ABSTRACT
Inulin-type fructans (ITF) and arabinoxylan oligosaccharides (AXOS) are broken down to different extents by various bifidobacterial strains present in the human colon. To date, phenotypic heterogeneity in the consumption of these complex oligosaccharides at the strain level remains poorly studied. To examine mechanistic variations in ITF and AXOS constituent preferences present in one individual, ITF and AXOS consumption by bifidobacterial strains isolated from the simulator of the human intestinal microbial ecosystem (SHIME) after inoculation with feces from one healthy individual was investigated. Among the 18 strains identified, four species-independent clusters displaying different ITF and AXOS degradation mechanisms and preferences were found. Bifidobacterium bifidum B46 showed limited growth on all substrates, whereas B. longum B24 and B. longum B18 could grow better on short-chain-length fractions of fructooligosaccharides (FOS) than on fructose. B. longum B24 could cleave arabinose substituents of AXOS extracellularly, without using the AXOS-derived xylose backbones, whereas B. longum B18 was able to consume oligosaccharides (up to xylotetraose) preferentially and consumed AXOS to a limited extent. B. adolescentis B72 degraded all fractions of FOS simultaneously, partially degraded inulin, and could use xylose backbones longer than xylotetraose extracellularly. The strain-specific degradation mechanisms were suggested to be complementary and indicated resource partitioning. Specialization in the degradation of complex carbohydrates by bifidobacteria present on the individual level could have in vivo implications for the successful implementation of ITF and AXOS, aiming at bifidogenic and/or butyrogenic effects. Finally, this work shows the importance of taking microbial strain-level differences into account in gut microbiota research.
IMPORTANCE It is well known that bifidobacteria degrade undigestible complex polysaccharides, such as ITF and AXOS, in the human colon. However, this process has never been studied for strains coexisting in the same individual. To examine strain-dependent mechanistic variations in ITF and AXOS constituent preferences present in one individual, ITF and AXOS consumption by bifidobacterial strains isolated from the SHIME after inoculation with feces from one healthy individual was investigated. Among the 18 bifidobacterial strains identified, four species-independent clusters displaying different ITF and AXOS degradation mechanisms and preferences were found, indicating that such strains can coexist in the human colon. Such specialization in the degradation of complex carbohydrates by bifidobacteria present on the individual level could have in vivo implications for the successful implementation of ITF and AXOS, aiming at bifidogenic and/or butyrogenic effects.
KEYWORDS: bifidobacteria, colon fermentation, inulin-type fructans, arabinoxylan oligosaccharides, prebiotics
INTRODUCTION
The human colon contains >1011 bacteria per ml of luminal content and is actually an extra organ within the human body (1–3). Despite the expanding body of knowledge demonstrating the importance of the gut microbiota for normal development and functioning of the human body and behavior (3–5), only a few studies underline the importance of individual bacterial strains within the human colon (6–8). For instance, it has been shown that strains of the same bacterial species can display different probiotic effects (9, 10), differ in their antioxidant capacities (11), have different pathogenic effects (12), differ in their abilities to consume carbohydrates (7, 13–18), and interact differently with other bacterial species through cross-feeding (7, 19).
Bifidobacteria are some of the most studied bacteria of the human colon. Despite their low prevalence (<5%) in the adult colon (20), they are indispensable for gut homeostasis (21–23) and cross-feeding interactions with other colon bacteria through the production of acetate and lactate from nondigestible carbohydrates (6, 7, 24, 25). However, it is well known that bifidobacterial strains differ in their ability to degrade inulin-type fructans (ITF) (13), arabinoxylan oligosaccharides (AXOS) (16), xylooligosaccharides (XOS) (16), and galactooligosaccharides (14, 26). For instance, a comparative study of 18 bifidobacterial strains belonging to 10 different species and coming from the feces of different donors and from bovine rumen showed that four clusters of strains could be distinguished, differing in their capabilities and mechanisms for degradation of ITF (13). Some strains consume only fructose, some consume both fructose and fructooligosaccharides (FOS) (mainly short-chain-length fractions of FOS with a degree of polymerization [DP] of up to 7), some degrade FOS fractions of all chain lengths simultaneously, whereby fructose consumption is not faster than FOS degradation, and still others also consume inulin. In the case of AXOS, five species-independent clusters of strains with different AXOS degradation mechanisms and preferences were identified among 36 bifidobacterial strains tested, belonging to 11 different species and coming from the feces of different donors and from various origins (yogurt, bovine rumen, and sewage) (16). Some strains consume only arabinose and/or xylose monosaccharides and others consume the arabinose substituents of AXOS, followed or not by consumption of the xylose backbones, either only short chains (DP of 2 to 4) or also longer backbones, either with or without the release of arabinose and xylose into the extracellular medium. It has been suggested that these kinds of differences within bifidobacterial communities might help different strains to live together in the colon, by avoiding competition for the same substrate. This yields cross-feeding opportunities among bifidobacterial strains (7, 16). For instance, it was shown that a triculture of Bifidobacterium longum ATCC 15707, Bifidobacterium adolescentis ATCC 15703, and Bifidobacterium breve ATCC 15700 could consume the whole A2XX molecule [α-l-Araf-(1→2)-β-d-Xylp-(1→4)-β-d-Xylp-(1→4)-d-Xyl] when the strains were grown together but not when they were grown separately (27). However, it is not yet known whether bifidobacterial strains with different AXOS and ITF degradation mechanisms, and thus preferences, actually occur within the colon of the same human individuals. In addition, to link the capacity of bifidobacteria to degrade ITF and AXOS in such individuals with specific colon regions, strains from different colon regions must be investigated. The aim of this study was to elucidate the potential to ferment ITF-derived monosaccharides (fructose), AXOS-derived monosaccharides (arabinose and xylose), and xylose backbones (XOS), as well as the complete FOS, inulin, and AXOS molecules, by performing laboratory fermentations with bifidobacterial strains isolated from the colon vessels of the simulator of the human microbial ecosystem (SHIME) that had been inoculated with the feces of one healthy individual.
RESULTS
Identification of bifidobacteria isolated from the SHIME.
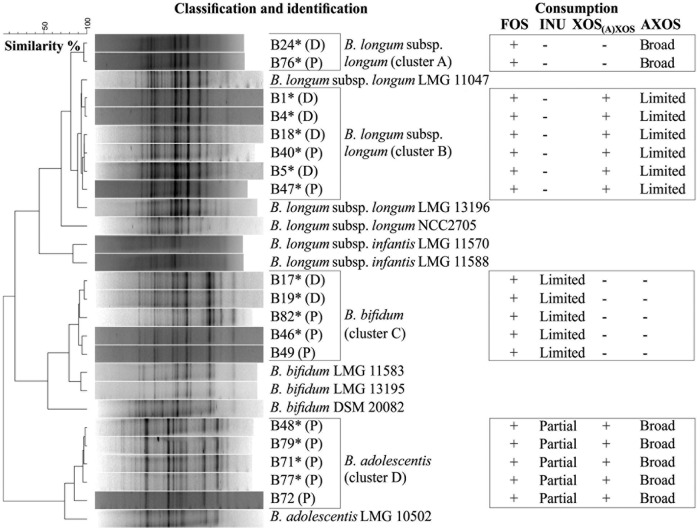
Of a total of 86 colonies picked up from different selective agar media, 28 isolates produced more acetate than lactate from fructose; among those isolates, 18 were identified as different Bifidobacterium species through 16S rRNA gene sequencing of genomic DNA (see Table S1 in the supplemental material). The dendrogram based on the numerical analysis of the digitized (GTG)5-PCR fingerprints of these 18 bifidobacterial strains consisted of four clusters (Fig. 1). Clusters A and B included strains belonging to the B. longum species, cluster C harbored strains identified as Bifidobacterium bifidum, and cluster D strains were allocated to the B. adolescentis species (Fig. 1). The (GTG)5-PCR fingerprints of clusters A and B were different, indicating the presence of different types of B. longum strains. Eleven bifidobacterial strains originated from the proximal colon vessel of the SHIME, and seven came from the distal colon vessel (Fig. 1). Cluster D was the sole cluster that contained only strains originating from the proximal colon vessel. The other clusters contained strains from both the proximal and distal colon vessels of the SHIME.
FIG 1.
Dendrogram based on the (GTG)5-PCR fingerprints of genomic DNA from 19 bifidobacterial isolates (designated with a B number, with the SHIME colon vessel origin indicated in parentheses) from the proximal (P) and distal (D) colon vessels of the SHIME and 9 reference bifidobacterial strains (designated with the culture collection number). The identity of the isolates marked with an asterisk was verified by 16S rRNA gene sequencing (see Table S1 in the supplemental material). The four columns on the right present the results of the screening experiments for the degradation of ITF and (A)XOS [consisting of XOS(A)XOS and AXOS] in MCB. +, consumption; −, no consumption; broad, several AXOS peaks in the HPAEC-PAD chromatogram disappeared; partial, consumption of inulin up to a particular chain length; limited, consumption of FOS (DP of 3 to 10) present in the commercial inulin substrate when the strain was grown on inulin or only one AXOS peak in the HPAEC-PAD chromatogram disappeared when the strain was grown on (A)XOS. Details of the results of the screening experiments can be found in Fig. S1 to S8 in the supplemental material.
Screening experiments with ITF and (A)XOS.
Differences in degradation capabilities for the ITF and (A)XOS [XOS(A)XOS and AXOS] substrates were found among the 18 bifidobacterial strains tested, corresponding to clusters A, B, C, and D in the (GTG)5-PCR fingerprint dendrogram (Fig. 1; also see Fig. S1 to S4 in the supplemental material). Within each cluster, no large differences in carbohydrate consumption between the strains were found.
Fermentation experiments with fructose, FOS, inulin, arabinose-xylose mixture, XOS, and (A)XOS.
Laboratory fermentation experiments with representative bifidobacterial strains of each cluster showed that differences occurred with regard to bacterial growth, carbohydrate consumption, and metabolite production (ratio of acetate to lactate) with fructose, FOS, inulin, an arabinose-xylose (A-X) mixture, XOS, or (A)XOS as the added energy source (Fig. 2 to 8). Also, different degradation mechanisms were found when strains were grown on FOS, inulin, or XOS (Fig. 3 to 8). Similarly, strain-specific differences were found in the case of the B. longum strains (Fig. 6A and B, 7A and B, and 8A and B).
FIG 2.
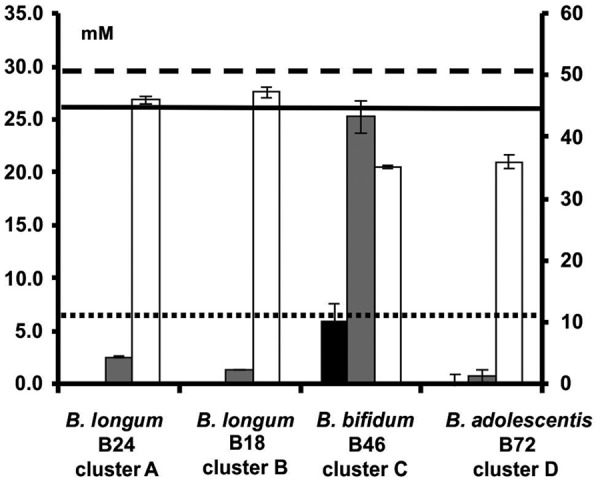
Residual concentrations of fructose (white bars; right axis), arabinose (gray bars; left axis), and xylose (black bars; left axis) after 24 h or 48 h of fermentation with bifidobacterial strains representing clusters A to D, in MCB supplemented with 9.0 g liter−1 (51.7 mM) of fructose or 1.0 g liter−1 (6.7 mM) of arabinose and 4.0 g liter−1 (26.6 mM) of xylose. The horizontal lines represent the initial concentrations of fructose (dashed line), arabinose (dotted line), and xylose (solid line).
FIG 3.
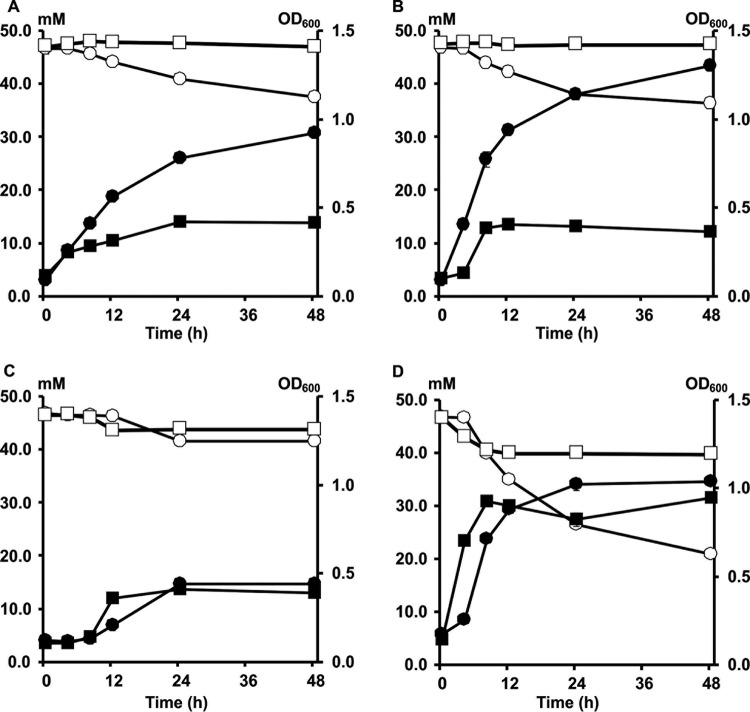
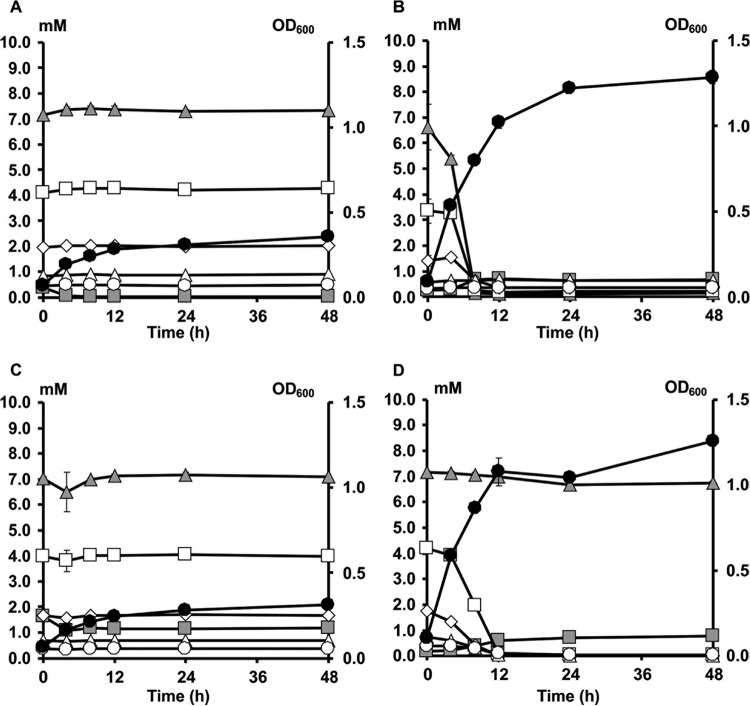
Growth and degradation of FOS and inulin by Bifidobacterium longum B24 (A), B. longum B18 (B), B. bifidum B46 (C), and B. adolescentis B72 (D), strains representing clusters A, B, C, and D, respectively, in MCB supplemented with 9.0 g liter−1 of FOS or 9.0 g liter−1 of inulin, as a function of time. Growth on FOS (black circles) and inulin (black squares) is represented as OD600 measurements. Consumption of FOS (white circles) and inulin (white squares) is represented as residual concentrations of fructose equivalents.
FIG 4.
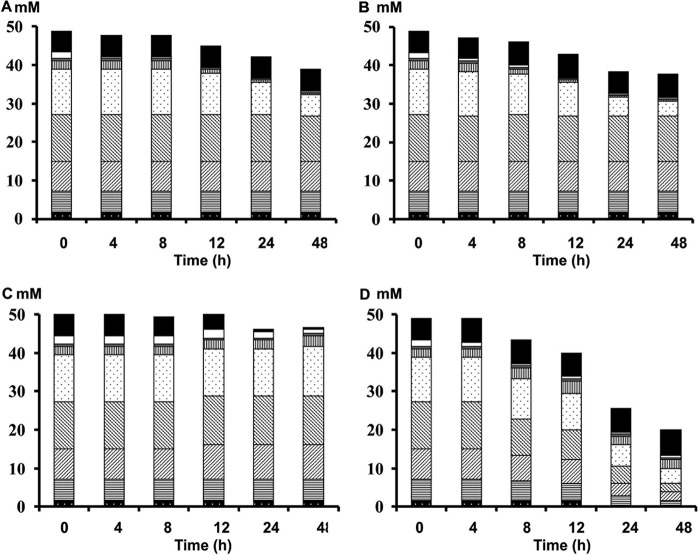
FOS degradation by Bifidobacterium longum B24 (A), B. longum B18 (B), B. bifidum B46 (C), and B. adolescentis B72 (D), strains representing clusters A, B, C, and D, respectively, in MCB supplemented with 9.0 g liter−1 of FOS, as a function of time. Consumption of fructose (black), glucose (white), sucrose (gray), inulibiose (vertically lined), GF2 +F3 (dotted), GF3 +F4 (right-leaning diagonally lined), GF4 +F5 (left-leaning diagonally lined), GF5 +F6 (horizontally lined), and GF6 +F7 (black with white dots) is represented as residual concentrations of fructose equivalents.
FIG 5.
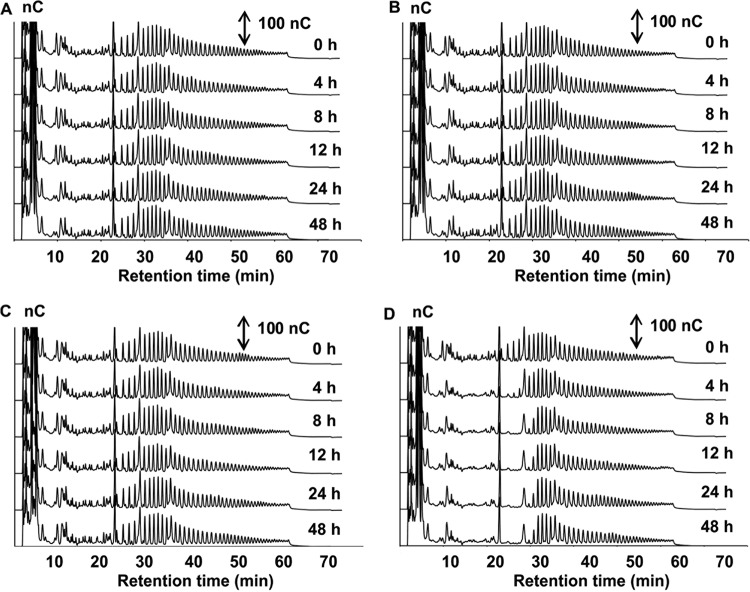
Inulin degradation by Bifidobacterium longum B24 (A), B. longum B18 (B), B. bifidum B46 (C), and B. adolescentis B72 (D), strains representing clusters A, B, C, and D, respectively, in MCB supplemented with 9.0 g liter−1 of inulin, as a function of time. HPAEC-PAD chromatograms of inulin indicate signal intensity (in nanocoulombs) at each retention time. Double-headed arrows represent the scale of the PAD response.
FIG 6.
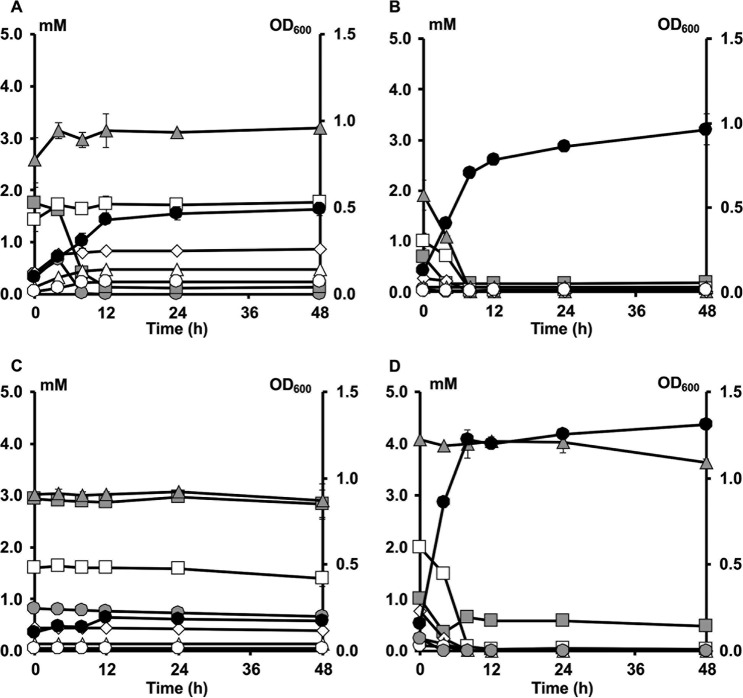
Growth and XOS degradation by Bifidobacterium longum B24 (A), B. longum B18 (B), B. bifidum B46 (C), and B. adolescentis B72 (D), strains representing clusters A, B, C, and D, respectively, in MCB supplemented with 5.0 g liter−1 of XOS, as a function of time. Growth is represented as OD600 measurements (black circles). Monosaccharide and XOS consumption are represented as residual concentrations of xylose (gray squares), xylobiose (gray triangles), xylotriose (white squares), xylotetraose (white diamonds), xylopentaose (white triangles), and xylohexaose (white circles).
FIG 7.
Growth and AXOS degradation by Bifidobacterium longum B24 (A), B. longum B18 (B), B. bifidum B46 (C), and B. adolescentis B72 (D), strains representing clusters A, B, C, and D, respectively, in MCB supplemented with 5.0 g liter−1 of (A)XOS, as a function of time. Growth is represented as OD600 measurements (black circles). Monosaccharide and XOS(A)XOS consumption is represented as residual concentrations of arabinose (gray circles), xylose (gray squares), xylobiose (gray triangles), xylotriose (white squares), xylotetraose (white diamonds), xylopentaose (white triangles), and xylohexaose (white circles).
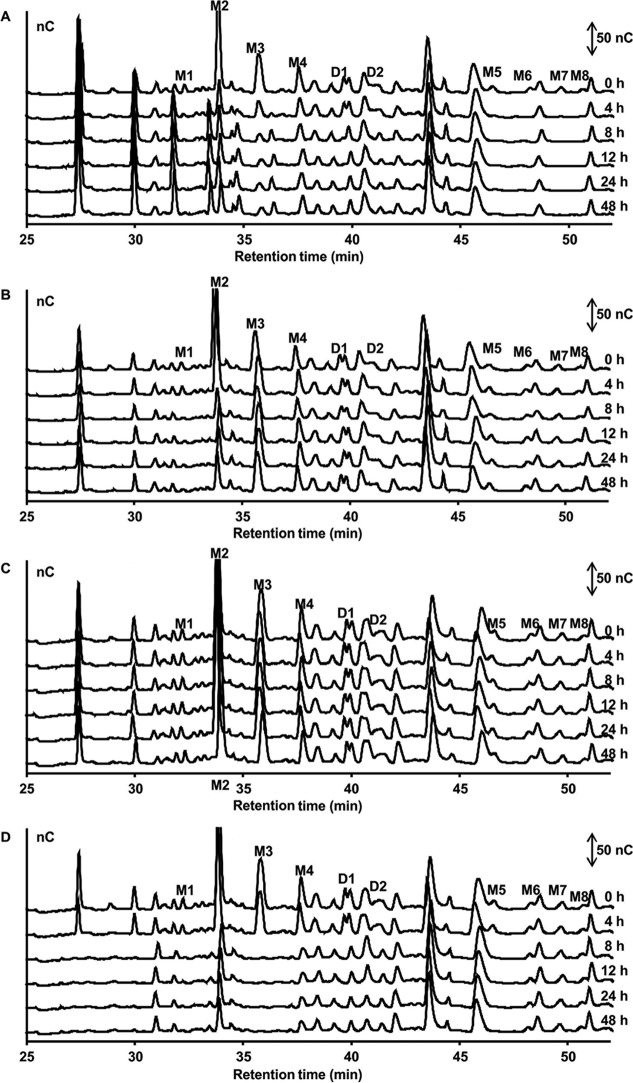
FIG 8.
AXOS degradation by Bifidobacterium longum B24 (A), B. longum B18 (B), B. bifidum B46 (C), and B. adolescentis B72 (D), strains representing clusters A, B, C, and D, respectively, in MCB supplemented with 5.0 g liter−1 of (A)XOS, as a function of time. HPAEC-PAD chromatograms of AXOS indicate signal intensity (in nanocoulombs) at each retention time. Double-headed arrows represent the scale of the PAD response. Peaks D1 and D2 represent disubstituted AXOS molecules containing at least one doubly substituted xylose residue; peaks M1 to M8 represent monosubstituted AXOS molecules containing at least one monosubstituted xylose residue.
Both B. longum B24 and B. longum B18, belonging to cluster A and cluster B, respectively, consumed FOS, i.e., 9.04 ± 1.24 mM fructose equivalents (FE) and 10.55 ± 0.93 mM FE, respectively, after 48 h of fermentation (Fig. 3A and B). For both strains, degradation of fructose, glucose, inulibiose, kestose, and kestotriose was found (Fig. 4A and B). Better growth was obtained with B. longum B18 (optical density at 600 nm [OD600] of 1.32 ± 0.03) than with B. longum B24 (OD600 of 0.92 ± 0.03) (Fig. 3A and B), but similar acetate/lactate ratios after 48 h of fermentation were found for the two strains (ratios of 5.2 and 4.5, respectively). Besides acetate and lactate, FOS consumption resulted in the production of formate and ethanol (see Fig. S5A and B in the supplemental material). Although inulin could not be degraded by B. longum B24 and B. longum B18 (Fig. 5A and B), limited growth took place with short-chain-length fractions present in the commercial inulin substrate used (maximum OD600 values of 0.42 ± 0.01 and 0.37 ± 0.01, respectively) (Fig. 3A and B). Both strains consumed arabinose and xylose monosaccharides (Fig. 2 and 6A and B). However, whereas B. longum B24 could not use XOS (Fig. 6A) or XOS(A)XOS (Fig. 7A), B. longum B18 consumed xylobiose, xylotriose, and xylotetraose (Fig. 6B and 7B) but not longer XOS or XOS(A)XOS molecules (Fig. 6B and 7B). Nevertheless, during growth on AXOS, B. longum B24 could use the arabinose substituents of almost all monosubstituted and disubstituted AXOS molecules (Fig. 8A), which resulted in increases in the xylose backbone concentrations (e.g., xylobiose, xylotriose, xylotetraose, xylopentaose, xylohexaose, and possibly also longer xylose backbones for which no standards were available) in the extracellular medium (Fig. 7A). Accordingly, better growth was measured for B. longum B18 grown on XOS or (A)XOS (maximum OD600 values of 1.23 ± 0.04 and 0.96 ± 0.09, respectively), compared to B. longum B24 (maximum OD600 values of 0.36 ± 0.01 and 0.49 ± 0.03, respectively) (Fig. 6A and B and 7A and B). However, B. longum B18 could use only certain monosubstituted AXOS molecules (corresponding to one peak [M2] in the high-performance anion-exchange chromatography with pulsed amperometric detection [HPAEC-PAD] chromatogram) (Fig. 8B). XOS and (A)XOS consumption by both strains resulted in the production of acetate, lactate, formate, and ethanol (see Fig. S7A and B and S8A and B in the supplemental material). In the case of B. longum B24, a higher acetate/lactate ratio after 48 h of fermentation was found when the strain was grown on XOS (ratio of 1.8), compared to (A)XOS (ratio of 1.3). Given the better growth of B. longum B18 on both XOS and (A)XOS, higher concentrations of acetate, lactate, formate, and ethanol were found, with comparable acetate/lactate ratios after 48 of fermentation (ratios of 1.3 and 1.4, respectively).
The cluster C representative, B. bifidum B46, did not degrade FOS or inulin (Fig. 4C and 5C), although consumption of 5.28 ± 0.18 mM FE and 2.70 ± 0.22 mM FE with the FOS and inulin substrates, respectively, supported limited growth (maximum OD600 values of 0.39 ± 0.01 and 0.44 ± 0.01, respectively) (Fig. 3C). Similarly, this strain could not use the arabinose and xylose monosaccharides, XOS, XOS(A)XOS, or the arabinose substituents of AXOS (Fig. 2, 6C, 7C, and 8C) but did consume fructose (Fig. 2). However, limited bacterial growth and production of low concentrations of acetate and lactate (no formate) were measured when the strain was grown in a medium for colon bacteria (MCB) supplemented with FOS, inulin, XOS, or (A)XOS (Fig. 3C, 6C, and 7C; also see Fig. S5C, S6C, S7C, and S8C in the supplemental material), indicating the presence of other energy sources in MCB and/or the commercial substrates used.
B. adolescentis B72, belonging to cluster D, consumed FOS (Fig. 4D). Degradation of fractions of all chain lengths resulted in consumption of 25.85 ± 0.50 mM FE, with a maximum OD600 value of 1.04 ± 0.03 (Fig. 3D). Moreover, B. adolescentis B72 was the only strain that performed partial inulin degradation (Fig. 5D), with the consumption of 6.89 ± 0.36 mM FE (Fig. 3D). Acetate, lactate, and ethanol were produced on both energy sources after 48 h of fermentation (see Fig. S5D and S6D in the supplemental material). A higher acetate/lactate ratio after 48 h of fermentation was found with inulin (ratio of 2.0), compared with FOS (ratio of 1.6). Also, this strain consumed arabinose and xylose (Fig. 2 and 6D), XOS and XOS(A)XOS (Fig. 6D and 7D), and several monosubstituted and disubstituted AXOS molecules (Fig. 8D), although fewer AXOS peaks in the chromatogram disappeared, compared to those for B. longum B24 (Fig. 8A). B. adolescentis B72 did not consume xylobiose but could use xylotriose, xylotetraose, xylopentaose, xylohexaose, and possibly also longer xylose backbones for which no standards were available (Fig. 6D and 7D). Moreover, the consumption of those xylose backbones was accompanied by increases in the xylose concentrations in the extracellular medium, indicating extracellular degradation. Similar maximum OD600 values were measured when the strain was grown on XOS (OD600 of 1.26 ± 0.02) (Fig. 6D) or (A)XOS (OD600 of 1.31 ± 0.01) (Fig. 7D), but higher concentrations of acetate and lactate (no formate) were produced when the strain was grown on XOS (see Fig. S7D and S8D in the supplemental material). Also, the acetate/lactate ratios after 48 h of fermentation were similar with the two substrates, i.e., 1.1 (XOS) versus 1.2 [(A)XOS].
DISCUSSION
Several studies have shown that the consumption of ITF and AXOS increases bifidobacterial numbers in the colon of healthy humans (28–33) and restores them in the cecum of mice with diet-induced obesity (34, 35). It has been suggested that the complementarity in ITF and AXOS degradation mechanisms among bifidobacterial strains, together with their potential for cell-associated or intracellular degradation of these polymers, could explain the bifidogenic effects (7, 13, 16). Indeed, in vitro cocultivation of strains with complementary degradation mechanisms resulted in transcriptional upregulation of the genes for carbohydrate metabolism and, consequently, survival in the case of starch and xylan (36), mucin glycoproteins (37), and AXOS (24).
The present study of monoculture fermentations with bifidobacterial strains isolated from colon vessels of the SHIME that had been inoculated with fecal material from one healthy individual, belonging to three different bifidobacterial species (B. longum, B. adolescentis, and B. bifidum), proved that, within an individual gut microbiota, different bifidobacterial strains with complementary ITF and AXOS degradation mechanisms are present. The bifidobacterial species identified are commonly encountered in the human gut microbiota of adults. Indeed, it has been shown that, on average, two or three different Bifidobacterium species are present in fecal samples from human adults, with B. longum (present in up to 96% of adults), B. adolescentis (present in up to 83% of adults), and B. bifidum (present in up to 40% of adults) being the most frequently detected (38, 39). In view of the stability of the gut microbiota, it has been estimated that 60% of all bacterial strains within an individual persist for 5 years (40). However, the diversity of the microbial communities within an individual can be reversible (41). The feature of generally low diversity of the microbial communities is related to many diseases, and enrichment of a consortium of strains in the gut requires the administration of the missing taxa in combination with dietary carbohydrates (41). To this end, development of tailor-made food supplements is required.
The four strains tested, representing four different phenotypic clusters (clusters A, B, C, and D), displayed different ITF and AXOS degradation mechanisms. The ITF degradation mechanisms characterized in the present study were comparable to three of the four clusters (clusters A, B, and D) in a previous fermentation study with 18 bifidobacterial strains performed by Falony et al. (13). Their AXOS degradation mechanisms corresponded to the degradation mechanisms of four of the five clusters (clusters I, II, III, and V) encountered previously in a comparative fermentation study with 36 bifidobacterial strains performed by Rivière et al. (16). Like all other B. bifidum strains (belonging to cluster I [16] and cluster A [13]), B. bifidum B46 (cluster C) could not use FOS, inulin, arabinose, xylose, XOS, or (A)XOS. Moreover, since B. bifidum strains display the lowest genetic potential within the Bifidobacterium genus to use plant-derived carbohydrates, it is likely that this species does not contribute to the bifidogenic effects of ITF and AXOS in the human colon. This species can use fructose (13), however, and has the highest genetic potential within the Bifidobacterium genus to use host-associated carbohydrates such as human milk oligosaccharides and mucin (37, 42, 43). The degradation mechanisms of B. longum B24 (cluster A) and B. longum B18 (cluster B) corresponded to those of cluster B strains described by Falony et al. (13), which were characterized by the uptake and subsequent intracellular degradation of short-chain-length fractions of oligofructose. This preferential degradation was shown to be a competitive advantage of bifidobacterial strains toward extracellular FOS-degrading colon bacteria (encompassing Bacteroides spp. and Clostridium cluster IV and XIVa species) during coculture batch fermentations (7, 19, 25). This selective oligosaccharide degradation may be explained by the presence of multiple FOS transport systems with different specificities (17).
The strain-dependent differences in XOS and AXOS degradation by B. longum B24 and B. longum B18 are due to the cellular localization of their enzymes. The phenotype of B. longum B24 is characterized by extracellular cleavage of arabinose substituents of AXOS without using the AXOS-derived xylose backbones (cluster II) (16). In previous work, such a degradation mechanism was linked to the presence of genes encoding an extracellular β-endoxylanase and an extracellular β-xylosidase (BL1543 and BL1544, respectively), which were identified as extracellular membrane-associated α-arabinofuranosidases in B. longum subsp. longum NCC2705 (16, 44, 45). Another B. longum strain (B. longum B18, in cluster B) showed the same degradation mechanisms as the bifidobacterial strains belonging to cluster III, which are characterized by the preferential uptake of short oligosaccharides (up to xylotetraose) and limited consumption of AXOS (16). Like other cluster D bifidobacteria (13), B. adolescentis B72 could use all oligofructose fractions and could partially break down inulin. Moreover, B. adolescentis B72 could also use xylose backbones longer than xylotetraose; this was accompanied by increases in xylose concentrations in the medium. Such a breakdown profile was described in the case of Bifidobacterium catenulatum LMG 11043T, belonging to cluster V, and indicated extracellular degradation (6, 16). In contrast to B. catenulatum LMG 11043T, B. adolescentis B72 did not use xylobiose. The inability to use xylobiose has also been found for bifidobacterial strains belonging to cluster III (16) and has been ascribed in Bifidobacterium animalis subsp. lactis Bl-04 to the presence of the binding protein BlAXBP (part of an ATP-binding cassette transporter), which has very low affinity for xylobiose, compared to xylotriose and xylotetraose (46). Furthermore, xylose-backbone-degrading enzymes with weak activity for xylobiose, compared to longer xylose backbones, have been characterized in B. adolescentis LMG 10502T (47, 48). The coexistence of these bifidobacterial strains with complementary ITF and AXOS degradation mechanisms, and thus preferences, within the gut microbiota of one individual suggests cooperation among bifidobacteria for the degradation of ITF and AXOS.
Apart from differences in carbohydrate consumption, the representative strains tested showed differences in metabolite production depending on the type of substrate consumed [FOS versus inulin and XOS versus (A)XOS] and the degradation capabilities of the strains. Arabinose and xylose resulting from XOS and (A)XOS degradation can enter the bifid shunt through conversion to xylulose 5-phosphate (7, 49). Theoretically, a bifid shunt produces an acetate/lactate molar ratio of 1.0 in the case of pentoses, instead of a ratio of 1.5 for hexoses (e.g., fructose). However, this molar ratio was not observed for the representative strains of clusters A and B when the strains were grown on XOS or (A)XOS, due to the production of additional acetate, formate, and ethanol, instead of lactate, from pyruvate. In the case of ITF fermentation, this shift in metabolism from lactate production to mixed-acid production was found for all strains tested and can be explained by the need for additional ATP production by cells growing on complex carbohydrates, to improve their competitiveness because of their slower growth (7, 19, 50–52). In contrast, B. adolescentis B72, which grew well on all of the substrates tested, produced equimolar concentrations of acetate and lactate, without measurable concentrations of formate and ethanol. A trend of increasing acetate/lactate ratios characterized all bifidobacterial strains tested in the following sequence: XOS, AXOS, FOS, and inulin.
Previous SHIME experiments have shown that the primary site of inulin fermentation (DP of 2 to 60, with an average DP of 10) is the ascending colon (53), whereas AXOS (AXOS-15-0.27 and AXOS-29-0.30) are gradually fermented along the transverse and distal colon vessels of the SHIME (53, 54); inulin and long-chain arabinoxylans (LC-AX) (LC-AX-60-0.70) stimulate bifidobacterial growth (especially B. longum) in all colon vessels of the SHIME (55). Indeed, the B. longum strains belonging to clusters A and B originated from both the proximal and distal colon vessels of the SHIME. Also, the B. bifidum strains occurred in both the proximal and distal colon vessels, whereas the B. adolescentis strains originated only from the proximal colon vessel. Similarly, it was recently shown that bifidobacterial strains with different ITF degradation mechanisms occur within the same colon region (56). This finding, together with the present results, supports the cooperation hypothesis and resource partitioning of bifidobacteria to degrade prebiotics together in the human colon. Resource partitioning is niche differentiation that enables species coexistence (2).
In this work, it has been demonstrated that different bifidobacterial strains can coexist in the human colon by having different carbohydrate preferences and applying complementary carbohydrate degradation mechanisms. This may explain the lack of outcompetition. Indeed, since the bifidobacterial strains used in the present study originated from one individual, they may use the available resources together, possibly through cooperation. More studies have to be performed to determine whether such bifidobacterial strains are also present in other individuals and to investigate the precise set of enzymes that are required to act together at a precise physical location (57). Moreover, specialization in the degradation of complex carbohydrates by bifidobacteria present on the individual level could have in vivo implications for the successful implementation of ITF and AXOS, aiming at bifidogenic and/or butyrogenic effects.
MATERIALS AND METHODS
Substrates.
Different substrates were used in this study. Concerning ITF, fructose (VWR International, Darmstadt, Germany), FOS (OraftiP95; BENEO-Orafti, Tienen, Belgium), and inulin (OraftiHP; BENEO-Orafti) were used. OraftiP95 consists mainly of FOS (≥93.2% [mass/mass]), with the DP ranging from 2 to 8 and an average DP of 4, and minor concentrations of glucose, fructose, and sucrose. OraftiHP contains inulin (≥99.5% [mass/mass]) and minor concentrations of glucose, fructose, and sucrose (<0.5% [mass/mass]). The DP of inulin varies between 12 and 65, with an average of ≥23. Concerning AXOS, arabinose (Sigma-Aldrich, Steinheim, Germany), xylose (Sigma-Aldrich), an A-X mixture (1.0 g liter−1 of arabinose and 4.0 g liter−1 of xylose, to mimic the typical A/X ratio of 0.25 in AXOS), XOS 95P (Shandong Longlive Bio-Technology, Shandong, China), LC-AX (BioActor, Ghent, Belgium), and AXOS-5-0.27 (Fugeia, Leuven, Belgium) were used. XOS 95P (referred to as XOS) consists mainly of XOS molecules with a DP of 2 to 7 (XOS2–7, ≥95.0% [mass/mass]; XOS2–4, ≥65.0% [mass/mass]) and minor concentrations of xylose, arabinose, and dextrose (≤0.5% [mass/mass]). LC-AX has an average DP of 60, an A/X ratio of 0.7, and a purity of 60%. AXOS-5-0.27 has an average DP of 5 and an A/X ratio of 0.27 and contains 1% (mass/mass) ferulic acid. Because it has been shown that AXOS-5-0.27 contains XOS as well (16, 58), this substrate is referred to as (A)XOS and its individual components as AXOS and XOS(A)XOS.
Stock solutions of fructose, A-X mixture, and LC-AX were sterilized by autoclaving (121°C at 210 kPa for 20 min). Stock solutions of FOS and inulin were sterilized by membrane filtration (Millex-GP filters with 0.22-μm pores; Millipore, Molsheim, France). Stock solutions of XOS and (A)XOS were sterilized by membrane filtration using Minisart filters (0.2-μm pores; Sartorius, Göttingen, Germany) and MediaKap-50 Plus high-performance hollow fiber membrane filters (0.2-μm pores; Microgon, Rancho Dominguez, CA), respectively.
Isolation of bifidobacteria from the SHIME.
A long-term fermentation experiment was performed in the SHIME. The SHIME is a dynamic in vitro model mimicking the human gastrointestinal tract by carrying a complex, stable, individual-specific, and colon region-specific microbial community (59). The SHIME used consisted of one vessel simulating the stomach, one vessel simulating the small intestine, and two vessels simulating the colon, namely, one representing the proximal colon (pH 5.6 to 5.9) and one the distal colon (pH 6.6 to 6.9), all kept at 37°C. The vessels were made anaerobic by flushing with 100% nitrogen gas (Air Liquide, Paris, France). The SHIME medium used was composed of the following compounds: pectin (Sigma-Aldrich), 2.0 g liter−1; xylan (Sigma-Aldrich), 1.0 g liter−1; glucose (Sigma-Aldrich), 0.4 g liter−1; potato starch (Anco, Roeselare, Belgium), 2.0 g liter−1; yeast extract (Oxoid, Basingstoke, Hampshire, UK), 3.0 g liter−1; peptone (Oxoid), 1.0 g liter−1; type II mucin (Sigma-Aldrich), 0.4 g liter−1; cysteine-HCl (Sigma-Aldrich), 0.5 g liter−1. All compounds were dissolved in demineralized water and autoclaved at 121°C and 210 kPa for 20 min. A sterilized stock solution of LC-AX was added aseptically to the SHIME medium at a final concentration of 10.0 g liter−1. Pancreatic juice was composed of the following compounds: NaHCO3 (Merck, Darmstadt, Germany), 12.5 g liter−1; ox gall (Becton Dickinson, Le Pont de Claix, France), 6.0 g liter−1; pancreatin (Sigma-Aldrich), 0.9 g liter−1. All compounds were dissolved in autoclaved (121°C at 210 kPa for 20 min) demineralized water. Three times per day, 140 ml of medium (pH 2.0) and 60 ml of pancreatic juice were added to the stomach and small intestine vessels, respectively, from which 200 ml flowed to the proximal and distal colon vessels three times a day. The colon vessels were inoculated with the human fecal microbiota from a healthy male individual 29 years of age, who had no history of antibiotic intake for 6 months prior to the study. Inoculum preparation was performed as described previously (60). Briefly, 20 g of freshly voided feces was diluted and homogenized with 100 ml of anaerobic phosphate-buffered saline containing 0.5 g liter−1 of cysteine-HCl. After removal of the particulate material by centrifugation (500 × g for 1 min), 40 ml of supernatant was added to the colon vessels. Two weeks after inoculation, the microbial community was stable, and samples were taken from the colon vessels of the SHIME to isolate bifidobacterial strains. Samples were immediately diluted in anaerobic phosphate-buffered saline containing 0.5 g liter−1 of cysteine-HCl. Platings were performed on different selective agar media for bifidobacteria, as follows: Wilkins-Chalgren (WC) agar (Oxoid) supplemented with 0.5 g liter−1 of cysteine-HCl and 50 mg liter−1 of mupirocin (Applichem, Darmstadt, Germany) (61, 62); de Man-Rogosa-Sharpe (MRS) agar (Oxoid) supplemented with 0.5 g liter−1 of cysteine-HCl and 50 mg liter−1 of mupirocin; raffinose-Bifidobacterium (RB) agar (63); and transoligosaccharide (TOS)-propionate agar (Merck) supplemented with 0.5 g liter−1 of cysteine-HCl and 50 mg liter−1 of mupirocin. Plates were incubated at 37°C for 48 h under anaerobic conditions in a MG anaerobic workstation (Don Whitley Scientific, Shipley, West Yorkshire, UK), which was continuously sparged with a mixture of 80% nitrogen gas, 10% carbon dioxide, and 10% hydrogen gas (Air Liquide). Next, 10% of the grown colonies of the highest dilutions were purified by streaking on the appropriate agar medium and growth of the isolates in the corresponding liquid medium for 24 h at 37°C, under anaerobic conditions. For storage of the isolates at −80°C, the media were supplemented with 25% (vol/vol) glycerol (Sigma-Aldrich), as cryoprotectant.
Identification of bifidobacterial isolates.
The identity of the bifidobacterial isolates was first assessed through a phenotypic test for the presence of a bifid shunt that results in the production of a theoretical molar ratio of acetate to lactate of 3:2 when the isolates are grown on hexoses. Therefore, incubations with the isolates were carried out in test tubes containing 10 ml of a slightly modified version of MCB (51), which did not contain Tween 80 and was supplemented with 9.0 g liter−1 of fructose as the sole added energy source. All compounds were dissolved in demineralized water, and the medium was adjusted to pH 6.3. After autoclaving at 121°C and 210 kPa for 20 min, the test tubes were put immediately under anaerobic conditions. Next, a sterilized stock solution of fructose was added aseptically to the test tubes. For inoculum preparation, the strains were inoculated from stock cultures stored at −80°C into 10 ml of the appropriate liquid medium (WC, MRS, RB, or TOS-propionate medium) and were incubated anaerobically at 37°C for 12 h. Next, 5.0% (vol/vol) of the first subculture was grown in 10 ml of MCB at 37°C for 48 h, under anaerobic conditions. Acetate and lactate concentrations were determined through high-performance liquid chromatography with refractive index detection (HPLC-RI), as described below.
Selected isolates, together with nine reference bifidobacterial strains (B. longum subsp. longum LMG 11047, B. longum subsp. longum LMG 13196, B. longum subsp. longum NCC2705, B. longum subsp. infantis LMG 11570, B. longum subsp. infantis LMG 11588, B. bifidum LMG 11583, B. bifidum LMG 13195, B. bifidum DSM 20082, and B. adolescentis LMG 10502), were classified and identified by (GTG)5-PCR fingerprinting of genomic DNA. For the isolation of genomic DNA, cell pellets of overnight cultures grown in the appropriate medium were collected by microcentrifugation at 21,036 × g for 15 min at 4°C and were stored at −20°C for at least 1 h. DNA was extracted from the cell pellets as described previously (64). Genomic DNA was amplified with the (GTG)5 primer (65, 66). After electrophoresis, image analysis of the resulting amplicon fingerprints was performed with the BioNumerics 5.10 software package (Applied Maths, Sint-Martens-Latem, Belgium). Similarities between the digitized profiles of the isolates and reference bifidobacterial strains were quantified using the Pearson correlation coefficient, and dendrograms were constructed using the unweighted pair group method with arithmetic average clustering algorithm. For identification and verification of the clusters assigned, the 16S rRNA genes of representative isolates from each cluster were amplified and sequenced using the genus-specific primers Bif164 and Bif662 (67). The sequence results were aligned and assembled with BioEdit software (68) and were evaluated through comparison with the nucleotide collection database (69) using the BLASTn algorithm (70) (see Table S1 in the supplemental material).
Screening experiments.
Initial screening experiments with bifidobacterial strains were performed in duplicate in 10-ml test tubes containing MCB (pH 6.3) supplemented with 5.0 g liter−1 of AXOS-5-0.27, 9.0 g liter−1 of FOS, or 9.0 g liter−1 of inulin. These concentrations of the energy sources were according to a previous study with bifidobacterial strains isolated from feces (13, 16).
First, the MCB-containing test tubes were autoclaved (121°C at 210 kPa for 20 min) and put immediately under anaerobic conditions. Next, sterilized stock solutions of the substrates were added aseptically to the sterile MCB. The inocula (5.0% [vol/vol]) were prepared after transfer of the bifidobacterial strains from −80°C stock cultures to 10 ml of reinforced clostridial medium (Oxoid) or MRS medium (in the case of the B. bifidum strains), and the cultures were incubated at 37°C for 12 h, under anaerobic conditions (13, 16). To obtain bacterial cells in the exponential growth phase, 0.5 ml of these cultures was transferred into 10 ml of MCB supplemented with fructose or A-X mixture, and the cultures were incubated anaerobically at 37°C for 12 h. Then, 5.0% (vol/vol) of these cultures was transferred into 10 ml of MCB supplemented with FOS, inulin, or (A)XOS, and the cultures were incubated anaerobically at 37°C. Samples were taken after 48 h of fermentation to analyze the ITF and (A)XOS degradation capacities of the bifidobacterial strains, as described above.
Fermentation experiments.
Laboratory fermentations were carried out with four representative bifidobacterial strains (B. longum B24, B. longum B18, B. bifidum B46, and B. adolescentis B72), each representing a different ITF and (A)XOS degradation phenotype, in duplicate. Fermentations were performed in 10-ml test tubes containing MCB (pH 6.3) supplemented with 5.0 g liter−1 of A-X mixture or in 100-ml Schott flasks containing MCB (pH 6.3) supplemented with 9.0 g liter−1 of fructose, FOS, or inulin, 5.0 g liter−1 of XOS 95P, or 5.0 g liter−1 of (A)XOS (13, 16). Sterilized stock solutions of these energy sources were added aseptically to sterile MCB. The inocula were prepared via two subcultivations at 37°C under anaerobic conditions, as described above. Fermentations with fructose and the A-X mixture were sampled after inoculation (0 h) and after 24 h (A-X mixture) or 48 h (fructose) of fermentation, to monitor monosaccharide consumption. Fermentations with XOS and (A)XOS were sampled after inoculation (0 h) and after 4, 8, 12, 24, and 48 h of fermentation, to monitor bacterial growth, carbohydrate consumption, and metabolite production as a function of time.
Fermentation analyses. (i) Bacterial growth.
To monitor bacterial growth, OD600 values were determined with a visible-wavelength spectrophotometer (Genesys 20; Thermo Scientific, Waltham, MA). Ultrapure water was used as a blank. Each measurement was performed in triplicate.
(ii) Carbohydrate consumption.
Residual concentrations of arabinose, xylose, XOS, and XOS(A)XOS were determined using HPAEC-PAD, as described previously (16). Briefly, samples were deproteinized with Carrez A solution (36 g liter−1 of K2[Fe(CN)6]·3H2O [Sigma-Aldrich]) and Carrez B solution (72 g liter−1 of ZnSO4·7H2O [VWR International]) and injected (10 μl) into a CarboPac PA100 analytical column (Thermo Scientific) on an ICS-3000 chromatograph (Thermo Scientific). A mixture of arabinose (Sigma-Aldrich), xylose (Sigma-Aldrich), xylobiose, xylotriose, xylotetraose, xylopentaose, and xylohexaose (all from Megazyme International, Bray, Ireland) was used as an external standard to perform quantifications. All samples were analyzed in triplicate. The results are represented as averages of three independent measurements.
The breakdown of AXOS was analyzed through HPAEC-PAD as described previously (16, 58). Briefly, samples were deproteinized with Carrez A and B solutions and injected (10 μl) into a CarboPac PA200 analytical column (Thermo Scientific) on an ICS-3000 chromatograph (Thermo Scientific). To interpret the chromatograms, enzymatic reference degradation chromatograms, which were generated through degradation of (A)XOS in solution by α-arabinofuranosidases with known specificities (58), were used.
Residual concentrations of fructose, FOS, and inulin were determined through HPLC-RI, as described previously (13). Briefly, samples were deproteinized with Carrez A and B solutions and injected (30 μl) into an ICSep ICE ORH-801 column (Transgenomic North America, Omaha, NE) on a Waters chromatograph (Waters, Milford, MA), as described previously. For complete acid hydrolysis of the glycosidic bonds of FOS and inulin, samples were treated with equal volumes of 20% (mass/vol) trichloroacetic acid (deproteinization) for 24 h at room temperature. All samples were analyzed in triplicate. Calibration was performed with external standards. The results (expressed as FE) are represented as averages of three independent measurements.
Oligofructose fractions were analyzed quantitatively by gas chromatography with flame ionization detection (GC-FID), using a 5300-HT high-resolution gas chromatograph (Carlo Erba, Rodina, Italy) equipped with a SGE HT-5 aluminum-clad capillary column (Achrom, Zulte, Belgium) and applying as external standards oligofructose (RaftiloseP95X; BENEO-Orafti), glucose, fructose, and sucrose, as described previously (13). Briefly, the isooctane phase of derivatized samples was injected (1 μl) into the column (71). Concentrations of fructose (F), glucose (G), sucrose (GF), the oligosaccharides inulobiose (F2) up to inuloheptaose (F7), and kestose (GF2) up to kestoheptaose (GF6) were determined separately. In the figures concerning the breakdown of FOS, the concentrations of the different-chain-length fractions of oligofructose with the same DP (starting from a DP of 3) are represented as the sum of the concentrations of the different types of fractions, i.e., GFX and FX+1, expressed as FE. Each sample was analyzed once, which was possible because of the high reproducibility and reliability of the method (13, 71).
The breakdown of inulin was analyzed with HPAEC-PAD, using a DX500 chromatograph equipped with a CarboPac PA100 analytical column, as described previously (19). For the mobile phase, three different solutions were used, at a flow rate of 1 ml min−1, namely, 0.1 M NaOH (eluent A), 0.1 M NaOH and 0.4 M CH3COONa (eluent B), and 1 M NaOH (eluent C). The following gradient was applied: 0 min, 96% A, 4% B, and 0% C; 15 min, 60% A, 40% B, and 0% C; 30 min, 30% A, 70% B, and 0% C; 50 min, 10% A, 90% B, and 0% C; 60 min, 0% A, 0% B, and 100% C; 70 min, 0% A, 96% B, and 4% C. Samples that had been deproteinized with Carrez A and B solutions were filtered (0.2-μm filters; Whatman, GE Healthcare Life Sciences, Buckinghamshire, UK) and injected (50 μl) into the column.
Metabolite production.
Acetate, lactate, formate, and ethanol concentrations were determined through HPLC-RI as described above. A mixture of acetate (Merck), lactate (Sigma-Aldrich), formate (Merck), and ethanol (Merck) was used as an external standard to perform quantifications. All samples were analyzed in triplicate. The results are represented as averages of three independent measurements.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the financial support of the Research Council of the Vrije Universiteit Brussel (SRP7, IRP2, and IOF342 projects) and the Hercules Foundation (project UABR 09/004). A.R. was the recipient of a PhD fellowship from the Research Foundation-Flanders (FWO-Vlaanderen). M.S. was the recipient of a PhD fellowship from the Vrije Universiteit Brussel, in the framework of a bilateral agreement with the University of Ljubljana.
We thank Christophe Courtin (KU Leuven) for providing AXOS. Filip Timmermans (formerly of Thermo Scientific) is acknowledged for kindly providing the CarboPac PA200 column. We also thank the analytical laboratory team of BENEO-Orafti (Tienen, Belgium) for the oligofructose analyses.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02893-17.
REFERENCES
- 1.Whitman WB, Coleman DC, Wiebe WJ. 1998. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A 95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE, Peterson DA, Gordon JI. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 3.Sommer F, Bäckhed F. 2013. The gut microbiota: masters of host development and physiology. Nat Rev Microbiol 11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 4.O'Hara AM, Shanahan F. 2006. The gut flora as a forgotten organ. EMBO Rep 7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinan TG, Stilling RM, Stanton C, Cryan JF. 2015. Collective unconscious: how gut microbes shape human behavior. J Psychiatr Res 63:1–9. doi: 10.1016/j.jpsychires.2015.02.021. [DOI] [PubMed] [Google Scholar]
- 6.De Vuyst L, Leroy F. 2011. Cross-feeding between bifidobacteria and butyrate-producing colon bacteria explains bifidobacterial competitiveness, butyrate production, and gas production. Int J Food Microbiol 149:73–80. doi: 10.1016/j.ijfoodmicro.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 7.De Vuyst L, Moens F, Selak M, Rivière A, Leroy F. 2014. Summer meeting 2013: growth and physiology of bifidobacteria. J Appl Microbiol 116:477–491. doi: 10.1111/jam.12415. [DOI] [PubMed] [Google Scholar]
- 8.Greenblum S, Carr R, Borenstein E. 2015. Extensive strain-level copy-number variation across human gut microbiome species. Cell 160:583–594. doi: 10.1016/j.cell.2014.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fåk F, Bäckhed F. 2012. Lactobacillus reuteri prevents diet-induced obesity, but not atherosclerosis, in a strain dependent fashion in Apoe−/− mice. PLoS One 7:e46837. doi: 10.1371/journal.pone.0046837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wall R, Marques TM, O'Sullivan O, Ross RP, Shanahan F, Quigley EM, Dinan TG, Kiely B, Fitzgerald GF, Cotter PD, Fouhy F, Stanton C. 2012. Contrasting effects of Bifidobacterium breve NCIMB 702258 and Bifidobacterium breve DPC 6330 on the composition of murine brain fatty acids and gut microbiota. Am J Clin Nutr 95:1278–1287. doi: 10.3945/ajcn.111.026435. [DOI] [PubMed] [Google Scholar]
- 11.Gagnon M, Savard P, Rivière A, Lapointe G, Roy D. 2015. Bioaccessible antioxidants in milk fermented by Bifidobacterium longum subsp. longum strains. BioMed Res Int 2015:169381. doi: 10.1155/2015/169381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 13.Falony G, Lazidou K, Verschaeren A, Weckx S, Maes D, De Vuyst L. 2009. In vitro kinetic analysis of fermentation of prebiotic inulin-type fructans by Bifidobacterium species reveals four different phenotypes. Appl Environ Microbiol 75:454–461. doi: 10.1128/AEM.01488-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watson D, O'Connell Motherway M, Schoterman MHC, Joost van Neerven RJ, Nauta A, van Sinderen D. 2013. Selective carbohydrate utilization by lactobacilli and bifidobacteria. J Appl Microbiol 114:1132–1146. doi: 10.1111/jam.12105. [DOI] [PubMed] [Google Scholar]
- 15.Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. 2013. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain dependent. PLoS One 8:e76341. doi: 10.1371/journal.pone.0076341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rivière A, Moens F, Selak M, Maes D, Weckx S, De Vuyst L. 2014. The ability of bifidobacteria to degrade arabinoxylan oligosaccharide constituents and derived oligosaccharides is strain dependent. Appl Environ Microbiol 80:204–217. doi: 10.1128/AEM.02853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLaughlin HP, O'Conell Motherway M, Lakshminarayanan B, Stanton C, Ross RP, Brulc J, Menon R, O'Toole PW, van Sinderen D. 2015. Carbohydrate catabolic diversity of bifidobacteria and lactobacilli of human origin. Int J Food Microbiol 203:109–121. doi: 10.1016/j.ijfoodmicro.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Odamaki T, Horigome A, Sugahara H, Hashikura N, Minami Xiao J-Z, Abe F. 2015. Comparative genomics revealed genetic diversity and species/strain-level differences in carbohydrate metabolism of three probiotic bifidobacterial species. Int J Genomics 2015:567809. doi: 10.1155/2015/567809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falony G, Calmeyn T, Leroy F, De Vuyst L. 2009. Coculture fermentations of Bifidobacterium species and Bacteroides thetaiotaomicron reveal a mechanistic insight into the prebiotic effect of inulin-type fructans. Appl Environ Microbiol 75:2312–2319. doi: 10.1128/AEM.02649-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Mérieux A, Melo Minardi R, M'rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. 2011. Enterotypes of the human gut microbiome. Nature 473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leahy SC, Higgins DG, Fitzgerald GF, van Sinderen D. 2005. Getting better with bifidobacteria. J Appl Microbiol 98:1303–1315. doi: 10.1111/j.1365-2672.2005.02600.x. [DOI] [PubMed] [Google Scholar]
- 22.Di Gioia D, Aloisio I, Mazzola G, Biavati B. 2014. Bifidobacteria: their impact on gut microbiota composition and their applications as probiotics in infants. Appl Microbiol Biotechnol 98:563–577. doi: 10.1007/s00253-013-5405-9. [DOI] [PubMed] [Google Scholar]
- 23.Grimm V, Westermann C, Riedel CU. 2014. Bifidobacteria-host interactions: an update on colonisation factors. BioMed Res Int 2014:960826. doi: 10.1155/2014/960826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rivière A, Gagnon M, Weckx S, Roy D, De Vuyst L. 2015. Mutual cross-feeding interactions between Bifidobacterium longum NCC2705 and Eubacterium rectale ATCC 33656 explain the bifidogenic and butyrogenic effects of arabinoxylan-oligosaccharides. Appl Environ Microbiol 81:7767–7781. doi: 10.1128/AEM.02089-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moens F, Weckx S, De Vuyst L. 2016. Bifidobacterial inulin-type fructan degradation capacity determines cross-feeding interactions between bifidobacteria and Faecalibacterium prausnitzii. Int J Food Microbiol 231:76–85. doi: 10.1016/j.ijfoodmicro.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 26.Garrido D, Ruiz-Moyano S, Jimenez-Espinoza R, Eom HJ, Block DE, Mills DA. 2013. Utilization of galactooligosaccharides by Bifidobacterium longum subsp. infantis isolates. Food Microbiol 33:262–270. doi: 10.1016/j.fm.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pastell H, Westermann P, Meyer AS, Tuomainen P, Tenkanen M. 2009. In vitro fermentation of arabinoxylan-derived carbohydrates by bifidobacteria and mixed faecal microbiota. J Agric Food Chem 57:8598–8606. doi: 10.1021/jf901397b. [DOI] [PubMed] [Google Scholar]
- 28.Cloetens L, Broekaert WF, Delaedt Y, Ollevier F, Courtin CM, Delcour JA, Rutgeerts P, Verbeke K. 2010. Tolerance of arabinoxylan-oligosaccharides and their prebiotic activity in healthy subjects: a randomised, placebo-controlled cross-over study. Br J Nutr 103:703–713. doi: 10.1017/S0007114509992248. [DOI] [PubMed] [Google Scholar]
- 29.Maki KC, Gibson GR, Dickmann RS, Kendall CW, Chen CY, Costabile A, Comelli EM, McKay DL, Almeida NG, Jenkins D, Zello GA, Blumberg JB. 2012. Digestive and physiologic effects of a wheat bran extract, arabino-xylan-oligosaccharide, in breakfast cereal. Nutrition 28:1115–1121. doi: 10.1016/j.nut.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 30.Walton GE, Lu C, Trogh I, Arnaut F, Gibson GR. 2012. A randomised, double-blind, placebo controlled cross-over study to determine the gastrointestinal effects of consumption of arabinoxylan-oligosaccharides enriched bread in healthy volunteers. Nutr J 11:36. doi: 10.1186/1475-2891-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouhnik Y, Raskine L, Champion K, Andrieux C, Penven S, Jacobs H, Simoneau G. 2007. Prolonged administration of low-dose inulin stimulates the growth of bifidobacteria in humans. Nutr Res 27:187–193. doi: 10.1016/j.nutres.2007.01.013. [DOI] [Google Scholar]
- 32.Gibson G, Beatty E, Wang X, Cummings J. 1995. Selective stimulation of bifidobacteria in the human colon by FOS and inulin. Gastroenterology 108:975–982. doi: 10.1016/0016-5085(95)90192-2. [DOI] [PubMed] [Google Scholar]
- 33.Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. 2009. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr 101:541–550. doi: 10.1017/S0007114508019880. [DOI] [PubMed] [Google Scholar]
- 34.Neyrinck AM, Van Hée VF, Piront N, De Backer F, Toussaint O, Cani PD, Delzenne NM. 2012. Wheat-derived arabinoxylan oligosaccharides with prebiotic effect increase satietogenic gut peptides and reduce metabolic endotoxemia in diet-induced obese mice. Nutr Diabetes 2:e28. doi: 10.1038/nutd.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM, Gibson GR, Delzenne NM. 2007. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia 50:2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- 36.Turroni F, Özcan E, Milani C, Mancabelli L, Viappiani A, van Sinderen D, Sela DA, Ventura M. 2015. Glycan cross-feeding activities between bifidobacteria under in vitro conditions. Front Microbiol 6:1030. doi: 10.3389/fmicb.2015.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egan M, O'Connell Motherway M, Kilcoyne M, Kane M, Joshi L, Ventura M, van Sinderen D. 2014. Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiol 14:282. doi: 10.1186/s12866-014-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Junick J, Blaut M. 2012. Quantification of human fecal Bifidobacterium species by use of quantitative real-time PCR analysis targeting the groEL gene. Appl Environ Microbiol 78:2613–2622. doi: 10.1128/AEM.07749-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishikawa E, Matsuki T, Kubota H, Makino H, Sakai T, Oishi K, Kushiro A, Fujimoto J, Watanabe K, Watanuki M, Tanaka R. 2013. Ethnic diversity of gut microbiota: species characterization of Bacteroides fragilis group and genus Bifidobacterium in healthy Belgian adults, and comparison with data from Japanese subjects. J Biosc Bioeng 116:265–270. doi: 10.1016/j.jbiosc.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 40.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. 2013. The long-term stability of the human gut microbiota. Science 341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. 2016. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duranti S, Milani C, Lugli GA, Turroni F, Mancabelli L, Sanchez B, Ferrario C, Viappiani A, Mangifesta M, Mancino W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2015. Insights from genomes of representatives of the human gut commensal Bifidobacterium bifidum. Environ Microbiol 17:2515–2531. doi: 10.1111/1462-2920.12743. [DOI] [PubMed] [Google Scholar]
- 43.Turroni F, Duranti S, Bottacini F, Guglielmetti S, van Sinderen D, Ventura M. 2014. Bifidobacterium bifidum as an example of a specialized human gut commensal. Front Microbiol 5:437. doi: 10.3389/fmicb.2014.00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lagaert S. 2013. Study of the arabinoxylan and arabinoxylan oligosaccharide-degrading enzymes of Bifidobacterium adolescentis and Bifidobacterium longum. PhD thesis Katholieke Universiteit Leuven, Leuven, Belgium. [Google Scholar]
- 45.Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen MC, Desiere F, Bork P, Delley M, Pridmore RD, Arigoni F. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci U S A 99:14422–14427. doi: 10.1073/pnas.212527599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ejby M, Fredslund F, Vujicic-Zagar A, Svensson B, Slotboom DJ, Abou Hachem M. 2013. Structural basis for arabinoxylo-oligosaccharide capture by the probiotic Bifidobacterium animalis subsp. lactis Bl-04. Mol Microbiol 90:1100–1112. doi: 10.1111/mmi.12419. [DOI] [PubMed] [Google Scholar]
- 47.Lagaert S, Van Campenhout S, Pollet A, Bourgois TM, Delcour JA, Courtin CM, Volckaert G. 2007. Recombinant expression and characterization of a reducing end xylose-releasing exo-oligoxylanase from Bifidobacterium adolescentis. Appl Environ Microbiol 73:5374–5377. doi: 10.1128/AEM.00722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lagaert S, Pollet A, Delcour JA, Lavigne R, Courtin CM, Volckaert G. 2011. Characterization of two β-xylosidases from Bifidobacterium adolescentis and their contribution to the hydrolysis of prebiotic xylooligosaccharides. Appl Microbiol Biotechnol 92:1179–1185. doi: 10.1007/s00253-011-3396-y. [DOI] [PubMed] [Google Scholar]
- 49.Pokusaeva K, Fitzgerald GF, van Sinderen D. 2011. Carbohydrate metabolism in bifidobacteria. Genes Nutr 6:285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van der Meulen R, Avonts L, De Vuyst L. 2004. Short fractions of oligofructose are preferentially metabolized by Bifidobacterium animalis DN-173 010. Appl Environ Microbiol 70:1923–1930. doi: 10.1128/AEM.70.4.1923-1930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van der Meulen R, Makras E, Verbrugghe K, Adriany T, De Vuyst L. 2006. In vitro kinetic analysis of oligofructose consumption by Bacteroides and Bifidobacterium spp. indicates different degradation mechanisms. Appl Environ Microbiol 72:1006–1012. doi: 10.1128/AEM.72.2.1006-1012.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van der Meulen R, Adriany T, Verbrugghe K, De Vuyst L. 2006. Kinetic analysis of bifidobacterial metabolism reveals a minor role for succinic acid in the regeneration of NAD+ through its growth-associated production. Appl Environ Microbiol 72:5204–5210. doi: 10.1128/AEM.00146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grootaert C, Van den Abbeele P, Marzorati M, Broekaert WF, Courtin CM, Delcour JA, Verstraete W, Van de Wiele T. 2009. Comparison of prebiotic effects of arabinoxylan oligosaccharides and inulin in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol Ecol 69:231–242. doi: 10.1111/j.1574-6941.2009.00712.x. [DOI] [PubMed] [Google Scholar]
- 54.Sanchez JI, Marzorati M, Grootaert C, Baran M, Van Craeyveld V, Courtin CM, Broekaert WF, Delcour JA, Verstraete W, Van de Wiele T. 2009. Arabinoxylan-oligosaccharides (AXOS) affect the protein/carbohydrate fermentation balance and microbial population dynamics of the simulator of human intestinal microbial ecosystem. Microb Biotechnol 2:101–113. doi: 10.1111/j.1751-7915.2008.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van den Abbeele P, Venema K, Van de Wiele T, Verstraete W, Possemiers S. 2013. Different human gut models reveal the distinct fermentation patterns of arabinoxylan versus inulin. J Agric Food Chem 61:9819–9827. doi: 10.1021/jf4021784. [DOI] [PubMed] [Google Scholar]
- 56.Selak M, Rivière A, Moens F, Van den Abbeele P, Geirnaert A, Rogelj I, Leroy F, De Vuyst L. 2016. Inulin-type fructan fermentation by bifidobacteria depends on the strain rather than the species and region in the human intestine. Appl Microbiol Biotechnol 100:4097–4107. doi: 10.1007/s00253-016-7351-9. [DOI] [PubMed] [Google Scholar]
- 57.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. 2013. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol 11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 58.Rivière A, Eeltink S, Pierlot C, Balzarini T, Moens F, Selak M, De Vuyst L. 2013. A high-resolution ion-exchange chromatography method for monitoring of prebiotic arabinoxylan-oligosaccharide degradation in a complex fermentation medium. Anal Chem 85:4982–4990. doi: 10.1021/ac400187f. [DOI] [PubMed] [Google Scholar]
- 59.Van den Abbeele P, Grootaert C, Marzorati M, Possemiers S, Verstraete W, Gérard P, Rabot S, Bruneau A, El Aidy S, Derrien M, Zoetendal E, Kleerebezem M, Smidt H, Van de Wiele T. 2010. Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for Bacteroidetes and Clostridium cluster IX. Appl Environ Microbiol 76:5237–5246. doi: 10.1128/AEM.00759-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Possemiers S, Verthe K, Uyttendaele S, Verstraete W. 2004. PCR-DGGE-based quantification of stability of the microbial community in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol Ecol 49:495–507. doi: 10.1016/j.femsec.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 61.Ferraris L, Aires J, Waligora-Dupriet A-J, Butel M-J. 2010. New selective medium for selection of bifidobacteria from human feces. Anaerobe 16:469–471. doi: 10.1016/j.anaerobe.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 62.Serafini F, Bottacini F, Viappiani A, Baruffini E, Turroni F, Foroni E, Lodi T, van Sinderen D, Ventura M. 2011. Insights into physiological and genetic mupirocin susceptibility in bifidobacteria. Appl Environ Microbiol 77:3141–3146. doi: 10.1128/AEM.02540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hartemink R, Kok B, Weenk G, Rombouts F. 1996. Raffinose-Bifidobacterium (RB) agar, a new selective medium for bifidobacteria. J Microbiol Methods 27:33–43. doi: 10.1016/0167-7012(96)00926-8. [DOI] [Google Scholar]
- 64.Braem G, De Vliegher S, Supré K, Haesebrouck F, Leroy F, De Vuyst L. 2011. (GTG)5-PCR fingerprinting for the classification and identification of coagulase-negative Staphylococcus species from bovine milk and teat apices: a comparison of type strains and field isolates. Vet Microbiol 147:67–74. doi: 10.1016/j.vetmic.2010.05.044. [DOI] [PubMed] [Google Scholar]
- 65.Gevers D, Huys G, Swings J. 2001. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol Lett 205:31–36. doi: 10.1111/j.1574-6968.2001.tb10921.x. [DOI] [PubMed] [Google Scholar]
- 66.Masco L, Huys G, Gevers D, Verbrugghen L, Swings J. 2003. Identification of Bifidobacterium species using rep-PCR fingerprinting. Syst Appl Microbiol 26:557–563. doi: 10.1078/072320203770865864. [DOI] [PubMed] [Google Scholar]
- 67.Kok RG, De Waal A, Schut F, Welling GW, Weenk G, Hellingwerf KJ. 1996. Specific detection and analysis of a probiotic Bifidobacterium strain in infant faeces. Appl Environ Microbiol 62:3668–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
- 69.NCBI Resource Coordinators. 2014. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 42:D7–D17. doi: 10.1093/nar/gkt1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 71.Joye D, Hoebregs H. 2000. Determination of oligofructose, a soluble dietary fiber, by high-temperature capillary gas chromatography. J AOAC Int 83:1020–1025. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.