ability to sequester viral particles in infected cells(Pichlmair et al. 2011), and initiate transcription(Hui et al. 2003), being therefore an important component in infection. In the other hand, it has been described as a predictor of cancer(Weichselbaum et al. 2008). Interestingly, IFIT1, together with IRF7 and IFNb, has been suggested to be upregulated by aging in correlation with negative brain performance(Baruch et al. 2014). The binding of Sirt-1 onto the IFIT1 promoter was completely lost in SIV infection, and was also not observed in samples from aged macaques’ frontal cortex. As expected, we observed the transcriptional upregulation and the presence of IFIT1-positive cells, with myelomonocytic morphology, both in SIV and in the brain of aged macaques.
We have confirmed the regulation of IRF7 and of AIF1 by Sirt-1 as a silencer in vitro, using inhibition of Sirt-1 deacetylase activity by Sirtinol, in human macrophages. The upregulation of IRF7 and AIF1 in the presence of Sirt-1 inhibitor was higher than upon stimulation with the positive control CpG ODN, which suggests that the inhibition of Sirt-1 unrepresses genes of the inflammatory response to reach a maximum transcription threshold. On the other hand, IRF1 and IFIT1 were not affected by the blockage of Sirt-1 activity, suggesting that other control layers may be operating.
The question that remains to be examined relates to the specificity of Sirt-1 to histone substrates, given that the randomness of the Sirt-1 binding and regulation does not seem to be as high as expected. While NAD-dependent, Sirt-1 activation is susceptible to changes in the energy balance and in the redox environment(Circu and Aw 2010; Imai et al. 2000a; Imai et al. 2000b). In addition, DNA damage is a potent recruiter of Sirt-1 binding. These could explain the increased number of genes experiencing Sirt-1 binding in microglia from infected macaques, in spite of the transcriptional decrease. The Sirt-1 binding towards target promoters seems to be restricted to histones that bear lysine residues at specific positions, such as H1 (K26), H3 (K9, K14), and H4 (K16)(Imai et al. 2000a; Imai et al. 2000b; Vaquero et al. 2004). Conversely, H3K9 and H4K16 acetylation is increased in Sirt−/− MEF cells(Wang et al. 2008). On the other hand, loss of H4K16 acetylation is commonly observed in human cancers(Fraga et al. 2005), leading to the question whether this is due to Sirt-1 activity or failure on the acetylation process that provides substrate for Sirt-1 silencing activity. In yeast, increase in H4K16 acetylation is concomitant to a decrease in Sir2 (SIRT1) protein, causing reduced transcriptional silencing(Dang et al. 2009). A similar mechanism may be associated to the severe encephalitis associated to disease progression. Other Sirt-1 substrate is the H3K56 acetylation, mediated by p300, and induced by DNA damage(Vempati and Haldar 2012; Vempati et al. 2010), particularly at the Bclaf1 promoter, thereby reducing T cell activation and promoting immune tolerance(Kong et al. 2011; Vempati et al. 2010; Zhang et al. 2009). Thus, H3K56 deacetylation may play a role in immune function.
Attempts to identify Sirt-1 substrate specificity have been made in vitro without success (Blander et al. 2005). However, stereospecific catalytic preference for L-AcK versus the D-isomer has been demonstrated(Jamonnak et al. 2010). In fact, crystal structures of several sirtuins have revealed a highly conserved catalytic domain(Sanders et al. 2010), where NAD+ and acetyl-Lys enter from a cleft between a Rossmann fold domain and a smaller, structurally more variable Zn2+-binding domain. The peptide binding grooves are architecturally conserved but exhibit isoform-specific features such as charge distribution and shape details. Although certain sirtuins appear to lack sequence specificity, most studies find sirtuin selectivity for substrates with certain residue types around the deacetylation site (Cosgrove et al. 2006; Garske and Denu 2006; Smith et al. 2011). Sirt-1, for instance, has a preference for polar, mainly positively charged residues surrounding the deacetylation site, with non-charged residues inserted at positions +2, +3 and −2, correlated with the hydrophilic Sirt1 peptide binding groove, negatively charged on the bottom(Rauh et al. 2013). Thus, there is some specificity to the choice of silenced target genes, especially in uninfected controls, but other layers of biochemical histone modifications in particular positions, acetylations and methylations, which precede the regulation by this molecule, may be enhanced in the infected brain, particularly in cells that become activated such as microglia, and explaining a spectrum of outcomes. Curiously, a relative consistency of targets is observed, likely due to the consistency of the cell activation process and receptor-triggered pathways that precede transcription.
A role for Sirt-1 in the brain has been proposed, in controlling blood brain barrier proteins and HIV infection in pericytes (Castro et al. 2016). On the other hand, a potential ability of microglial Sirt-1 to control the onset of cognitive decline by affecting microglia and brain inflammatory components has been observed (Chen et al. 2005; Cho et al. 2015). Our results endorse a role for Sirt-1 in brain homeostasis, and offer a mechanism to explain HIV neuropathogenesis. It is important to acknowledge that cell preparations such as the ones used in this study, contain a small (<10%) number of cells of subtypes other than microglia, such as CD8 T cells and Natural killer cells, as reported by us in previous studies(Marcondes et al. 2001; Marcondes et al. 2010; Marcondes et al. 2015; Marcondes et al. 2006; Marcondes et al. 2003; Marcondes et al. 2008; Marcondes et al. 2013). However, changes in these minor populations, although meaningful, are also only detectable in the isolated subsets. Conversely, the changes that we have observed in this study by enriching the microglial/myeloid cell pool were not observed in whole brain tissue preparations (not shown). This suggests that subset-specific changes can be missed by whole tissue approaches. This also points to the necessity of a future examination of Sirt-1 binding patterns in minor immune cell subpopulations from the brain. It is also important to acknowledge that animals with SIVE have a higher viral load and faster disease progression compared to SIV animals(Bortell et al. 2015). Whether differences in the epigenetic control of gene expression by Sirt-1 and its associated factors, in other pathways could contribute to this effect, remains to be established. Our analysis suggests that innate immune activity, viral control and disease progression are tightly linked to aging.
Here, we have identified changes in Sirt-1 binding to chromatin of brain microglia and myeloid cells caused by SIV infection that can explain important transcriptional changes in the brain of infected subjects, including the development of inflammation. We found Sirt-1-regulated genes that reportedly participate in inflammatory processes, as negative regulators of viral entry, but that can be involved in neuropathogenesis in the chronically infected brain. The upregulation of these same inflammatory genes in the brain of aging macaques suggests that there are common mechanisms between the inflammatory process during infection and aging, with a potential link established both by the recruitment of Sirt-1 to other sites upon chromatin remodeling and DNA damage, and loss of the ability of Sirt-1 to repress key targets. The molecules described here to be under Sirt-1 control may be confirmed as markers of CNS aging induced by the infection, in macaques and in humans. The results suggest that the molecules regulated by Sirt-1, are critical in the development of an inflammatory environment in the CNS and may be important markers of the CNS aging-like phenotypes induced by SIV.
The changes in Sirt-1 activity, and the meaning of the dynamic rearrangement of Sirt-1 binding to specific gene promoter histone targets, suggests a mechanism for controlling phenotype upon microglia/macrophage activation, through a mechanism that is similar in aging. This study offers evidence of a role for Sirt-1, a molecule involved in aging, specifically in microglia/myeloid-rich cell populations and CNS inflammation following HIV infection of the brain, triggering changes that resemble and may be common to the ones occurring in the aging brain. The implications of these findings for the increasingly aging HIV+ population remain to be examined.
Methods
Monkeys and SIV Infection
All experiments had the approval from the Institutional Animal Care and Use Committee of The Scripps Research Institute and University of Nebraska, and followed National Institutes of Health guidelines. SIV-negative, simian retrovirus type D-negative, and herpes B virus-free rhesus macaques with and average of four years old, were purchased from Valley Biosystems (West Sacramento, CA). From the animals infected with a cell-free SIV stock derived from SIVmac251(Lane et al. 1995; Watry et al. 1995), four exhibited a regular course of infection (SIV), and were sacrificed between 180 and 200 days after infection. The other four exhibited signs of progression to AIDS and were sacrificed between 56 and 82 days after infection. After terminal anesthesia, animals were intracardially perfused with sterile PBS containing 1 U/ml heparin. Brain tissue samples were taken for cell isolation, virus quantification, and formalin fixation for histology. Brain samples from six older, uninfected macaques, with age between 18 and 23 years old, were kindly donated by the NIH National Institute of Aging Non Human Primate Tissue Repository, at the Wisconsin National Primate Center.
SIV Viral load in the brain
Brain SIV RNA was calculated in frontal cortical brain fragments by using a quantitative branched DNA signal amplification assay, performed by Siemens Clinical Laboratory (Emeryville, CA).
Microglial Cell isolation
The brain tissue was removed at necropsy after intravascular perfusion. For isolation of cells from the brain, the brain was carefully freed of meninges. Brain immune cells were isolated from all representative areas of the brain, by enzymatic digestion of minced tissue, followed by Percoll (Sigma-Aldrich) gradient, as described previously(Marcondes et al. 2001). The isolated cells were quantified in a Z2 Coulter Counter (Beckman Coulter, Brea, CA), and further characterized by flow cytometry, using anti-CD11b (clone M1/70, BD Bioscience, San Diego, CA), anti-CD45LCA (clone 2B11, BD Biosciences, San Diego, CA), anti- human CD14 (clone M5E2, BioLegend, San Diego, CA), anti- human CD16 (clone 3G8, BD Bioscience) anti-monkey CD3-biotin (clone FN-18, Invitrogen Biosource, Carlsbad, CA) followed by streptavidin-PerCP or -APC (BD Bioscience), and anti-human CD8-PE, -FITC, or -PeCy5 (clone DK25, Dako, Carpinteria, CA), or isotype controls (BD Bioscience). Stained cells were acquired by a FACSCalibur (BD Bioscience, San Jose, CA) flow cytometer, and analyzed in FlowJo 6.2.1 software (Tree Star Inc., Ashland, OR), as previously described( ; Marcondes et al. 2001).
Samples, Labeling and Gene array
Gene analysis was performed on cryopreserved microglial cells, by custom Miltenyi Biotec Array Services. All samples were individually performed in duplicate. RNA was isolated from all macaques using standard RNA extraction protocols (NucleoSpin RNA II, Macherey-Nagel). Quality of the samples was checked via Agilent 2100 Bioanalyzer platform (Agilent technologies). A RNA integrity number (RIN) was calculated by a proprietary algorithm that takes several QC parameters into account, such as 28S/18S RNA peak area ratios and unexpected peaks in the 5S RNA region, and RIN number of 10 indicates high quality, and 1 low quality. A RIN >6 is of sufficient quality for gene expression profiling experiments (Fleige and Pfaffl 2006; Fleige et al. 2006). All samples, except for 1 (animal 492 – Normal control) showed values above 6. That animal was excluded from the analysis, leaving the healthy control group with an n=4. For the linear T7-based amplification step, 100ng of each total RNA sample was used. To produce Cy3-labeled cRNA, the RNA samples were amplified and labeled using the Agilent Low Input Quick Amp Labeling kit (Agilent technologies) following the manufacturer’s protocol. Yields of cRNA and the dye incorporation rate were measured with the ND-1000 spectrophotometer (Nanodrop Technologies). The hybridization procedure was performed according to Agilent 60-mer 0ligo microarray processing protocol using the Agilent gene expression hybridization kit (Agilent Technologies). Briefly, 1.65ug Cy3-labeled fragmented cRNA in hybridization buffer was hybridized overnight (17 hours, 65oC) to Agilent Whole Rhesus monkey Genome oligo microarrays 4×44K (one-color) using Agilent’s recommended hybridization chamber and oven. Finally, the microarrays were washed once with the Agilent Gene expression wash buffer 1 for 1 min at room temperature, followed by a second wash with pre-heated Agilent Gene expression wash buffer 2 (37oC) for 1 min. The last washing step was performed with acetonitrile. Fluorescent signals of the hybridized microarrays were detected using Agilent’s microarray Scanner System (Agilent Technologies). The Agilent Feature extraction software (FES) was used to read out and process the microarray image files, determining feature intensities (including background subtraction), rejecting outliers and calculating statistical confidences. For determination of differential gene expression FES derived output data files were further analyzed using Rosetta Resolver gene expression data analysis system (Rosetta Biosoftware), for comparing two single intensity profiles in a ratio experiment.
Systems Analysis
Pathway assignments and functional annotations were analyzed using DAVID Bioinformatics Database(Huang da et al. 2009). To complete the bioinformatics analysis, two knowledge base resources were queried: the Ingenuity Knowledge Base (Calvano et al. 2005) and an interaction repository, which is based on cpath (Cerami et al. 2006; Cerami et al. 2011; Cline et al. 2007) and includes interactions that have been curated by GeneGo (http://www.genego.com), the Kyoto Encyclopedia of Genes and Genomes (KEGG - http://www.genome.jp/kegg/), and Ingenuity. Benjamini False Discovery Rate (FDR) adjusted values <0.01 and p values < 0.05 (provided by DAVID) were utilized as conservative filters for identification of true values. Cluster analysis and networks were obtained and visualized using Cytoscape (Shannon et al. 2003).
Quantitative RT-PCR
First Strand kit (Qiagen) was used for cDNA synthesis. Primers were purchased from Qiagen (Valencia, CA) for detection of relative levels using SyBrGreen/ROX mix (Qiagen) in an ABI HT7900 machine. Data was analyzed with Sequence Detection System software and normalized with GAPDH and 18S expression.
ChIP-Seq
A ChIP reaction was carried out with 30ug of microglia chromatin and anti-Sirt-1 antibody (Millipore). The ChIP DNA was processed into an Illumina ChIP-Seq library and sequenced to generate >2 million reads, which were aligned to the M.mulatta genome annotation (MacaM/Oct 2014 assembly) and 6.2 million unique aligns (removed duplicates) were obtained. A signal map showing fragment densities along the genome was visualized in the Integrated Genome Browser (IGB) and MACS peak finding was used to identify peaks. Control data was derived from 5.1 million (positive control) and 5.8 million (negative control) alignments. With default settings, 2141 Sirt-1 meaningful peaks genome-wide were identified. Raw data and metadata are available at GEO GSE95793.
Immunohistochemistry (IHC)
Formalin-fixed, paraffin embedded brain tissue was sectioned and the detection of molecular markers was performed using antibodies against Iba-1 (AIF1 - WAKO, Richmond, VA), CD163 (Vector Labs, Burlingame, CA), Mac3 (BioLegend, San Diego, CA) and glial fibrillary acidic protein (GFAP, Dako, Carpinteria, CA), as well as IRF1 (Novus Biologicals, Littleton, CO), IRF7 (Novus), and IFIT1 (Novus), using standard procedures(Bortell et al. 2015). The secondary antibodies were biotinilated (Vector Labs, Burlingame, CA), and were followed by Streptavidin-HRP, and were developed using NovaRed (Vector Labs).
THP1 cell culture and in vitro treatments
THP-1 cells, a pro-monocytic cell line, were cultured in RPMI 1640 (Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum (Gemini Bio-Products, West Sacramento, CA), 10 mM Hepes, 0.1 mM MEM non-essential amino acids, 1 mM sodium pyruvate, and 100 nM penicillin/ streptomycin (Life Technologies). The cells were maintained at a low density (2.5 × 105/ml) with media being refreshed every three days and were maintained at 5% CO2, 37°C. The basal cells were stimulated with 6ug/ml of CpG ODN, and/or the 100uM of the Sirt-1 inhibitor Sirtinol for 24 hrs. RNA was extracted from the cell pellets for determination of AIF1, IFIT1, IRF1 and IRF7, using primers purchased from Qiagen, and using SyBr Green qRT-PCR (Qiagen). Values were normalized againt the expression of GAPDH.
Human blood samples
Human blood from 7 adult individuals, 4 males and 3 females, was obtained through the Scripps Research Institute Normal Blood Donor Service, under the IRB number 17-6927 approved to MCGM, and under informed consent. Fifty ml of EDTA-treated blood from each individual donor was centrifuged at 1500 rpm at 4oC for separation of plasma and isolation of leukocyte buffy coats. Leukocytes were resuspended in complete RPMI and layered over the same volume of Histopaque 1077 (Sigma Aldrich), then centrifuged at 2000 rpm for 20 minutes at room temperature. The leukocyte ring was washed twice at 1200 rpm, then twice more at 900 rpm for platelet separation. The cell pellet was incubated with CD14 magnetic beads (Myltenyi Biotec, Auburn, CA, USA), and separated using magnetic columns, following manufacturer’s protocols. The cell purity was confirmed using PE-labelled anti- human CD11b (clone HI11b, Biosource, San Diego, CA). The cells were plated at 2.5×106/ml cell density and let rest overnight, before stimulation. The stimulation was performed with 6ug/ml of CpG ODN, or with 1, 10 and 30 ng/ml of HIV-1 IIIB Tat recombinant peptide (10ng/ml is shown in Figure 8; Catalog 2222, NIH AIDS Reagent Program), and/or the 100uM of the Sirt-1 inhibitor Sirtinol for 24 hrs. RNA was extracted from the cell pellets for determination of AIF1, IFIT1, IRF1, IRF7, as well as Sirt1, in triplicates, using primers purchased from Qiagen, and using SyBr Green qRT-PCR (Qiagen). Values were normalized against the expression of GAPDH.
Statistical analysis
Statistical analysis was performed using Prism software (GraphPad, La Jolla, CA). One-way ANOVA followed by Bonferroni’s multiple comparisons were performed as applicable. Human samples were analyzed using repeated measures ANOVA, followed by Tukey test.
Acknowledgments
The authors would like to thank Steven A. Totusek (University of Nebraska Medical Center, Omaha, NE) for the help on depositing the ChIP data into the Gene Expression Omnibus (GEO) system. We thank Dr. Douglas Galasko (Department of Neurosciences, University of California, San Diego - UCSD, and Director of the UCSD Shiley-Marcos Alzheimer's Disease Research Center (ADRC), for important discussions and critical guidance throughout the study. We want to deeply thank the NIH National Institute of Aging Non Human Primate Tissue Repository, at the Wisconsin National Primate Center, for the amazing resource it provides. We also want to thank Dr. Lindsay Whitton for the use of his microscope. This work has been funded by NIH/NIDA grant R01DA036164 and by NIH/NIA grant R21AG054240 to MCGM.
Footnotes
Authors’ contributions
NB prepared cells for ChIP procedures, analyzed the data, performed most PCRs, and helped write the manuscript.
LB performed all the in vitro assays, the PCRs related to those experiments, and helped write the methods session of the manuscript.
JAN performed cell fixations and participated in discussions.
BM provided paraffin sections from SIV-infected macaques and controls.
HSF was present in all the necropsies, provided samples from macaques, participated in all discussions, and proofread the manuscript.
MCGM designed the study, was present in necropsies, performed cell and tissue isolation, analyzed data, performed systems analysis, performed PCRs, obtained funding and wrote the manuscript.
The authors declare that there are no competing financial interests.
References
- Baruch K, et al. Aging. Aging-induced type I interferon response at the choroid plexus negatively affects brain function. Science. 2014;346:89–93. doi: 10.1126/science.1252945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander G, Olejnik J, Krzymanska-Olejnik E, McDonagh T, Haigis M, Yaffe MB, Guarente L. SIRT1 shows no substrate specificity in vitro. The Journal of biological chemistry. 2005;280:9780–9785. doi: 10.1074/jbc.M414080200. [DOI] [PubMed] [Google Scholar]
- Boily G, He XH, Pearce B, Jardine K, McBurney MW. SirT1-null mice develop tumors at normal rates but are poorly protected by resveratrol. Oncogene. 2009;28:2882–2893. doi: 10.1038/onc.2009.147. [DOI] [PubMed] [Google Scholar]
- Boily G, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PloS one. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortell N, Morsey B, Basova L, Fox HS, Marcondes MC. Phenotypic changes in the brain of SIV-infected macaques exposed to methamphetamine parallel macrophage activation patterns induced by the common gamma-chain cytokine system. Frontiers in microbiology. 2015;6:900. doi: 10.3389/fmicb.2015.00900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch-Presegue L, Vaquero A. The dual role of sirtuins in cancer. Genes & cancer. 2011;2:648–662. doi: 10.1177/1947601911417862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch-Presegue L, Vaquero A. Sirtuins in stress response: guardians of the genome. Oncogene. 2013 doi: 10.1038/onc.2013.344. [DOI] [PubMed] [Google Scholar]
- Burudi EM, Marcondes MC, Watry DD, Zandonatti M, Taffe MA, Fox HS. Regulation of indoleamine 2,3-dioxygenase expression in simian immunodeficiency virus-infected monkey brains. J Virol. 2002;76:12233–12241. doi: 10.1128/JVI.76.23.12233-12241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvano SE, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- Carvallo L, Lopez L, Fajardo JE, Jaureguiberry-Bravo M, Fiser A, Berman JW. HIV-Tat regulates macrophage gene expression in the context of neuroAIDS. PloS one. 2017;12:e0179882. doi: 10.1371/journal.pone.0179882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro V, et al. Occludin controls HIV transcription in brain pericytes via regulation of SIRT-1 activation. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2016;30:1234–1246. doi: 10.1096/fj.15-277673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami EG, Bader GD, Gross BE, Sander C. cPath: open source software for collecting, storing, and querying biological pathways. BMC bioinformatics. 2006;7:497. doi: 10.1186/1471-2105-7-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami EG, et al. Pathway Commons, a web resource for biological pathway data. Nucleic acids research. 2011;39:D685–690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri AD, Yelamanchili SV, Marcondes MC, Fox HS. Up-regulation of microRNA-142 in simian immunodeficiency virus encephalitis leads to repression of sirtuin1. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27:3720–3729. doi: 10.1096/fj.13-232678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, et al. SIRT1 protects against microglia-dependent amyloid-beta toxicity through inhibiting NF-kappaB signaling. The Journal of biological chemistry. 2005;280:40364–40374. doi: 10.1074/jbc.M509329200. [DOI] [PubMed] [Google Scholar]
- Cho SH, et al. SIRT1 deficiency in microglia contributes to cognitive decline in aging and neurodegeneration via epigenetic regulation of IL-1beta. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2015;35:807–818. doi: 10.1523/JNEUROSCI.2939-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Circu ML, Aw TY. Reactive oxygen species, cellular redox systems, and apoptosis. Free radical biology & medicine. 2010;48:749–762. doi: 10.1016/j.freeradbiomed.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, et al. Integration of biological networks and gene expression data using Cytoscape. Nature protocols. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove MS, Bever K, Avalos JL, Muhammad S, Zhang X, Wolberger C. The structural basis of sirtuin substrate affinity. Biochemistry. 2006;45:7511–7521. doi: 10.1021/bi0526332. [DOI] [PubMed] [Google Scholar]
- Dang W, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danish HH, Goyal S, Taunk NK, Wu H, Moran MS, Haffty BG. Interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) as a prognostic marker for local control in T1–2 N0 breast cancer treated with breast-conserving surgery and radiation therapy (BCS + RT) The breast journal. 2013;19:231–239. doi: 10.1111/tbj.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Molecular aspects of medicine. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Fleige S, Walf V, Huch S, Prgomet C, Sehm J, Pfaffl MW. Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnology letters. 2006;28:1601–1613. doi: 10.1007/s10529-006-9127-2. [DOI] [PubMed] [Google Scholar]
- Fraga MF, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nature genetics. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- Franceschi C. Inflammaging as a major characteristic of old people: can it be prevented or cured? Nutrition reviews. 2007;65:S173–176. doi: 10.1111/j.1753-4887.2007.tb00358.x. [DOI] [PubMed] [Google Scholar]
- Franceschi C, et al. Inflammaging and anti-inflammaging: a systemic perspective on aging and longevity emerged from studies in humans. Mechanisms of ageing and development. 2007;128:92–105. doi: 10.1016/j.mad.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Frankel S, Rogina B. Sir2, caloric restriction and aging. Pathologie-biologie. 2006;54:55–57. doi: 10.1016/j.patbio.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Gabay O, et al. Sirt1-deficient mice exhibit an altered cartilage phenotype Joint, bone, spine : revue du rhumatisme. 2013;80:613–620. doi: 10.1016/j.jbspin.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagarina V, Gabay O, Dvir-Ginzberg M, Lee EJ, Brady JK, Quon MJ, Hall DJ. SirT1 enhances survival of human osteoarthritic chondrocytes by repressing protein tyrosine phosphatase 1B and activating the insulin-like growth factor receptor pathway. Arthritis and rheumatism. 2010;62:1383–1392. doi: 10.1002/art.27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garske AL, Denu JM. SIRT1 top 40 hits: use of one-bead, one-compound acetyl-peptide libraries and quantum dots to probe deacetylase specificity. Biochemistry. 2006;45:94–101. doi: 10.1021/bi052015l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersten M, Alirezaei M, Marcondes MC, Flynn C, Ravasi T, Ideker T, Fox HS. An integrated systems analysis implicates EGR1 downregulation in simian immunodeficiency virus encephalitis-induced neural dysfunction. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:12467–12476. doi: 10.1523/JNEUROSCI.3180-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greter M, Lelios I, Croxford AL. Microglia Versus Myeloid Cell Nomenclature during Brain Inflammation. Frontiers in immunology. 2015;6:249. doi: 10.3389/fimmu.2015.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greter M, Merad M. Regulation of microglia development and homeostasis. Glia. 2013;61:121–127. doi: 10.1002/glia.22408. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes & development. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Herranz D, Serrano M. Impact of Sirt1 on mammalian aging. Aging. 2010a;2:315–316. doi: 10.18632/aging.100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz D, Serrano M. SIRT1: recent lessons from mouse models. Nature reviews Cancer. 2010b;10:819–823. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, et al. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 2005a;434:1035–1040. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- Honda K, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005b;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- Honda K, Yanai H, Takaoka A, Taniguchi T. Regulation of the type I IFN induction: a current view. International immunology. 2005c;17:1367–1378. doi: 10.1093/intimm/dxh318. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang DW, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic acids research. 2007;35:W169–175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui DJ, Bhasker CR, Merrick WC, Sen GC. Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2.GTP.Met-tRNAi. The Journal of biological chemistry. 2003;278:39477–39482. doi: 10.1074/jbc.M305038200. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000a;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Imai S, Johnson FB, Marciniak RA, McVey M, Park PU, Guarente L. Sir2: an NAD-dependent histone deacetylase that connects chromatin silencing, metabolism, and aging. Cold Spring Harbor symposia on quantitative biology. 2000b;65:297–302. doi: 10.1101/sqb.2000.65.297. [DOI] [PubMed] [Google Scholar]
- Jamonnak N, Hirsch BM, Pang Y, Zheng W. Substrate specificity of SIRT1-catalyzed lysine Nepsilon-deacetylation reaction probed with the side chain modified Nepsilon-acetyl-lysine analogs. Bioorganic chemistry. 2010;38:17–25. doi: 10.1016/j.bioorg.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & development. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorooshi R, Owens T. Injury-induced type I IFN signaling regulates inflammatory responses in the central nervous system. Journal of immunology. 2010;185:1258–1264. doi: 10.4049/jimmunol.0901753. [DOI] [PubMed] [Google Scholar]
- Kierdorf K, et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nature neuroscience. 2013;16:273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- Kierdorf K, Prinz M. Factors regulating microglia activation. Frontiers in cellular neuroscience. 2013;7:44. doi: 10.3389/fncel.2013.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Kukkonen S, Martinez-Viedma Mdel P, Gupta S, Aldovini A. Tat engagement of p38 MAP kinase and IRF7 pathways leads to activation of interferon-stimulated genes in antigen-presenting cells. Blood. 2013;121:4090–4100. doi: 10.1182/blood-2012-10-461566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolthur-Seetharam U, Teerds K, de Rooij DG, Wendling O, McBurney M, Sassone-Corsi P, Davidson I. The histone deacetylase SIRT1 controls male fertility in mice through regulation of hypothalamic-pituitary gonadotropin signaling. Biology of reproduction. 2009;80:384–391. doi: 10.1095/biolreprod.108.070193. [DOI] [PubMed] [Google Scholar]
- Kong S, Kim SJ, Sandal B, Lee SM, Gao B, Zhang DD, Fang D. The type III histone deacetylase Sirt1 protein suppresses p300-mediated histone H3 lysine 56 acetylation at Bclaf1 promoter to inhibit T cell activation. The Journal of biological chemistry. 2011;286:16967–16975. doi: 10.1074/jbc.M111.218206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan S, et al. Evidence for innate immune system activation in HIV type 1-infected elite controllers. The Journal of infectious diseases. 2014;209:931–939. doi: 10.1093/infdis/jit581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, et al. Age-associated changes in gene expression in human brain and isolated neurons. Neurobiology of aging. 2013;34:1199–1209. doi: 10.1016/j.neurobiolaging.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane TE, Buchmeier MJ, Watry DD, Jakubowski DB, Fox HS. Serial passage of microglial SIV results in selection of homogeneous env quasispecies in the brain. Virology. 1995;212:458–465. doi: 10.1006/viro.1995.1503. [DOI] [PubMed] [Google Scholar]
- Lemieux ME, et al. The Sirt1 deacetylase modulates the insulin-like growth factor signaling pathway in mammals. Mechanisms of ageing and development. 2005;126:1097–1105. doi: 10.1016/j.mad.2005.04.006. [DOI] [PubMed] [Google Scholar]
- London A, Cohen M, Schwartz M. Microglia and monocyte-derived macrophages: functionally distinct populations that act in concert in CNS plasticity and repair. Frontiers in cellular neuroscience. 2013;7:34. doi: 10.3389/fncel.2013.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcondes MC, et al. Highly activated CD8(+) T cells in the brain correlate with early central nervous system dysfunction in simian immunodeficiency virus infection. Journal of immunology. 2001;167:5429–5438. doi: 10.4049/jimmunol.167.9.5429. [DOI] [PubMed] [Google Scholar]
- Marcondes MC, Flynn C, Watry DD, Zandonatti M, Fox HS. Methamphetamine increases brain viral load and activates natural killer cells in simian immunodeficiency virus-infected monkeys. Am J Pathol. 2010;177:355–361. doi: 10.2353/ajpath.2010.090953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcondes MC, Morsey B, Emanuel K, Lamberty BG, Flynn CT, Fox HS. CD8+ T cells maintain suppression of simian immunodeficiency virus in the central nervous system. The Journal of infectious diseases. 2015;211:40–44. doi: 10.1093/infdis/jiu401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcondes MC, Penedo MC, Lanigan C, Hall D, Watry DD, Zandonatti M, Fox HS. Simian immunodeficiency virus-induced CD4+ T cell deficits in cytokine secretion profile are dependent on monkey origin. Viral Immunol. 2006;19:679–689. doi: 10.1089/vim.2006.19.679. [DOI] [PubMed] [Google Scholar]
- Marcondes MC, Phillipson CA, Fox HS. Distinct clonal repertoire of brain CD8+ cells in simian immunodeficiency virus infection. Aids. 2003;17:1605–1611. doi: 10.1097/01.aids.0000072648.21517.c9. [DOI] [PubMed] [Google Scholar]
- Marcondes MC, Sopper S, Sauermann U, Burdo TH, Watry D, Zandonatti M, Fox HS. CD4 deficits and disease course acceleration can be driven by a collapse of the CD8 response in rhesus macaques infected with simian immunodeficiency virus. Aids. 2008;22:1441–1452. doi: 10.1097/QAD.0b013e3283052fb5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcondes MC, Spina C, Bustamante E, Fox H. Increased toll-like receptor signaling pathways characterize CD8+ cells in rapidly progressive SIV infection. BioMed research international. 2013;2013:796014. doi: 10.1155/2013/796014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott JE, Vartanian KB, Mitchell H, Stevens SL, Sanfilippo A, Stenzel-Poore MP. Identification and validation of Ifit1 as an important innate immune bottleneck. PloS one. 2012;7:e36465. doi: 10.1371/journal.pone.0036465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning S, Pagano JS, Barber GN. IRF7: activation, regulation, modification and function. Genes and immunity. 2011;12:399–414. doi: 10.1038/gene.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios JA, Herranz D, De Bonis ML, Velasco S, Serrano M, Blasco MA. SIRT1 contributes to telomere maintenance and augments global homologous recombination. The Journal of cell biology. 2010;191:1299–1313. doi: 10.1083/jcb.201005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, et al. IFIT1 is an antiviral protein that recognizes 5'-triphosphate RNA. Nature immunology. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- Postler E, Rimner A, Beschorner R, Schluesener HJ, Meyermann R. Allograft-inflammatory-factor-1 is upregulated in microglial cells in human cerebral infarctions. Journal of neuroimmunology. 2000;104:85–91. doi: 10.1016/s0165-5728(99)00222-2. [DOI] [PubMed] [Google Scholar]
- Prinz M. Microglia and monocytes: molecularly defined. Acta neuropathologica. 2014;128:317–318. doi: 10.1007/s00401-014-1331-x. [DOI] [PubMed] [Google Scholar]
- Prinz M, Tay TL, Wolf Y, Jung S. Microglia: unique and common features with other tissue macrophages. Acta neuropathologica. 2014;128:319–331. doi: 10.1007/s00401-014-1267-1. [DOI] [PubMed] [Google Scholar]
- Rauh D, et al. An acetylome peptide microarray reveals specificities and deacetylation substrates for all human sirtuin isoforms. Nature communications. 2013;4:2327. doi: 10.1038/ncomms3327. [DOI] [PubMed] [Google Scholar]
- Roberts ES, et al. Acute SIV infection of the brain leads to upregulation of IL6 and interferon-regulated genes: expression patterns throughout disease progression and impact on neuroAIDS. Journal of neuroimmunology. 2004a;157:81–92. doi: 10.1016/j.jneuroim.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Roberts ES, Masliah E, Fox HS. CD163 identifies a unique population of ramified microglia in HIV encephalitis (HIVE) Journal of neuropathology and experimental neurology. 2004b;63:1255–1264. doi: 10.1093/jnen/63.12.1255. [DOI] [PubMed] [Google Scholar]
- Roberts ES, Zandonatti MA, Watry DD, Madden LJ, Henriksen SJ, Taffe MA, Fox HS. Induction of pathogenic sets of genes in macrophages and neurons in NeuroAIDS. Am J Pathol. 2003;162:2041–2057. doi: 10.1016/S0002-9440(10)64336-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders BD, Jackson B, Marmorstein R. Structural basis for sirtuin function: what we know and what we don't. Biochimica et biophysica acta. 2010;1804:1604–1616. doi: 10.1016/j.bbapap.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab JM, Frei E, Klusman I, Schnell L, Schwab ME, Schluesener HJ. AIF-1 expression defines a proliferating and alert microglial/macrophage phenotype following spinal cord injury in rats. Journal of neuroimmunology. 2001;119:214–222. doi: 10.1016/s0165-5728(01)00375-7. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Chen ZJ. Antiviral innate immunity pathways. Cell research. 2006;16:141–147. doi: 10.1038/sj.cr.7310019. [DOI] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirois M, Robitaille L, Allary R, Shah M, Woelk CH, Estaquier J, Corbeil J. TRAF6 and IRF7 control HIV replication in macrophages. PloS one. 2011;6:e28125. doi: 10.1371/journal.pone.0028125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BC, Settles B, Hallows WC, Craven MW, Denu JM. SIRT3 substrate specificity determined by peptide arrays and machine learning. ACS chemical biology. 2011;6:146–157. doi: 10.1021/cb100218d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song NY, Surh YJ. Janus-faced role of SIRT1 in tumorigenesis. Annals of the New York Academy of Sciences. 2012;1271:10–19. doi: 10.1111/j.1749-6632.2012.06762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Molecular cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Vempati RK, Haldar D. DNA damage in the presence of chemical genotoxic agents induce acetylation of H3K56 and H4K16 but not H3K9 in mammalian cells. Molecular biology reports. 2012;39:303–308. doi: 10.1007/s11033-011-0739-9. [DOI] [PubMed] [Google Scholar]
- Vempati RK, Jayani RS, Notani D, Sengupta A, Galande S, Haldar D. p300-mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals. The Journal of biological chemistry. 2010;285:28553–28564. doi: 10.1074/jbc.M110.149393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nature cell biology. 2006;8:1025–1031. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- Wang RH, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watry D, Lane TE, Streb M, Fox HS. Transfer of neuropathogenic simian immunodeficiency virus with naturally infected microglia. The American journal of pathology. 1995;146:914–923. [PMC free article] [PubMed] [Google Scholar]
- Weichselbaum RR, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley CA, et al. Damage and repair of DNA in HIV encephalitis. Journal of neuropathology and experimental neurology. 2000;59:955–965. doi: 10.1093/jnen/59.11.955. [DOI] [PubMed] [Google Scholar]
- Yan N, Chen ZJ. Intrinsic antiviral immunity. Nature immunology. 2012;13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, et al. The type III histone deacetylase Sirt1 is essential for maintenance of T cell tolerance in mice. The Journal of clinical investigation. 2009;119:3048–3058. doi: 10.1172/JCI38902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Kraus WL. SIRT1-dependent regulation of chromatin and transcription: linking NAD(+) metabolism and signaling to the control of cellular functions. Biochimica et biophysica acta. 2010;1804:1666–1675. doi: 10.1016/j.bbapap.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]