ABSTRACT
Selective packaging is a mechanism used by multiple virus families to specifically incorporate genomic RNA (gRNA) into virions and exclude other types of RNA. Lineage A betacoronaviruses incorporate a 95-bp stem-loop structure, the packaging signal (PS), into the nsp15 locus of ORF1b that is both necessary and sufficient for the packaging of RNAs. However, unlike other viral PSs, where mutations generally resulted in viral replication defects, mutation of the coronavirus (CoV) PS results in large increases in subgenomic RNA packaging with minimal effects on gRNA packaging in vitro and on viral titers. Here, we show that selective packaging is also required for viral evasion of the innate immune response and optimal pathogenicity. We engineered two distinct PS mutants in two different strains of murine hepatitis virus (MHV) that packaged increased levels of subgenomic RNAs, negative-sense genomic RNA, and even cellular RNAs. All PS mutant viruses replicated normally in vitro but caused dramatically reduced lethality and weight loss in vivo. PS mutant virus infection of bone marrow-derived macrophages resulted in increased interferon (IFN) production, indicating that the innate immune system limited the replication and/or pathogenesis of PS mutant viruses in vivo. PS mutant viruses remained attenuated in MAVS−/− and Toll-like receptor 7-knockout (TLR7−/−) mice, two well-known RNA sensors for CoVs, but virulence was restored in interferon alpha/beta receptor-knockout (IFNAR−/−) mice or in MAVS−/− mice treated with IFNAR-blocking antibodies. Together, these data indicate that coronaviruses promote virulence by utilizing selective packaging to avoid innate immune detection.
KEYWORDS: coronavirus, interferon response, murine hepatitis virus, RNA packaging, packaging signal, selective packaging
IMPORTANCE
Coronaviruses (CoVs) produce many types of RNA molecules during their replication cycle, including both positive- and negative-sense genomic and subgenomic RNAs. Despite this, coronaviruses selectively package only positive-sense genomic RNA into their virions. Why CoVs selectively package their genomic RNA is not clear, as disruption of the packaging signal in MHV, which leads to loss of selective packaging, does not affect genomic RNA packaging or virus replication in cultured cells. This contrasts with other viruses, where disruption of selective packaging generally leads to altered replication. Here, we demonstrate that in the absence of selective packaging, the virulence of MHV was significantly reduced. Importantly, virulence was restored in the absence of interferon signaling, indicating that selective packaging is a mechanism used by CoVs to escape innate immune detection.
INTRODUCTION
Coronaviruses (CoVs), like all positive-single-stranded RNA (+ssRNA) viruses, replicate in the cytoplasm of the host cell on restructured membranes (1, 2). In the case of coronaviruses, replication/transcription complexes (RTCs) assemble on endoplasmic reticulum (ER) membranes. The RTC, a multiprotein complex, replicates genomic RNA (gRNA) and transcribes subgenomic RNA (sgRNA) during viral infection. During replication, different viral RNAs are replicated, transcribed, translated, or assembled into virions. In RNA virus infections, replication and assembly are often coordinated by cis-acting RNA elements, which may undergo intramolecular RNA interactions to form secondary structures important for recognition. Concordantly, during coronavirus infection, gRNA and sgRNAs must be distinguishable for the purposes of replication, transcription, translation, and assembly.
CoVs are very adept at packaging only gRNA into virions, while sgRNAs are almost completely excluded from virions, despite having identical 5′ and 3′ ends as the gRNA. In lineage A betacoronaviruses (β-CoVs), such as murine hepatitis virus (MHV), the selective packaging of gRNA is dependent on the packaging signal (PS) and its interaction with structural proteins. Early studies of the MHV PS identified a 190-bp region within the nsp15 locus as the PS (3). In both infection and virus-like particle systems, the 190-bp PS was sufficient for packaging of nonviral RNA (4). Selective packaging is intimately tied to interactions with the structural proteins N and M. The selective packaging of nonviral RNA was correlated with M protein binding (5). Furthermore, N protein binds to RNA indiscriminately and was not required for packaging of nonviral RNA. However, when the N protein C-terminal domain (CTD) from MHV was swapped with sudden acute respiratory syndrome-associated CoV (SARS-CoV), significant increases in packaged sgRNAs were observed (6). These two studies propose two different models for selective packaging with M, or alternatively N, protein playing the dominant role in conferring packaging selectivity.
A subsequent report on the PS identified a 95-bp stem-loop structure within the previously described 190-bp region (7). Structural study of the PS from MHV, human coronavirus HKU1 (HCoV-HKU1), bovine CoV, and HCoV-OC43 revealed several conserved elements within the 95-bp stem-loop structure, including 2-nucleotide (nt) bulges, AGC/GUAAU motifs, and a pentaloop (Fig. 1A) (7). More recent studies utilizing 20 synonymous mutations predicted to abolish the PS stem-loop structure demonstrated that the secondary structure of the PS was necessary for selective packaging during MHV infection (8). In this instance, significantly higher levels of sgRNA were packaged into PS mutants than in wild-type MHV. Remarkably, this increase in sgRNA did not accompany equivalent decreases in packaging of gRNA or in viral titers (8). Thus, these PS mutants had no defect in virus production in tissue culture cells compared to wild-type MHV (8). This is in contrast to the role of the alphavirus packaging signal, where PS mutants also lead to increased sgRNA packaging, but at the expense of gRNA packaging and viral fitness in cultured cells (9). Retrovirus packaging signal mutants also lead to greatly decreased genomic RNA packaging with a concomitant increase in cellular mRNA packaging (10).
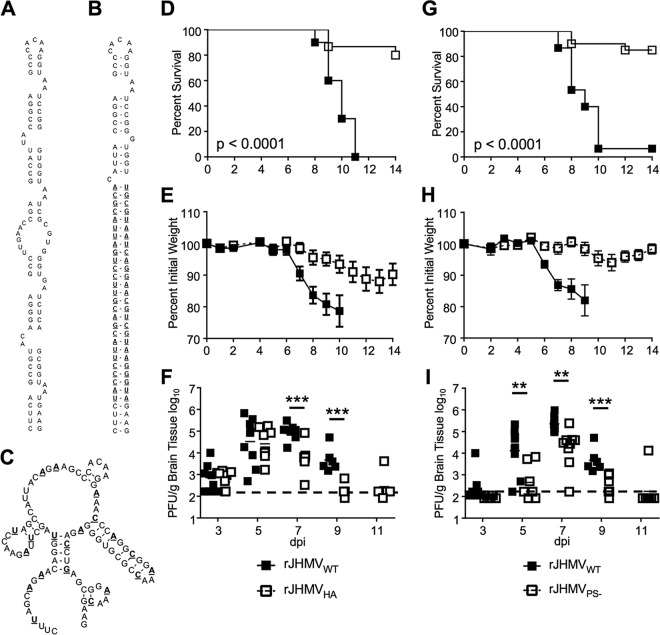
FIG 1 .
rJHMV packaging mutants are attenuated in vivo. (A) The RNA secondary structure of the MHV packaging signal (7). (B) Mfold predicted RNA secondary structure of rA59Nsp15-HA. HA epitope coding sequence and complement are boldfaced and underlined. (C) Mfold predicted RNA secondary structure of MHV PS−. Synonymous mutations are boldfaced and underlined. (D to F) Five- to 8-week-old male B6 mice were infected with 3 × 104 PFU of rJHMVNsp15-HA or rJHMVWT by intranasal inoculation. Infected mice were monitored for survival (D) and weight loss (E) for 14 dpi (rJHMVWT, n = 10; rJHMVNsp15-HA, n = 15). (F) Infected brains were harvested from mice at the indicated day postinfection. Brains were homogenized in PBS, and titers of infectious virus were determined by plaque assay on HeLa-MHVR cells. (G to I) Five- to 8-week-old male B6 mice were infected with 3 × 104 PFU of either rJHMVPS− or rJHMVWT by intranasal inoculation. Infected mice were monitored for survival (G) and weight loss (H) for 14 dpi (rJHMVWT, n = 15; rJHMVPS−, n = 20). (I) Infected brains were harvested from mice at the indicated day postinfection. Brains were homogenized in PBS, and titers of infectious virus were determined by plaque assay on HeLa-MHVR cells. The dashed line in panels F and I represents the limit of detection for the plaque assay. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Mann-Whitney test.
Since PS mutations in non-CoVs result in large decreases in gRNA incorporation into virions and diminished replication, infection with lineage A betacoronaviruses provides a unique opportunity to study the effect of selective viral packaging in vivo. Here, using two different PS mutants, we demonstrate that disrupting the selective packaging of MHV resulted in highly attenuated viruses in vivo, and this was true for A59 and JHMV, two different strains of MHV. Furthermore, the PS was important for repressing the type I interferon (IFN-I) response during MHV infection of bone marrow-derived macrophages (BMMs). Finally, IFN-I signaling was essential for the attenuation of selective packaging mutants as the virulence of P/S mutants was restored in interferon alpha/beta receptor-knockout (IFNAR−/−) mice or mice treated with IFNAR-blocking antibody. This study establishes the PS as a novel virulence factor for MHV, and likely other coronaviruses.
RESULTS
MHV packaging signal mutants are attenuated in vivo.
Previously, we inserted the RNA sequence encoding a hemagglutinin (HA) tag within the PS of the nsp15 locus (shown in Fig. 1A and B). This virus, termed rA59Nsp15-HA, replicated like wild-type virus but had increased levels of packaged sgRNA, indicating that selective packaging of gRNA was disrupted (11). Based on these observations, we next investigated the effect of selective packaging on virulence. B6 male mice were infected with rA59Nsp15-HA by intracranial injection and monitored for morbidity and mortality for 12 days postinfection (dpi). rA59Nsp15-HA-infected mice had decreased weight loss and increased survival compared to wild-type rA59 (rA59WT) (see Fig. S1A and B in the supplemental material). To test whether this phenotype was strain specific, we introduced the HA sequence into nsp15 of the rJHMV strain of MHV (rJHMVNsp15-HA), using a previously described full-length pBAC-based reverse genetics system (12). rJHMVNsp15-HA had growth kinetics in vitro equivalent to that of rJHMVWT (Fig. S2A), as previously described for rA59Nsp15-HA-infected cells (11). Mice infected with rJHMVNsp15-HA had substantially decreased weight loss and increased survival (Fig. 1D and E). While these data suggest that selective packaging is important for MHV virulence, we cannot rule out subtle effects of the HA tag on the function of nsp15 endoribonuclease, a known IFN-I antagonist. Further, addition of the HA sequence resulted in a 31-bp double-stranded RNA (dsRNA) stem, which could conceivably induce an innate immune response (13, 14).
rA59 packaging mutants are attenuated in vivo. (A and B) Five- to 8-week-old male B6 mice were infected by intracranial inoculation of 700 PFU of rA59Nsp15-HA or rA59WT and monitored for survival (A) and weight loss (B) for 10 dpi (rA59WT, n = 10; rA59Nsp15-HA, n = 10). (C and D) Five- to 8-week-old B6 mice were infected by intracranial inoculation of 700 PFU of rA59PS− or rA59WT and monitored for weight loss (C) and survival (D) for 12 dpi (rA59WT, n = 10; rA59PS−, n = 10). Download FIG S1, TIF file, 0.4 MB (371.6KB, tif) .
Copyright © 2018 Athmer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
rJHMV packaging mutants replicate the same as wild-type virus in tissue culture cells. (A and B) 17Cl-1 cells were infected at an MOI of 0.1 PFU/cell with rJHMVWT, rJHMVNsp15-HA (A), or rJHMVPS− (B), and viral progeny were collected at the indicated time points. Input virus was collected following the adsorption step. Virus titers were determined by plaque assay on HeLa-MHVR cells. The average and range for three biological replicates are plotted from a single experiment. Data are representative of two independent experiments. 3.2 and 4.2 represent two separate clones of rJHMVPS−. Download FIG S2, TIF file, 0.3 MB (280KB, tif) .
Copyright © 2018 Athmer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To separate the roles of nsp15 protein and of the PS in virulence, we constructed a second set of viruses with packaging defects in the context of both rA59 and rJHMV (termed PS−), as previously described (8). These mutants have synonymous mutations throughout the PS, which are predicted to alter the stem-loop structure of the PS without changing the amino acid sequence of nsp15 (Fig. 1C). The growth kinetics of two independent rJHMVPS− clones (3.2 and 4.2) was nearly identical to that of rJHMVWT in vitro (Fig. S2B). These data are consistent with previous results using rA59PS− and indicate that the PS has few, if any, effects on replication in vitro (8). After confirming the fitness of rJHMVPS− in vitro, we next assessed the virulence of rJHMVPS− in infected mice. B6 mice were infected intranasally with either rJHMVWT or rJHMVPS− and monitored for morbidity and mortality for 14 dpi. Intranasal infection of rJHMVWT resulted in nearly 100% mortality (Fig. 1G) and substantial weight loss (Fig. 1H). Mice infected with rJHMVPS− had significantly increased survival (Fig. 1G) and reduced weight loss (Fig. 1H). In agreement with results using rJHMV, mice infected with rA59PS− also had decreased weight loss and increased survival compared to mice infected with rA59WT (Fig. S1C and D).
We next measured the viral load of rJHMVWT, rJHMVPS−, and rJHMVNsp15-HA in the brains of infected mice. Both rJHMVWT and rJHMVNsp15-HA invaded the central nervous system (CNS) within 3 dpi and replicated to similar levels up to 5 dpi (Fig. 1F). By 7 dpi, titers in rJHMVNsp15-HA-infected mice were decreased in the CNS compared to those in mice infected with rJHMVWT. rJHMVNsp15-HA was cleared by 9 dpi. rJHMVPS− also invaded the CNS, albeit with a delay, and reached similar peak titers as rJHMVNsp15-HA but not rJHMVWT (Fig. 1I). rJHMVPS− titers decreased on 9 dpi and were cleared by 11 dpi (Fig. 1I). Thus, both PS mutant viruses did not reach the same peak viral titers as, and were cleared more rapidly than, wild-type JHMV. These data, utilizing two distinct PS mutant viruses, indicate that selective packaging of viral genomic RNA is essential for optimal virulence in multiple models of MHV infection.
rA59PS− and rA59Nsp15-HA have increased sgRNA and cellular RNA packaged into virions.
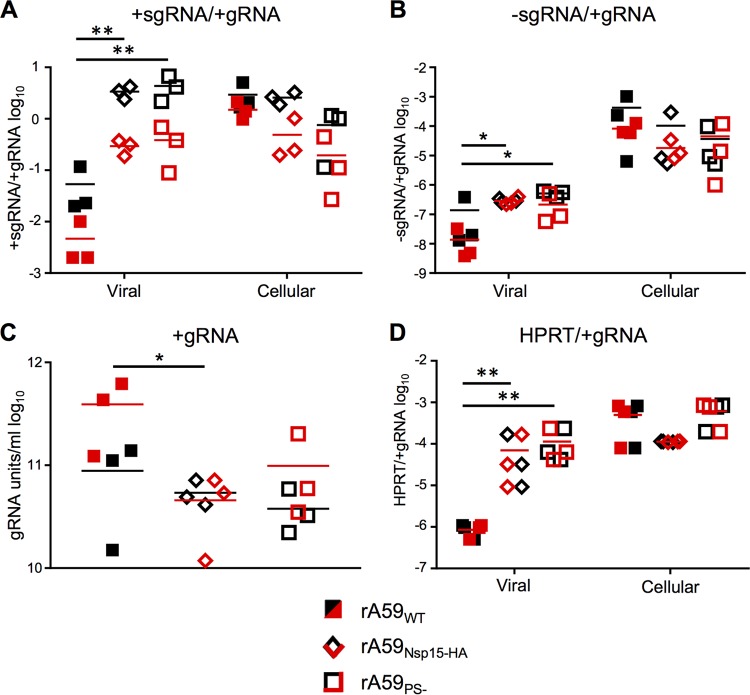
We next assessed the degree of the selective packaging defect observed in rA59PS− and rA59Nsp15-HA virions (Fig. 2). To accomplish this goal, the levels of positive- and negative-strand sgRNA7 (+sgRNA7 and –sgRNA7, respectively) and positive-strand gRNA (gRNA) were determined using concentrated virus samples prepared by velocity centrifugation of infected-cell supernatants. In agreement with previous publications (8, 11), both rA59PS− and rA59Nsp15-HA virions had higher levels of packaged +sgRNA7 than gRNA compared to rA59WT virions (Fig. 2A). Furthermore, the ratio of +sgRNA to gRNA in rA59PS− and rA59Nsp15-HA virions was nearly equivalent to the intracellular ratio in rA59PS−-infected and rA59Nsp15-HA-infected cells. This enrichment of +sgRNA7 to gRNA in rA59PS− and rA59Nsp15-HA virions contrasts with rA59WT, where the +sgRNA7-to-gRNA ratio was dramatically decreased compared to the intracellular ratio (Fig. 2A). An increased ratio of −sgRNA7 to gRNA was also observed in rA59PS− and rA59Nsp15-HA compared to rA59WT virions (Fig. 2B) but was not equivalent to the intracellular ratio of these RNAs. Finally, these increases in sgRNA7 incorporation had little effect on the levels of gRNA (Fig. 2C), as neither rA59PS− nor rA59Nsp15-HA virions had more than an ~2-fold decrease in packaged gRNA compared to rA59WT virions.
FIG 2 .
rA59PS− and rA59Nsp15-HA have increased sgRNA and cellular RNA packaged into virions. (A to C) Supernatants from rA59PS−-, rA59Nsp15-HA-, and rA59WT-infected 17Cl-1 cells were collected, and cell debris was removed. Virions were pelleted by ultracentrifugation through a 30% sucrose cushion, and viral RNA was isolated. Intracellular RNA was isolated from the 17Cl-1 monolayers using Trizol. The levels of +sgRNA7, −sgRNA7, and gRNA in concentrated virus and intracellular RNA were measured using RT-qPCR standard curves. The ratios of +sgRNA7 (A) and −sgRNA7 (B) to gRNA were calculated and plotted for viral and intracellular RNA. (C) The number of gRNA units per milliliter was calculated and plotted for each sample. (D) HPRT and gRNA were measured by RT-qPCR. Six biological replicates from two independent experiments are plotted; each experiment is plotted as either red or black. The ratio of HPRT to gRNA was calculated and plotted for viral and intracellular RNA. *, P < 0.05; **, P < 0.01 by Mann-Whitney test.
In retroviruses, the loss of gRNA packaging results in robust packaging of cellular RNA (10). To determine the extent of cellular RNA incorporation by MHV selective packaging mutants, we measured the ratio of hypoxanthine-guanine phosphoribosyltransferase (HPRT) RNA to gRNA in concentrated virus and intracellular RNA samples. We found a significant increase in the HPRT-to-gRNA ratio in both rA59PS− and rA59Nsp15-HA (Fig. 2D) compared to rA59WT virions. However, the ratio of HPRT to gRNA in rA59PS− and rA59Nsp15-HA virions was not equivalent to the intracellular ratio of HPRT to gRNA. These data establish that rA59PS− and rA59Nsp15-HA were similarly defective in selective packaging and suggest that cellular RNA can be packaged into virions at increased levels in selective packaging mutants.
rA59PS− induces increased levels of IFN-I in BMMs.
Because the two selective packaging mutants have similar phenotypes in vivo and similar packaging defects, we utilized only rA59PS− and rJHMVPS− to further investigate the mechanism of PS-mediated virulence. The elevated levels of sgRNA, especially –sgRNA, in virions may act as pathogen-associated molecular patterns (PAMPs), which may be sensed during entry, replication, or packaging of PS mutants. To initially test this hypothesis, we utilized bone marrow-derived macrophages (BMMs) because they are easily culturable, are highly susceptible to MHV, and are known to express IFN-I following infection (15). BMMs infected with rA59PS− had an ~2- to 3-fold increase in beta interferon (IFN-β) at 12 h postinfection (hpi) compared to rA59WT-infected BMMs (Fig. 3A). The increased IFN-I response was not due to increased replication of rA59PS− as rA59PS− infection resulted in a small (~1.5- to 2-fold) reduction in gRNA (Fig. 3B). In agreement with previous reports, we found that IFN-I production was dependent on MDA5 for both rA59WT and rA59PS− (Fig. 3C). Furthermore, there was still a small difference (~1.5-fold) in replication of rA59PS− in MDA5−/− cells (Fig. 3D). These results indicate that alteration of the packaging signal in MHV can lead to an enhanced innate immune response and perhaps a modest defect in replication in primary cells.
FIG 3 .

The PS is required to suppress IFN production in bone marrow-derived macrophages (BMDMs). Cultured BMDMs from B6 or MDA5−/− mice were infected at an MOI of 1 PFU/cell with either rA59WT or rA59PS−, and RNA was isolated at the indicated time points. The levels of IFN-I (A and C) and gRNA (B and D) were measured by qRT-PCR and normalized to HPRT levels. The average and standard error of the mean for six biological replicates are plotted from two independent experiments.
IFN-I signaling is required for rJHMVPS− attenuation.
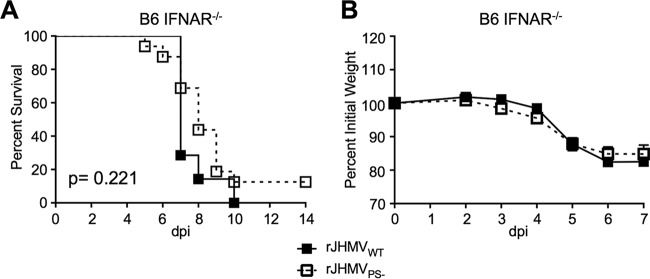
Increased levels of IFN-I in BMMs following rA59PS− infection and the well-established protective role of IFN-I signaling in MHV infection (16) suggested that IFN-I signaling would be important for rA59PS− or rJHMVPS− attenuation. Since rJHMV was more virulent than rA59, rJHMVPS− was used to test this hypothesis. IFNAR−/− mice were infected with rJHMVPS− or rJHMVWT and monitored for morbidity and mortality for 14 dpi. rJHMVWT-infected IFNAR−/− mice had substantial weight loss and succumbed to infection earlier than rJHMVWT-infected wild-type mice (Fig. 4A and B compared to Fig. 1D and G). IFNAR−/− mice infected with rJHMVPS− at the standard dose of 3 × 104 PFU succumbed to infection in a manner similar to infection with rJHMVWT (Fig. 4A and B). However, at a low dose of virus, 1 × 103 PFU, the virulence of rJHMVPS− was not completely restored (Fig. S3). The increased levels of IFN-β in rA59PS−-infected BMMs and rescued virulence of rJHMVPS− in IFNAR−/− mice suggest that IFN-I signaling plays an important role in the attenuation of rJHMVPS−. Additionally, other factors, such as delayed neuroinvasion, may also be important for the attenuation of PS− viruses.
FIG 4 .
IFN-I signaling is required for rJHMVPS− attenuation. (A and B) Five- to 8-week-old male IFNAR−/− B6 mice were infected with 3 × 104 PFU of rJHMVPS− or rJHMVWT by intranasal inoculation. Infected mice were monitored for survival (A) and weight loss (B) for 14 dpi (rJHMVWT, n = 7; rJHMVPS−, n = 14). *, P < 0.05 by Mann-Whitney test.
rJHMVPS− virulence is not fully restored in IFNAR−/− mice after infection with a lower dose of virus. (A and B) Five- to 8-week-old male IFNAR−/− B6 mice were infected with 1 × 103 PFU of rJHMVPS− or rJHMVWT by intranasal inoculation. Infected mice were monitored for survival (A) and weight loss (B) for 14 dpi (rJHMVWT, n = 6; rJHMVPS−, n = 10). Download FIG S3, TIF file, 0.3 MB (291.8KB, tif) .
Copyright © 2018 Athmer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Previous work has demonstrated protective roles for MDA5 and Toll-like receptor 7 (TLR7) during coronavirus infection (15, 17). Next, we utilized MAVS−/− mice to determine the role of MDA5-mediated signaling in attenuating rJHMVPS−. rJHMVWT caused a lethal infection in MAVS−/− mice, but rJHMVPS− remained attenuated, causing moderate weight loss and no decrease in survival (Fig. 5A and B). MHV induces significant levels of IFN-I in plasmacytoid dendritic cells via TLR7 signaling, contributing to protection (17). To determine the role of TLR7 signaling in protection, we infected TLR7−/− mice with rJHMVPS−. Since the TLR7−/− mice that were used were on a BALB/c background, we first confirmed that BALB/c mice were protected from rJHMVPS−-mediated lethality. rJHMVWT infection caused complete lethality and substantial weight loss, while rJHMVPS−-infected BALB/c mice had a phenotype similar to that observed in rJHMVPS−-infected B6 mice, with increased survival and less weight loss (Fig. 5C and D). TLR7−/− mice infected with rJHMVWT succumbed to infection, while we observed no increase in weight loss or lethality after infection with rJHMVPS− (Fig. 5E and F). These results suggest that neither signaling through MAVS nor that through TLR7 is solely responsible for attenuation of packaging signal mutants and suggest that initiation of IFN-I signaling occurs via alternate sensors or is multifactorial.
FIG 5 .
IFN-I signaling during rJHMVPS− is not solely dependent upon either TLR7- or MAVS-mediated signaling. (A and B) Five- to 8-week-old male MAVS−/− mice were infected with 3 × 104 PFU of either rJHMVPS− or rJHMVWT by intranasal injection. Infected mice were monitored for survival (A) and weight loss (B) (rJHMVWT, n = 7; rJHMVPS−, n = 12). (C and D) Five- to 8-week-old male BALB/c mice were infected with 1 × 104 PFU of either rJHMVPS− or rJHMVWT by intranasal inoculation. Infected mice were monitored for survival (C) and weight loss (D) (rJHMVWT, n = 5; rJHMVPS−, n = 5). (E and F) Five- to 8-week-old male TLR7−/− (BALB/c) mice were infected with 1 × 104 PFU of either rJHMVPS− or rJHMVWT by intranasal inoculation. Infected mice were monitored for survival (E) and weight loss (F) (rJHMVWT, n = 5; rJHMVPS−, n = 5). (G and H) Five- to 8-week-old male MAVS−/− (B6) mice were infected with 3 × 104 PFU of rJHMVPS− by intranasal injection. At the time of inoculation and at 3 dpi, 1.5 µg and 0.5 µg of IFNAR-blocking antibody were administered, respectively, by the i.p. route to infected mice, which were monitored for survival (G) and weight loss (H) (rJHMVWT, n = 7; rJHMVPS−, n = 7).
While neither TLR7 nor MAVS signaling was solely responsible for rJHMVPS− attenuation, it is possible that a protective level of IFN-I was still produced in infected MAVS−/− or TLR7−/− mice. IFNAR−/− mice are incapable of IFN-I signaling and therefore lack expression of interferon-stimulated genes (ISGs). In contrast, MDA5−/− mice infected with MHV had decreased but detectable levels of IFN-I, and the expression of many ISGs was not altered during MHV infection of MDA5−/− mice (18). Next, to determine if IFN-I signaling during infection was responsible for protecting mice or if basal levels of ISGs were playing a significant role, we treated MAVS−/− mice with an IFNAR-blocking antibody (19). In these experiments, IFNAR-blocking antibody was injected intraperitoneally 6 h prior to infection and again 3 days later. Control MAVS−/− mice administered rat IgG remained protected from rJHMVPS−; conversely, MAVS−/− mice administered IFNAR-blocking antibody displayed increased weight loss and succumbed to infection (Fig. 5G and H). These data suggest that IFN-I signaling during infection is important for the attenuation of rJHMVPS−.
DISCUSSION
Due to the largely unaltered growth of packaging mutants in vitro, the lineage A betacoronaviruses, specifically MHV, provide a unique opportunity to study the effect of viral packaging on virulence. This is not possible in both alphaviruses and retroviruses since packaging mutants have significant decreases in gRNA incorporation into virions and in infectivity (9, 10). Our data identify the lineage A betacoronavirus PS as a novel cis-acting virulence factor in MHV.
While the function of the lineage A betacoronavirus packaging signal is well understood in vitro, its relevance in vivo has not been addressed. The location of the stem-loop structure within the nsp15 locus allows these coronaviruses to selectively package gRNA and not coterminal sgRNA or host RNA. In this work, we quantified the packaging defect in rA59Nsp15-HA and rA59PS−. The defects present in these two different PS mutants were strikingly similar, despite rA59Nsp15-HA maintaining the overall structure, the pentaloop, and one of the 2-nucleotide (nt) bulges, elements hypothesized to be important for packaging (Fig. 1A and B). In agreement with previous publications (8), we found a significant increase in sgRNA packaging in both rA59PS− and rA59Nsp15-HA virions. We further divided the sgRNA into +sgRNA and −sgRNA and found increased levels of both in the virions of mutant viruses compared to parental viruses. Furthermore, we found increased levels of host cellular RNA in both packaging mutant virions. Interestingly, while the ratios of +sgRNA7 to gRNA in the virions of both rA59PS− and rA59Nsp15-HA were equivalent to the intracellular ratio, this was not true for ratios of –sgRNA7 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) to gRNA.
Packaging occurs through an interaction between the PS stem-loop and coronavirus structural proteins. A direct interaction between the M protein and the PS was previously reported and correlated with the packaging of nonviral RNA (20). It can be hypothesized that in the absence of M-PS interactions, packaging of RNA is dependent on N protein. Since N protein binds to RNA indiscriminately (5), levels of packaged RNA would be predicted to closely mimic those present in the intracellular environment. However, the ratio of both –sgRNA7 and GAPDH to gRNA was increased in virions but did not reach intracellular ratios, suggesting that RNA binding by N is not the sole determinant of RNA packaging in rA59PS− and rA59Nsp15-HA. RNA localization, secondary structure in other regions of the genome, or other proteins may influence RNA packaging. The relative lack of –sgRNA7 packaging may reflect its localization to membranous structures associated with replication while +sgRNA7 would be exported to the cytosol.
All packaging mutants tested were attenuated in mice. rJHMVPS− and rJHMVNsp15-HA, and rA59PS− and rA59Nsp15-HA, were attenuated in the context of lethal encephalitis (Fig. 1) and hepatitis (see Fig. S1 in the supplemental material), respectively. It can be hypothesized that packaged sgRNA and/or host RNAs behave as PAMPs, eliciting an increased IFN-I response in rJHMVPS−-infected and rJHMVNsp15-HA-infected cells. Consistent with this hypothesis, IFN-I signaling was essential for this attenuation in vivo (Fig. 3). Furthermore, BMMs infected with rJHMVPS− expressed higher levels of IFN-I than rJHMVWT (Fig. 3A). These data indicate that packaging mutants are capable of inducing an increased IFN-I response, which may be protective in vivo. We investigated the two best-described sensors for IFN-I signaling during coronavirus infection, MDA5 and TLR7. We found that MAVS−/− and TLR7−/− mice were both resistant to rJHMVPS−. These data indicate that neither TLR7- nor MAVS-mediated signaling was solely responsible for the attenuation of rJHMVPS−. Furthermore, MAVS−/− mice treated with IFNAR-blocking antibody were more susceptible to rJHMVPS− than control antibody-treated mice, suggesting that IFN-I was still responsible for rJHMVPS− attenuation in these mice. Together, these data support a role for IFN-I signaling in rJHMVPS− attenuation and suggest that TLR7 and MDA5 have a redundant function(s) or that other sensors/inflammatory pathways play a role in MHV packaging mutant attenuation.
The sgRNAs could act as PAMPs during the replication and/or entry of MHV packaging mutants. The presence of −sgRNAs in virions, albeit in small amounts, might be most important for the induction of IFN-I since negative-strand viral RNA is presumably not capped but, rather, contains a triphosphate at the 5′ end. This would facilitate recognition by the intracellular helical receptor RIG-I. Negative-sense sgRNA is expected to be present as double-stranded RNA (dsRNA), which could be detected by RIG-I, TLR3, and protein kinase R (PKR), in addition to MDA5. It should be noted that while our data suggest that IFN-I signaling and perhaps other changes in the innate immune response are largely responsible for rJHMVPS− attenuation, defects in virus replication in cells in the infected brain may also contribute to attenuation. We observed no differences between the replication kinetics of rJHMVPS− or rJHMVNsp15-HA and those of rJHMVWT in vitro (Fig. S1), but this does not preclude subtle differences in primary cells, such as BMMs (Fig. 3D).
In conclusion, our results demonstrate that the selective packaging of gRNA plays an important role in the virulence of MHV and probably other lineage A betacoronaviruses. This work uncovers a unique method by which an RNA virus shields its RNA products from the host to prevent an innate immune response. Selective packaging is conserved among coronaviruses and therefore is likely important for the virulence of most, if not all, coronaviruses. However, the localization of the packaging signal for most coronaviruses is unknown. Thus, an important goal for future research will be to identify the packaging signals in pathogenic human and animal coronaviruses and determine their roles in virulence.
MATERIALS AND METHODS
Cell culture.
17Cl-1 cells, HeLa cells expressing MHV receptor carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1a) (HeLa-MHVR), and baby hamster kidney cells expressing CEACAM1a (BHK-MHVR) were grown in Dulbecco’s modified Eagle’s medium (DMEM), with 10% fetal calf serum, 1% penicillin-streptomycin (Pen-Strep), 2% sodium bicarbonate, 2% l-glutamine, 1% nonessential amino acids, and 1% sodium pyruvate (D10). Low-serum medium is the same as D10 with 2% fetal calf serum (D2).
Mice.
Pathogen-free C57BL/6 (B6) and BALB/c mice were purchased from Charles River Laboratories. B6 interferon alpha/beta receptor-knockout (IFNAR−/−) mice were obtained from the Jackson Laboratory, and Toll-like receptor 7-knockout (TLR7−/−) mice were obtained from Shizuo Akira (Osaka University) via Westley Reeves (University of Florida). Mice were bred in the animal care facility at the University of Iowa. Animal studies were approved by the University of Iowa Animal Care and Use Committee (IACUC).
Generation of recombinant viruses.
pBAC-rJHMVPS− and pBAC-rJHMVNsp15-HA were created using pBAC-rJHM.IA, by Red recombination, and using the primers listed in Table S1 in the supplemental material. Using gBlock fragments with the corresponding mutations and listed primers, a PCR product was created with the HA or PS− substitution, kanamycin followed by an ISCE-I restriction site (KanI), and homologous arms. Either the KanI-HA- or KanI-PS−-containing product was transformed into competent GS1783 cells harboring the rJHMV.IA pBAC. Chloramphenicol-positive (CML+) Kan+ colonies were screened for intermolecular recombination by PCR and restriction enzyme digestion. Colonies with the correct intermediates were then used for intramolecular recombination of the homologous arms. Correct colonies were amplified, and the ISCE-I enzyme was induced with 1% arabinose for 2 h. The Lambda Red recombination enzymes were induced by heat shock at 42°C for 30 min, and cells were then incubated at room temperature for 4 h. CML+ Kan− colonies were screened for correct final products by PCR, restriction enzyme digestion, and sequencing. pBAC.IAPS− or pBAC.IANsp15-HA was cotransfected with plasmid expressing N protein into BHK-MHVR cells using Lipofectamine 2000 (Fisher Scientific) according to the manufacturer’s protocol. Transfected BHK cells were collected at the time of maximal cytopathic effect and lysed by freeze-thawing. rJHMVPS− and rJHMVNsp15-HA were then passaged 3 times to obtain working stocks. Two independent clones of the PS− virus (Fig. 1C), rJHMVPS−, were isolated, and the sequence of each clone was confirmed. No coding mutations were present in either clone; however, rJHMVPS−3.2 did contain a synonymous mutation in nsp15 3′ of the PS. It is unlikely that this mutation significantly alters the secondary structure in this region, and this isolate behaved identically to rJHMVPS−4.2. rJHMVNsp15-HA was sequenced after passaging with no mutations found.
Primers used to engineer recombinant viruses. Download TABLE S1, DOCX file, 0.2 MB (253.8KB, docx) .
Copyright © 2018 Athmer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To introduce the previously published PS− mutation (8) into rA59, the published mutations were introduced into the F plasmid of the rA59 system (21). The F plasmid was linearized, and homologous arms, with the PS− mutations, were added by PCR using the primers listed in Table S1. The linearized plasmid was recombined using the homologous arms and the In-Fusion cloning kit (Clontech) according to the manufacturer’s protocol. The PS− mutations were screened for by PCR and sequencing. The F plasmid and other fragments were digested with BsmBI and ligated together using T4 DNA ligase. Ligated DNAs and N protein transcript were transcribed in vitro using the mMessage mMachine T7 transcription kit (Fisher Scientific) according to the manufacturer’s protocol. BHK-MHVR cells were then transfected by electroporation with transcribed RNAs. Transfected BHK-MHVR cells were plated in D10 and monitored for cytopathic effects. Rescued recombinant viruses were collected, their titers were determined as described above, and they were termed rA59PS− P0. rA59PS− P0 stocks were used to generate rA59PS− P1 stocks by infecting 17Cl-1 cells with virus at a multiplicity of infection (MOI) of 0.1 PFU/cell and collecting cells at times of maximal cytopathic effects. Titers of P1 stocks of rA59WT and rA59PS− were determined, and the stocks were utilized for all experiments with rA59.
Tissue culture cell infection.
Unless otherwise noted, all infections were completed as follows. Semiconfluent monolayers of cells were infected on either 12-well or 24-well plates. Virus was adsorbed in DMEM at 37°C for 30 min at the indicated MOI. After adsorption, remaining virus was removed, and the cells were returned to growth medium for the indicated times.
Mouse infection.
For rJHMV infections, 5- to 8-week-old male mice were sedated using isoflurane and infected by the intranasal route with 3 × 104 PFU, unless otherwise stated, in 12 µl. In IFNAR-blocking experiments, 1.5 µg of InVivoMAb (Bio X Cell; MAR1-5A3) IFNAR-blocking antibody or rat IgG (MP Biomedical) was administered by intraperitoneal (i.p.) injection 6 h prior to infection and 0.5 µg was administered 3 days postinfection. For rA59 infections, 5- to 8-week-old male mice were sedated and infected intracranially with 700 PFU. All infected mice were weighed and monitored for 2 weeks and euthanized if the mouse dropped below 70% of initial body weight or became dehydrated, in accordance with University of Iowa IACUC-approved animal protocols.
RT-qPCR.
For cellular RNA experiments, RNA was isolated from cultured cells using Trizol (Fisher Scientific). cDNAs for all RNAs except sgRNAs (see below) were prepared using Moloney murine leukemia virus (MMLV) reverse transcriptase with random hexamers according to the manufacturer’s instructions. Real-time quantitative PCR (RT-qPCR) was performed using Applied Biosystems QuantStudio 3 and PowerUp Sybr green master mix (Applied Biosystems). All primers used for qPCR are listed in Table S2. Target gene levels were normalized to hypoxanthine-guanine phosphoribosyltransferase (HPRT) by the threshold cycle (CT) equation: ΔCT = CT of the gene of interest − CT of HPRT. All results are shown as a ratio to HPRT calculated as 2−ΔCT.
Primers used for qPCR. Download TABLE S2, DOCX file, 0.1 MB (140.2KB, docx) .
Copyright © 2018 Athmer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Selective viral RNA packaging.
Infected 17Cl-1 supernatants were collected at 20 hpi, when maximal cytopathic effects were observed but before cell lysis. Supernatants were clarified by centrifugation at 1,000 × g and filtered through a 0.45-µm filter. Clarified supernatants were overlaid on 30% sucrose in Na-Tris buffer (100 mM NaCl, 10 mM Tris-Cl, pH 8.0) and centrifuged at 27,000 rpm for 4 h at 4°C. Pelleted virions were resuspended in DMEM. Viral RNA and cellular RNA from infected monolayers were isolated using Trizol (Fisher Scientific) according to the manufacturer’s protocol. cDNA for viral RNAs was generated using Superscript IV (Fisher Scientific) and strand-specific primers (Table S2) according to the manufacturer’s protocol, while cDNA for HPRT was generated using random hexamers as described above. Quantification for viral RNAs was performed utilizing a standard curve, which was generated from reverse-transcribed PCR-derived gRNA and sgRNA products. Relative quantification was performed using ΔCT with normalization to gRNA. Data are presented as a ratio of +/−sgRNA7 RNA to gRNA, gRNA units per milliliter, or HPRT RNA to gRNA.
Viral titers.
Viral titers were determined by infecting HeLa-MHVR cells as described above. Following rA59 infection, a 1:1 ratio of 1.2% agarose and D2 was applied to cells and the cells were incubated for 24 h at 37°C. Following incubation, cell monolayers were fixed with 4% formaldehyde in phosphate-buffered saline (PBS) and stained with 0.1% crystal violet in 2% methanol. Following rJHMV infection, cells were returned to D2 medium for 16 h at 37°C. After incubation, a 1:1 mixture of 1.2% agarose and D2 with 1% neutral red was applied for 4 h at 37°C before counting plaques.
BMM cultures.
Male and female B6 mice were euthanized, and bone marrow was isolated from each femur and tibia. The bone marrow was treated with ammonium-chloride-potassium lysing buffer for 30 s and then washed three times with RPMI containing 10% fetal calf serum. Bone marrow cells were then strained through 70-µm strainers and counted. Bone marrow cells (5 × 105) were plated in BMM medium (RPMI, 10% fetal calf serum, 10% L929 cell supernatant, 1% Pen-Strep, 1% sodium pyruvate, 1% l-glutamine) and cultured for 4 days. Following the 4th day, medium was changed daily until the day of experiment (day 7). BMMs were infected as described above at an MOI of 1 PFU/cell.
Statistics.
Student’s unpaired t test or the Mann-Whitney U test was used to analyze differences in mean values between groups. All results are expressed as mean ± range or ±standard error of the mean where indicated. P values of ≤0.05 were considered statistically significant. Survival data were analyzed using Mantel-Cox tests (*, P < 0.05).
ACKNOWLEDGMENTS
This work was supported in part by grants from the NIH (RO1 NS36592 to S.P., R01 AI108197 to M.R.D., RO1 AI104002 to M.G., T32-GM007347 to K.W.G., and F30-AI129229 to K.W.G.) and the National Multiple Sclerosis Society (RG 5340-A-7 to S.P.).
Footnotes
Citation Athmer J, Fehr AR, Grunewald ME, Qu W, Wheeler DL, Graepel KW, Channappanavar R, Sekine A, Aldabeeb DS, Gale M, Jr, Denison MR, Perlman S. 2018. Selective packaging in murine coronavirus promotes virulence by limiting type I interferon responses. mBio 9:e00272-18. https://doi.org/10.1128/mBio.00272-18.
REFERENCES
- 1.Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snijder EJ, van der Meer Y, Zevenhoven-Dobbe J, Onderwater JJ, van der Meulen J, Koerten HK, Mommaas AM. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J Virol 80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fosmire JA, Hwang K, Makino S. 1992. Identification and characterization of a coronavirus packaging signal. J Virol 66:3522–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woo K, Joo M, Narayanan K, Kim KH, Makino S. 1997. Murine coronavirus packaging signal confers packaging to nonviral RNA. J Virol 71:824–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narayanan K, Makino S. 2001. Cooperation of an RNA packaging signal and a viral envelope protein in coronavirus RNA packaging. J Virol 75:9059–9067. doi: 10.1128/JVI.75.19.9059-9067.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo L, Koetzner CA, Masters PS. 2016. A key role for the carboxy-terminal tail of the murine coronavirus nucleocapsid protein in coordination of genome packaging. Virology 494:100–107. doi: 10.1016/j.virol.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen SC, van den Born E, van den Worm SH, Pleij CW, Snijder EJ, Olsthoorn RC. 2007. New structure model for the packaging signal in the genome of group IIa coronaviruses. J Virol 81:6771–6774. doi: 10.1128/JVI.02231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuo L, Masters PS. 2013. Functional analysis of the murine coronavirus genomic RNA packaging signal. J Virol 87:5182–5192. doi: 10.1128/JVI.00100-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim DY, Firth AE, Atasheva S, Frolova EI, Frolov I. 2011. Conservation of a packaging signal and the viral genome RNA packaging mechanism in alphavirus evolution. J Virol 85:8022–8036. doi: 10.1128/JVI.00644-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rulli SJ Jr, Hibbert CS, Mirro J, Pederson T, Biswal S, Rein A. 2007. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J Virol 81:6623–6631. doi: 10.1128/JVI.02833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Athmer J, Fehr AR, Grunewald M, Smith EC, Denison MR, Perlman S. 2017. In situ tagged nsp15 reveals interactions with coronavirus replication/transcription complex-associated proteins. mBio 8:e02320-16. doi: 10.1128/mBio.02320-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fehr AR, Athmer J, Channappanavar R, Phillips JM, Meyerholz DK, Perlman S. 2015. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J Virol 89:1523–1536. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kindler E, Gil-Cruz C, Spanier J, Li Y, Wilhelm J, Rabouw HH, Züst R, Hwang M, V’kovski P, Stalder H, Marti S, Habjan M, Cervantes-Barragan L, Elliot R, Karl N, Gaughan C, van Kuppeveld FJ, Silverman RH, Keller M, Ludewig B, Bergmann CC, Ziebuhr J, Weiss SR, Kalinke U, Thiel V. 2017. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog 13:e1006195. doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng X, Hackbart M, Mettelman RC, O’Brien A, Mielech AM, Yi G, Kao CC, Baker SC. 2017. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci U S A 114:E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roth-Cross JK, Bender SJ, Weiss SR. 2008. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J Virol 82:9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ireland DD, Stohlman SA, Hinton DR, Atkinson R, Bergmann CC. 2008. Type I interferons are essential in controlling neurotropic coronavirus infection irrespective of functional CD8 T cells. J Virol 82:300–310. doi: 10.1128/JVI.01794-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cervantes-Barragan L, Züst R, Weber F, Spiegel M, Lang KS, Akira S, Thiel V, Ludewig B. 2007. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 109:1131–1137. doi: 10.1182/blood-2006-05-023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zalinger ZB, Elliott R, Rose KM, Weiss SR. 2015. MDA5 is critical to host defense during infection with murine coronavirus. J Virol 89:12330–12340. doi: 10.1128/JVI.01470-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinto AK, Daffis S, Brien JD, Gainey MD, Yokoyama WM, Sheehan KC, Murphy KM, Schreiber RD, Diamond MS. 2011. A temporal role of type I interferon signaling in CD8+ T cell maturation during acute West Nile virus infection. PLoS Pathog 7:e1002407. doi: 10.1371/journal.ppat.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narayanan K, Chen CJ, Maeda J, Makino S. 2003. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J Virol 77:2922–2927. doi: 10.1128/JVI.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yount B, Denison MR, Weiss SR, Baric RS. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J Virol 76:11065–11078. doi: 10.1128/JVI.76.21.11065-11078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
rA59 packaging mutants are attenuated in vivo. (A and B) Five- to 8-week-old male B6 mice were infected by intracranial inoculation of 700 PFU of rA59Nsp15-HA or rA59WT and monitored for survival (A) and weight loss (B) for 10 dpi (rA59WT, n = 10; rA59Nsp15-HA, n = 10). (C and D) Five- to 8-week-old B6 mice were infected by intracranial inoculation of 700 PFU of rA59PS− or rA59WT and monitored for weight loss (C) and survival (D) for 12 dpi (rA59WT, n = 10; rA59PS−, n = 10). Download FIG S1, TIF file, 0.4 MB (371.6KB, tif) .
Copyright © 2018 Athmer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
rJHMV packaging mutants replicate the same as wild-type virus in tissue culture cells. (A and B) 17Cl-1 cells were infected at an MOI of 0.1 PFU/cell with rJHMVWT, rJHMVNsp15-HA (A), or rJHMVPS− (B), and viral progeny were collected at the indicated time points. Input virus was collected following the adsorption step. Virus titers were determined by plaque assay on HeLa-MHVR cells. The average and range for three biological replicates are plotted from a single experiment. Data are representative of two independent experiments. 3.2 and 4.2 represent two separate clones of rJHMVPS−. Download FIG S2, TIF file, 0.3 MB (280KB, tif) .
Copyright © 2018 Athmer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
rJHMVPS− virulence is not fully restored in IFNAR−/− mice after infection with a lower dose of virus. (A and B) Five- to 8-week-old male IFNAR−/− B6 mice were infected with 1 × 103 PFU of rJHMVPS− or rJHMVWT by intranasal inoculation. Infected mice were monitored for survival (A) and weight loss (B) for 14 dpi (rJHMVWT, n = 6; rJHMVPS−, n = 10). Download FIG S3, TIF file, 0.3 MB (291.8KB, tif) .
Copyright © 2018 Athmer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used to engineer recombinant viruses. Download TABLE S1, DOCX file, 0.2 MB (253.8KB, docx) .
Copyright © 2018 Athmer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used for qPCR. Download TABLE S2, DOCX file, 0.1 MB (140.2KB, docx) .
Copyright © 2018 Athmer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.