ABSTRACT
In soil, the link between microbial diversity and carbon transformations is challenged by the concept of functional redundancy. Here, we hypothesized that functional redundancy may decrease with increasing carbon source recalcitrance and that coupling of diversity with C cycling may change accordingly. We manipulated microbial diversity to examine how diversity decrease affects the decomposition of easily degradable (i.e., allochthonous plant residues) versus recalcitrant (i.e., autochthonous organic matter) C sources. We found that a decrease in microbial diversity (i) affected the decomposition of both autochthonous and allochthonous carbon sources, thereby reducing global CO2 emission by up to 40%, and (ii) shaped the source of CO2 emission toward preferential decomposition of most degradable C sources. Our results also revealed that the significance of the diversity effect increases with nutrient availability. Altogether, these findings show that C cycling in soil may be more vulnerable to microbial diversity changes than expected from previous studies, particularly in ecosystems exposed to nutrient inputs. Thus, concern about the preservation of microbial diversity may be highly relevant in the current global-change context assumed to impact soil biodiversity and the pulse inputs of plant residues and rhizodeposits into the soil.
IMPORTANCE With hundreds of thousands of taxa per gram of soil, microbial diversity dominates soil biodiversity. While numerous studies have established that microbial communities respond rapidly to environmental changes, the relationship between microbial diversity and soil functioning remains controversial. Using a well-controlled laboratory approach, we provide empirical evidence that microbial diversity may be of high significance for organic matter decomposition, a major process on which rely many of the ecosystem services provided by the soil ecosystem. These new findings should be taken into account in future studies aimed at understanding and predicting the functional consequences of changes in microbial diversity on soil ecosystem services and carbon storage in soil.
KEYWORDS: microbial diversity, soil organic matter, priming effect, functional redundancy, carbon mineralization
INTRODUCTION
Biodiversity enhances ecosystem stability and productivity. This assumption has been broadly verified for plant communities, thanks to the vast body of evidence from more than 200 years of studies, most of which were based on the manipulation of taxonomic diversity and/or diversity of functional groups (1–4). Compared to plant ecology, microbial ecology is still lacking demonstrations of the relationship between biodiversity and function (2, 5). Consequently, while it is widely recognized that microorganisms perform a crucial role in many key ecosystem functions involved in soil fertility and environmental and water quality (6), the importance of microbial diversity is still debated (7, 8). However, this question is critical when the impact of climatic changes (9) and land management (10–12) on microbial diversity in soil ecosystems is considered.
With hundreds of thousands of taxa per gram of soil, microbial diversity dominates soil biodiversity (8). Due to this enormous diversity, functional redundancy within the soil microbial component is assumed to be very high, so high, in fact, that it is still widely assumed that community diversity and composition are decoupled from functioning (8, 13). Nevertheless, functional redundancy may vary between the numerous processes driven by soil microorganisms (14). More precisely, processes carried out by small pools of microbial species might be more affected by changes in diversity than processes involving large numbers of microbial taxa (15, 16). Within the soil microbial community, the decomposition of organic matter is one of the most redundant functions since most soil microorganisms can be broadly grouped as heterotrophs. Such high redundancy was evidenced by several studies which reported that microbial diversity had no influence on a global estimation of organic matter decomposition based on total CO2 release (17, 18). However, total CO2 release integrates a large range of emission sources, ranging from labile to recalcitrant C-organic compounds, and the enzymatic capacities required to degrade recalcitrant compounds, compared to more labile C compounds, are provided by only a small pool of microbial species (16, 19, 20). This suggests that functional redundancy may decrease with increasing carbon source recalcitrance and that coupling of diversity with C cycling may change accordingly. As a result, modifications in microbial diversity may not necessarily affect the intensity of the broad CO2 flux itself (17, 18) but, rather, the composition of this flux in terms of the relative contributions of CO2 emitted from labile versus recalcitrant C sources. Studies providing such evidence should be highly relevant to predicting how much the current impact of anthropogenic activities and of climate changes on soil microbial diversity is likely to affect the carbon balance in soil.
The aim of this study was to investigate the impact of changes in microbial diversity (bacteria and fungi) on the decomposition of different C substrates in soil. We hypothesized that a decrease in microbial diversity would have less effect on the decomposition of allochthonous, easily degradable C sources than on more recalcitrant autochthonous C sources. To test this hypothesis, we took advantage of the particularities of the soil microbial component, i.e., microscopic size and high growth rates, which make it a highly suitable model that is easy to manipulate for testing a diversity function hypothesis under controlled conditions. We simulated an erosion of soil microbial diversity by using a dilution-to-extinction approach in a microcosm experiment (16, 21). This approach has advantages over other possible ways of manipulating diversity, such as community construction (22, 23), in that (i) it allows manipulation irrespective of the cultivability of the microbial species and (ii) it leads to the establishment of highly complex communities composed of several hundreds of different species, which is realistic with regard to the huge genetic diversity that characterizes the soil microbial world. 13C-labeled wheat residues were used as allochthonous carbon input to distinguish the relative contributions of mineralization from allochthonous (13C-CO2) and autochthonous (12C-CO2) carbon sources according to microbial diversity.
RESULTS
Microbial biomass.
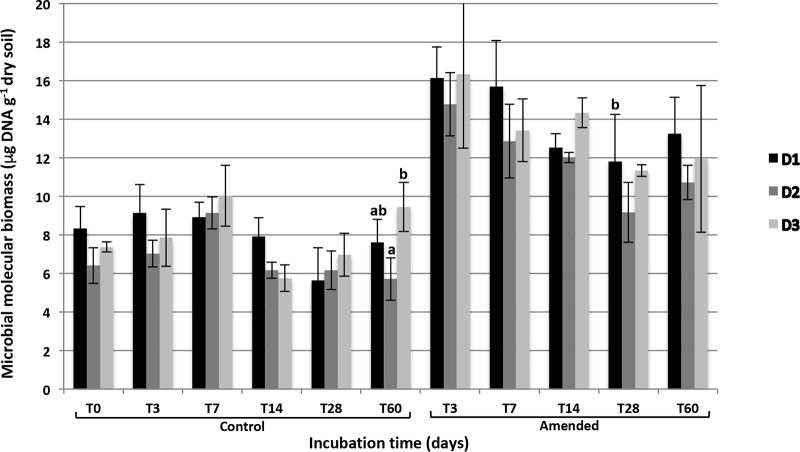
Microbial molecular biomass levels at time zero (T0; i.e., after 6 weeks of preincubation) were similar at all the three levels of diversity (D1 to D3) (Fig. 1). In the control microcosms, microbial biomass levels did not evolve with time and remained similar between D1, D2, and D3 at each sampling date throughout the 60 days of incubation (P > 0.05). Likewise, similar bacterial and fungal densities were observed at the three diversity levels (by quantitative PCR [qPCR]) (see Fig. S1 in the supplemental material). Incorporation of wheat residues increased microbial biomass in the amended microcosms, with significantly higher values than in controls occurring up to 14 days of incubation (P < 0.05). However, the increases in D1, D2, and D3 were similar, and no difference of biomass was observed according to the diversity level irrespective of the time of incubation time. Similarly, the bacterial and fungal densities estimated by quantitative PCR of the 16S and 18S rRNA genes did not differ between D1, D2, and D3 irrespective of the time of incubation (Fig. S1).
FIG 1.
Soil microbial biomass in control and wheat-amended microcosms for each of the three levels of diversity (D1, D2, and D3) during the incubation. Error bars denote standard deviations of biological replicates (n = 3).
Diversity of bacterial and fungal communities.
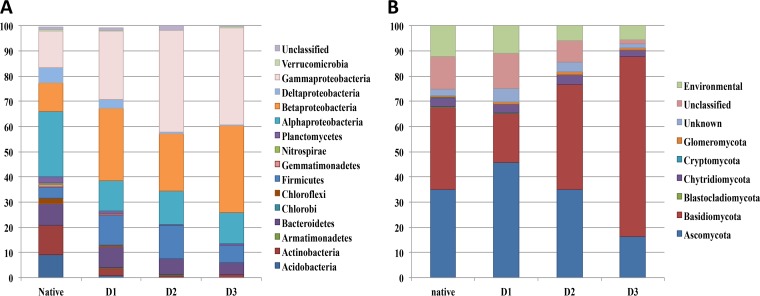
The numbers of sequences retained for each sample after the bioinformatics filters were 5,630 for bacteria and 4,763 for fungi. The number of sequences between the samples was standardized to allow comparison of the diversity metrics between the three levels of diversity (24). For bacterial communities, the operational taxonomic unit (OTU) richness was decreased by 30% in D1 compared to the level in the native community. This decrease was associated with a higher dominance of OTUs affiliated to Firmicutes and Beta- and Gammaproteobacteria and a decrease of in the levels of Acidobacteria, Actinobacteria, Planctomycetes, Chloroflexi, Gemmatimonadetes, and Deltaproteobacteria (Fig. 2A). When the diversity gradient was considered, the average OTU richness decreased significantly (P < 0.05), that is, by 30% and 52% in D2 and D3, respectively, compared to the level of D1 (Table 1). In the same way, the Shannon and evenness diversity indices evidenced a decrease in bacterial diversity along the dilution gradient and better evenness in the most species-rich communities. As observed between D1 and the native community, the decrease of evenness along the diversity gradient was associated with an increase in the dominance of OTUs affiliated to Beta- and Gammaproteobacteria and a decrease of Acidobacteria, Actinobacteria, Planctomycetes, Gemmatimonadetes, Chloroflexi, and Deltaproteobacteria (Fig. 2A).
FIG 2.
Relative abundances of bacterial (A) and fungal (B) phyla for the native community and each level of diversity (D1, high; D2, medium; D3, low) at T0. Only phylogenetic groups with a relative abundance of >1% are shown. Abundance of each phylum was determined from n = 3 biological replicates.
TABLE 1.
Diversity and microbial cooccurrence network indices of the microbial communitiesa
| Level of diversityb | Index of diversity by community |
Microbial cooccurrenced |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria |
Fungi |
|||||||||||
| Richnessc | Shannon index | Evenness index | Richness | Shannon index | Evenness index | Nb nodes | Total no. of links | No. of bacterial links | No. of fungal links | B/F | P/N | |
| Native | 1,004 ± 98.8 A | 5.48 ± 0.13 A | 0.79 ± 0.01 A | 449 ± 54.8 A | 3.87 ± 0.21 AB | 0.63 ± 0.07 A | 446 | 16,822 | 8,099 | 1,668 | 4.86 | 1.09 |
| D1 | 659 ± 19.4 B | 4.20 ± 0.09 B | 0.65 ± 0.02 B | 462 ± 51.8 A | 3.95 ± 0.31 A | 0.64 ± 0.04 A | 331 | 9,524 | 3,222 | 1,718 | 1.88 | 1.12 |
| D2 | 435 ± 110.7 C | 3.62 ± 0.54 B | 0.59 ± 0.06 B | 356 ± 51.5 A | 3.32 ± 0.29 AB | 0.57 ± 0.04 A | 186 | 3,718 | 1,499 | 532 | 2.89 | 1.07 |
| D3 | 313 ± 56.6 C | 2.97 ± 0.40 C | 0.51 ± 0.05 C | 194 ± 91.0 B | 2.51 ± 1.42 B | 0.47 ± 0.22 A | 107 | 1,367 | 593 | 219 | 2.71 | 1.09 |
Values were calculated from the sequencing data for the native community and for each level of diversity at T0 (i.e., the end of the 6 weeks of preincubation). For each level of diversity, values with different letters differ significantly (P < 0.05). Values are expressed as means ± standard deviation (n = 3 biological replicates).
D1, high; D2, medium; D3, low.
Richness refers to the number of OTUs defined at the genus level.
Nb nodes, number of taxa connected in the network; bacterial links and fungal links, links involving, respectively, bacteria only and fungi only; B/F, ratio between the numbers of bacterial and fungal links; P/N, ratio between the numbers of positive and negative links in the cooccurrence network.
The diversity metrics of the fungal communities in the native and the most-species-rich community, D1, were similar (Table 1). Composition at the phylum level was slightly different, mainly due to an increase of Ascomycota and a decrease of Basidiomycota in D1 (Fig. 2B). As described above for bacteria, the diversity of the fungal community also decreased with increased dilution, as evidenced by (i) the average OTU richness, which decreased significantly (P < 0.05) by 23% and 58% in D2 and D3, respectively, compared to the level in D1, and (ii) the values of the Shannon index (Table 1). Evenness of the fungal communities also decreased with decreasing diversity. This was mainly due to the increased dominance of OTUs affiliated to Basidiomycota over Ascomycota along the diversity gradient (Fig. 2B).
The complexity of the microbial community (bacteria and fungi) was lower in D1 than in the native community, as evidenced by the values of the microbial cooccurrence indices (Table 1). This was due to the decreased number of bacterial links, but not of fungal links, contributing to the network. As indicated above for the diversity metrics, the complexity of the microbial network decreased with dilution in terms of the number of nodes and links. However, values for the balance between positive and negative links were similar in all communities of the gradient at the level of the native community.
Soil and wheat residue C mineralization.
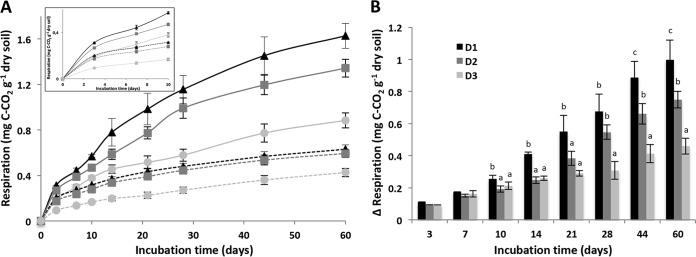
In the control microcosms, cumulative soil respiration (Rs) levels were similar for D1 and D2 but significantly decreased in D3, the lowest diversity level (P < 0.05) (Fig. 3A). This lower level was mainly ascribed to a lower respiration during the first 7 days of incubation than the rates for D1 and D2 (Fig. 3A). After 7 days, the respiration rates no longer differed between the three diversity levels. At the end of the incubation, 0.42 ± 0.04 mg g−1 soil C-CO2 had been emitted in D3, which was significantly lower, by 33% and 29%, than the values recorded for D1 (0.63 ± 0.04 mg g−1) and D2 (0.59 ± 0.03 mg g−1), respectively (P < 0.05).
FIG 3.
(A) Cumulative total soil respiration over 60 days in control (dashed lines) and wheat-amended (solid lines) microcosms for the three levels of diversity. Black, high diversity (D1); dark gray, medium diversity (D2); light gray, low diversity (D3). (B) Difference between respiration levels of control and wheat-amended microcosms for the three levels of diversity. For each incubation time and level of diversity, values with different letters differ significantly (P < 0.05). The inset in panel A represents a focus on the first 10 days to visualize the early shifts. Error bars denote standard deviations of biological replicates (n = 3).
Wheat residue incorporation significantly increased respiration irrespective of the soil diversity level (Fig. 3A). However, the stimulation of CO2 release increased with increasing microbial diversity (P < 0.05), leading to a clear gradient of respiration, with D1 > D2 > D3. More precisely, at the end of incubation the total C-CO2 emitted from the amended microcosms (Rt) ranged from 1.63 ± 0.109 (D1) to 1.34 ± 0.08 (D2) and 0.88 ± 0.07 (D3) mg g−1 soil, corresponding to decreases in C-CO2 release of 46% and 18%, respectively, in D3 and D2, compared to the D1 level. Analysis of the difference between the respiration levels in control and wheat-amended microcosms for each of the three levels of diversity (Fig. 3B) evidenced three distinct periods as follows: (i) an initial early phase represented by the first week following wheat incorporation and characterized by similar increases in respiration for each of the three diversity levels; (ii) a second phase between 10 and 21 days during which the respiration increase was significantly higher in D1 than in D2 and D3; and (iii) a third and later phase from 21 to 60 days characterized by a gradient in the respiration increase which followed the gradient of diversity.
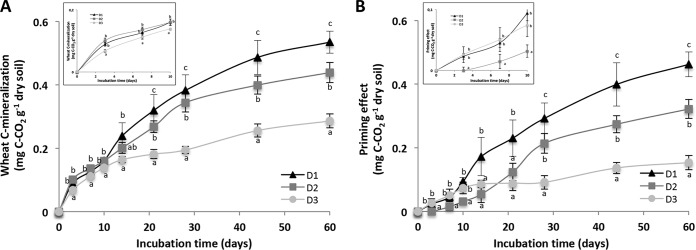
The increased C-CO2 release in amended microcosms was explained in part by wheat residue decomposition (Rr), which occurred in all three diversity levels (Fig. 4A). However, wheat mineralization increased significantly with increasing soil microbial diversity (P < 0.05), and at T60 days about 63%, 50%, and 35% of the wheat residues had been mineralized, respectively, in the high-, medium-, and low-diversity levels (D1, D2, and D3, respectively). Interestingly, the difference in residue mineralization between the three diversity levels increased with time (Fig. 4A).
FIG 4.
Cumulative respiration of wheat residues (A) and cumulative priming effect (B) were measured during 60 days of incubation for the three levels of diversity: D1, high diversity; D2, medium diversity; D3, low diversity. For each incubation time and level of diversity, values with different letters differ significantly (P < 0.05). Insets represent a focus on the first 10 days to visualize the early shifts. Error bars denote standard deviations of biological replicates (n = 3).
The release of 12C-CO2 (i.e., C-CO2 emitted through mineralization of the autochthonous carbon source) was also significantly stimulated, compared to the control level, following wheat residue incorporation. This pointed to a priming effect (PE) of native soil organic matter, which also contributed significantly to the increased C-CO2 release observed in amended microcosms (Fig. 4B). This PE occurred at all the three diversity levels, but its intensity was significantly and positively related to the diversity of the soil microbial community (P < 0.05) (Fig. 4B). At the end of the incubation, PE represented 0.46, 0.32, and 0.15 mg C g−1 soil, corresponding to increases of 73% ± 4%, 52% ± 3%, and 40% ± 1% in autochthonous carbon decomposition for D1, D2, and D3, respectively. In addition to the absolute amount of C-CO2 emitted through PE, the amount of C-CO2 emitted by PE per unit of wheat organic carbon (C-wheat) residue mineralized was also dependent on the diversity level, ranging from 0.53 ± 0.04 mg g−1 for D3 to 0.73 ± 0.02 mg g−1 for D2 and 0.86 ± 0.04 mg g−1 for D1. This indicated that, for a given quantity of C-wheat residue mineralized, the quantity of C-CO2 released through PE increased with increasing soil microbial diversity.
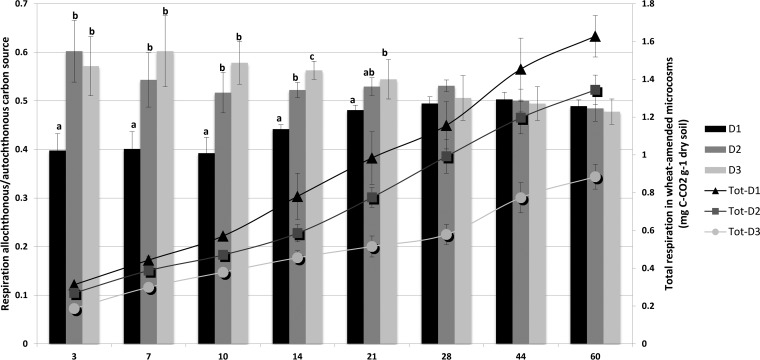
In amended microcosms, microbial diversity also impacted the composition of total CO2 flux in terms of the relative contribution of C-CO2 emitted from allochthonous wheat residues versus an autochthonous carbon source (Fig. 5 and Table S1). Most markedly, the contribution of C-CO2 emitted from wheat residues was significantly higher in the species-poor communities in D2 and D3 than in D1 up to 3 weeks.
FIG 5.
Relative contribution of allochthonous versus autochthonous carbon sources to total CO2 emission (bars) and cumulative total soil respiration in wheat-amended microcosms (curves) during 60 days of incubation for the three levels of diversity (D1, high; D2, medium; D3, low). For each incubation time and level of diversity, values with different letters differ significantly (P < 0.05). Error bars denote standard deviations of biological replicates (n = 3).
DISCUSSION
Community manipulation decreased microbial diversity in soil.
The aim of our study was to evaluate the effect of microbial diversity on carbon transformations in soil. To this end, we manipulated experimentally the diversity of a soil microbial community using a dilution-to-extinction approach (16, 18, 21).
Compared to the native community, the fungal community that established in the highest-diversity level, D1, harbored similar characteristics in terms of richness and complexity. In contrast, and despite the high values of diversity indices, the diversity of the bacterial community was lower in D1 than in the native community. This decrease was explained by a lower abundance of phyla reported to be mainly composed of slow-growing organisms, such as Acidobacteria, Actinobacteria, Planctomycetes, Chloroflexi, and Gemmatimonadetes (25–27), and an increased dominance of particular populations described as fast-growing r-strategists, mainly affiliated to Proteobacteria (mostly the Alphaproteobacteria and Gammaproteobacteria subclasses of Proteobacteria), and Firmicutes (25–27). Altogether, the shifts in diversity observed between the D1 and the native communities highlighted the finding that colonization impacted the bacterial community more than the fungal community. The observed changes are in agreement with the commonly accepted idea that bacteria are generally more opportunistic than fungi due to differences in physiology and metabolic capacities (19, 28, 29).
The dilution-to-extinction approach was successful in establishing gradients of bacterial and fungal diversity among the three diversity levels, with D1 > D2 > D3 (Table 1). It was clearly apparent that the decreased diversity could be ascribed to a loss of the least abundant (i.e., rare) species at the moment of dilution, as evidenced by the richness index, which was decreased by 34% and 52% for bacteria and by 23% and 58% for fungi in D2 and D3, respectively, compared to levels in D1. However, the ecological attributes of the inoculated populations may also have determined the fitness of these populations during soil colonization and, hence, determined the diversity of the finally established community (30). This may explain why the differences between the three diversity levels did not simply result from the arithmetic loss of microbial populations due to dilution (31). More precisely, the decrease of evenness observed for bacterial and fungal communities along the diversity gradient highlighted the finding that dilution led to the increased dominance of a particular subset of microbial populations at the end of colonization, in agreement with previous studies (30). However, the composition of the established communities suggested that the mechanisms that determined the success of the microbial populations in colonizing the soil differed between bacteria and fungi.
For bacteria, evenness was decreased along the gradient due to the increased dominance of the most opportunistic fast-growing r-strategists over the slower-growing populations (interestingly, the same phyla were involved as the phyla, discussed above, that differed between the native and D1 communities). This suggests that the growth rate may have been the main determinant of the success of bacterial populations during soil colonization. In contrast, for fungi the decrease of evenness was mainly associated with an increased dominance of Basidiomycota over Ascomycota. Both phyla are reported to be major contributors to C cycling in soil. However, Basidiomycota are assumed to possess improved metabolic capacities over Ascomycota for decomposing more recalcitrant soil C compounds (19, 32). Accordingly, our results suggest that the success of the fungal populations during the colonization phase may have been determined more by their metabolic abilities than by their growth rates. It is noticeable that this difference in colonization strategy between bacteria and fungi led to an increase in the relative importance of fungi in determining the properties of the microbial network in the inoculated soils, as evidenced by the decrease in the ratio of the numbers of bacterial-to-fungal links in D1 (1.88), D2 (2.89), and D3 (2.71) compared to the ratio in the native community (4.86). However, in our study this did not impact labile carbon availability in the three diversity levels, based on the size of the dissolved organic carbon pool as estimated by applying the method of Guigue et al. (33). At T0 the carbon pools were similar for D1 (737 ± 103 μg g−1 soil), D2 (532 ± 207 μg g−1 soil), and D3 (724 ± 28 μg g−1 soil).
In spite of large differences in terms of the levels of bacterial and fungal diversity, the microbial biomasses and densities at T0 were similar for all three diversity levels (Fig. 1). This indicated that after inoculation, the microbial communities developed until the soil carrying capacity was attained (16, 34), which was not dependent on the diversity of the soil microbial community. The microbial biomass in the controls remained stable throughout the experiment but was similarly increased by the addition of wheat residues in all three diversity levels. The possibility that any further observed shifts in respiration rates with dilution would be due to a community biomass effect could therefore be excluded.
Decrease in microbial diversity affects the decomposition of allochthonous and autochthonous carbon sources.
High microbial diversity generally stimulated the decomposition of both autochthonous and allochthonous carbon sources (Fig. 3 and 4). In the controls, basal respiration (i.e., autochthonous organic matter decomposition) was not affected by the decrease of microbial diversity at the two highest diversity levels, D1 and D2 (Fig. 3A), in agreement with the functional redundancy hypothesis (18). However, respiration decreased by 33% in the most species-poor community, D3, compared to that of D1, highlighting the limits of functional redundancy to buffer the effect of diversity loss on ecosystem processes (30, 35). This was unexpected, however, since previous studies had not shown any impact of an even greater decrease in microbial diversity on basal respiration (18, 36). This discrepancy with previously published data may be attributed to our experimental design, which included mechanical mixing of the soil in the controls to mimic the disturbance associated with the incorporation of wheat residues in amended microcosms. Mechanical disturbance is known to release additional C substrates due to the disruption of soil aggregates and hence to increase nutrient availability for microbes, particularly during the first days following the perturbation (37). Such a transitory increase of nutrient availability may have contributed to the functional dissimilarity observed in the lowest diversity level (D3) during the first week of incubation, in agreement with van der Heijden et al. (6), who proposed that the relationship between microbial diversity and ecosystem functioning may differ between nutrient-poor and nutrient-rich ecosystems. The importance of nutrient availability was verified in recent studies that reported an increase of the diversity effect with increasing nutrient availability for denitrification or of microbial stability following disturbance (30, 34).
In our study, an increase of the functional significance of microbial diversity with increasing nutrient availability was further evidenced by the results obtained in wheat-amended microcosms. As expected, C-CO2 release was increased following wheat residue input, corresponding to the input of an easily degradable C substrate (38), which stimulated microbial biomass during the first 2 weeks of incubation (Fig. 1). However, whereas the increases in microbial biomass were similar at all three diversity levels, decreasing microbial diversity had a strong and highly significant effect on decreasing C-CO2 emission. In contrast to the above-mentioned results obtained for basal respiration, no saturation effect was observed in amended microcosms since the total C-CO2 released after 60 days was decreased, respectively, by 18% and 44% in D2 and D3 compared to the D1 level (Fig. 3A). Altogether, our results indicate that functional redundancy may have been overestimated and that C transformations in soil may be more sensitive to microbial diversity changes than previously expected, particularly in ecosystems exposed to nutrient inputs.
Interestingly, the difference in C-CO2 (delta C-CO2) amounts released between the controls and their respective amended treatments for each of the three diversity levels increased with time (Fig. 3B). We suggest that this increase of functional dissimilarity between the three diversity levels with time may in part be explained by (i) the chemical and structural complexity of the allochthonous C source and (ii) the sequential decomposition of the different components of wheat residues that occurred during the degradation process from labile (i.e., sugars) to more recalcitrant (i.e., lignin) compounds (39). While the labile fraction of wheat residues provides easily available C and energy sources for many microorganisms (i.e., highly redundant function), the cometabolic nature of lignin breakdown depends on energy-providing processes and requires a specific set of enzymes and microbial populations (i.e., weakly redundant function) (19, 20). Accordingly, during the first week of decomposition, similar values of delta C-CO2 were observed at all three diversity levels, reflecting the availability and decomposition of the most degradable wheat compounds. After 7 days, the decrease of delta C-CO2 observed in D3 and D2 compared to that of D1 may reflect the increased recalcitrance of residues due to prior decomposition of the most degradable compounds, hence requiring more specialized functions carried out by a less redundant community and more likely to be represented in the most species-rich community. Finally, the gradient observed of D1 > D2 > D3 after 28 days may be explained by the subsequent increased recalcitrance of the remaining residues. Altogether, these results show that functional redundancy decreases with increasing carbon source recalcitrance, leading to an increased effect of biodiversity with time following allochthonous C source inputs. To our knowledge, the dynamic of the diversity effect following an allochthonous C source input has not been reported previously. However, it may be highly relevant in the current global-change context assumed to impact the pulse inputs of plant residues and rhizodeposits into the soil (40, 41).
In addition to CO2 emitted from wheat decomposition, the stimulation of autochthonous carbon mineralization after wheat residue addition to the soil, i.e., the priming effect, also contributed significantly to the increased CO2 release in amended microcosms (Fig. 4B). This was in agreement with numerous studies that reported a PE following the incorporation of plant residues into the soil (26, 42–44). However, here we provide the proof of concept that PE is positively dependent on microbial diversity since it decreased along the diversity gradient (Fig. 4B). This diversity effect was highly significant since autochthonous C mineralization was 30% higher in D1 than in D3. In addition to the amount of C-CO2 emitted through PE, the efficiency of C-wheat to produce C-CO2 through PE increased with increasing microbial diversity (0.86, 0.73, and 0.53 C unit were emitted per unit of C-wheat mineralized in D1, D2, and D3, respectively). Different microbial mechanisms have been proposed to explain the PE (27, 45, 46). However, in our study involving an erosion of microbial diversity, the differences in PEs and better efficiency observed according to diversity may be mainly explained by an increased cometabolism process in species-rich communities: with autochthonous C decomposers (i.e., K-strategist populations carrying a specific set of enzymes) more likely to be present in species-rich communities and degrading recalcitrant soil organic matter (SOM) compounds by using the allochthonous C source as energy source (47).
The dynamics of the ratio between C-CO2 released from allochthonous sources to that from autochthonous C sources provided additional interesting information in this sense (Fig. 5). According to our hypothesis, the relative contribution of C-CO2 emitted from the allochthonous C source was increased in the most-depauperate communities (D2 and D3) up to 3 weeks after wheat residue incorporation into the soil, indicating that species-poor communities preferentially decomposed the more degradable allochthonous C sources rather than the autochthonous C source. However, after 3 weeks, similar values of this ratio were observed at all three diversity levels, probably resulting from the establishment of limiting conditions due to nutrient depletion and a higher recalcitrance of the remaining residues (39), which were becoming close to the recalcitrance of the autochthonous C sources.
In our study based on the use of the dilution-to-extinction approach to manipulate bacterial and fungal diversity, it must be kept in mind that higher trophic-level organisms, i.e., specialized predators like protozoa, are likely to have been influenced most by dilution because these organisms are less abundant in soil food webs (48). More precisely, considering that protozoa are present at an average density of 106 g−1 soil in most soils, they may have been weakly present in D3 (dilution 10−5) compared to their levels in D1 and D3. Given the importance of these organisms for ecosystem functions (for a review, see reference 49), the loss of protozoa, in addition to the erosion of the fungal and bacterial diversity discussed above, may have contributed to the changes in C-CO2 emissions observed among the three diversity levels. The loss of protozoa may, indeed, possibly have decreased the availability of macronutrients in D3 through the decrease of the release of N by bacterivores and/or the immobilization of N in the microbial biomass due to lower stimulation of microbial activity and turnover, hence limiting the mineralization of organic carbon (49).
This food web hypothesis may explain the slight decrease of mineralization observed in D3 compared to that of the two other diversity levels in the controls. However, it cannot explain the gradient of mineralization observed following wheat addition between D1, D2, and D3, despite the presence of protozoa in D1 and D2. In addition, it was globally estimated in a recent meta-analysis that grazing decreases the soil microbial biomass and bacterial abundance by 16 and 17%, respectively (49). In our study, neither the microbial biomass nor the bacterial density was decreased between the three diversity levels, suggesting that the grazing pressures were similar in the three diversity levels. In another respect, our results show that the priming effect decreased with the decrease in microbial diversity. Again, this is not in agreement with the food web hypothesis since a decrease in macronutrient availability would, on the contrary, have increased the intensity of the PE due to stimulation of the nutrient-mining mechanism (44, 50, 51) that has been demonstrated to take place in cases of nutrient depletion. Finally, the hypothesis of a decrease of nutrient availability is not in agreement with the decrease of the decomposition of lignin following the erosion of diversity previously reported in the context of the same experiment (16). Limiting-nitrogen conditions were, on the contrary, shown to induce ligninolytic activity (52). Altogether, these elements highlight the finding that the decrease of mineralization observed between D1, D2, and D3 was ascribed to the decrease of microbial diversity rather than to the loss of higher trophic levels.
In conclusion, our results demonstrate that C cycling in soil may be more vulnerable to microbial diversity changes than expected from previous studies. They indicate mainly (i) that a decrease of soil microbial diversity affected the decomposition of both autochthonous and allochthonous carbon sources, hence reducing global CO2 emission (i.e., emission from allochthonous and autochthonous sources) by up to 40%, (ii) that this decrease in diversity modified the source of CO2 emission toward preferential decomposition of allochthonous C-substrates, and (iii) that the significance of the diversity effect increases with nutrient availability. Altogether, these highly relevant new findings should be taken into account in future studies aiming to understand and predict the functional consequences of decreased microbial diversity on soil ecosystem services and carbon storage in soil, particularly in the current context of global changes assumed to impact both microbial diversity and the pulse inputs of plant residues and rhizodeposits into the soil (i.e., the functional significance of the microbial diversity variable in predictive models of C turnover in soil will be increased with the increase of C substrate availability). The coupling of community diversity with ecosystem functioning also implies that concerns about the need to preserve biodiversity are also relevant for soil microbial communities.
MATERIALS AND METHODS
Plant culture and 13C-labeling.
Seeds of wheat (Triticum aestivum cv. Caphorn) were germinated at 4°C in darkness. Plantlets were grown in a mix of sand (1/4) and perlite (3/4) in an airtight chamber which allowed accurate regulation of atmospheric gas composition and environmental parameters (GRAP, CEA Cadarache, France). The plants were watered with half-diluted Hoagland's nutrient solution, and CO2 concentration was maintained at 380 μl liter−1. The partial pressures of both 13CO2 and 12CO2 in the chamber were continuously monitored by near-infrared spectroscopy to determine 13C enrichment of the CO2. Regulation was achieved by automatic injection of pure (>99 atom% 13C) 13CO2 (purchased from CortecNet, Paris, France). The 13C isotope excess in the chamber was fixed at >80 atom% during the first 10 days and at >90 atom% thereafter. Plants were grown for 196 days (i.e., to maturity). The roots and shoots were then separated and dried before further use. The wheat was thus labeled with 13C at 96 atom% and characterized by a C/N ratio of 78.2. Only the shoots (aerial parts) were used for the experiment; the roots were discarded.
Soil.
Soil samples were taken from the top 10 cm of a Cambisol on a temporary grassland site, part of a long-term observatory (LTO) for environmental research (Lusignan LTO-ACBB, INRA), located in the southwest of France (46°25′12.91″ N, 0°07′29.35″ E). The soil pH in water (pHH2O) was 6.6, and the soil contained 17.5% clay, 36.9% fine silt, 30.4% coarse silt, 15.2% sand, <1 g kg−1 CaCO3, 2.29% organic C, and 0.13% total N. The C/N ratio was 9.9. The soil was sieved at 4 mm and homogenized prior to gamma ray sterilization (35 kGy; Conservatome, Dagneux, France). The sterility of the irradiated soil was verified by spreading serial dilutions of the soil onto nutrient agar plates.
Experimental design.
Microcosms were set up by placing 40 g of dry sterile soil in 150-ml plasma flasks. Sterile soil microcosms were inoculated with suspensions of the same nonsterile soil (16, 30, 34). Briefly, an initial soil suspension was prepared by mixing 100 g of native soil (equivalent dry mass) with 300 ml of sterile distilled water using a Waring blender. After the soil suspension was subjected to blending for 5 min at maximum speed, it was serially diluted. Three levels of dilution of the soil suspension were used as inocula to create a gradient of diversity, i.e., undiluted (100; D1), 1/103 dilution (D2), and 1/105 dilution (D3); the soil moisture was fixed at 60% water-holding capacity. After inoculation, the microcosms were closed to avoid air contamination and preincubated for 6 weeks at 20°C (i.e., average autumn temperature at the sampling site) with weekly aeration and verification of soil water content to allow colonization and stabilization of the inoculated communities in terms of density and structure. On day 0 (T0; after 6 weeks of preincubation), for each diversity level, half of the microcosms were amended with 13C-labeled wheat residues (dried, ground to <1 mm, 96 atom% 13C labeled), incorporated at a rate of 5 mg g−1, while the other half were not amended (control). The wheat residues were homogeneously incorporated into the soil by mixing. The soil in the control microcosms was also mixed at T0 to avoid any bias due to the mixing step. The soil microcosms were then incubated in the dark at 20°C for 60 days. During incubation the soil moisture was maintained by replacing weight loss with autoclaved reverse-osmosis water when necessary. In total, there were 90 microcosms corresponding to 2 treatments (wheat-amended versus control) times 3 levels of diversity times 5 sampling dates times 3 replicates. At T0, labile carbon availability was determined in triplicate microcosms for each diversity level by quantifying the size of the dissolved organic carbon pool after extraction of the water-extractable organic matter by percolation at high pressure and temperature (33).
Total and 13C-CO2 measurements.
At T0, T3, T7, T10, T14, T21, T28, T44, and T60 days of incubation, the gaseous phase of the microcosm was sampled in 10-ml airtight flasks to measure the CO2 concentration and in 12-ml airtight flasks to determine the 13C-CO2 enrichment. CO2 concentration was determined on an Agilent 7890 GC system equipped with a Pora-Plot Q column and a thermal conductivity detector coupled to an automatic sampler (G1888 headspace sampler; Agilent). Carbon isotopic enrichment was determined by gas chromatography isotope ratio mass spectrometry (GC-IRMS) using a trace gas interface coupled in continuous flow with a VG Isochrom mass spectrometer (Micromass, Manchester, England). For isotopic CO2, a pure bottle (CO2 of >99.999% [N48; Air Liquide, France]), previously calibrated against a certified isotopic standard (δ13C = −25.5‰ ± 0.2‰ versus the Pee Dee belemnite [PDB] standard; Isotop, Air Liquide), was selected as internal standard. Gas samples were manually injected into the trace gas with a gas-tight syringe.
By using 13C-labeled plant residues, the autochthonous soil C (Rs) and plant residue C (Rr) respiration (milligrams of C-CO2 kilogram−1 soil) could be calculated from the following mass balance equations: Rs + Rr = Rt and Rs × As13 + Rr × Ar13 = Rt × At13, where As13 is the 13C abundance (dimensionless) of soil carbon, Ar13 is the 13C abundance of plant residue, Rt is the total CO2 emitted by soil with plant residue, and At13 is its 13C abundance.
The priming effect (PE; milligrams of C-CO2 kilogram−1 soil) induced by the addition of plant residue was calculated as PE = (Rs of soil with plant residue) − (Rs of control soil), where (Rs of control soil) was the CO2 emitted by control soil.
DNA extraction from soil and molecular microbial biomass determination.
At T0, T7, T14, T28, and T60 days of incubation, DNA was extracted from 2 g of soil from all triplicate microcosms of each treatment and diversity level and quantified as previously described (11). DNA was also extracted from triplicates of native soil samples. DNA concentrations of crude extracts were determined by electrophoresis in a 1% agarose gel using a calf thymus DNA standard curve and used as estimates of microbial molecular biomass (53). After quantification, DNA was purified using a MinElute gel extraction kit (Qiagen, Courtaboeuf, France).
Pyrosequencing of 16S and 18S rRNA genes.
Bacterial and fungal diversity was determined by 454 pyrosequencing of ribosomal genes in the three replicates of each of the three diversity levels at T0, i.e., at the end of the 6 weeks of preincubation, and in three replicates of the native soil. For bacterial communities, a 16S rRNA gene fragment with sequence variability and the appropriate size (about 450 bases) was amplified using the primers F479 and R888. For fungal communities, an 18S rRNA gene fragment of about 350 bases was amplified using the primers FR1 and FF390 (54). Primers and PCR conditions were as described previously (11). Finally, the PCR products were purified and quantified, and pyrosequencing was carried out on a GS FLX Titanium 454 sequencing system (Roche).
Taxonomic assignments and clustering of 16S and 18S rRNA gene fragments.
Bioinformatics analyses were done with GnS-PIPE developed by the GenoSol platform (INRA, Dijon, France) and described previously (11). First, all reads were sorted according to the chosen multiplex identifiers. Then, raw reads were filtered and deleted as follows: (i) if the exact primers were not found both at the beginning and end of the sequence, (ii) if the sequences contained any ambiguity, or (iii) if the sequence length was below 350 and 300 bases for 16S and 18S reads, respectively. A PERL program was then applied to obtain strict dereplication. The dereplicated reads were then aligned using Infernal alignments (55) and clustered into operational taxonomic units (OTU) at 95% sequence similarity using a PERL program that clusters rare reads to abundant ones and does not count differences in homopolymer lengths (main bias of pyrosequencing technologies). Another homemade filtering step was then applied to eliminate potential sources of error (e.g., PCR chimeras, sequencing errors, or OTU overestimation) based on taxonomic results. To efficiently compare the data sets and avoid biased community comparisons, high-quality reads were rarefied by random selection closed to the lowest data sets (5.630 and 4.763 reads for 16S and 18S rRNA gene sequences, respectively). Kept reads were then compared for taxonomy-based analysis against the Silva database (version r111) using similarity approaches (USEARCH). Finally, diversity indexes were determined using the detected taxonomic groups at the genus level. The numbers of OTUs and the Shannon (H′) and evenness (J) indexes were used as indicators of soil microbial richness, diversity, and structure, respectively.
Statistical analysis.
Molecular biomass and diversity indexes were compared between the three levels of diversity by applying a nonparametric Kruskal-Wallis test. For gaseous data, statistical analyses were conducted using two-way analysis of variance (ANOVA), and differences between means were tested with the Fisher test (P ≤ 0.05). These statistical calculations were carried out using XLSTAT software (Addinsoft, Paris, France). Microbial cooccurrence networks were built using a previously described method (56). For each dilution level and for the native soil, the core community of OTUs in the three replicates was used to identify the significant Spearman correlations (corrected with the false discovery rate method; P < 0.05) that were interpreted as nonrandom cooccurrences. The bacterial, fungal, and combined networks were computed to estimate the proportion of links involving bacteria only, fungi only, or both bacteria and fungi. The calculation and definition of the network metrics were detailed previously in Karimi et al. (57).
Accession number(s).
The nucleotide sequences determined in this study have been submitted to the European Bioinformatics Institute (EBI) database under accession number PRJEB19513.
Supplementary Material
ACKNOWLEDGMENTS
This study was financially supported by the French National Research Agency (ANR) under the framework of the ANR Systerra project DIMIMOS (ANR-08-STRA-06).
We thank D. Warwick for correction and improvement of the English in the manuscript.
P.-A.M. and L.R. designed the study; A.S., A.K., J.L., O.M., A.C., and J.G. performed the research; S.T., L.B., B.K., and S.D. performed bioinformatic and statistical analyses; P.-A.M. wrote the first draft of the manuscript, and all authors contributed substantially to the revision.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02738-17.
REFERENCES
- 1.Lawes JB, Gilbert JH, Master MT. 1882. Agricultural, chemical, and botanical results of experiments on the mixed herbage of permanent grasslands, conducted for more than twenty years in succession on the same land. Part II. The botanical results. Philos Trans R Soc Lond 173:1181–1423. [Google Scholar]
- 2.Balvanera P, Pfisterer AB, Buchmann N, He JS, Nakashizuka T, Raffaelli D, Schmid B. 2006. Quantifying the evidence for biodiversity effects on ecosystem functioning and services. Ecol Lett 9:1146–1156. doi: 10.1111/j.1461-0248.2006.00963.x. [DOI] [PubMed] [Google Scholar]
- 3.Campbell V, Murphy G, Romanuk TN. 2011. Experimental design and the outcome and interpretation of diversity-stability relations. Oikos 120:399–408. doi: 10.1111/j.1600-0706.2010.18768.x. [DOI] [Google Scholar]
- 4.Delgado-Baquerizo M, Maestre FT, Reich PB, Jeffries TC, Gaitan JJ, Encinar D, Berdugo M, Campbell CD, Singh BK. 2016. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat Commun 7:10541. doi: 10.1038/ncomms10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reed HE, Martiny JBH. 2007. Testing the functional significance of microbial composition in natural communities. FEMS Microbiol Ecol 62:161–170. doi: 10.1111/j.1574-6941.2007.00386.x. [DOI] [PubMed] [Google Scholar]
- 6.van der Heijden MGA, Bardgett RD, van Straalen NM. 2008. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett 11:296–310. doi: 10.1111/j.1461-0248.2007.01139.x. [DOI] [PubMed] [Google Scholar]
- 7.Setälä H, McLean MA. 2004. Decomposition rate of organic substrates in relation to the species diversity of soil saprophytic fungi. Oecologia 139:98–107. doi: 10.1007/s00442-003-1478-y. [DOI] [PubMed] [Google Scholar]
- 8.Allison SD, Martiny JBH. 2008. Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci U S A 105:11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruess L, Michelsen A, Schmidt IK, Jonasson S. 1999. Simulated climate change affecting microorganisms, nematode density and biodiversity in subarctic soils. Plant Soil 212:63–73. doi: 10.1023/A:1004567816355. [DOI] [Google Scholar]
- 10.Tardy V, Spor A, Mathieu O, Leveque J, Terrat S, Plassart P, Régnier T, Bardgett RD, van der Putten W, Roggero PP, Seddaiu G, Bagella S, Lemanceau P, Ranjard L, Maron PA. 2015. Shifts in microbial diversity through land use intensity as drivers of carbon mineralization in soil. Soil Biol Biochem 90:204–213. doi: 10.1016/j.soilbio.2015.08.010. [DOI] [Google Scholar]
- 11.Tardy V, Chabbi A, Charrier X, de Berranger C, Reignier T, Dequiedt S, Faivre-Primot C, Terrat S, Ranjard L, Maron PA. 2015. Land use history shifts in situ fungal and bacterial successions following wheat straw input into the soil. PLoS One 10:e0130672. doi: 10.1371/journal.pone.0130672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arcand MM, Helgason BI, Lemke RI. 2016. Microbial crop residue decomposition dynamics in organic and conventionally managed soils. Appl Soil Ecol 107:347–359. doi: 10.1016/j.apsoil.2016.07.001. [DOI] [Google Scholar]
- 13.Bardgett RD, van der Putten WH. 2014. Belowground biodiversity and ecosystem functioning. Nature 515:505–511. doi: 10.1038/nature13855. [DOI] [PubMed] [Google Scholar]
- 14.Schimel J. 1995. Ecosystem consequences of microbial diversity and community structure, p 239–254. In Chapin F, Koerner C (ed), Arctic and alpine biodiversity: patterns, causes, and ecosystem consequences. Springer-Verlag, New York, NY. [Google Scholar]
- 15.Peter H, Beier S, Bertilsson S, Lindström ES, Langenheder S, Tranvik LJ. 2011. Function-specific response to depletion of microbial diversity. ISME J 5:351–361. doi: 10.1038/ismej.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baumann K, Dignac MF, Rumpel C, Bardoux G, Sarr A, Steffens M, Maron PA. 2012. Soil microbial diversity affects soil organic matter decomposition in a silty grassland soil. Biogeochemistry 114:201–212. doi: 10.1007/s10533-012-9800-6. [DOI] [Google Scholar]
- 17.Griffiths BS, Ritz K, Bardgett RD, Cook R, Christensen S, Ekelund F, Sørensen SJ, Bååth E, Bloem J, De Ruiter PC, Dolfing J, Nicolardot B. 2000. Ecosystem response of pasture soil communities to fumigation-induced microbial diversity reductions: an examination of the biodiversity-ecosystem function relationship. Oikos 2:279–294. doi: 10.1034/j.1600-0706.2000.900208.x. [DOI] [Google Scholar]
- 18.Wertz S, Degrange V, Prosser JI, Poly F, Commeaux C, Freitag T, Guillaumaud N, Le Roux X. 2006. Maintenance of soil functioning following erosion of microbial diversity. Environ Microbiol 8:2162–2169. doi: 10.1111/j.1462-2920.2006.01098.x. [DOI] [PubMed] [Google Scholar]
- 19.de Boer W, Folman LB, Summerbell RC, Boddy L. 2005. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev 29:795–811. doi: 10.1016/j.femsre.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 20.Niemenmaa O, Uusi-Rauva A, Hatakka A. 2008. Demethoxylation of [O14CH3]-labelled lignin model compounds by the brown-rot fungi Gloeophyllum trabeum and Poria (Postia) placenta. Biodegradation 19:555–565. doi: 10.1007/s10532-007-9161-3. [DOI] [PubMed] [Google Scholar]
- 21.Roger F, Bertilsson S, Langenheder S, Osman OA, Gamfeldt L. 2016. Effects of multiple dimensions of bacterial diversity on functioning, stability and multifunctionality. Ecology 97:2716–2728. doi: 10.1002/ecy.1518. [DOI] [PubMed] [Google Scholar]
- 22.Liebich J, Schloter M, Schäffer A, Vereecken H, Burauel P. 2007. Degradation and humification of maize straw in soil microcosms inoculated with simple and complex microbial communities. Eur J Soil Sci 58:141–151. doi: 10.1111/j.1365-2389.2006.00816.x. [DOI] [Google Scholar]
- 23.Bell T, Newman JA, Silverman BW, Turner SL, Lilley AK. 2005. The contribution of species richness and composition to bacterial services. Nature 436:1157–1160. doi: 10.1038/nature03891. [DOI] [PubMed] [Google Scholar]
- 24.Gihring TM, Green SJ, Schadt CW. 2012. Massively parallel rRNA gene sequencing exacerbates the potential for biased community diversity comparisons due to variable library sizes. Environ Microbiol 14:285–290. doi: 10.1111/j.1462-2920.2011.02550.x. [DOI] [PubMed] [Google Scholar]
- 25.Fierer N, Bradford MA, Jackson RB. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 26.DeBruyn JM, Nixon LT, Fawaz MN, Johnson AM, Radosevich M. 2011. Global biogeography and quantitative seasonal dynamics of Gemmatimonadetes in soil. Appl Environ Microbiol 77:6295–6300. doi: 10.1128/AEM.05005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pascault N, Ranjard L, Kaisermann A, Bachar D, Christen R, Terrat S, Mathieu O, Lévêque J, Mougel C, Henault C, Lemanceau P, Péan M, Boiry S, Fontaine F, Maron PA. 2013. Stimulation of different functional groups of bacteria by various plant residues as a driver of soil priming effect. Ecosystems 16:810–822. doi: 10.1007/s10021-013-9650-7. [DOI] [Google Scholar]
- 28.Paterson E, Osler G, Dawson LA, Gebbing T, Sim A, Ord B. 2008. Labile and recalcitrant plant fractions are utilised by distinct microbial communities in soil: independent of the presence of roots and mycorrhizal fungi. Soil Biol Biochem 40:1103–1113. doi: 10.1016/j.soilbio.2007.12.003. [DOI] [Google Scholar]
- 29.Poll C, Marhan S, Ingwersen J, Kandeler E. 2008. Dynamics of litter carbon turnover and microbial abundance in a rye detritusphere. Soil Biol Biochem 40:1306–1321. doi: 10.1016/j.soilbio.2007.04.002. [DOI] [Google Scholar]
- 30.Tardy V, Mathieu O, Leveque J, Terrat S, Chabbi A, Lemanceau P, Ranjard L, Maron PA. 2014. Stability of soil microbial structure and activity depends on microbial diversity. Environ Microbiol Rep 6:173–183. doi: 10.1111/1758-2229.12126. [DOI] [PubMed] [Google Scholar]
- 31.Franklin RB, Garland JL, Bolster CH, Mills AL. 2001. Impact of dilution on microbial community structure and functional potential: comparison of numerical simulations and batch culture experiments. Appl Environ Microbiol 67:702–712. doi: 10.1128/AEM.67.2.702-712.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voříšková J, Baldrian P. 2013. Fungal community on decomposing leaf litter undergoes rapid successional changes. ISME J 7:477–486. doi: 10.1038/ismej.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guigue J, Mathieu O, Leveque J, Mounier S, Laffont R, Maron PA, Navarro N, Château C, Amiotte-Suchet P, Lucas Y. 2014. A comparison of extraction procedures for water-extractable organic matter in soils. Eur J Soil Sci 65:520–530. doi: 10.1111/ejss.12156. [DOI] [Google Scholar]
- 34.Philippot L, Spor A, Hénault C, Bru D, Bizouard F, Jones CM, Sarr A, Maron PA. 2013. Loss in microbial diversity affects nitrogen cycling in soil. ISME J 7:1609–1619. doi: 10.1038/ismej.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Juarez S, Nunan N, Duday AC, Pouteau V, Chenu C. 2013. Soil carbon mineralisation responses to alterations of microbial diversity and soil structure. Biol Fertil Soils 49:939–948. doi: 10.1007/s00374-013-0784-8. [DOI] [Google Scholar]
- 36.Griffiths BS, Ritz K, Wheatley R, Kuan HL, Boag B, Christensen S, Ekelund F, Sørensen SJ, Muller S, Bloem J. 2001. An examination of the biodiversity-ecosystem function relationship in arable soil microbial communities. Soil Biol Biochem 33:1713–1722. doi: 10.1016/S0038-0717(01)00094-3. [DOI] [Google Scholar]
- 37.Lienhard P, Terrat S, Mathieu O, Levêque J, Chemidlin Prévost-Bouré N, Nowak V, Régnier T, Faivre C, Sayphoummie S, Panyasiri K, Tivet F, Ranjard L, Maron PA. 2013. Soil microbial diversity and C turnover modified by tillage and cropping in Laos tropical grassland. Environ Chem Lett 11:391–398. doi: 10.1007/s10311-013-0420-8. [DOI] [Google Scholar]
- 38.Cayuela ML, Sinicco T, Mondini C. 2009. Mineralization dynamics and biochemical properties during initial decomposition of plant and animal residues in soil. Appl Soil Ecol 41:118–127. doi: 10.1016/j.apsoil.2008.10.001. [DOI] [Google Scholar]
- 39.Pascault N, Nicolardot B, Bastian F, Thiébeau P, Ranjard L, Maron PA. 2010. In situ dynamics and spatial heterogeneity of soil bacterial communities under different crop residue management. Microb Ecol 60:291–303. doi: 10.1007/s00248-010-9648-z. [DOI] [PubMed] [Google Scholar]
- 40.Bardgett RD, Freeman C, Ostle NJ. 2008. Microbial contributions to climate change through carbon cycle feedbacks. ISME J 2:805–814. doi: 10.1038/ismej.2008.58. [DOI] [PubMed] [Google Scholar]
- 41.Roy J, Picon-Cochard C, Augusti A, Benot ML, Thiery L, Darsonville O, Landais D, Piel C, Defossez M, Devidal S, Escape C, Ravel O, Fromin N, Volaire F, Milcu A, Bahn M, Soussana JF. 2016. Elevated CO2 maintains grassland net carbon uptake under a future heat and drought extreme. Proc Natl Acad Sci U S A 113:6224–6229. doi: 10.1073/pnas.1524527113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fontaine S, Barot S, Barré P, Bdioui N, Mary B, Rumpel C. 2007. Stability of organic carbon in deep soil layers controlled by fresh carbon supply. Nature 450:277–280. doi: 10.1038/nature06275. [DOI] [PubMed] [Google Scholar]
- 43.Blagodatskaya E, Khomyakov N, Myachina O, Bogomolova I, Blagodatsky S, Kuzyakov Y. 2014. Microbial interactions affect sources of priming induced by cellulose. Soil Biol Biochem 74:39–49. doi: 10.1016/j.soilbio.2014.02.017. [DOI] [Google Scholar]
- 44.Rousk J, Hill PW, Davey LJ. 2015. Priming of the decomposition of ageing soil organic matter: concentration dependence and microbial control. Funct Ecol 29:285–296. doi: 10.1111/1365-2435.12377. [DOI] [Google Scholar]
- 45.Chen R, Senbayram M, Blagodatsky S, Myachina O, Dittert K, Lin X, Blagodatskaya E. 2014. Soil C and N availability determine the priming effect: microbial N mining and stoichiometric decomposition theories. Glob Chang Biol 20:2356–2367. doi: 10.1111/gcb.12475. [DOI] [PubMed] [Google Scholar]
- 46.Perveen N, Barot S, Alvarez G, Klumpp K, Martin R, Rapaport A, Herfurth D, Louault F, Fontaine S. 2014. Priming effect and microbial diversity in ecosystem functioning and response to global change: a modeling approach using the SYMPHONY model. Glob Chang Biol 20:1174–1190. doi: 10.1111/gcb.12493. [DOI] [PubMed] [Google Scholar]
- 47.Fontaine S, Barot S. 2005. Size and functional diversity of microbe populations control plant persistence and carbon accumulation. Ecol Lett 8:1075–1087. doi: 10.1111/j.1461-0248.2005.00813.x. [DOI] [Google Scholar]
- 48.Hunt HW, Wall DH. 2002. Modelling the effects of loss of soil biodiversity on ecosystem function. Glob Chang Biol 8:33–50. doi: 10.1046/j.1365-2486.2002.00425.x. [DOI] [Google Scholar]
- 49.Trap J, Bonkowski M, Plassard C, Villenave C, Blanchart E. 2016. Ecological importance of soil bacterivores for ecosystem functions. Plant Soil 398:1–24. doi: 10.1007/s11104-015-2671-6. [DOI] [Google Scholar]
- 50.Fontaine S, Henault C, Aamor A, Bdioui N, Bloor JMG, Maire V, Mary B, Revaillot S, Maron PA. 2011. Fungi mediate long term sequestration of carbon and nitrogen in soil through their priming effect. Soil Biol Biochem 43:86–96. doi: 10.1016/j.soilbio.2010.09.017. [DOI] [Google Scholar]
- 51.Razanamalala K, Razafimbelo T, Maron PA, Ranjard L, Chemidlin N, Lelièvre M, Dequiedt S, Ramaroson VH, Marsden C, Becquer T, Trap J, Blanchart E, Bernard L. 2018. Soil microbial diversity drives the priming effect along climate gradients: a case study in Madagascar. ISME J 12:451–462. doi: 10.1038/ismej.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sterjiades R, Erikson KEL. 1993. Biodegradation of lignins, p 115–126. In Scalbert A. (ed), Polyphenolic phenomena. INRA Editions, Paris, France. [Google Scholar]
- 53.Dequiedt S, Saby NPA, Lelievre M, Jolivet C, Thioulouse J, Toutain B, Arrouays D, Bispo A, Lemanceau P, Ranjard L. 2011. Biogeographical patterns of soil molecular microbial biomass as influenced by soil characteristics and management. Glob Ecol Biogeogr 20:641–652. doi: 10.1111/j.1466-8238.2010.00628.x. [DOI] [Google Scholar]
- 54.Vainio EJ, Hantula J. 2000. Direct analysis of wood-inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Mycol Res 104:927–936. doi: 10.1017/S0953756200002471. [DOI] [Google Scholar]
- 55.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohidee AS, McGarrell DM, March T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for r RNA analysis. Nucleic Acids Res 37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karimi B, Meyer C, Gilbert D, Bernard N. 2016. Air pollution below WHO levels decreases by 40% the links of terrestrial microbial networks. Environ Chem Lett 14:467–475. doi: 10.1007/s10311-016-0589-8. [DOI] [Google Scholar]
- 57.Karimi B, Maron PA, Chemidlin-Prevost Boure N, Bernard N, Gilbert D, Ranjard L. 2017. Microbial diversity and ecological networks as indicators of environmental quality. Environ Chem Lett 15:265–281. doi: 10.1007/s10311-017-0614-6. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.