Abstract
Capillary zone electrophoresis (CZE)-tandem mass spectrometry (MS/MS) has been recognized as a useful tool for top-down proteomics. However, its performance for deep top-down proteomics is still dramatically lower than widely used reversed-phase liquid chromatography (RPLC)-MS/MS. We present an orthogonal multi-dimensional separation platform that couples size exclusion chromatography (SEC) and RPLC based protein pre-fractionation to CZE-MS/MS for deep top-down proteomics of Escherichia coli. The platform generated high peak capacity (~4 000) for separation of intact proteins, leading to the identification of 5 700 proteoforms from the Escherichia coli proteome. The data represents a 10-fold improvement in the number of proteoform identifications compared with previous CZE-MS/MS studies and represents the largest bacterial top-down proteomics dataset reported to date. The performance of the CZE-MS/MS based platform is comparable to the state-of-the-art RPLC-MS/MS based systems in terms of the number of proteoform identifications and the instrument time.
Graphical Abstract
In top-down proteomics, intact proteins extracted from cells are typically fractionated by liquid chromatography (LC) or electrophoresis, followed by reversed-phase LC-tandem mass spectrometry (RPLC-MS/MS) analysis. The resulting MS/MS spectra are compared with a protein database derived from the genome sequence for proteoform identifications (IDs). [1–3] The state-of-the-art RPLC-MS/MS based workflows have approached 3 000–5 000 proteoform IDs corresponding to around 1 000 proteins. [4–7]
Capillary zone electrophoresis (CZE)-MS/MS has been recognized as a useful tool for top-down proteomics due to the high resolution of CZE for separation of intact proteins and the high sensitivity of CZE-MS/MS for detection of intact proteins. [8–14] However, the performance of CZE-MS/MS based platforms are still far below that of RPLC-MS/MS based platforms in terms of the number of proteoform IDs. Several groups have made some effort to improve CZE-MS/MS for top-down proteomics.[15–19] Li et al. identified 30 large proteins (30–80 kDa) from P. aeruginosa PA01 cell lysate using CZE-MS/MS, indicating the potential of CZE-MS/MS for top-down identification of large proteins from a complex proteome. [15] Han et al. coupled RPLC fractionation to CZE-MS/MS for top-down proteomics of Pyrococcus furiosus and identified nearly 300 proteoforms corresponding to 134 proteins, demonstrating the capability of CZE-MS/MS for large-scale top-down proteomics. [16] Zhao et al. combined high-resolution RPLC fractionation and CZE-MS/MS for large-scale top-down proteomics of yeast and observed nearly 600 proteoform and 200 protein IDs. [18] The data represents the state of the art of CZE-MS/MS for top-down proteomics.
Two major issues have limited the number of proteoform IDs from complex proteomes using CZE-MS/MS. One issue is the low sample loading capacity of CZE. The other one is the low peak capacity of CZE for separation of intact proteins. The sample loading capacity and peak capacity of CZE was 200 nL or lower and less than 100, respectively, in the reports mentioned in the previous paragraph. Recently, we boosted the sample loading capacity and peak capacity of CZE-MS/MS to 1 μL and 280, respectively, using dynamic pH junction based sample stacking [20–22] for analysis of complex mixtures of intact proteins. [19] Duplicate CZE-MS/MS analyses of an Escherichia coli (E. coli) proteome generated 586 ±38 proteoform IDs with a 1% spectrum-level false discovery rate (FDR). [19] We compared the identified proteoforms from the duplicate CZE-MS/MS analyses and revealed that on average, about 76% of the proteoform IDs were the same in each CZE-MS/MS run, suggesting the good reproducibility of the CZE-MS/MS system.
Based on the previous work, we report a multi-dimensional platform with high peak capacity for separation of intact proteins in complex proteomes, Figure 1. The proteins in an E. coli lysate were first fractionated with size exclusion chromatography (SEC) into five fractions based on their size, Figure 1A. The proteins in each SEC fraction were further fractionated with RPLC into 20 fractions based on their hydrophobicity, resulting in 100 RPLC fractions (5×20) in total, Figure 1B. The proteins in those fractions were separated by the dynamic pH junction based CZE based on their size-to-charge ratios, followed by electrospray ionization (ESI)-MS/MS analysis, Figure 1C. The proteins in each RPLC fraction were dissolved in 5 μL of 50 mM ammonium bicarbonate (pH 8.0) for CZE-MS/MS. The background electrolyte (BGE) of CZE was 10% (v/v) acetic acid (pH 2.2). The electro-kinetically pumped sheath flow interface was employed to couple CZE to MS.[23,24] About 10% of the sample (500 nL) was injected into the separation capillary for CZE-MS/MS. The SEC-RPLC-CZE platform produced orthogonal and high-capacity separation of intact proteins. The peak capacity of the platform was estimated to be around 4 000 based on the full width at half maximum (FWHM) of protein peaks. The acquired MS/MS spectra of proteins were subjected to a database search using TopPIC (Top-down mass spectrometry based Proteoform Identification and Characterization) software for identification and characterization of proteoforms, [25,26] Figure 1D. Experimental details are shown in the Supporting Information I.
Figure 1.

The multi-dimensional platform with high peak capacity for separation of intact proteins in complex proteomes. (A) Chromatogram of an E. coli lysate after SEC separation. (B) Chromatogram of an SEC fraction of the E. coli lysate after RPLC separation. (C) Total ion current (TIC) electropherogram of an RPLC fraction of the E. coli lysate after CZE-MS/MS analysis. (D) Fragmentation pattern of one identified proteoform from gene hdeB after database search with TopPIC (Top-down mass spectrometry based Proteoform Identification and Characterization) software. AU=absorbance units, mAU=milli- absorbance units, NL=normalized level.
We identified over 58 000 proteoform-spectrum matches (PrSMs), 5 705 proteoforms and 850 proteins from the E. coli proteome using the SEC-RPLC-CZE-MS/MS platform with a 1% spectrum-level FDR. We observed reasonable protein signal from 43 RPLC fractions using CZE-MS/MS and the proteoform/protein IDs were from those 43 CZE-MS/MS runs. The corresponding electropherograms are shown in Figures S1–S9 in Supporting Information I. The dataset represents an order of magnitude improvement in the number of proteoform IDs compared with previous CZE-MS/MS studies (5 700 vs. 300–600 proteoforms).[16,18,19] The dataset also represents the largest bacterial top-down proteomics dataset reported to date. The details of the identified PrSMs and proteoforms are listed in Supporting Information II.
We attribute the dramatic improvement in the number proteoform IDs to two major reasons. First, the SEC-RPLC-CZE platform produced high peak capacity (~4 000) for separation of intact proteins. The peak capacity is at least 4 times higher than that in previous top-down proteomics studies using CZE-MS/MS. [16,18,19] Second, the dynamic pH junction based CZE-MS/MS system had high sample loading capacity. About 10% of the proteins in each RPLC fraction (500 nL vs. 5 μL) was injected into the capillary for CZE-MS/MS, and the sample loading volume is 2–5 times higher than previous top-down proteomics studies using LC-CZE-MS/MS.[16,18] Both the high peak capacity and high sample loading capacity benefit the identification of relatively low abundant proteins and proteoforms.
We then performed various analyses of the proteoforms and proteins that were identified from the E. coli proteome using the SEC-RPLC-CZE-MS/MS platform. Single-shot CZE-MS/MS produced nearly 500 proteoform IDs from two of the 43 RPLC fractions and yielded 200–400 proteoform IDs from most of the RPLC fractions, Figure 2A. The number of cumulative proteoform IDs increased steadily with the increase of the number of RPLC fraction or SEC fraction, indicating the efficient pre-fractionation performance of SEC and RPLC, Figure 2A. SEC fractions 3–5 made greater contribution to the proteoform IDs than SEC fraction 1 and we did not observe significant protein signal from SEC fraction 2, Figure 2A. The majority of the identified proteoforms had mass in a range of 10–20 kDa and 52 proteoforms with mass bigger than 30 kDa were identified, indicating the potential of the platform for top-down characterization of large proteins, Figure 2B. The number of proteoforms per gene ranged from 1 to 345, Figure S10 in Supporting Information I. The detected mass shifts from the identified proteoforms ranged from −600 Da to 600 Da, corresponding to various modifications, e.g., protein truncations, cysteine carbamidomethylation (57 Da), methylation (14 Da), acetylation (42 Da), and oxidation (16 Da), Figure S11 in Supporting Information I. We also detected N-terminal methionine excision and signal peptide removal.
Figure 2.
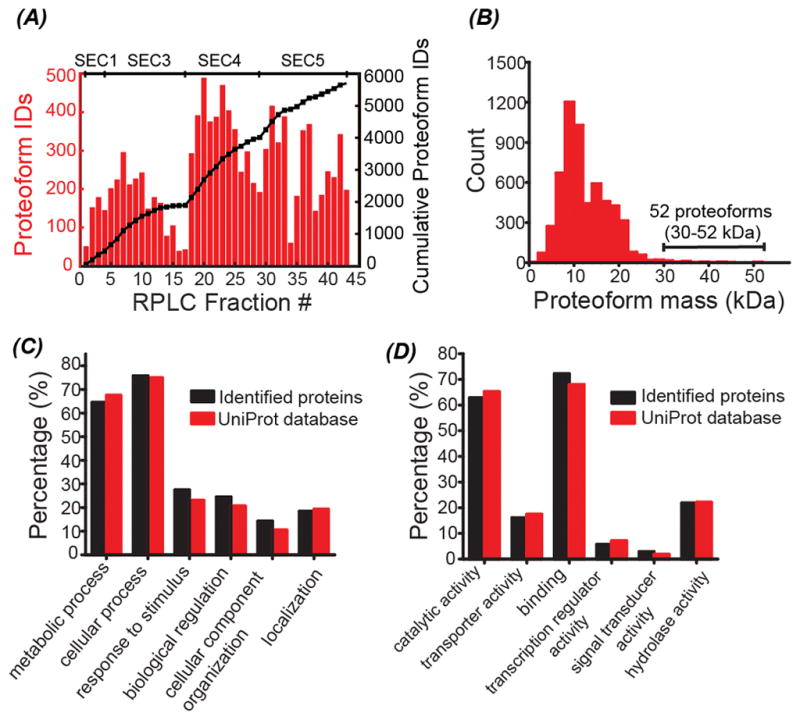
Summary of the identified proteins and proteoforms. (A) The number of proteoform IDs in each RPLC fraction (the red colored bars); the cumulative proteoform IDs vs. the number of RPLC fractions (the black line with squares). The SEC fraction to which the RPLC fractions belong was labelled on the top of the figure. (B) Distribution of the mass of the identified proteoforms. (C) Distribution of the biological process of identified proteins in this work and the proteins in the UniProt E. coli database. (D) Distribution of the molecular function of identified proteins in this work and the proteins in the UniProt E.coli database. The “Retrieve/ID mapping” tool in the UniProt website was used to obtain the gene ontology (GO) information of proteins.
We observed good linear correlation between the number of PrSMs and the abundance (ppm) of 20 randomly selected proteins in a mass range of 6–20 kDa, Table S1 and Figure S12 in Supporting Information I. The data suggested that the number of PrSMs of proteins (<20 kDa) could be used to roughly estimate their abundance in cells, which is similar to the spectral count idea used in bottom-up proteomics.[27] Similarly, we used the number of PrSMs to estimate the relative abundance of various proteoforms derived from a same gene and we took two genes, hdeA and hdeB, as the examples. We identified 345 proteoforms (6 634 PrSMs) and 47 proteoforms (1 084 PrSMs) for hdeA and hdeB, respectively, Figure S10. For hdeA, 62% of the identified proteoforms (214 out of the 345) related to various truncations at the termini of the protein molecules, and 131 proteoforms had no truncations. The data suggest that protein truncation is one major reason for the large number of identified proteoforms of hdeA. The 131 proteoforms of hdeA that were not truncated corresponded to 87% of all the PrSMs of hdeA, and the 214 truncated proteoforms only accounted for 13% of the total PrSMs of hdeA. For hdeB, only 10% of the proteoforms (5 out of the 47) related to various truncations and those proteoforms only represented 1% of the total PrSMs. The data clearly indicate that the majority of the hdeA and hdeB protein molecules in the E. coli cells have no truncations. As shown in Table S2 in Supporting Information I, the majority of the hdeA and hdeB protein molecules in E. coli cells had the mass shift as 0 Da based on their PrSM data. A small percentage of the hdeB protein molecules had methylation (mass shift as 14 Da), dimethylation (mass shift as 28 Da), acetylation (mass shift as 42 Da), or combination of methylation and acetylation (mass shift as 56 Da), Table S2. Those PTMs of hdeB detected here agreed well with that in one E. coli PTM database established recently by the Smith group using bottom-up proteomics.[28] Similarly, we also identified some hdeA proteoforms with the same mass shifts as the hdeB proteoforms, e.g., 14 Da, 28 Da, and 42 Da. However, we could not find any PTM information about hdeA from UniProt database (http://www.uniprot.org/uniprot/P0AES9) and the E. coli PTM database in reference 28. The results here highlight the capability of the CZE-MS/MS based top-down proteomics for accurate characterization of proteins in cells.
We further compared the identified proteins (850) with the proteins in UniProt E. coli database (~4 000 proteins) in terms of the gene ontology (GO) information, Figures 2C–2D and Figure S13 in Supporting Information I. Our SEC-RPLC-CZE-MS/MS platform had no obvious bias in protein ID with respect to the biological process and molecular function distributions. About 36% of the identified proteins were membrane proteins and this percentage was only slightly lower than that in the UniProt database (43%). The data indicated that our platform was efficient for identification of membrane proteins. The percentage of proteins that located in the intracellular part, cytosol or ribosomal subunit was higher in the identified protein pool than that in the UniProt database.
We also compared our work with recent deep top-down proteomics studies that employed RPLC as the final dimension for separation of intact proteins prior to MS and MS/MS analysis. In our work, 5 705 proteoform and 850 protein IDs were observed from the 43 CZE-MS/MS runs, corresponding to roughly 4 680 minutes of instrument time. Tran et al. combined solution isoelectric focusing (sIEF), gel elution liquid fraction entrapment electrophoresis (GELFrEE), and RPLC-MS/MS for top-down analysis of a human cell line, resulting in over 3 000 proteoform IDs from 1 063 proteins with 3 825 minutes of instrument time.[4] Anderson et al. identified 3 238 proteoforms and 684 proteins from human colorectal cancer cells using GELFrEE prefractionation followed by RPLC-MS/MS.[7] Overall, the data acquisition took roughly 4 960 minutes. Catherman et al. combined subcellular fractionation, sIEF, GELFrEE, and RPLC-MS/MS for deep top-down proteomics of the transformed human cell line H1299 proteome.[5] Over 5 000 proteoforms and 1 220 proteins were identified, representing the largest top-down proteomic dataset of the human proteome reported to date. Hundreds of RPLC-MS/MS runs (~90 min per run) were performed in that study. In summary, our SEC-RPLC-CZE-MS/MS platform is comparable with the state-of-the-art RPLC-MS/MS based systems for deep top-down proteomics in terms of the number of proteoform IDs and the total instrument time.
It is noteworthy that the total CZE-MS/MS analysis time can be easily reduced via boosting the electric field across the separation capillary. In this work, 20 kV was applied across the capillary for separation, and increasing the voltage to 30 kV will improve the throughput by 1.5 fold theoretically. In addition, in this work, we did not fully use the instrument time for proteoform IDs, and there was significant dead time in each CZE-MS/MS run. For instance, all of the identified PrSMs concentrated in a 10-min window for the RPLC fraction 15, and the dead time of this CZE-MS/MS run was 110 min, Figure S14A in Supporting Information I. As another example, the identified PrSMs spread over an 80-min window for the RPLC fraction 19, and about 40 PrSMs/min was approached across a 35-min window, Figure S14B. The dead time of that CZE-MS/MS run was still 40 min. We believe the sequential sample injection method that has been tested for high-throughput bottom-up proteomics using CZE-MS/MS recently will allow us to reduce the dead time in each CZE-MS/MS run. [29–31] Those improvements will be very helpful to increase the throughput of our SEC-RPLC-CZE-MS/MS platform for deep top-down proteomics.
We speculate that the number of proteoform and protein IDs from the SEC-RPLC-CZE-MS/MS platform can be significantly boosted via several improvements. First, the SEC separation can be further improved via simply increasing the length of the SEC column and employing the serial SEC method developed recently by the Ge group.[6] Second, the RPLC separation can be improved via investigating different RP beads and employing longer columns.[32,33] Third, the performance of CZE can be improved with longer separation capillaries (i.e., 1.5 meters) and higher separation voltage (i.e., 60 kV). Fourth, the improvement in mass resolution and scan speed of mass spectrometers definitely will benefit large-scale top-down proteomics of complex proteomes. In addition, combination of different protein fragmentation techniques, e.g., high energy collision dissociation (HCD),[34] electron transfer dissociation (ETD),[35–37] and ultraviolet photodissociation (UVPD),[38,39] will be invaluable for boosting the scale of top-down proteomics and improving the quality of proteoform characterization.
Supplementary Material
Acknowledgments
We thank Prof. Heedeok Hong’s group at Michigan State University (Department of Chemistry) for kindly providing the Escherichia coli cells for our research. We thank the support from the National Institute of General Medical Sciences, National Institutes of Health (NIH), through Grant R01GM118470 (X. Liu) and R01GM125991 (L. Sun and X. Liu).
Footnotes
Notes
The authors declare no competing financial interest.
Experimental details; summary of the number of PrSMs and abundance of the selected 20 proteins; summary of the number of PrSMs of various proteoforms of hdeA and hdeB with different mass shifts; base peak electropherograms of the 43 CZE-MS/MS runs; distribution of the number of identified proteoforms from each E. coli gene; distribution of the detected mass shifts from the identified proteoforms; correlation between the number of PrSMs and the abundance of the 20 randomly selected proteins; and distribution of the cellular component of the identified proteins and the proteins in the UniProt E. coli database (PDF)
The identified PrSMs and proteoforms (XLSX)
References
- 1.Toby TK, Fornelli L, Kelleher NL. Annu Rev Anal Chem. 2016;9:499–519. doi: 10.1146/annurev-anchem-071015-041550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelleher NL, Lin HY, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. J Am Chem Soc. 1999;121:806–812. [Google Scholar]
- 3.Ge Y, Lawhorn BG, ElNaggar M, Strauss E, Park JH, Begley TP, McLafferty FW. J Am Chem Soc. 2002;124:672–678. doi: 10.1021/ja011335z. [DOI] [PubMed] [Google Scholar]
- 4.Tran JC, Zamdborg L, Ahlf DR, Lee JE, Catherman AD, Durbin KR, Tipton JD, Vellaichamy A, Kellie JF, Li M, Wu C, Sweet SM, Early BP, Siuti N, LeDuc RD, Compton PD, Thomas PM, Kelleher NL. Nature. 2011;480:254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Catherman AD, Durbin KR, Ahlf DR, Early BP, Fellers RT, Tran JC, Thomas PM, Kelleher NL. Mol Cell Proteomics. 2013;12:3465–3473. doi: 10.1074/mcp.M113.030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai W, Tucholski T, Chen B, Alpert AJ, McIlwain S, Kohmoto T, Jin S, Ge Y. Anal Chem. 2017;89:5467–5475. doi: 10.1021/acs.analchem.7b00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson LC, DeHart CJ, Kaiser NK, Fellers RT, Smith DF, Greer JB, LeDuc RD, Blakney GT, Thomas PM, Kelleher NL, Hendrickson CL. J Proteome Res. 2017;16:1087–1096. doi: 10.1021/acs.jproteome.6b00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jorgenson JW, Lukacs KD. Science. 1983;222:266–272. doi: 10.1126/science.6623076. [DOI] [PubMed] [Google Scholar]
- 9.Valaskovic GA, Kelleher NL, McLafferty FW. Science. 1996;273:1199–1202. doi: 10.1126/science.273.5279.1199. [DOI] [PubMed] [Google Scholar]
- 10.Sun L, Knierman MD, Zhu G, Dovichi NJ. Anal Chem. 2013;85:5989–5995. doi: 10.1021/ac4008122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haselberg R, de Jong GJ, Somsen GW. Anal Chem. 2013;85:2289–2296. doi: 10.1021/ac303158f. [DOI] [PubMed] [Google Scholar]
- 12.Bush DR, Zang L, Belov AM, Ivanov AR, Karger BL. Anal Chem. 2016;88:1138–1146. doi: 10.1021/acs.analchem.5b03218. [DOI] [PubMed] [Google Scholar]
- 13.Sarg B, Faserl K, Kremser L, Halfinger B, Sebastiano R, Lindner HH. Mol Cell Proteomics. 2013;12:2640–2656. doi: 10.1074/mcp.M112.024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han X, Wang Y, Aslanian A, Fonslow B, Graczyk B, Davis TN, Yates JR., III J Proteome Res. 2014;13:6078–6086. doi: 10.1021/pr500971h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Compton PD, Tran JC, Ntai I, Kelleher NL. Proteomics. 2014;14:1158–1164. doi: 10.1002/pmic.201300381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han X, Wang Y, Aslanian A, Bern M, Lavallée-Adam M, Yates JR., III Anal Chem. 2014;86:11006–11012. doi: 10.1021/ac503439n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y, Sun L, Champion MM, Knierman MD, Dovichi NJ. Anal Chem. 2014;86:4873–4878. doi: 10.1021/ac500092q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, Sun L, Zhu G, Dovichi NJ. J Proteome Res. 2016;15:3679–3685. doi: 10.1021/acs.jproteome.6b00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lubeckyj RA, McCool EN, Shen X, Kou Q, Liu X, Sun L. Anal Chem. 2017;89:12059–12067. doi: 10.1021/acs.analchem.7b02532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Britz-McKibbin P, Chen DDY. Anal Chem. 2000;72:1242–1252. doi: 10.1021/ac990898e. [DOI] [PubMed] [Google Scholar]
- 21.Zhu G, Sun L, Yan X, Dovichi NJ. Anal Chem. 2014;86:6331–6336. doi: 10.1021/ac5004486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen D, Shen X, Sun L. Analyst. 2017;142:2118–2127. doi: 10.1039/c7an00509a. [DOI] [PubMed] [Google Scholar]
- 23.Wojcik R, Dada OO, Sadilek M, Dovichi NJ. Rapid Commun Mass Spectrom. 2010;24:2554–2560. doi: 10.1002/rcm.4672. [DOI] [PubMed] [Google Scholar]
- 24.Sun L, Zhu G, Zhao Y, Yan X, Mou S, Dovichi NJ. Angew Chem Int Ed. 2013;52:13661–13664. doi: 10.1002/anie.201308139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kou Q, Xun L, Liu X. Bioinformatics. 2016;32:3495–3497. doi: 10.1093/bioinformatics/btw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, Inbar Y, Dorrestein PC, Wynne C, Edwards N, Souda P, Whitelegge JP, Bafna V, Pevzner PA. Mol Cell Proteomics. 2010;9:2772–2782. doi: 10.1074/mcp.M110.002766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H, Sadygov RG, Yates JR., III Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 28.Dai Y, Shortreed MR, Scalf M, Frey BL, Cesnik AJ, Solntsev S, Schaffer LV, Smith LM. J Proteome Res. 2017;16:4156–4165. doi: 10.1021/acs.jproteome.7b00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faserl K, Sarg B, Sola L, Linder HH. Proteomics. 2017;17 doi: 10.1002/pmic.201700310. doi.org/10.1002/pmic.201700310. [DOI] [PubMed] [Google Scholar]
- 30.Boley DA, Zhang Z, Dovichi NJ. J Chromatogr A. 2017;1523:123–126. doi: 10.1016/j.chroma.2017.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garza S, Moini M. Anal Chem. 2006;78:7309–7316. doi: 10.1021/ac0612269. [DOI] [PubMed] [Google Scholar]
- 32.Ansong C, Wu S, Meng D, Liu X, Brewer HM, Deatherage Kaiser BL, Nakayasu ES, Cort JR, Pevzner P, Smith RD, Heffron F, Adkins JN, Pasa-Tolic L. Proc Natl Acad Sci USA. 2013;110:10153–10158. doi: 10.1073/pnas.1221210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen Y, Tolić N, Piehowski PD, Shukla AK, Kim S, Zhao R, Qu Y, Robinson E, Smith RD, Paša-Tolić L. J Chromatogr A. 2017;1498:99–110. doi: 10.1016/j.chroma.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 34.Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Nat Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 35.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc Natl Acad Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JE, Coon JJ. Anal Chem. 2007;79:477–485. doi: 10.1021/ac061457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia Y, Han H, McLuckey SA. Anal Chem. 2008;80:1111–1117. doi: 10.1021/ac702188q. [DOI] [PubMed] [Google Scholar]
- 38.Shaw JB, Li W, Holden DD, Zhang Y, Griep-Raming J, Fellers RT, Early BP, Thomas PM, Kelleher NL, Brodbelt JS. J Am Chem Soc. 2013;135:12646–12651. doi: 10.1021/ja4029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Brien JP, Li W, Zhang Y, Brodbelt JS. J Am Chem Soc. 2014;136:12920–12928. doi: 10.1021/ja505217w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


