Abstract
Background
Multiple antigen miniarrays used for detecting autoantibodies to tumor-associated antigens (TAAs) can be a useful approach for cancer detection and diagnosis. We here address a very specific question: might there be autoimmune responses to TAAs which precede clinical detection of hepatocellular carcinoma (HCC) in HBV and HCV chronic liver disease patients under continuous medical surveillance, and if so, could these anti-TAAs be added to the armamentarium of diagnostic tests?
Methods
We here examine the utility of a panel of 12 TAAs for the diagnosis of hepatocellular carcinoma (HCC). We derived a predictive rule for the presence of HCC based on the panel, from a cohort comprising 160 HCC patients and 90 normals. We then applied this rule to sequential anti-TAA data from a cohort of 17 HCC patients, from whom this information was available prior to diagnosis.
Results
The predictors (autoantibodies to HCC1, P16, P53, P90, and survivin) indicated the presence of HCC prior to diagnosis in 16 of the 17 patients, at a median lead time of 0.75 year.
Conclusions
We believe these findings warrant further study of anti-TAA profiles as biomarkers for primary or early diagnosis of HCC.
Electronic supplementary material
The online version of this article (10.1007/s00262-018-2135-y) contains supplementary material, which is available to authorized users.
Keywords: Autoantibody profiles, Tumor-associated antigens, Classification, Hepatocellular carcinoma
Introduction
In 2012, liver cancer was the fifth most common cancer in men, and the ninth in women, in terms of estimated incidence worldwide [1]. It is a pernicious disease, and is the second most common cause of death from cancer worldwide. The prognosis for liver cancer is quite poor, with overall ratio of mortality to incidence of about 0.95. In the United States, 1 year survival is less than 40%, and 5 year survival is less than 15% [2, 3]. Definitive diagnosis is typically achieved with either CT scanning or MRI, but these modalities may well be too costly or unavailable in many regions with high prevalence of liver cancer. Techniques for early detection and diagnosis that involve “liquid biopsy” [4] may be viable alternatives in these situations, and our study is an initial step toward this goal.
A long-standing research interest of ours has been the investigation of the utility of autoantibody profiles to tumor-associated antigens (TAAs) for discriminating between cancer patients and controls [5–7]. We found that these multiple antigen miniarrays could provide accurate tools for cancer detection and diagnosis, and suggested that performance of the miniarrays might be enhanced by other combinations of TAAs appropriately selected for different cancer cohorts. From our previous studies, we had amassed a collection of serial serum samples from individuals with HBV and HCV-associated liver diseases who developed hepatocellular carcinoma (HCC) and came from liver disease clinics in three different countries, China, Japan, and South Korea. This presented us the opportunity to ask the important, clinically relevant question: Would these patients develop autoantibodies to tumor-associated antigens prior to clinically detectable HCC? We address this question directly in the present paper, where we examine the utility of an expanded panel of 12 autoantibody profiles for cancer diagnosis of HCC, based on serum samples from newly-diagnosed HCC patients and normal controls. In particular, we hypothesized that a predictive tool for HCC derived from a panel of TAAs could serve as an early warning system for HCC.
Materials and methods
Sera samples
In all, serum samples from 177 HCC patients and 90 normal controls were amassed, as follows. Sera from 76 patients with HCC from Xiamen in China were obtained from the serum bank of the Cancer Autoimmunity Research Laboratory at the University of Texas (El Paso, Texas, USA), which were originally provided by a collaborator in Sun Yat-sen University (Guangzhou, China). 84 HCC patients’ sera were collected from South Korea (Yonsei University, Seoul) and 17 from Japan (Shinshu University Hospital, Matsumoto). Ninety normal human sera (NHS) were originally obtained from the serum bank of the Autoimmune Disease Center at the Scripps Research Institute (La Jolla, CA, USA). These controls were actively working individuals who had no obvious signs or symptoms of viral hepatitis or any other forms of hepatitis. All cancer patients were diagnosed according to established criteria; their serum samples were collected at the time of initial cancer diagnosis, when the patients had not received treatment with any chemotherapy or radiation therapy. In addition, sera from the 17 patients from Japan had also been collected periodically for varying lengths of time prior to diagnosis. This study was approved by the Institutional Review Board of the University of Texas at El Paso and collaborating institutions.
Expression and purification of recombinant proteins
The 12 antigens HCC1, IMP1, Koc, MDM2, NPM1, p16, p53, p62, p90, RalA, survivin, and 14-3-3 ζ, were expressed as recombinant proteins. The sources of the 12 genes’ cDNA clones were described in detail in a previous study [7]. cDNAs of HCC1, IMP1, Koc, MDM2, NPM1, p53, p62, survivin, RalA, survivin, and 14-3-3 ζ genes were subcloned into pET28a vector producing a fusion protein with NH-terminal 6x histidine and T7 epitope tags [8–14]. The recombinant proteins expressed in Escherichia coli BL21 (DE3) were purified using nickel column chromatography. cDNAs of p16 and p90 were subcloned into pEGX vector expressing protein with glutathione S transferase (GST) fusion partner. The GST gene fusion system was utilized for the expression and purification of p16 and p90 recombinant protein [15, 16]. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining were utilized to establish that expression products with expected molecular masses were produced. In addition, western blotting analysis was used to confirm that the bands seen in SDS-PAGE were reactive with reference antibodies.
Enzyme-linked immunosorbent assay (ELISA)
Autoantibodies against 12 TAAs were determined by ELISA using purified recombinant proteins as described in our previous study [7]. Briefly, the recombinant proteins were coated into 96 well plates at a concentration of 0.5 µg/mL. The human serum samples, diluted 1:200, were incubated in the antigen-coated wells for 90 min, followed by horseradish peroxidase (HRP)-conjugated goat anti-human IgG used as the secondary antibody at 1:4,000 dilution. The substrate 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS) (Sigma, St. Louis, MO, USA) was used as the detecting agent. The optical density (OD) value was read at 405 nm for each well. Each sample was tested in duplicate, and the average OD value was used for all analyses. Each run of ELISA included eight normal human sera (NHS), representing a range of absorbance above and below the mean of the original 90 NHS, and the average OD value of eight NHS was used to normalize all absorbance values to the standard mean of the entire 90 normal samples. All of the sera with positive results in ELISA were verified by Western Blotting with purified recombinant proteins, as described previously [7].
Statistical methods
Logistic regression was used to derive a predictor rule for the presence or absence of HCC, based on anti-TAA levels in the set of 90 normals and 160 cases (84 from South Korea, 76 from China). In this regard, the minimum Bayes information criterion (BIC [17]) was used to identify the optimal subset of TAAs for the logistic regression classifier, using an all possible subsets algorithm. (The BIC is based on the likelihood function, but with a penalty for overfitting.) This subset included autoantibodies to HCC1, P16, P53, P90, and survivin.
Operating characteristics of the logistic regression classifier based on these 5 TAAs were derived from 1000 cross validation runs for tenfold cross validation, and also leave one out cross validation (LOOCV). For each tenfold cross-validation run, the entire data set was randomly divided into ten equally sized subsets, and the classifiers were trained on nine subsets and tested on the remaining subset (the validation subset). This cross validation was repeated nine additional times (so that each of the ten subsets served as the validation subset once only), and the results on the test sets combined to calculate the predictive accuracy and error rates of each classifier. We initially performed tenfold cross validation on the original data set of 250 individuals. We then refined the cross-validation procedure, by stratifying the random division (into ten subsets) separately by location (90 normals, 76 cases from China, and 84 cases from South Korea). For LOOCV, each observation in the training sample is omitted in turn, and the logistic classifier is trained on the remaining observations; this classifier is used to predict the status (class) of the omitted observation. The LOOCV estimates of the classification rates are based on the fractions of the omitted observations that are correctly classified.
The data set comprising the anti-TAA data from the 90 normals and 160 HCC cases (76 from China, 84 from South Korea) was used solely to derive the predictor rule for HCC. We then took the prediction rule from the logistic regression classifier, and applied it to the data consisting of the sequential anti-TAA levels in 17 HCC patients from Japan, the goal here being to determine whether, or when, HCC would be suspected or considered in these patients based on the prediction rule, prior to “official” diagnosis of HCC.
There were no missing data in either the 250 subject data set or the sequential data from Japan. Descriptive statistics were calculated in SPSS v22 (IBM Corp., 2013), and the logistic regression analyses were undertaken in JMP 11.0 (SAS Institute Inc., 2013) and Matlab R2014b (The MathWorks Inc., 2014).
Results
Descriptive statistics
Descriptive statistics relating to anti-TAA levels in the first data set (160 HCC patients, and 90 controls) are given in Table 1. In general, autoantibody levels are not normally distributed in either cases or controls, but tend to be characterized by positive skewness and heavy kurtosis. In addition, anti-TAA levels tend to be higher in cases than controls, with varying degrees of overlap. This can be seen in Supplementary Fig. 1, which depicts dual histograms with each TAA, for the patients and the controls. Generally, anti-TAA levels appear to be somewhat elevated in cases relative to controls, but occasional extreme values might well distort the perceived separation between the cohorts. Although discrimination between cases and controls on the basis of some of the anti-TAAs appears feasible, it is interesting that occasionally, anti-TAA levels in a few of the controls are consistent with a diagnosis of HCC [e.g., 14-3-3ζ, P16, P90]; and, with RalA, anti-TAA levels are noticeably less in some cases than in the controls.
Table 1.
Descriptive statistics for serum autoantibody levels to 12 tumor-associated antigens
| 14-3-3ζ | HCC1 | IMP1 | KOC | MDM2 | NPM1 | P16 | P53 | P62 | P90 | RalA | survivin | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases (N = 160) | ||||||||||||
| Minimum | 0.000 | 0.000 | 0.028 | 0.003 | 0.002 | 0.007 | − 0.010 | 0.000 | 0.000 | 0.001 | 0.000 | 0.027 |
| Maximum | 0.493 | 0.580 | 4.031 | 2.193 | 0.602 | 1.706 | 1.166 | 3.743 | 3.347 | 0.660 | 0.750 | 2.760 |
| Range | 0.493 | 0.580 | 4.003 | 2.190 | 0.600 | 1.699 | 1.176 | 3.743 | 3.347 | 0.659 | 0.750 | 2.733 |
| Interquartile range | 0.069 | 0.093 | 0.120 | 0.080 | 0.080 | 0.222 | 0.165 | 0.095 | 0.184 | 0.089 | 0.146 | 0.128 |
| Median | 0.082 | 0.099 | 0.140 | 0.088 | 0.127 | 0.163 | 0.184 | 0.114 | 0.226 | 0.124 | 0.194 | 0.149 |
| Arithmetic mean | 0.107 | 0.123 | 0.221 | 0.126 | 0.153 | 0.256 | 0.235 | 0.176 | 0.274 | 0.140 | 0.214 | 0.203 |
| Standard deviation | 0.083 | 0.099 | 0.400 | 0.193 | 0.105 | 0.257 | 0.176 | 0.347 | 0.315 | 0.087 | 0.128 | 0.257 |
| Skewness | 2.305 | 1.757 | 6.811 | 8.183 | 1.968 | 2.803 | 2.060 | 8.276 | 6.477 | 2.411 | 1.160 | 6.999 |
| Kurtosis | 7.422 | 3.880 | 55.932 | 83.437 | 4.436 | 10.581 | 6.224 | 77.763 | 58.267 | 9.838 | 1.959 | 63.834 |
| Controls (N = 90) | ||||||||||||
| Minimum | 0.012 | 0.004 | 0.024 | 0.012 | 0.005 | 0.000 | 0.017 | 0.014 | 0.015 | 0.011 | 0.042 | 0.018 |
| Maximum | 0.260 | 0.236 | 0.383 | 0.271 | 0.217 | 0.616 | 0.685 | 0.224 | 0.347 | 0.452 | 0.272 | 0.376 |
| Range | 0.248 | 0.232 | 0.359 | 0.259 | 0.212 | 0.616 | 0.668 | 0.210 | 0.332 | 0.441 | 0.230 | 0.358 |
| Interquartile range | 0.038 | 0.044 | 0.071 | 0.039 | 0.054 | 0.083 | 0.111 | 0.033 | 0.090 | 0.031 | 0.063 | 0.044 |
| Median | 0.060 | 0.043 | 0.090 | 0.047 | 0.076 | 0.057 | 0.110 | 0.056 | 0.106 | 0.042 | 0.107 | 0.061 |
| Arithmetic mean | 0.067 | 0.055 | 0.110 | 0.058 | 0.078 | 0.099 | 0.142 | 0.059 | 0.120 | 0.053 | 0.118 | 0.075 |
| Standard deviation | 0.037 | 0.045 | 0.068 | 0.051 | 0.043 | 0.114 | 0.107 | 0.029 | 0.069 | 0.049 | 0.049 | 0.057 |
| Skewness | 2.144 | 1.997 | 1.822 | 2.497 | 0.632 | 2.466 | 2.199 | 2.219 | 0.907 | 6.177 | 0.993 | 3.479 |
| Kurtosis | 7.781 | 4.845 | 4.086 | 7.146 | 0.299 | 6.789 | 7.157 | 10.689 | 0.621 | 48.773 | 0.857 | 16.097 |
14-3-3ζ: tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta
HCC1: RNA binding motif protein 39
IMP1: insulin like growth factor 2 mRNA binding protein 1
KOC: insulin like growth factor 2 mRNA binding protein 3
MDM2: MDM2 proto-oncogene
NPM1: nucleophosmin
P16/CDKN2A: cyclin dependent kinase inhibitor 2A
P53/TP53: tumor protein p53
P62/IGF2BP2: insulin like growth factor 2 mRNA binding protein 2
P90/CIP2A: KIAA1524
RalA: RAS like proto-oncogene A
survivin: baculoviral IAP repeat-containing protein 5.1-A-like
Classifier: first data set
As described above, logistic regression was used to derive a classifier for HCC vs. normal from the data set comprising 90 normals and 160 HCC cases (76 from China, 84 from South Korea). The optimal classifier, based on a minimum Bayes information criterion, is based on 5 of the 12 TAAs, namely, HCC1, P16, P53, P90, and survivin. To implement the logistic regression classifier for an individual, a numerical score for that individual is calculated, based on that individual’s OD values for those TAAs:
Then, the classification rule is:
If score > 0, the individual is classified as having HCC.
If score < 0, the individual is classified as not having HCC.
The operating characteristics of the classifier rule based on these 5 TAAs were assessed by cross validation, as detailed in the methods. Interestingly, operating characteristics for leave one out cross-validation (LOOCV) and both methods of tenfold cross-validation were virtually identical: sensitivity of the 5-TAA classifier was estimated as 0.880 from the original tenfold cross-validation compared to 0.881 with stratified tenfold cross-validation and LOOCV; specificity was estimated as 0.841 with the original tenfold cross-validation compared to 0.833 with both stratified tenfold cross-validation and LOOCV.
Classifier: sequential data
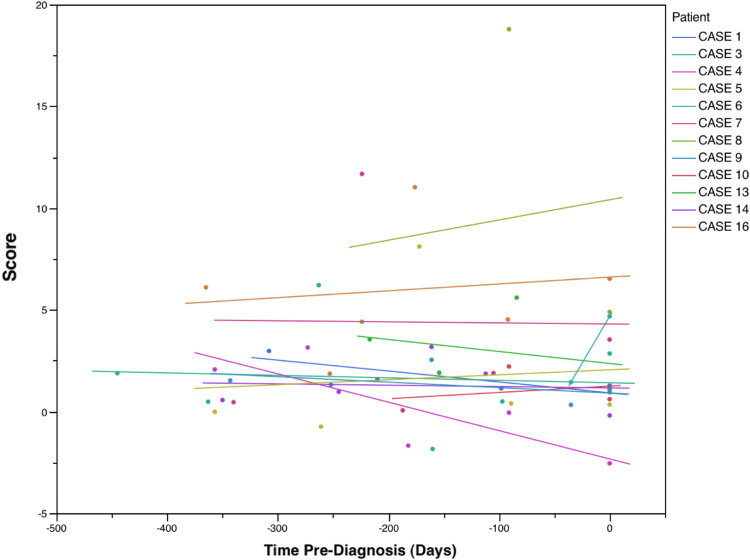
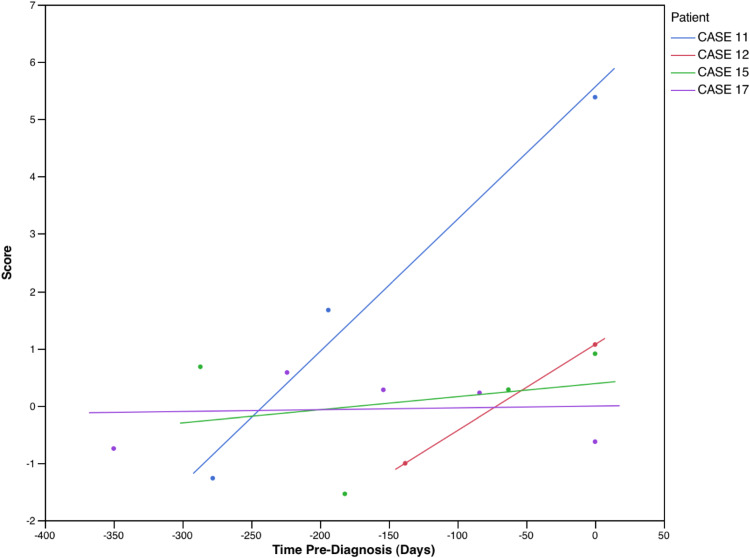
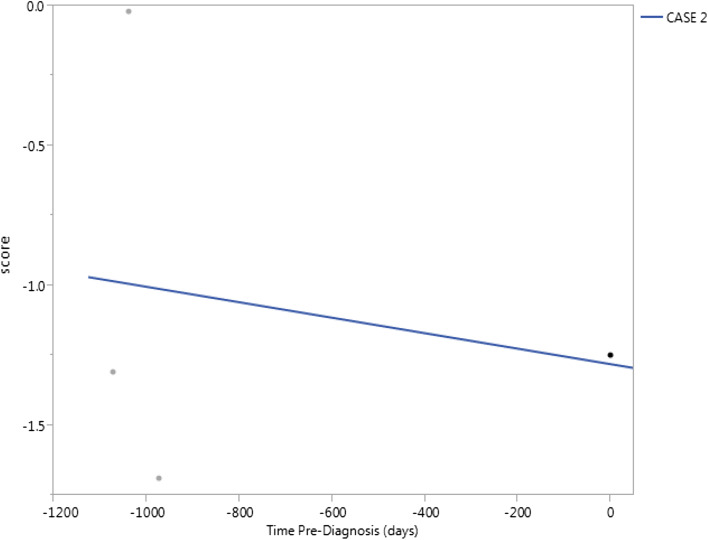
As noted previously, there were sequential anti-TAA data available for 17 HCC patients, prior to diagnosis of HCC. The classifier rule was applied to the sequential data, the purpose being to identify the earliest preclinical stage at which the presence of HCC would be indicated with the TAA classifier rule. In 12 patients (Fig. 1), HCC would have been indicated (i.e., score > 0) at the earliest time at which anti-TAA values were recorded; in 4 patients (Fig. 2), HCC would have first been indicated subsequent to the initial time of availability of anti-TAA levels, but prior to time of diagnosis; and in 1 patient (Fig. 3), the classifier would not have indicated the presence of HCC prior to time of diagnosis. Among the 16 patients for whom HCC was flagged by the classifier, the time when flagged ranged from 0.1 to 1.2 years, median 0.75 year (mean 0.72 year, standard deviation 0.31 year).
Fig. 1.
Classification scores for 12 HCC patients with sequential anti-TAA data, prior to time of diagnosis (time = 0). A score > 0 would indicate presence of HCC. In these 12 patients, the classification score would have indicated presence of HCC at initial evaluation
Fig. 2.
Classification scores for four HCC patients with sequential anti-TAA data, prior to time of diagnosis (time = 0). A score > 0 would indicate presence of HCC. In these four patients, the classification score would have indicated presence of HCC subsequent to initial evaluation, but prior to formal diagnosis
Fig. 3.
Classification scores for 1 HCC patient with sequential anti-TAA data, prior to time of diagnosis (time = 0). A score > 0 would indicate presence of HCC. In this patient, the classification score would not have indicated presence of HCC prior to time of diagnosis
Discussion
In a previous study [5], we reported that multiple antigen miniarrays could serve as useful tools for cancer detection and diagnosis. The utility of autoantibodies in cancer diagnosis was demonstrated there because of the typical absence of elevated or depressed levels of particular autoantibodies in normal individuals. In addition, we had previously proposed [18, 19] that autoantibodies might successfully be used as indicators of aberrant cellular mechanisms in tumorigenesis. In the oncologic setting, we further suggested that autoantibody panels might be adopted as predictive markers, that is, they might provide early warning of the onset of cancer.
The current study investigates this concept: we examined the utility of an expanded panel of 12 anti-TAA profiles for diagnosis of hepatocellular carcinoma. We first derived a classifier for HCC from a training set of 160 HCC patients and 90 normals, based on 5 of the 12 TAAs in our panel; then, using this classifier on a test set comprising sequential data from 17 HCC patients prior to diagnosis, we found that the classifier signals HCC in 16 of the 17 subjects, at a median of 9 months prior to diagnosis. This finding raises the notion that some autoantibodes may be detectable before overt signs and symptoms of HCC and might serve as an early warning system of malignant conversion in high risk patients such as those with with HBV/HCC hepatitis. Early detection of HCC should lead to improved survival rates, since tumors should be smaller and at an earlier stage in tumor evolution, and metastatic spread minimized, leading to improvements in overall survival [20]. Nevertheless, incorporation of a panel of TAAs into a systematic program of HCC surveillance for high-risk patients ought to be preceded by evaluation of the panel with a defined disease control group (e.g., autoimmune liver disease, obstructive biliary disease, and cirrhosis) as comparator.
We believe that a strength of our approach is the incorporation of a multivariate classifier for the presence of HCC. We have shown in the previous studies [e.g., 5] that panels of TAAs provide improved operating characteristics [e.g., sensitivity, specificity] over individual TAAs for diagnosis, and reliance on fixed cutoffs for positivity in ELISAs [e.g., mean + 2 standard deviations for ODs] does not derive full information from these assays [5, 21]. [From a statistical perspective, judicious combinations of variables tend to outperform individual variables as classifiers; and, one typically derives more information from a “continuous” variable than from a “dichotomous” version of it].
Nevertheless, our study has limitations. From a statistical perspective, our classifier could be improved on. Our use of logistic regression as a comparator is motivated by its ease of interpretation, and relatively good performance in our previous studies; as well, both cases and controls in our training set are numerous and not totally unbalanced, and our explanatory variables are not highly correlated. Still, classifiers based on other criteria or methods might enjoy gains in performance. For example, we could have used a different criterion to BIC for variable selection. In this regard, we did examine classifiers chosen on the basis of Akaike’s information criterion: this resulted in an expansion of the number of variables in our classifier [while maintaining the core group of HCC1, P16, P53, P90, and survivin], but with no noticeable improvements in performance. In addition, in the current setting, we have previously investigated classifiers based on restricted Boltzmann machines [22], and were encouraged by their relatively good performance as classifiers. Perhaps, more pertinent would be optimal selection of TAAs for any classification rule: we might expect improvements in the operating characteristics of any classifier through inclusion of judiciously selected TAAs that are not part of our panel.
We acknowledge that our study is retrospective, and nothing is known about the clinical characteristics of the subjects in either the original data set of 250 subjects (China, South Korea, US) or the sequential data set from Japan. Serum samples were provided anonymously, with no additional information, reflective of the national laws and confidentiality and ethical standards of the institutions from which the samples were obtained. Furthermore, our normal controls were not matched for age, gender, or ethnicity with the cases, though anti-TAA levels should be negligible among normal individuals regardless of demographics. Still, it is quite possible that clinical or demographic information could have profitably been incorporated into the classifier, or perhaps identification of high-risk subjects for whom frequent monitoring of anti-TAA levels would be warranted.
In addition, a reviewer has commented that “it would be important to have as controls viral hepatitis patients that do not progress to malignancy. The effects of hepatitis viruses, especially HBV, are well known and it is important to determine what is malignancy-related and what is virus-related, with an appreciation that there may be an overlap in these distinctions”. We agree with this assessment, which is congruent with our objective of drawing interest to the possibilities of utilizing anti-TAAs as tools for early detection of conversion from chronic liver disease to HCC.
Given our study limitations, we caution against interpreting this study to be a conclusive validation of the clinical value or utility of TAA panel-based testing for HCC. Nevertheless, our positive finding that some autoantibodies to TAAs do antedate clinically overt HCC provides some justification for well-controlled prospective studies with complete demographic and clinical data so as to either confirm or invalidate our observations. We, therefore, hope to raise interest in such prospective studies by clinics in countries with high prevalence of HBV- and HCV- associated liver diseases. We of course would be happy to encourage such studies by supplying cDNA clones encoding TAAs.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We wish to acknowledge the generous donation of HCC sera from Dr. David Lee.
Abbreviations
- BIC
Bayes information criterion
- ELISA
Enzyme-linked immunosorbent assay
- LOOCV
Leave one out cross validation
- NHS
Normal human ser
Author contributions
Conception and design: JAK and EMT. Development of methodology: HI, LD, J-YZ, and EMT. Acquisition of data: HI, LD, J-YZ, and EMT. Analysis and interpretation of data (computational and statistical analysis): JAK, LD, and EMT. Writing and revision of the manuscript: JAK, LD, EMT. Review of the manuscript: JAK, HI, LD, J-YZ, and EMT. Study supervision: EMT.
Funding
The authors would like to acknowledge funding received from the National Institutes of Health: 1 PO1 HL119165 (Koziol).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval and ethical standards
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committees and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. In particular, this study was approved by the Institutional Review Board of the University of Texas at El Paso and collaborating institutions. This study does not contain any studies with animals performed by any of the authors.
References
- 1.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F (2013) GLOBOCAN 2012 v1.0, cancer incidence and mortality worldwide: IARC CancerBase No. 11 [Internet]. Lyon, France: international agency for research on cancer. http://globocan.iarc.fr. Accessed 17 Oct 2015
- 2.National Cancer Institute, Surveillance, Epidemiology, and End Results Program, Fast Stats, SEER Mortality. http://seer.cancer.gov/faststats. Accessed 17 Oct 2015
- 3.El-Serag H, Kanwal F. Epidemiology of hepatocellular carcinoma in the United States: where are we? Where do we go? Hepatology. 2014;60:1767–1775. doi: 10.1002/hep.27222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pantel K, Alix-Panabières C. Real-time liquid biopsy in cancer patients: fact or fiction? Can Res. 2013;73:6384–6388. doi: 10.1158/0008-5472.CAN-13-2030. [DOI] [PubMed] [Google Scholar]
- 5.Koziol JA, Zhang JY, Casiano CA, Peng XX, Shi FD, Feng AC, Chan EK, Tan EM. Recursive partitioning as an approach to selection of immune markers for tumor diagnosis. Clin Cancer Res. 2003;9:5120–5126. [PubMed] [Google Scholar]
- 6.Zhang JY, Casiano CA, Peng XX, Koziol JA, Chan EK, Tan EM. Enhancement of antibody detection in cancer using panel of recombinant tumor-associated antigens. Cancer Epidemiol Biomarkers Prev. 2003;12:136–143. [PubMed] [Google Scholar]
- 7.Dai L, Ren P, Liu M, Imai H, Tan EM, Zhang JY. Using immunomic approach to enhance tumor-associated autoantibody detection in diagnosis of hepatocellular carcinoma. Clin Immunol. 2014;152:127–139. doi: 10.1016/j.clim.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang JY, Chan EKL, Peng XX, Tan EM. A novel cytoplasmic protein with RNA-binding motifs is an autoantigen in human hepatocellular carcinoma. J Exp Med. 1999;189:1101–1110. doi: 10.1084/jem.189.7.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Müeller-Pillasch F, Lacher U, Wallrapp C, Micha A, Zimmerhackl F, Hameister H, Varga G, Friess H, Büchler M, Beger HG, Vila MR, Adler G, Gress TM. Cloning of a gene highly overexpressed in cancer coding for a novel KH-domain containing protein. Oncogene. 1997;14:2729–2733. doi: 10.1038/sj.onc.1201110. [DOI] [PubMed] [Google Scholar]
- 10.Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19:1262–1270. doi: 10.1128/MCB.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imai H, Chan EK, Kiyosawa K, Fu XD, Tan EM. Novel nuclear autoantigen with splicing factor motifs identified with antibody from hepatocellular carcinoma. J Clin Invest. 1993;92:2419–2426. doi: 10.1172/JCI116848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu M, Varela-Ramirez A, Li J, Dai L, Aguilera RJ, Zhang JY. Humoral autoimmune response to nucleophosmin in the immunodiagnosis of hepatocellular carcinoma. Oncol Rep. 2015;33:2245–2252. doi: 10.3892/or.2015.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu M, Zheng SJ, Chen Y, Li N, Ren PF, Dai LP, Duan ZP, Zhang JY. Autoantibody response to murine double minute 2 protein in immunodiagnosis of hepatocellular carcinoma. J Immunol Res. 2014;2014:906532. doi: 10.1155/2014/906532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu M, Liu X, Ren P, Li J, Zheng SJ, Chen Y, Duan ZP, Li N, Zhang JY. A cancer-related protein 14-3-3ζ is a potential tumor-associated antigen in immunodiagnosis of hepatocellular carcinoma. Tumor Biol. 2014;35:4247–4256. doi: 10.1007/s13277-013-1555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Looi K, Megliorino R, Shi FD, Peng XX, Chen Y, Zhang JY. Humoral immune response to p16, a cyclin-dependent kinase inhibitor in human malignancies. Oncol Rep. 2006;16:1105–1110. [PubMed] [Google Scholar]
- 16.Looi KS, Nakayasu ES, Diaz RA, Tan EM, Almeida IC, Zhang JY. Using proteomic approach to identify tumor-associated antigens as markers inhepatocellular carcinoma. J Proteome Res. 2008;7:4004–4012. doi: 10.1021/pr800273h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarz G. Estimating the dimension of a model. Ann Stat. 1978;6:461–464. doi: 10.1214/aos/1176344136. [DOI] [Google Scholar]
- 18.Tan EM. Autoantibodies as reporters identifying aberrant cellular mechanisms in tumorigenesis. J Clin Invest. 2001;108:1411–1415. doi: 10.1172/JCI14451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan EM, Zhang JY. Autoantibodies to tumor-associated antigens: reporters from the immune system. Immunol Rev. 2008;222:328–340. doi: 10.1111/j.1600-065X.2008.00611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singal AG, Pillai A, Tiro J. Early detection, curative treatment, and survival rates for hepatocellular carcinoma surveillance in patients with cirrhosis: a meta-analysis. PLoS Med. 2014;11:e1001624. doi: 10.1371/journal.pmed.1001624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai L, Tsay JC, Li J, Yie TA, Munger JS, Pass H, Rom WN, Zhang Y, Tan EM, Zhang JY. Autoantibodies against tumor-associated antigens in the early detection of lung cancer. Lung Cancer. 2016;99:172–179. doi: 10.1016/j.lungcan.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 22.Koziol JA, Tan EM, Dai DL, Ren P, Zhang JY. Restricted Boltzmann machines for classification of hepatocellular carcinoma. Comput Biol J. 2014;2014:418069. doi: 10.1155/2014/418069. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.