ABSTRACT
Glucansucrases (GSs) in glycoside hydrolase family 70 (GH70) catalyze the synthesis of α-glucans from sucrose, a reaction that is widely seen in lactic acid bacteria (LAB). These enzymes have been implicated in many aspects of microbial life. Products of GSs have great commercial value as food supplements and medical materials; therefore, these enzymes have attracted much attention from both science and industry. Certain issues concerning the origin and evolution of GSs are still to be addressed, although an increasing number of GH70 enzymes have been characterized. This study describes a GS enzyme with the appearance of a branching sucrase (BrS). Structural analysis indicated that this GS enzyme produced a type of glucan composed of an α-(1→6) glucosidic backbone and α-(1→4) branches, as well as a considerable amount of α-(1→3) branches, distinguishing it from the GSs identified so far. Moreover, sequence-based analysis of the catalytic core of this enzyme suggested that it might be an evolutionary intermediate between the BrS and GS subgroups. These results provide an evolutionary link between these subgroups of GH70 enzymes and shed new light on the origination of GSs.
IMPORTANCE GH70 GSs catalyze the synthesis of α-glucans from sucrose, a reaction that is widely seen in LAB. Products of these enzymes have great commercial value as food supplements and medical materials. Moreover, these enzymes have attracted much attention from scientists because they have potential in tailored synthesis of α-glucans with desired structures and properties. Although more and more GSs have been characterized, the origin and evolution of these enzymes have not been well addressed. This study describes a GS with the appearance of a BrS (i.e., high levels of similarity to BrSs in sequence analysis). Further analysis indicated that this enzyme synthesized a type of insoluble glucan composed of an α-(1→6) glucosidic backbone and many α-(1→4)- and α-(1→3)-linked branches, the linkage composition of which has rarely been reported in the literature. This BrS-like GS enzyme might be an evolutionary intermediate between BrS and GS enzymes.
KEYWORDS: GH70 enzymes, glucansucrase, branching sucrase, insoluble glucan, Leuconostoc mesenteroides, intermediate
INTRODUCTION
Enzymes of glycoside hydrolase family 70 (GH70) are increasingly attracting attention for their potential in producing α-glucans with wide applications in food and feed products, as well as health care products (1–3). GH70 enzymes are structurally and mechanistically related to the GH13 (4, 5) and GH77 (4, 6) families, with which they form the GH-H clan (4, 6) of glycoside hydrolases (5, 7, 8). To date, a total of 66 GH70 enzymes have been biochemically characterized, most of which are distributed in the genera Lactobacillus, Leuconostoc, Streptococcus, and Weissella (http://www.cazy.org/GH70_characterized.html) (9). Crystallographic structures of these enzymes suggested that they adopt a unique U-shaped fold organized into five distinct structural domains, i.e., A, B, C, IV, and V (10–12), of which A, B, and C are shared by GH13 enzymes, suggesting an evolutionary relationship between these two families (5).
Historically, GH70 enzymes are mainly glucansucrases (GSs) that catalyze the synthesis of α-glucan homopolysaccharides from the sole substrate sucrose, and enzymes of this type have been widely investigated (5, 7, 13). According to the CAZy database (http://www.cazy.org/GH70.html; accessed 4 July 2017), most GH70 enzymes identified in lactic acid bacteria (LAB) are GSs (9). However, several other types of enzymes have been found recently in GH70. A truncated GH70 enzyme derived from DsrE, designated GBD-CD2, was found to be inactive on sucrose, whereas it was active when dextran was supplemented as a glucosyl acceptor, forming α-(1,2) linkages on linear dextran (11, 14, 15). Based on this enzyme characteristic, GBD-CD2 was identified as an α-(1→2) branching sucrase (BrS) (11, 15). Thereafter, several other branching sucrases have been identified, and these enzymes constitute a new subfamily of GH70 enzymes, referred to as BrS enzymes (7, 8, 11, 16, 17). Moreover, two new subfamilies, i.e., the glucosyltransferase B (GTFB)-like 4,6-α-glucanotransferases (4,6-α-GTs) and the glucosyltransferase C (GTFC)-like 4,6-α-GTs, have been established as GH70 enzymes (18, 19). Like the BrS enzymes, 4,6-α-GTs are inactive on sucrose; however, they show enzyme specificities distinct from those of BrS enzymes. As illustrated in previous studies (7, 18, 19), 4,6-α-GTs synthesize novel α-glucans but they use α-(1→4)-linked glucans (i.e., maltooligosaccharides and amylose), rather than α-(1→6)-linked linear glucan (substrate of BrS enzymes), as substrates.
A conserved catalytic core composed of A, B, and C domains was identified in GH70 enzymes (10). At the level of the primary sequence, the catalytic core contains seven conserved motifs in total, designated motifs I to VII (4, 12, 17, 19). There are seven strictly conserved residues distributed among these motifs (7, 20), and these residues were also conserved across the GH-H clan (4).
GH70 enzymes are generally thought to originate from GH13 enzymes; GH13 is presently one of the largest families of glycoside hydrolases (4, 6), and its members have been found to be widely distributed in organisms from all domains of life (9). An evolutionary pathway based on “permutation per duplication” was proposed to illustrate the evolution of GH13 enzymes to GH70 enzymes (7, 19, 21, 22). As depicted in this model, GH70 enzymes share with GH13 enzymes similar domains (i.e., domain A, B, and C) in the catalytic core, whereas these domains were rearranged in GH70 enzymes in a permutation per duplication manner (7, 22). Despite these rearrangements, there are several conserved motifs (e.g., motifs I to VII) in members of the two families, and these conserved motifs provide crucial information for analyzing the origin and evolution of GH70 enzymes.
Notably, members of GH70 without circular permutation have been identified recently. GtfC from Exiguobacterium sibiricum 255-15 (19) and GtfD from Azotobacter chroococcum NCIMB8003 (23) represent two GH70 subfamily members without the circular permutation events discussed above.
Previously, we isolated BD3749, a strain of Leuconostoc mesenteroides with a high capacity for insoluble exopolysaccharide (EPS) synthesis (24). To explore the mechanism of insoluble EPS synthesis, the inventory of GH70-encoding genes in BD3749 was investigated. In the present study, Gsy, a GH70 enzyme with the appearance of a BrS enzyme, was examined for its enzymatic characteristics. The results showed that, despite the appearance of a BrS enzyme, Gsy has the activity of de novo synthesis of insoluble glucan. Further, structural analysis indicated that the glucan product is composed mainly of α-(1→6)- and α-(1→4)-linked glucose, with a considerable amount of α-(1→3) linkages. Moreover, phylogenetic analysis of the catalytic core of GH70 enzymes identified so far suggested that Gsy might be an evolutionary intermediate between BrS and GS enzymes.
RESULTS
Gsy catalysis of de novo polymerization from sucrose.
Strains of the genus Leuconostoc are important sources of EPSs and GH70 enzymes. Previous studies demonstrated that L. mesenteroides BD3749 produces large amounts of EPSs, including water-soluble and insoluble ones. A recent study reported Gsy as the main enzyme responsible for production of insoluble EPSs (24). To investigate the enzyme characteristics of Gsy, the encoding gene gsy was PCR amplified from BD3749 genomic DNA and introduced into Escherichia coli after ligation with the vector pET-28a(+) (Invitrogen). The transformants were then streaked on Luria-Bertani (LB) plates supplemented with 0.1 mM isopropyl-β-d-thiogalactoside (IPTG) and 20 g/liter sucrose, for EPS synthesis. As shown in Fig. 1A, colonies of the E. coli strain carrying pET28a(+)-gsy showed slimy surfaces when plated on the plates, a conspicuous phenotype in comparison with the control strain, which does not carry gsy. Notably, the morphology of the E. coli strain carrying gsy is reminiscent of the appearance of EPS-producing L. mesenteroides.
FIG 1.
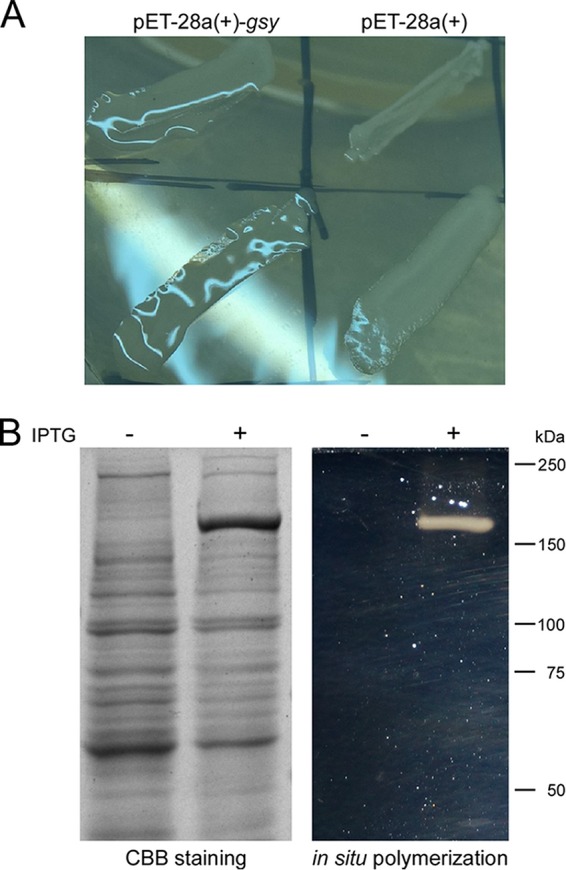
De novo polymerization activity of the recombinant Gsy. (A) Morphology of E. coli strains carrying pET28a(+)-gsy (left), compared to those carrying the empty vector (right). (B) SDS-PAGE analyses. (Left) Total protein extracts of E. coli cells were analyzed by SDS-PAGE, followed by staining with Coomassie brilliant blue (CBB). Note that a differentially expressed protein band was apparent in samples induced with IPTG. (Right) An equivalent gel was subjected to an in situ polymerization assay. At the site similar to the differential band in the CBB-stained gel, a polysaccharide-active band of ∼170 kDa was obvious.
Subsequently, the expression of the recombinant enzyme was confirmed by electrophoresis of the total protein extract of the E. coli strain carrying pET28a(+)-gsy. As shown by SDS-PAGE (Fig. 1B, left), a specific protein band with an apparent molecular mass of ∼170 kDa was obtained upon IPTG induction. This molecular mass is in accordance with that of the recombinant Gsy, confirming the accurate expression of Gsy. Moreover, to test whether the recombinant protein has the activity of a GS, an in situ polymerization assay was carried out with SDS-PAGE, as described previously (25, 26). As shown in Fig. 1B, right, the ectopically expressed enzyme was active in the transformation of sucrose into polysaccharides. This result showed that Gsy has the activity of de novo polymerization and is a GS.
Gsy synthesis of an insoluble glucan.
The recombinant enzyme was then purified through Ni2+-nitrilotriacetate (Ni-NTA) column affinity chromatography (see Fig. S1 in the supplemental material), and the polymer synthesized by purified Gsy was further analyzed. Purified recombinant Gsy produced an insoluble polymer from sucrose in sodium acetate buffer, with the release of fructose, as detected in the enzyme activity assay. The polymer remained insoluble under acidic conditions, whereas it was soluble in aqueous 0.5 M NaOH, giving a clear or slightly opalescent solution. Analysis of the monosaccharide composition by high-performance anion-exchange chromatography (HPAEC) demonstrated that the digested polymer displayed only one peak, at 5.875 min (Fig. S2), which accurately matched the retention time of glucose, indicating that the insoluble polymer contained glucose as the only monosaccharide. Therefore, the insoluble polymer produced by Gsy was a glucan.
Enzyme characterization.
Monosaccharide composition analysis demonstrated that Gsy is a GS. To study the enzyme characteristics of Gsy, an enzyme activity assay was performed as described previously (27–29). The purified Gsy showed an activity of 3.5 U/ml (16.5 U/mg of purified enzyme), as determined by the release of fructose.
Previous studies showed that L. mesenteroides GSs have optimal reaction temperatures and pH values ranging from 30°C to 35°C and from 5.0 to 6.0, respectively (13, 30). To determine the optimal reaction conditions for Gsy, the effects of temperature and pH on enzyme activity were examined. As shown in Fig. 2, the optimal pH for Gsy activity was 5.5 in 50 mM sodium acetate buffer; at higher pH values, the enzyme activity decreased rapidly. As for the temperature conditions, the enzyme activity increased with higher temperatures up to 30°C and remained at a high level to 35°C, after which it decreased sharply. These results showed that Gsy is a GS with typical GS enzyme characteristics.
FIG 2.
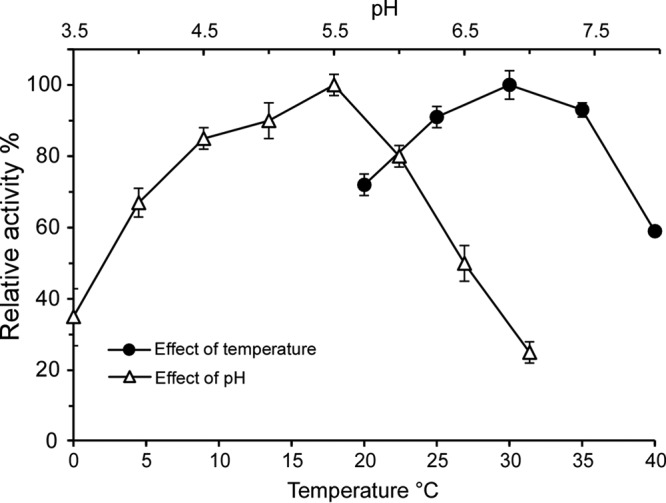
Effects of pH and temperature on the activity of the purified Gsy. The effect of pH was assayed in 50 mM sodium acetate (pH 3.5 to 7.5), and the effect of temperature (20°C to 40°C) was assayed in 50 mM sodium acetate (pH 5.4). Reactions were carried out for 15 min. The enzyme activity was reflected by the amount of fructose released. The highest activity was set as 100%, and the other values were calculated accordingly (relative activity). Shown are the average values of three independent experiments, and the error bars represent the standard errors (n = 3).
Structural characterization of the insoluble glucan.
The types of glycosidic linkages and the functional groups of the glucan were analyzed by using Fourier transform infrared (FTIR) spectroscopy. As shown in Fig. 3, the glucan produced by Gsy mainly contained α-(1→6) linkages, as demonstrated by the peak at 1,004 cm−1, which characterizes the considerable chain flexibility present in dextran around this type of glycosidic bond (31, 32). The α-glycosidic bond was also confirmed by the peak at 919 cm−1 and the band at 1,147 cm−1, caused by covalent vibrations of the C-O-C bond and glycosidic bridge. The peak at 1,102 cm−1 was due to vibration of the C-O bond at the C-4 position of the glucose residue (31). The hydroxyl and C-H stretching vibrations of the polysaccharide caused the bands in the regions of 3,281 cm−1 and 2,922 cm−1, respectively (33, 34). The band in the region of 1,645 cm−1 was due to bound water, according to previous analyses (32, 35).
FIG 3.
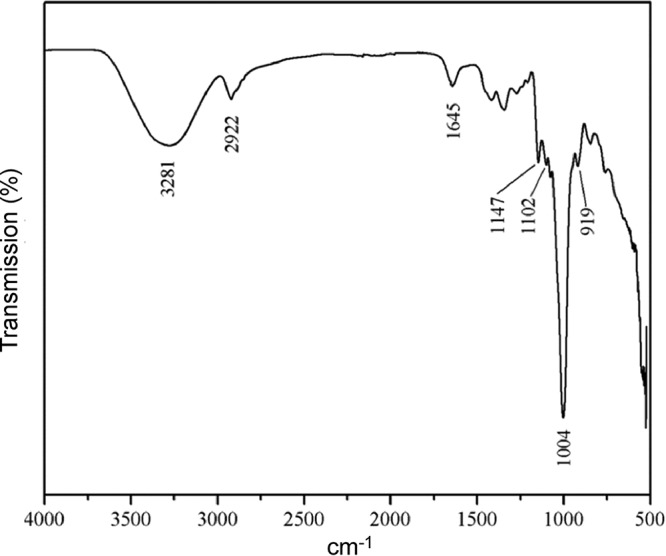
FTIR spectrum of the glucan synthesized by purified Gsy. The glucan sample was prepared as described in Materials and Methods. The FTIR spectrum was obtained using a Nicolet Nexus 470 spectrometer.
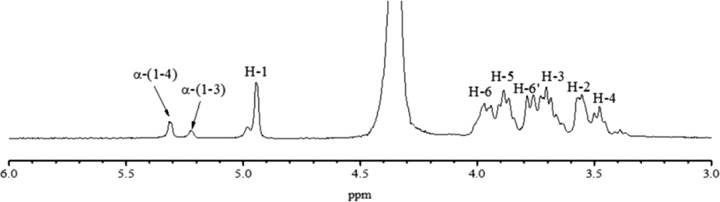
Subsequently, the types of glycosidic linkages were analyzed by 1H NMR spectroscopy. Since α-(1→6) linkages are flexible while α-(1→2), α-(1→3), and α-(1→4) linkages are more rigid, the insolubility of some α-glucans is usually the result of more rigid linkages in their structure (36). In the 1H NMR spectrum shown in Fig. 4, there are three anomeric protons, at 4.82 to 5.10, 5.22, and 5.31 ppm, which are ascribed to the anomeric protons of α-(1→6) d-Glcp, α-(1→3) d-Glcp, and α-(1→4) d-Glcp, respectively (37–39), demonstrating that the polymer is an α-glucan. As listed in Table 1, integration of the anomeric signals revealed that the mole percentages of the three corresponding linkages were 63.1%, 14.3%, and 22.6%, respectively. Due to its limited solubility, as well as the high concentration of NaOD in D2O, no ideal 13C NMR spectrum of the insoluble glucan has been obtained yet. Even so, based on the FTIR and 1H NMR data, it can be confirmed that this insoluble glucan contained an α-(1→6) glucosidic backbone and fairly considerable α-(1→4)- and α-(1→3)-linked branches.
FIG 4.
1H NMR spectrum of the glucan produced by purified Gsy. The 1H NMR spectrum was recorded at 500 MHz in D2O at 30°C. 1H chemical shifts are given using sodium 2,2,3,3-tetradeutero-3-trimethylsilylpropanoate (1H = 0 ppm) as an internal reference. The 1H NMR spectrum was obtained with a Varian Mercury Plus 400 spectrometer (Varian, USA).
TABLE 1.
Mole percentages of glycosidic linkages in different insoluble α-glucans, as determined by methylation or NMR
| Strain | Glycosidic linkage (mol %) |
Reference | |||
|---|---|---|---|---|---|
| α-(1→6) | α-(1→2) | α-(1→3) | α-(1→4) | ||
| L. mesenteroides R1510a | 67.7 | 12.5 | 19.8 | 0 | 49 |
| L. mesenteroides NRRLB-1299a,b | 65 | 30 | 5 | 0 | 50, 51 |
| L. mesenteroides NRRLB-1399a | 87.28 | 6.81 | 5.91 | 0 | 52 |
| L. mesenteroides NRRLB-1254a | 76.49 | 0 | 0 | 23.51 | 51 |
| L. mesenteroides NRRLB-1118a | 41.65 | 0 | 58.34 | 0 | 53 |
| L. mesenteroides NRRLB-742a | 86.63 | 0 | 0.71 | 12.65 | 54 |
| L. mesenteroides BD3749c | 63.1 | 0 | 14.3 | 22.6 | This study |
The mole percentages of glycosidic linkages were determined by methylation.
NRRLB-1299 was also classified as Leuconostoc citreum on the basis of repetitive-sequence-based PCR analysis and fermentation profiles (55).
The mole percentages of glycosidic linkages were determined by NMR.
Appearance of a branching sucrase.
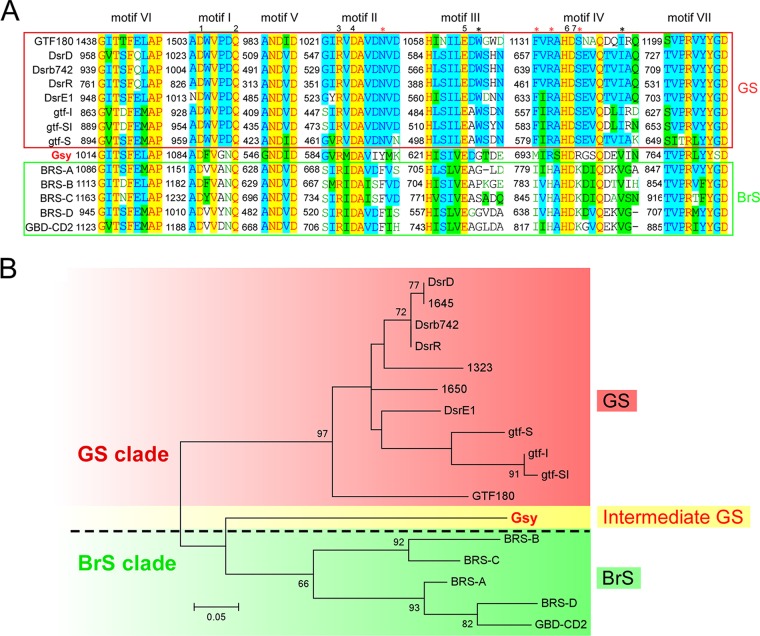
Although the biochemical studies described above demonstrated the enzyme activity of Gsy as a GS enzyme, sequence-based analysis showed that Gsy shares greater similarity (i.e., 50% identity) with the BrS enzyme GBD-CD2 (11, 15) than with any other GS enzyme identified so far. Besides the overall similarities, the seven functional sequence signatures (motifs I to VII) were also exploited in sequence analysis of GH70 members (4, 11, 12, 19). Vuillemin et al. compared the sequences of these conserved motifs of the BrS enzymes identified so far with those of the GSs and found several unique amino acids that appear exclusively in BrS enzymes (16). Accordingly, they suggested that BrS enzymes contain uniquely conserved residues in the catalytic core, compared to GSs. To obtain a detailed sequence feature of this peculiar enzyme, the sequence of the conserved motifs of Gsy was aligned with that of the representative GSs (7, 12, 19), as well as the BrS enzymes identified so far (8, 11, 16). As shown in Fig. 5A, although Gsy shared considerable similarities with GBD-CD2 and other BrS enzymes at some of the unique sites identified previously (marked with red asterisks in Fig. 5A) (16), it showed a pattern distinct from that of any of the BrS enzymes at other unique sites. For instance, at site 695 (numbered according to Gsy), an arginine was observed in most GSs, while a histidine was found in all of the BrS enzymes identified so far; interestingly, Gsy contains an arginine at this site, resembling a GS enzyme. At site 693, Gsy contains a methionine, distinct from either GS or BrS enzymes. At site 549, Gsy contains an isoleucine, similar to Gtf180 in Lactobacillus and gtf-S in Streptococcus and distinct from the BrS enzymes identified so far. Moreover, a serine appears at site 771 in Gsy, whereas a conserved glycine has been observed in the GS and BrS enzymes. Collectively, Gsy does not show the typical sequence characteristics identified by Vuillemin et al. for BrSs (16).
FIG 5.
Sequence-based analysis of motifs I to VII of the catalytic core of different GH70 enzymes. (A) Alignment of motifs I to VII from different GH70 enzymes. Sequence alignment was performed with Align X (Vector NTI, Invitrogen), and motifs I to VII are indicated above the sequences. GSs are shown in the upper lines enclosed in the red box, while BrSs are shown in the lower lines enclosed in the green box; Gsy is shown between the two subgroups. The numbers 1 to 7 indicate the highly conserved sites of GH70 enzymes, and the asterisks indicate the sites conserved exclusively in BrSs, as identified by Vuillemin et al. (16). The black asterisks indicate the conserved sites shared by Gsy, and the red asterisks indicate the sites at which Gsy showed a pattern distinct from that of the BrSs identified so far. (B) Phylogenetic analysis of GH70 enzymes, as indicated. Phylogenetic analysis was performed using MEGA 6.0, and the evolutionary tree was constructed based on the sequences of the core motifs shown in panel A. The scale bar represents a 5% difference in amino acid sequences. To illustrate the peculiar characteristics of Gsy, it is marked in red font to emphasize its enzyme function as a GS enzyme, but it is shaded yellow to indicate its appearance as a BrS enzyme as well as a GS enzyme. The dashed line indicates the separation of the GS and BrS clades.
Evolutionary intermediate between BrS and GS enzymes.
To obtain a clearer view of the relationship between BrS and GS enzymes, a phylogenetic analysis was performed using MEGA 6.0 and an evolutionary tree was constructed based on the sequence of the core motifs. As shown in Fig. 5B, the two subgroups (i.e., the GS subgroup, shaded in red, and the BrS subgroup, shaded in green) were separated from each other in the evolutionary tree. Interestingly, Gsy fell between these two subgroups in the evolutionary tree. This evolutionary position, together with GS activity under BrS appearance, suggested that Gsy might have significance in the separation of these two subgroups. As indicated in previous studies, the BrS and GS subgroups are in close relationship with each other in the evolutionary process (7, 11). Our analysis of the peculiar enzyme Gsy suggests that it might be an evolutionary intermediate linking these two subgroups.
Evolutionary aspects of GH70 subgroups.
As previous studies have illustrated, GH70 enzymes originated from GH13 (7, 10, 22), a large family of glycoside hydrolases with members widely distributed in all three domains of life (4). In GH70, four subgroups of enzymes with distinct enzyme characteristics, i.e., the GS subgroup, the BrS subgroup, and the GTFB-like and GTFC-like subgroups of 4,6-α-GTs, have been identified so far. According to Gangoiti et al., the 4,6-α-GT subgroup was formed in the process of the evolution of ancient GH13 enzymes into GSs (19, 40), and the BrS enzymes emerged after the separation of 4,6-α-GTs, as demonstrated in the previous study (7).
To investigate the position of Gsy in the evolutionary process, a phylogenetic analysis was performed with members of the four subgroups of GH70 and GH13. As shown in Fig. 6, along with the process of evolution from the ancient GH13 enzymes to modern GSs, three GH70 subgroups have emerged, in the following order: GTFC-like 4,6-α-GT subgroup, GTFB-like 4,6-α-GT subgroup (7, 19), and BrS subgroup (8, 16, 17). The enzyme Gsy emerged after the formation of the BrS subgroup and before that of the GS subgroup, indicating that Gsy might play an intermediate role in the divergence of the BrS and GS subgroups.
FIG 6.
Phylogenetic analysis of the four subgroups of GH70 enzymes and GH13 proteins. Alignment was performed with the GH70 and GH13 enzymes listed in Table S1 in the supplemental material. Overall, the GH70 enzymes were obtained by a BLAST analysis using the Gsy protein (GenBank accession number ANJ45894.1) as the query sequence. Notably, all of the characterized GH70 enzymes (http://www.cazy.org/GH70_characterized.html) were included in the alignment. Members of GH13 in the alignment were chosen according to previous studies (7, 19). The evolutionary tree is based on alignment of the full sequences of these enzymes. The biochemically characterized GSs and GTFB-like and GTFC-like 4,6-α-GTs of GH70, as well as members of GH13, are included. The evolutionary history was inferred using the maximum likelihood method, based on the JTT matrix-based model. The sequences are labeled with their GenBank accession numbers. The bar represents a genetic distance of 0.3 substitution per position. The BrS and GS subgroups of GH70 are indicated in green and red fonts, respectively. Gsy is shown in red font shaded with yellow, and its homologs that have not been biochemically characterized are shown in gray.
DISCUSSION
GH70 enzymes are transglucosylases produced by LAB, e.g., strains of Streptococcus, Leuconostoc, Weissella, or Lactobacillus (7, 12). Most of the enzymes classified in this family use sucrose as the d-glucopyranosyl donor to synthesize α-glucans of high molecular mass (>106 Da), with the release of fructose (12, 41). These enzymes catalyze the de novo synthesis of polysaccharide polymers from small molecules and thus are regarded as polymerases (11). In LAB, these enzymes are also referred to as glucosyltransferases (GTFs) (mainly used for Streptococcus) or glucansucrases (GSs). According to the differences in enzyme specificity and the type of glycosidic linkage formed, GSs can be classified into the following types: dextransucrase (EC 2.4.1.5), alternansucrase (EC 2.4.1.140), mutansucrase (EC 2.4.1.125), and reuteransucrase (EC 2.4.1.-) (7, 12). Apart from the polymerases described above, many GH70 enzymes with other enzyme characteristics have been identified recently (7, 12, 41). Overall, these enzymes do not have the activity of de novo polymerization (i.e., they are inactive on the substrate sucrose); instead, they can use oligosaccharides or polysaccharides as donor/acceptor substrates (8, 16, 18, 19, 40). BrSs (EC 2.4.1.-) are found in strains of Leuconostoc that produce exopolysaccharides with branching linkages (7). Although they are inactive on sucrose, they catalyze the transfer of glucosyl units from sucrose to dextrans, resulting in the synthesis of branching linkages onto dextrans (8, 11, 16). According to the type of branching linkage, BrS enzymes are classified into α-(1→2) and α-(1→3) branching sucrases, of which several members have been identified (11, 15, 16). α-4,6-Glucanotransferases (4,6-α-GTs) (EC 2.4.1.-) predominantly cleave an α-(1→4) glycosidic linkage from the nonreducing end of the donor substrate α-(1→4)-glucan and transfer the cleaved glucosyl unit to the nonreducing end of another α-(1→4)-glucan acceptor substrate, forming mainly α-(1→6) linkages, which results in the formation of isomaltooligosaccharide/maltooligosaccharide and polysaccharide mixtures with increasing percentages of α-(1→6) linkages (7). To date, two subgroups of 4,6-α-GTs have been implicated as GH70 enzymes, i.e., GTFB-like 4,6-α-GTs, with transglucosylase activity between α-(1→4)-glucans, and GTFC-like 4,6-α-GTs, with transglucosylase activity on maltodextrins and starch (7, 18, 19).
The origin and evolution of GH70 enzymes have been investigated extensively (7, 11, 20, 40, 42), and a generally accepted permutation-per-duplication-based model illustrated the evolution of the ancient GH13 to GH70. Moreover, in GH70, subgroups with distinct enzyme functions have been established (8, 11, 16, 18, 19, 40). Studies on the relationships of these subgroups have provided an evolutionary perspective on the structure-function relationships of GH70 enzymes (7, 18). However, certain issues regarding these models of evolution remain to be addressed. One of the remaining issues is the lack of intermediates in the conversion of enzyme activity. Based on bioinformatic data, Kralj et al. suggested that GTFB-like and GTFC-like 4,6-α-GTs represent the evolutionary intermediates between GH70 and GH13 enzymes (18). The BrS subgroup was established recently, and an increasing number of BrS members have been found and characterized (8, 11, 16, 17). Evolutionary analysis performed in a recent study suggested that BrSs formed a separate clade from the main cluster of GSs, reflecting their different enzyme specificity (7). Divergence from the BrS clade to the GS clade was accompanied by the conversion of enzyme specificity, i.e., from branching to polymerization (7). However, no intermediates have yet been observed in the conversion of BrS to GS enzymes, and the structural basis that underlies the conversion of enzyme specificity remains to be elucidated.
In this study, we identified Gsy, a GS with the appearance of a BrS enzyme. Sequence-based analysis (Fig. 5) suggested that it might be an intermediate between the BrS and GS clades of GH70 enzymes. Further study on the peculiar enzyme Gsy provided biochemical (Table 1 and Fig. 3 and 4) and bioinformatic (Fig. 6) data to support Gsy as an intermediate in the evolution of BrS to GS enzymes.
Moreover, this intermediate provides clues to the structural basis underlying the conversion of enzyme specificity and the acquisition of de novo polymerizing activity. As shown in Fig. 5A, alignment of the core motifs of Gsy with those of representative GS and BrS members identified several sites (including sites 695, 693, and 549, numbered according to Gsy) at which Gsy showed a pattern distinct from that of either GS or BrS enzymes, suggesting that these sites play important roles in the specificity of these enzymes and that residue changes at these sites might have led to the conversion of BrS enzymes to GS enzymes. Further studies, such as site-directed mutagenesis, will be required for full elucidation of the mechanism that underlies the conversion of the specificity of these enzymes and for better understanding of the structure-function relationships of GH70 GS and BrS enzymes.
MATERIALS AND METHODS
Cloning and recombinant expression of gsy in E. coli.
The sequence of gsy was obtained from GenBank under accession number KU306931. The gsy gene was cloned and transformed into E. coli as described previously (1). In brief, the gsy gene was PCR amplified from genomic DNA of L. mesenteroides BD3749. After purification and double digestion by BamHI and XhoI (Fermentas, USA), the fragment was ligated into the pET-28a(+) vector. The recombinant plasmid pET28a(+)-gsy was then transformed in Rosetta strains of E. coli, as described previously (1, 29). Transformants harboring the chimeric plasmid were selected on LB agar plates containing 20 μg/ml kanamycin. For induction of the recombinant Gsy, IPTG was added into the culture medium to a final concentration of 0.1 mM. For observation of EPSs on LB plates, 20 g/liter sucrose was included in the medium as a substrate.
Preparation and purification of the recombinant enzyme.
Recombinant Gsy was produced by growing the Rosetta strain harboring the recombinant plasmid pET28a(+)-gsy at 37°C in 20 ml of LB medium supplemented with 20 μg/ml kanamycin, with shaking. The overnight culture was then transferred into 500 ml of LB medium (as described above) and incubated until the optical density at 600 nm (OD600) reached 0.4 to 0.6. Subsequently, expression was induced by adding IPTG to a final concentration of 0.1 mM, and the culture was incubated overnight at 16°C, with constant shaking at 150 rpm. Subsequently, cells were harvested by centrifugation (10,000 × g for 10 min), resuspended in 40 ml of lysis buffer (5 mM imidazole, 20 mM Tris-HCl buffer [pH 8.0], 300 mM NaCl, 0.5 mg/ml lysozyme, 1 mM dithiothreitol, and one tablet of protease inhibitor cocktail [EDTA-free; Roche]), and sonicated for 10 min. After centrifugation (15,000 × g for 15 min), the recombinant enzyme was purified at 4°C by His-tag affinity chromatography using Ni-NTA as the column material (Sangon Biotech, Shanghai), according to the manufacturer's instructions. Elution was carried out with 200 mM imidazole buffer solution (200 mM imidazole, 20 mM Tris-HCl buffer [pH 8.0], and 300 mM NaCl), and the imidazole was removed by loading the sample onto a 10-DG column (Econo-Pac; Bio-Rad) that had been preequilibrated in 50 mM sodium acetate buffer (pH 5.6) with 0.45 mM CaCl2, 100 mM NaCl, and 0.1% (vol/vol) Tween 80, as described previously (16, 20). The purified enzyme was quantified with a Nanodrop 2000 spectrophotometer (Thermo Scientific) and detected by SDS-PAGE before further analysis.
SDS-PAGE.
E. coli cells were harvested and protein samples were prepared as described previously (29, 43). The protein concentrations of the crude extracts were determined by using the Bradford method. For each sample, an equivalent amount of total protein was suspended in SDS loading buffer (Sangon Biotech, Shanghai) and incubated at 37°C for 2 h. Each sample was then loaded and separated by 8% SDS-PAGE (120 V for 120 min). After electrophoresis, SDS was removed by washing the gel with distilled water, followed by 10 min of incubation at room temperature (repeated twice). The gel was then stained with Coomassie brilliant blue G250 (Sangon Biotech), according to the manufacturer's instructions.
In situ polymer synthesis.
In situ polymerization was carried out as described for previous studies (24, 43, 44). In brief, after electrophoresis and removal of SDS as described above, the SDS-PAGE gel was washed three times at room temperature with 50 mM sodium acetate buffer (pH 5.6) containing 2 mM CaCl2 and 0.1% (vol/vol) Triton X-100, for recovery of enzyme activity. Subsequently, the gel was incubated for 24 h at room temperature in the same buffer supplemented with 50 g/liter sucrose, and the active bands were detected by the appearance of glucan polymer, as described by Miller and Robyt (25).
Enzyme activity assays.
The activity of the purified Gsy enzyme was detected at 30°C in 50 mM sodium acetate containing 50 g/liter sucrose and 0.34 mM CaCl2 (buffered at pH 5.4), as described previously (24, 29). The enzyme activities were determined by measuring the amount of released fructose from 50 g/liter sucrose. The fructose concentrations were determined using the dinitrosalicylic acid method (8, 45). One unit was defined as the amount of enzyme that catalyzed the production of 1 μmol fructose per minute under the assay conditions. The protein concentration was measured with the bicinchoninic acid (BCA) protein assay kit (Pierce, Thermo Fisher Scientific), according to the manufacturer's instructions. The enzyme activity of the purified protein is described as units per milligram of total protein.
In order to examine the pH optimum of purified Gsy, the buffer described above was used for measurements in the pH range of 3.5 to 7.5, at intervals of 0.5 pH units. Assays were performed at 30°C. To determine the optimal temperature, the buffer (pH 5.4) described above was used and reactions were performed in the temperature range of 20°C to 40°C, at intervals of 5°C. All of the reaction mixtures were incubated for 15 min, and then the concentrations of fructose were determined as described above.
Preparation and purification of exopolysaccharides.
For polymer synthesis, enzymatic reactions were performed at 30°C for 24 h in the presence of 50 g/liter sucrose, 50 mM sodium acetate, and 0.34 mM CaCl2 (pH 5.4). One milliliter of purified enzyme (3.5 U/ml) was added, and the reaction was carried out overnight at 30°C. The insoluble product was harvested as described in previous studies (28, 32). In brief, the insoluble material was collected by centrifugation at 15,000 × g for 30 min at 4°C, washed with three rounds of suspension in 20 ml water and centrifugation, and then washed once in 50% ethanol and once in 95% ethanol before freeze-drying using a FreeZone 12-liter system (Labconco Corp., USA).
Structural analysis of the polysaccharide. (i) Monosaccharide composition.
To determine the monosaccharide composition, 3 mg of the polymer synthesized by recombinant Gsy was hydrolyzed with 4 M trifluoroacetic acid (TFA) at 100°C for 4 h. The excess TFA was removed by adding equal volumes of methanol to the digested sample and blowing the sample dry with a nitrogen purge. The hydrolyzed sample was then dissolved in deionized water, adjusted to its initial volume, and filtered through a 0.22-μm membrane (Millipore, USA). The monosaccharides were identified and quantified with a Dionex ICS-3000 HPAEC system equipped with a Dionex IonPac PA20 column (3 by 150 mm) and a pulsed amperometric detector (Thermo Fisher Scientific). The monosaccharides were eluted with a linear gradient of NaOH solution (6 mM to 300 mM) at a flow rate of 0.5 ml/min, according to the procedure described by Wood et al. (46). Rhamnose, arabinose, galactose, glucose, xylose, mannose, and fructose (Sigma, USA) were used as references. The percentages of the monosaccharides were calculated from the peak areas using a response factor.
(ii) FTIR analysis.
Five milligrams of the polymer was mixed with 150 mg of KBr, under pressure. The FTIR spectrum was obtained using a Nicolet Nexus 470 spectrometer (Nicolet Co., Waltham, MA). The sample was prepared as a KBr pellet and scanned against a blank KBr pellet background over a wavelength range of 4,000 to 500 cm−1, with a resolution of 4.0 cm−1.
(iii) 1H NMR spectroscopic analysis.
Ten milligrams of the lyophilized polymer was dissolved in 0.5 ml of 0.5 M NaOD and then placed into 5-mm NMR tubes. One-dimensional 1H NMR utilizing tetramethoxysilane as an internal standard was performed with a Varian Mercury Plus 400 spectrometer (Varian, USA) at 70°C. The 1H NMR data collection consisted of 256 acquisitions.
Sequence-based analysis of GH70 enzymes. (i) Sequence alignment.
The sequences of conserved motifs I to VII from representative GH70 enzymes, as indicated, were obtained through alignment of the GH70 enzymes with sequences of motifs I to VII identified in previous studies (6, 14, 19, 47). Alignments of the sequences of the conserved motifs were performed with Align X (Vector NTI, Invitrogen).
(ii) Phylogenetic analysis.
The full amino acid sequence of Gsy was used as the query sequence in BLASTp searches in NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Alignment of Gsy with the BLAST hits, other characterized GH70 enzymes (9), and representative GH13 enzymes (7, 19) was performed as described previously (31, 40). Sequence information for the GH70 enzymes was obtained from the CAZy database (http://www.cazy.org/GH70.html), with cross-reference to GenBank (see Table S1 in the supplemental material for details). Phylogenetic analysis of Gsy and the characterized GH70 enzymes was performed as in previous studies (5, 7, 24, 40). The phylogenetic tree was constructed with the maximum likelihood method, based on the JTT matrix-based model, using MEGA6 (48). Each protein was labeled with its GenBank accession number. The value on each branch is the estimated confidence limit for the position of the branches, as determined by bootstrap analysis. The scale bar represents the proportion of differences in amino acid sequences (42). Sequences of the core motifs (motifs I to VII) (Fig. 5B) or full sequences (Fig. 6) of the GH70 enzymes were used for phylogenetic analysis.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to colleagues from the Instrumental Analysis Center of Shanghai Jiao Tong University (Shanghai, China) for their technical assistance in structural analysis of the exopolysaccharides.
This work was supported by the National Science and Technology Pillar Program during the 12th Five-Year Plan Period (grant 2013BAD18B01) and the Engineering Center Capacity Improvement Project from the Science and Technology Commission of Shanghai Municipality (grant 16DZ2280600).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02810-17.
REFERENCES
- 1.Puanglek S, Kimura S, Enomoto-Rogers Y, Kabe T, Yoshida M, Wada M, Iwata T. 2016. In vitro synthesis of linear α-1,3-glucan and chemical modification to ester derivatives exhibiting outstanding thermal properties. Sci Rep 6:30479. doi: 10.1038/srep30479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freitas F, Alves VD, Reis MA. 2011. Advances in bacterial exopolysaccharides: from production to biotechnological applications. Trends Biotechnol 29:388–398. doi: 10.1016/j.tibtech.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Monsan P, Bozonnet S, Albenne C, Joucla G, Willemot RM, Remaud-Siméon M. 2001. Homopolysaccharides from lactic acid bacteria. Int Dairy J 11:675–685. doi: 10.1016/S0958-6946(01)00113-3. [DOI] [Google Scholar]
- 4.Janeček Š, Svensson B, MacGregor EA. 2014. α-Amylase: an enzyme specificity found in various families of glycoside hydrolases. Cell Mol Life Sci 71:1149–1170. doi: 10.1007/s00018-013-1388-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moulis C, André I, Remaud-Siméon M. 2016. GH13 amylosucrases and GH70 branching sucrases, atypical enzymes in their respective families. Cell Mol Life Sci 73:2661–2679. doi: 10.1007/s00018-016-2244-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janeček Š, Gabriško M. 2016. Remarkable evolutionary relatedness among the enzymes and proteins from the α-amylase family. Cell Mol Life Sci 73:2707–2725. doi: 10.1007/s00018-016-2246-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meng X, Gangoiti J, Bai Y, Pijning T, van Leeuwen SS, Dijkhuizen L. 2016. Structure-function relationships of family GH70 glucansucrase and 4,6-α-glucanotransferase enzymes, and their evolutionary relationships with family GH13 enzymes. Cell Mol Life Sci 73:2681–2706. doi: 10.1007/s00018-016-2245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Passerini D, Vuillemin M, Ufarté L, Morel S, Loux V, Fontagné-Faucher C, Monsan P, Remaud-Siméon M, Moulis C. 2015. Inventory of the GH70 enzymes encoded by Leuconostoc citreum NRRL B-1299: identification of three novel α-transglucosylases. FEBS J 282:2115–2130. doi: 10.1111/febs.13261. [DOI] [PubMed] [Google Scholar]
- 9.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vujicic-Zagar A, Pijning T, Kralj S, López CA, Eeuwema W, Dijkhuizen L, Dijkstra BW. 2010. Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes. Proc Natl Acad Sci U S A 107:21406–21411. doi: 10.1073/pnas.1007531107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brison Y, Pijning T, Malbert Y, Mourey L, Morel S, Monsan P, Tranier S, Dijkstra BW. 2012. Functional and structural characterization of α-(1→2) branching sucrase derived from DSR-E glucansucrase. J Biol Chem 287:7915–7924. doi: 10.1074/jbc.M111.305078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leemhuis H, Pijning T, Dobruchowska JM, van Leeuwen SS, Kralj S, Dijkstra BW, Dijkhuizen L. 2013. Glucansucrases: three-dimensional structures, reactions, mechanism, α-glucan analysis and their implications in biotechnology and food applications. J Biotechnol 163:250–272. doi: 10.1016/j.jbiotec.2012.06.037. [DOI] [PubMed] [Google Scholar]
- 13.van Hijum SA, Kralj S, Ozimek LK, Dijkhuizen L, van Geel-Schutten IG. 2006. Structure-function relationships of glucansucrase and fructansucrase enzymes from lactic acid bacteria. Microbiol Mol Biol Rev 70:157–176. doi: 10.1128/MMBR.70.1.157-176.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bozonnet S, Dols-Laffargue M, Fabre E, Pizzut S, Remaud-Siméon M, Monsan P, Willemot RM. 2002. Molecular characterization of DSR-E, an α-1,2 linkage-synthesizing dextransucrase with two catalytic domains. J Bacteriol 184:5753–5716. doi: 10.1128/JB.184.20.5753-5761.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabre E, Bozonnet S, Arcache A, Willemot RM, Vignon M, Monsan P, Remaud-Siméon M. 2005. Role of the two catalytic domains of DSR-E dextransucrase and their involvement in the formation of highly α-1,2 branched dextran. J Bacteriol 187:296–303. doi: 10.1128/JB.187.1.296-303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vuillemin M, Claverie M, Brison Y, Séverac E, Bondy P, Morel S, Monsan P, Moulis C, Remaud-Siméon M. 2016. Characterization of the first α-(1→3) branching sucrases of the GH70 family. J Biol Chem 291:7687–7702. doi: 10.1074/jbc.M115.688044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brison Y, Malbert Y, Czaplicki G, Mourey L, Remaud-Siméon M, Tranier S. 2016. Structural insights into the carbohydrate-binding ability of an α-(1→2) branching sucrase from glycoside-hydrolase family 70. J Biol Chem 291:7527–7540. doi: 10.1074/jbc.M115.688796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kralj S, Grijpstra P, van Leeuwen SS, Leemhuis H, Dobruchowska JM, van der Kaaij RM, Malik A, Oetari A, Kamerling JP, Dijkhuizen L. 2011. 4,6-α-Glucanotransferase, a novel enzyme that structurally and functionally provides an evolutionary link between glycoside hydrolase enzyme families 13 and 70. Appl Environ Microbiol 77:8154–8163. doi: 10.1128/AEM.05735-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gangoiti J, Pijning T, Dijkhuizen L. 2015. The Exiguobacterium sibiricum 255-15 GtfC enzyme represents a novel glycoside hydrolase 70 subfamily of 4,6-α-glucanotransferase enzymes. Appl Environ Microbiol 82:756–766. doi: 10.1128/AEM.03420-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gangoiti J, Lamothe L, van Leeuwen SS, Vafiadi C, Dijkhuizen L. 2017. Characterization of the Paenibacillus beijingensis DSM 24997 GtfD and its glucan polymer products representing a new glycoside hydrolase 70 subfamily of 4,6-α-glucanotransferase enzymes. PLoS One 12:e0172622. doi: 10.1371/journal.pone.0172622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peisajovich SG, Rockah L, Dan ST. 2006. Evolution of new protein topologies through multistep gene rearrangements. Nat Genet 38:168–174. doi: 10.1038/ng1717. [DOI] [PubMed] [Google Scholar]
- 22.MacGregor EA, Jespersen HM, Svensson B. 1996. A circularly permuted α-amylase-type α/β-barrel structure in glucan-synthesizing glucosyltransferases. FEBS Lett 378:263–266. doi: 10.1016/0014-5793(95)01428-4. [DOI] [PubMed] [Google Scholar]
- 23.Gangoiti J, van Leeuwen SS, Vafiadi C, Dijkhuizen L. 2016. The Gram-negative bacterium Azotobacter chroococcum NCIMB 8003 employs a new glycoside hydrolase family 70 4,6-α-glucanotransferase enzyme (GtfD) to synthesize a reuteran like polymer from maltodextrins and starch. Biochim Biophys Acta 1860:1224–1236. doi: 10.1016/j.bbagen.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Yan M, Han J, Xu X, Liu L, Gao C, Zheng H, Chen Y, Tao Y, Zhou H, Li Y. 2016. Gsy, a novel glucansucrase from Leuconostoc mesenteroides, mediates the formation of cell aggregates in response to oxidative stress. Sci Rep 6:38122. doi: 10.1038/srep38122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller AW, Robyt JF. 1986. Detection of dextransucrase and levansucrase on polyacrylamide gels by the periodic acid-Schiff stain: staining artifacts and their prevention. Anal Biochem 156:357–363. doi: 10.1016/0003-2697(86)90266-6. [DOI] [PubMed] [Google Scholar]
- 26.Dols M, Remaud-Siméon M, Willemot RM, Vignon M, Monsan P. 1998. Characterization of the different dextransucrase activities excreted in glucose, fructose, or sucrose medium by Leuconostoc mesenteroides NRRL B-1299. Appl Environ Microbiol 64:1298–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang HK, Kim YM, Kim DM. 2008. Functional, genetic, and bioinformatic characterization of dextransucrase (DSRBCB4) gene in Leuconostoc mesenteroides B-1299CB4. J Microbiol Biotechnol 18:1050–1058. [PubMed] [Google Scholar]
- 28.Côté GL, Skory CD. 2012. Cloning, expression, and characterization of an insoluble glucan-producing glucansucrase from Leuconostoc mesenteroides NRRL B-1118. Appl Microbiol Biotechnol 93:2387–2394. doi: 10.1007/s00253-011-3562-2. [DOI] [PubMed] [Google Scholar]
- 29.Rühmkorf C, Bork C, Mischnick P, Rübsam H, Becker T, Vogel RF. 2013. Identification of Lactobacillus curvatus TMW 1.624 dextransucrase and comparative characterization with Lactobacillus reuteri TMW 1.106 and Lactobacillus animalis TMW 1.971 dextransucrases. Food Microbiol 34:52–61. doi: 10.1016/j.fm.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 30.Monchois V, Willemot R, Monsan P. 1999. Glucansucrases: mechanism of action and structure–function relationships. FEMS Microbiol Rev 23:131–151. doi: 10.1111/j.1574-6976.1999.tb00394.x. [DOI] [PubMed] [Google Scholar]
- 31.Shingel KI. 2002. Determination of structural peculiarities of dexran, pullulan and γ-irradiated pullulan by Fourier-transform IR spectroscopy. Carbohydr Res 337:1445–1451. doi: 10.1016/S0008-6215(02)00209-4. [DOI] [PubMed] [Google Scholar]
- 32.Vettori MHPB, Franchetti SMM, Contiero J. 2012. Structural characterization of a new dextran with a low degree of branching produced by Leuconostoc mesenteroides FT045B dextransucrase. Carbohydr Polym 88:1440–1444. doi: 10.1016/j.carbpol.2012.02.048. [DOI] [Google Scholar]
- 33.Capek P, Hlavoňová E, Matulová M, Mislovicová D, Rùžička J, Koutný M, Keprdová L. 2011. Isolation and characterization of an extracellular glucan produced by Leuconostoc garlicum PR. Carbohydr Polym 83:88–93. doi: 10.1016/j.carbpol.2010.07.024. [DOI] [Google Scholar]
- 34.Liu C, Lin Q, Gao Y, Ye L, Xing Y, Xi T. 2007. Characterization and antitumor activity of a polysaccharide from Strongylocentrotus nudus eggs. Carbohydr Polym 67:313–318. doi: 10.1016/j.carbpol.2006.05.024. [DOI] [Google Scholar]
- 35.Parker FS. 1971. Application of infrared spectroscopy in biochemistry, biology and medicine. Springer, New York, NY. [Google Scholar]
- 36.Dowd MK, Zeng J, French AD, Reilly PJ. 1992. Conformational analysis of the anomeric forms of kojibiose, nigerose, and maltose using MM3. Carbohydr Res 230:223–244. doi: 10.1016/0008-6215(92)84035-Q. [DOI] [PubMed] [Google Scholar]
- 37.Olafsdottir ES, Ingolfsdottir K, Barsett H, Paulsen BS, Jurcic K, Wagner H. 1999. Immunologically active (1→3)-(1→4)-α-D-glucan from Cetraria islandica. Phytomedicine 6:33–39. doi: 10.1016/S0944-7113(99)80032-4. [DOI] [PubMed] [Google Scholar]
- 38.Kralj S, van Geel-Schutten IG, Rahaoui H, Leer RJ, Faber EJ, van der Maarel MJ, Dijkhuizen L. 2002. Molecular characterization of a novel glucosyltransferase from Lactobacillus reuteri strain 121 synthesizing a unique, highly branched glucan with α-(1→4) and α-(1→6) glucosidic bonds. Appl Environ Microbiol 68:4283–4291. doi: 10.1128/AEM.68.9.4283-4291.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Leeuwen SS, Kralj S, Gerwig GJ, Dijkhuizen L, Kamerling JP. 2008. Structural analysis of bioengineered α-d-glucan produced by a triple mutant of the glucansucrase GTF180 enzyme from Lactobacillus reuteri strain 180: generation of (α1→4) linkages in a native (1→3)(1–>6)-α-d-glucan. Biomacromolecules 9:2251–2258. doi: 10.1021/bm800410w. [DOI] [PubMed] [Google Scholar]
- 40.Gangoiti J, van Leeuwen SS, Gerwig GJ, Duboux S, Vafiadi C, Pijning T, Dijkhuizen L. 2017. 4,3-α-Glucanotransferase, a novel reaction specificity in glycoside hydrolase family 70 and clan GH-H. Sci Rep 7:39761. doi: 10.1038/srep39761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gangoiti J, Pijning T, Dijkhuizen L. 2018. Biotechnological potential of novel glycoside hydrolase family 70 enzymes synthesizing α-glucans from starch and sucrose. Biotechnol Adv 36:196–207. doi: 10.1016/j.biotechadv.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Hoshino T, Fujiwara T, Kawabata S. 2012. Evolution of cariogenic character in Streptococcus mutans: horizontal transmission of glycosyl hydrolase family 70 genes. Sci Rep 2:518. doi: 10.1038/srep00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanada N, Kuramitsu HK. 1988. Isolation and characterization of the Streptococcus mutans gtfC gene, coding for synthesis of both soluble and insoluble glucans. Infect Immun 56:1999–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monchois V, Remaud-Siméon M, Russell RR, Monsan P, Willemot RM. 1997. Characterization of Leuconostoc mesenteroides NRRL B-512F dextransucrase (DSRS) and identification of amino-acid residues playing a key role in enzyme activity. Appl Microbiol Biotechnol 48:465–472. doi: 10.1007/s002530051081. [DOI] [PubMed] [Google Scholar]
- 45.Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 46.Wood PJ, Weisz J, Blackwell BA. 1994. Structural studies of (1→3),(1→4)-β-d-glucans by 13C-nuclear magnetic resonance spectroscopy and by rapid analysis of cellulose-like regions using high-performance anion-exchange chromatography of oligosaccharides released by lichenase. Cereal Chem 71:301–307. [Google Scholar]
- 47.Janeček Š. 2002. How many conserved sequence regions are there in the α-amylase family? Biologia 57:29–41. [Google Scholar]
- 48.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cote GL, Ahlgren JA, Smith MR. 1999. Some structural features of an insoluble α-D-glucan from a mutant strain of Leuconostoc mesenteroides NRRL B-1355. J Industr Microbiol Biotechnol 23:656–660. doi: 10.1038/sj.jim.2900678. [DOI] [PubMed] [Google Scholar]
- 50.Kobayashi M, Shishido K, Kikuchi T, Matsuda K. 1973. Fractionation of the Leuconostoc mesenteroides NRRL B-1299 dextran and preliminary characterization of the fractions. Agric Biol Chem 37:357–365. doi: 10.1080/00021369.1973.10860671. [DOI] [Google Scholar]
- 51.Seymour FR, Slodki ME, Plattner RD, Jeanes A. 1977. Six unusual dextrans: methylation structural analysis by combined g.l.c.-m.s. of per-O-acetyl-aldononitriles. Carbohydr Res 53:153–166. doi: 10.1016/S0008-6215(00)88083-0. [DOI] [Google Scholar]
- 52.Jeans A, Seymour FR. 1979. The α-d-glucopyranosidic linkages of dextrans: comparison of percentages from structural analysis by periodate oxidation and by methylation. Carbohydr Res 74:31–40. doi: 10.1016/S0008-6215(00)84763-1. [DOI] [Google Scholar]
- 53.Shukla R, Shukla S, Bivolarski V, Iliev I, Ivanova I, Goyal A, Soccol CR, Pandey A, Larroche C, Thomazsoccol V. 2011. Structural characterization of insoluble dextran produced by Leuconostoc mesenteroides NRRL B-1149 in the presence of maltose. Food Technol Biotechnol 49:291–296. [Google Scholar]
- 54.Côté GL, Robyt JF. 1983. The formation of α-d-(1→3) branch linkages by an exocellular glucansucrase from Leuconostoc mesenteroides NRRL B-742. Carbohydr Res 119:141–156. doi: 10.1016/0008-6215(83)84053-1. [DOI] [Google Scholar]
- 55.Bounaix M-S, Gabriel V, Robert H, Morel S, Remaud-Siméon M, Gabriel B, Fontagné-Faucher C. 2010. Characterization of glucan-producing Leuconostoc strains isolated from sourdough. Int J Food Microbiol 144:1–9. doi: 10.1016/j.ijfoodmicro.2010.05.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.