ABSTRACT
To assess phenotypic bacterial antimicrobial resistance (AMR) in different strata (e.g., host populations, environmental areas, manure, or sewage effluents) for epidemiological purposes, isolates of target bacteria can be obtained from a stratum using various sample types. Also, different sample processing methods can be applied. The MIC of each target antimicrobial drug for each isolate is measured. Statistical equivalence testing of the MIC data for the isolates allows evaluation of whether different sample types or sample processing methods yield equivalent estimates of the bacterial antimicrobial susceptibility in the stratum. We demonstrate this approach on the antimicrobial susceptibility estimates for (i) nontyphoidal Salmonella spp. from ground or trimmed meat versus cecal content samples of cattle in processing plants in 2013-2014 and (ii) nontyphoidal Salmonella spp. from urine, fecal, and blood human samples in 2015 (U.S. National Antimicrobial Resistance Monitoring System data). We found that the sample types for cattle yielded nonequivalent susceptibility estimates for several antimicrobial drug classes and thus may gauge distinct subpopulations of salmonellae. The quinolone and fluoroquinolone susceptibility estimates for nontyphoidal salmonellae from human blood are nonequivalent to those from urine or feces, conjecturally due to the fluoroquinolone (ciprofloxacin) use to treat infections caused by nontyphoidal salmonellae. We also demonstrate statistical equivalence testing for comparing sample processing methods for fecal samples (culturing one versus multiple aliquots per sample) to assess AMR in fecal Escherichia coli. These methods yield equivalent results, except for tetracyclines. Importantly, statistical equivalence testing provides the MIC difference at which the data from two sample types or sample processing methods differ statistically. Data users (e.g., microbiologists and epidemiologists) may then interpret practical relevance of the difference.
IMPORTANCE Bacterial antimicrobial resistance (AMR) needs to be assessed in different populations or strata for the purposes of surveillance and determination of the efficacy of interventions to halt AMR dissemination. To assess phenotypic antimicrobial susceptibility, isolates of target bacteria can be obtained from a stratum using different sample types or employing different sample processing methods in the laboratory. The MIC of each target antimicrobial drug for each of the isolates is measured, yielding the MIC distribution across the isolates from each sample type or sample processing method. We describe statistical equivalence testing for the MIC data for evaluating whether two sample types or sample processing methods yield equivalent estimates of the bacterial phenotypic antimicrobial susceptibility in the stratum. This includes estimating the MIC difference at which the data from the two approaches differ statistically. Data users (e.g., microbiologists, epidemiologists, and public health professionals) can then interpret whether that present difference is practically relevant.
KEYWORDS: Escherichia coli, NARMS, Salmonella, assessment of antimicrobial resistance, bacterial antimicrobial resistance, cattle, fecal sampling, sample processing, sample types, statistical equivalence
INTRODUCTION
Informative assessment of bacterial antimicrobial resistance (AMR) within and among strata is the basic block in any investigation of AMR epidemiology or control approaches (1, 2). Such assessments are critical for identifying influential factors and mitigation strategies for AMR (1, 2). Examples of strata are animal or human populations, food products, environmental areas, manure effluents from food animal farms, and human sewage effluents. To assess AMR of a target bacterial species in a stratum, isolates of the bacteria are obtained from the sampling units (e.g., animal hosts or environmental area segments) in the stratum. Each isolate's phenotypic susceptibility to each target antimicrobial drug is measured as the drug's MIC inhibiting visible overnight growth of the isolate culture (3). The data for all the obtained isolates provide the distribution of the tested antimicrobial's MIC as an estimate for the target bacteria in the stratum. Descriptive statistics (e.g., elemental features of the data such as means, percentiles, or ranges) have been used extensively for the MIC distributions due to the ease of interpretation (the statistics used had been reviewed in detail by Wagner et al. [4]). Such distributions, however, could be subjected to statistical analyses to identify patterns and dynamics of the bacterial antimicrobial susceptibility in the stratum, compare sampling approaches or microbiological sample processing methods for the susceptibility assessments, or contrast the susceptibilities between strata. Analyzing the MIC distributions bears numerous challenges because the distributions tend to have complex shapes (e.g., do not follow the probability distributions commonly assumed for parametric statistical tests) and are inherently censored (i.e., all the isolates with MIC less than or equal to the smallest drug concentration tested are in one category in the beginning of the distribution, and all the isolates with MIC greater than the largest drug concentration tested are in one category in the end of the distribution) (4–7). Thus far, the analytical approaches have included comparing the histograms of relative frequency of the isolates with the specific MICs of the antimicrobial (7) and the cumulative frequency of the isolates over the increasing MICs of the antimicrobial (8, 9) in a stratum over time and between strata. The cumulative frequency distributions have been also used for comparing the antimicrobial susceptibility estimates between the isolate sets from different sampling approaches in a stratum (10). Survival analysis has been adapted to compare the probabilities of isolates with the specific MICs of the antimicrobial (the time to event is replaced by the concentration achieving bacterial growth inhibition, MIC) in a stratum over time and between strata defined by experimental factors (6, 11). Linear regression on the log2(MIC) has been used to compare the susceptibility to the antimicrobial in a stratum over time and between strata in the probabilistic framework (12) and to compare the MIC measurements obtained for the same strain set by different microbiological laboratories in the Bayesian framework (5). It has been suggested that a power analysis should be included for the statistical tests of tendencies in the MIC/log2(MIC) distributions to support interpretation of the results (12).
Different sampling approaches can be used to assess AMR in a target bacterial species in a stratum. For example, different sample types can be collected, from which the bacteria are then isolated. In other situations, once the samples have been collected, those can be subjected to different sample processing methods for bacterial isolation. For example, an aliquot of the sample can be plated on a bacteriological agar and a different number of the bacterial colonies tested for susceptibility to antimicrobials, or multiple aliquots of the sample can be plated and the bacterial colonies from each aliquot tested. The same analytical need arises in both of these scenarios: sampling a stratum by different sample types and applying different sample processing methods to the samples of one type. The need is to determine whether the sampling or the sample processing approaches yield similar estimates of phenotypic antimicrobial susceptibility in the target bacteria in the stratum. This question can be formulated as to whether the approaches yield equivalent estimates of the antimicrobial's MIC distribution for the bacteria in the stratum. This can be addressed by statistical equivalence testing (13, 14). This technique also provides a flexibility for the data users to interpret whether the existing differences between the bacterial susceptibility estimates for the stratum between the sampling or sample processing approaches are practically relevant (as shown below). The objective of this study was to demonstrate the utility of the statistical equivalence testing as a method to compare the bacterial antimicrobial susceptibility estimates for a stratum between sampling approaches (e.g., different sample types or sampling schemes) or sample processing methods.
RESULTS
Interpretation of statistical equivalence testing for MIC data from different sampling or sample processing approaches.
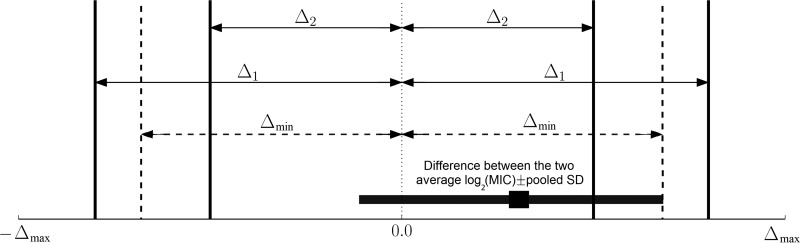
The most commonly used measurement of susceptibility of a bacterial isolate to an antimicrobial is the drug's MIC. When the MIC is measured using the broth microdilution assay based on serial 2-fold dilutions of the drug, the measurement is transformed to log2(MIC) for statistical analyses (12, 15). The measurements for all the target bacterial species' isolates obtained via a given sampling or sample processing approach from the target stratum yield the antimicrobial's MIC distribution for the species in the stratum. Such distributions from two sampling or sample processing approaches can be compared, and the minimum difference between the average log2(MIC) estimates from the two approaches at which the estimates are still statistically equivalent can be determined. We designate that difference Δmin. This threshold difference value can be found by performing the statistical equivalence testing on the log2(MIC) data from the two approaches starting from a large value of the difference Δ ≫ 1 ≥ Δmin and then reducing it until finding Δ→Δmin below which the hypothesis of a statistically significant difference between the average log2(MIC) cannot be rejected. This leads to the estimate of Δmin obtained from the confidence interval (CI) of the difference between the average log2(MIC) estimates from the two sampling or sample processing approaches (see Materials and Methods for details). The estimate of Δmin can be interpreted by data users as illustrated in Fig. 1. If a practically relevant difference Δ1 is outside Δmin, i.e., Δ1 ≥ Δmin, the two approaches yield statistically equivalent data. In contrast, if a practically relevant difference Δ2 is inside Δmin, i.e., Δ2 < Δmin, the two approaches yield statistically nonequivalent data. Thus, data users could apply their perspectives of which difference between the average log2(MIC) estimates from the two sampling or sample processing approaches is practically relevant and compare that to the existing statistically significant difference, Δmin. Summarizing the results as illustrated in Fig. 2 to 4 enables evaluating the differences in the average log2(MIC) estimates between the two sampling or sample processing approaches for individual antimicrobials tested (within and between the drug classes).
FIG 1.
Schematic representation of testing statistical equivalence of the bacterial antimicrobial susceptibility estimates from two sampling or sample processing approaches used in a stratum. If a practically relevant difference between the average log2(MIC) estimates from the two approaches is equal to or larger than Δmin (e.g., Δ1), the hypothesis of statistical nonequivalence of the estimates will be rejected, signaling equivalence of the MIC data from the two approaches. If a practically relevant difference is smaller than Δmin (e.g., Δ2), the hypothesis of statistical nonequivalence of the estimates will be accepted. The maximum possible difference between the log2(MIC) values from the two approaches is Δmax for the isolate sets Y1 and Y2).
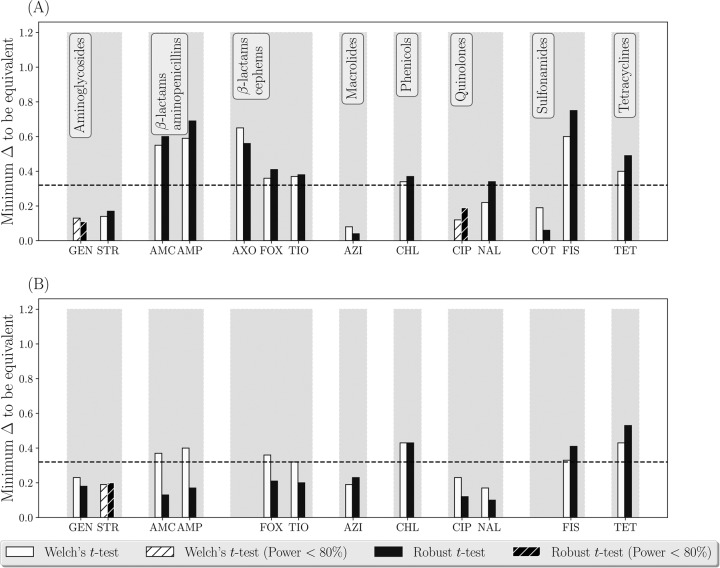
FIG 2.
Testing statistical equivalence of the estimates of phenotypic antimicrobial susceptibility of nontyphoidal Salmonella enterica subsp. enterica isolates from the ground- or trimmed-meat samples (n = 310 for 2013 and n = 344 for 2014) versus cecal content samples (n = 435 for 2013 and n = 318 for 2014) from cattle in the processing plants in the United States in 2013 (A) and 2014 (B). The data were collected by the NARMS. Aminoglycosides: GEN, gentamicin; STR, streptomycin. β-Lactam aminopenicillins: AMC, amoxicillin-clavulanic acid; AMP, ampicillin. β-Lactam cephems: AXO, ceftriaxone; FOX, cefoxitin; TIO, ceftiofur. Macrolides: AZI, azithromycin. Phenicols: CHL, chloramphenicol. Quinolones: CIP, ciprofloxacin; NAL, nalidixic acid. Sulfonamides: COT, trimethoprim sulfamethoxazole; FIS, sulfisoxazole. Tetracyclines: TET, tetracycline.
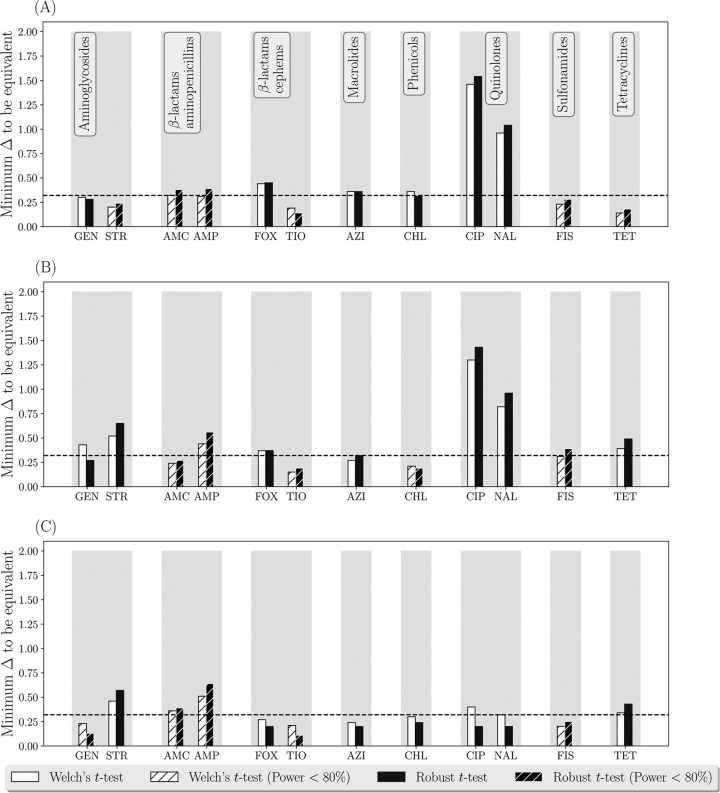
FIG 3.
Testing statistical equivalence of the estimates of phenotypic antimicrobial susceptibility of nontyphoidal Salmonella enterica subsp. enterica isolates from urine (n = 144), fecal (n = 1,495), and blood (n = 181) samples of humans in the United States in 2015. (A) Urine versus blood isolates; (B) fecal versus blood isolates; (C) urine versus fecal isolates. The data were collected by the NARMS. Aminoglycosides: GEN, gentamicin; STR, streptomycin. β-Lactam aminopenicillins: AMC, amoxicillin-clavulanic acid; AMP, ampicillin. β-Lactam cephems: FOX, cefoxitin; TIO, ceftiofur. Macrolides: AZI, azithromycin. Phenicols: CHL, chloramphenicol. Quinolones: CIP, ciprofloxacin; NAL, nalidixic acid. Sulfonamides: FIS, sulfisoxazole. Tetracycline: TET, tetracycline.
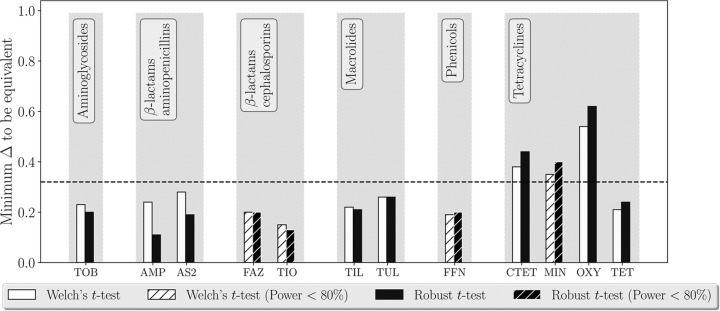
FIG 4.
Testing statistical equivalence of the estimates of phenotypic antimicrobial susceptibility of E. coli in cattle fecal pads (n = 32). The sample processing approaches compared were testing susceptibility of 4 E. coli isolates obtained from one aliquot of the pad versus testing susceptibility of 4 E. coli isolates each obtained from a different aliquot of the pad, with the aliquots collected from locations spread along the longest axis of the pad. Aminoglycosides: TOB, tobramycin. β-Lactam aminopenicillins: AMP, ampicillin; AS2, ampicillin-sulbactam (2:1 ratio). β-Lactam cephems: FAZ, cefazolin; TIO, ceftiofur. Macrolides: TIL, tilmicosin; TUL, tulathromycin. Phenicols: FFN, florfenicol. Tetracyclines: CTET, chlortetracycline; MIN, minocycline; OXY, oxytetracycline; TET, tetracycline.
Here we provide a suggestion on how Δmin could be interpreted systematically. When bioequivalence of two drug preparations is investigated based on a biological drug response variable for which logarithmic transformations are appropriate, the preparations are considered equivalent if the difference Δ in the variable values is such that 2Δ is ≤1.25 (16). This corresponds to ≤0.32 on the log2(MIC) scale. If none of the two sampling or sample processing approaches compared is a reference for the bacterial antimicrobial susceptibility assessment, data users can consider the absolute value of the difference between the average log2(MIC) estimates. They could interpret that the two approaches yield nonequivalent estimates of the average log2(MIC) if Δmin is >0.32. Such values of Δmin suggest that the estimates differ beyond the biological variation expected if the two approaches were gathering the isolates from the same subpopulation of the target bacteria in the sampled stratum. Note that the statistical determination of Δmin accounts for variability in the data from the two approaches (see Materials and Methods for details).
Case study 1: ground- or trimmed-meat versus cecal content samples from cattle in processing plants for assessing antimicrobial susceptibility of nontyphoidal Salmonella enterica subsp. enterica in cattle.
Monitoring of AMR in the U.S. food chain is conducted by the National Antimicrobial Resistance Monitoring System (NARMS) (17). In cattle processing plants, both samples of ground or trimmed meat and of cecal contents of cattle carcasses were collected in 2013-2014 (17, 18). Nontyphoidal Salmonella enterica subsp. enterica isolates of diverse serovars were obtained from both these sample types (17, 18). Phenotypic susceptibility of the isolates to antimicrobials representing major antimicrobial drug classes was tested (Table 1) (17, 18) (the data can be found here: https://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/ucm416741.htm). As the first case study, we investigated statistical equivalence of the average log2(MIC) estimates of each tested antimicrobial (Table 1) for S. enterica yielded by the ground- or trimmed-meat samples (n = 310 for 2013 and n = 344 for 2014) versus cecal content samples (n = 435 for 2013 and n = 318 for 2014). In both 2013 and 2014, the equivalence testing was used to determine for each antimicrobial Δmin, i.e., the difference between the average log2(MIC) from the two sampling approaches at which the approaches still yielded statistically equivalent data. The distributions of the log2(MIC) for all the tested antimicrobials in each 2013 and 2014 did not follow a normal distribution (Wilk-Shapiro test, P value < 0.05 for each of the two distributions from the sampling approaches). Because of this, the robust t test was used for the equivalence testing (see Materials and Methods for details). The values of Δmin tended to be larger for phenicols, sulfonamides, and tetracyclines in both 2013 and 2014 (Fig. 2A and B). Relatively large Δmin values were also estimated for β-lactams, both aminopenicillins and cephems, in 2013 (Fig. 2A). The statistical nonequivalence of the log2(MIC) data for these drug classes highlighted that the cecal content sampling gauged S. enterica subpopulations that differ in their phenotypic AMR from S. enterica subpopulations gauged via the ground- or trimmed-meat sampling in the cattle processing plants. This interpretation was based on considering the data for each antimicrobial from the two sampling approaches nonequivalent if Δmin was >0.32.
TABLE 1.
Antimicrobial drug susceptibilities tested for nontyphoidal Salmonella enterica subsp. enterica isolates from ground- or trimmed-meat samples and from cecal content samples collected from cattle in processing plants in the United States in 2013 and 2014a
| Antimicrobial drug class | Subclass | Combinatory formulation | Drug |
|---|---|---|---|
| Aminoglycosides | -Micins | No | Gentamicin |
| -Mycins | No | Kanamycin (tested in 2013 only) | |
| No | Streptomycin | ||
| β-Lactams | Aminopenicillins with β-lactamase inhibitors | Yes | Amoxicillin with clavulanic acid |
| Aminopenicillins | No | Ampicillin | |
| Cephems | No | Ceftriaxone | |
| No | Cefoxitine | ||
| No | Ceftiofur | ||
| Macrolides | Azalides | No | Azithromycin |
| Phenicols | NA | No | Chloramphenicol |
| Quinolones | Fluoroquinolones | No | Ciprofloxacin |
| Quinolones | No | Nalidixic acid | |
| Sulfonamides | NA | Yes | Sulfamethoxazole with trimethoprim |
| No | Sulfisoxazole | ||
| Tetracyclines | Tetracyclines | No | Tetracycline |
The data were collected by the U.S. National Antimicrobial Resistance Monitoring System. Note that due to a low variability in the data for kanamycin in 2013 and ceftriaxone in 2014, the equivalence testing could not be performed. NA, not applicable.
Case study 2: antimicrobial susceptibility of nontyphoidal S. enterica subsp. enterica from urine versus fecal versus blood samples of humans.
Monitoring of AMR in enteric pathogens of humans in the United States is also a part of the NARMS activities (17). Nontyphoidal Salmonella enterica subsp. enterica isolates of diverse serovars were obtained from urine, fecal, and blood samples of humans in the United States in 2015 (the analyzed data set is limited to those states that have permitted the U.S. Centers for Disease Control and Prevention to share the data with the public; the data set can be found at https://wwwn.cdc.gov/narmsnow/). Phenotypic susceptibility of the isolates to antimicrobials representing major antimicrobial drug classes was tested (Table 2). As the second case study, we applied the statistical equivalence testing to compare the estimates of antimicrobial susceptibility of nontyphoidal S. enterica isolates from urine (n = 144), fecal (n = 1,495), and blood (n = 181) samples of humans in the United States in 2015. The distributions of the log2(MIC) for all the tested antimicrobials for the S. enterica isolates from each urine, fecal, or blood samples did not follow a normal distribution (Wilk-Shapiro test, P value < 0.05 for each distribution). Thus, the robust t test was used for the equivalence testing. The results demonstrated that susceptibilities of the S. enterica isolates from human urine versus from blood to cephems (which are β-lactams), macrolides, phenicols, and quinolones were statistically nonequivalent (Fig. 3A). Further, susceptibilities of the S. enterica isolates from human feces versus from blood to aminoglycosides, cephems, quinolones, and tetracyclines were nonequivalent (Fig. 3B). Susceptibilities of the S. enterica isolates from human urine versus from feces differed to a lesser extent but still were nonequivalent for aminoglycosides, quinolones, and tetracyclines (Fig. 3C). These interpretations for each antimicrobial in a pairwise comparison of the isolate sources were based on considering the data nonequivalent if Δmin was >0.32. The largest differences were found for the fluoroquinolone ciprofloxacin and the older quinolone nalidixic acid (Fig. 3A and B), to which the isolates from human blood had lower susceptibilities (higher ciprofloxacin and nalidixic acid MICs) than the isolates from either urine or feces. The difference between the average log2(MIC) of ciprofloxacin for the isolates from blood versus urine was 1.08 (95% CI: 0.74, 1.42), and for the isolates from blood versus feces it was 1.20 (95% CI: 0.86, 1.54). The difference between the average log2(MIC) of nalidixic acid for the isolates from blood versus urine was 0.71 (95% CI: 0.46, 0.95), and for the isolates from blood versus feces it was 0.80 (95% CI: 0.57, 0.96).
TABLE 2.
Antimicrobial drug susceptibilities tested for nontyphoidal Salmonella enterica subsp. enterica isolates from urine, fecal, and blood samples of humans in the United States in 2015a
| Antimicrobial drug class | Subclass | Combinatory formulation | Drug |
|---|---|---|---|
| Drugs for which variability in the log2(MIC) data from urine, fecal, and blood samples was sufficient to perform the statistical equivalence testing | |||
| Aminoglycosides | -Micins | No | Gentamicin |
| -Mycins | No | Streptomycin | |
| β-Lactams | Penicillins, including amino-, carboxy-, and ureido-, with β-lactamase inhibitors | Yes | Amoxicillin with clavulanic acid |
| Penicillins, including amino-, carboxy-, and ureido- | No | Ampicillin | |
| Cephems | No | Cefoxitin | |
| No | Ceftiofur | ||
| Macrolides | Azalides | No | Azithromycin |
| Phenicols | NA | No | Chloramphenicol |
| Quinolones | Fluoroquinolones | No | Ciprofloxacin |
| Quinolones | No | Nalidixic acid | |
| Sulfonamides | NA | No | Sulfisoxazole |
| Tetracyclines | Tetracyclines | No | Tetracycline |
| Drugs for which variability in the log2(MIC) data from urine, fecal, and blood samples was insufficient to perform the statistical equivalence testing | |||
| β-Lactams | Cephems | No | Ceftriaxone |
| Sulfonamides | NA | Yes | Sulfamethoxazole with trimethoprim |
| Drugs for which statistical equivalence testing could not be performed because of a low no. of each urine and blood samples (n < 10) tested with the antimicrobials | |||
| β-Lactams | Penicillins, including amino-, carboxy-, and ureido-, with β-lactamase inhibitors | Yes | Piperacillin with tazobactam constant |
| Carbapenems | No | Imipenem | |
| Cephems with β-lactamase inhibitors | Yes | Cefotaxime with clavulanic acid Ceftazidime with clavulanic acid | |
| Cephems | No | Cefepime | |
| No | Cefotaxime | ||
| No | Cefquinome | ||
| No | Ceftazidime | ||
| Monobactams | No | Aztreonam |
The data were collected by the U.S. National Antimicrobial Resistance Monitoring System.
Case study 3: processing a fecal sample for assessing antimicrobial susceptibility of fecal Escherichia coli using multiple bacterial isolates from one aliquot versus one isolate from each of multiple aliquots of the sample.
Assessment of AMR in a target culturable bacterial species in a fecal sample customary involves using a single aliquot from the sample (19, 20). The aliquot is diluted (the dilution is chosen based on the expected bacterial density) and the dilution(s) is plated on a bacteriological agar (19, 20). One or more of the bacterial colonies with typical morphology for the species on the agar are selected, each of the colonies is replated for isolation, and the isolate's phenotypic susceptibility to antimicrobials is tested (19, 20). However, the population of commensal bacteria such as Escherichia coli in feces of an animal or human could consist of genetically diverse subpopulations (21–25). We have chosen to investigate for cattle fecal pads if testing susceptibility of E. coli obtained from a single aliquot of the pad (conventional approach) versus from multiple aliquots taken over the longest axis of the pad yield equivalent estimates of phenotypic antimicrobial susceptibility of fecal E. coli. Fresh fecal pads (n = 32) were collected from beef and dairy cattle at research facilities at Kansas State University during May to June 2016. The animals that were sampled had not received antimicrobial drugs in the preceding week, but 1 to 3 weeks prior they had received either antimicrobials or feed supplemented with copper (Cu) or zinc (Zn) (which can coselect AMR in the animal fecal bacteria [11, 26–29]). This animal selection was done to ensure that the cattle fecal E. coli would have detectable but not uniformly high levels of phenotypic AMR, to facilitate statistical analyses of the data. Four aliquots were taken equidistantly along the longest axis of each pad. One of the four aliquots was randomly selected, and four E. coli isolates were obtained from that aliquot. From each of the other three aliquots from the pad, one E. coli isolate was obtained. All the isolates were tested for phenotypic susceptibility to antimicrobials, which were chosen to represent most of the existing antimicrobial drug classes (Table 3). As the third case study, we investigated for each of these antimicrobials the statistical equivalence of the average log2(MIC) estimates for fecal E. coli yielded by testing four bacterial isolates from a single aliquot of the fecal pad versus testing one bacterial isolate from each of four aliquots taken from locations spread over the longest axis of the fecal pad. The results demonstrated detectable differences between the two sample processing methods in the estimates of fecal E. coli susceptibility to aminopenicillins (which are β-lactams), aminoglycosides, macrolides, and tetracyclines (Fig. 4). Only for tetracyclines did the two methods yield statistically nonequivalent estimates of fecal E. coli susceptibility. The interpretation for each tested antimicrobial was based on considering the data nonequivalent if Δmin was >0.32. The observed variability in the log2(MIC) for antimicrobials of newer classes (Table 3) was insufficient for the testing; the E. coli isolates were predominantly susceptible to these antimicrobials.
TABLE 3.
Antimicrobial drug susceptibilities tested for Escherichia coli isolates from cattle fecal pads
| Antimicrobial drug class | Subclass | Combinatory formulation | Drug |
|---|---|---|---|
| Drugs for which variability in the log2(MIC) data from the two sample processing approaches was sufficient to perform the statistical equivalence testing | |||
| Aminoglycosides | -Mycins | No | Tobramycin |
| β-Lactams | Penicillins, including amino-, carboxy-, and ureido- | No | Ampicillin |
| Penicillins, including amino-, carboxy-, and ureido-, with β-lactamase inhibitors | Yes | Ampicillin with sulbactam, 2:1 ratio | |
| Cephems | No | Cefazolin | |
| No | Ceftiofur | ||
| Macrolides | Macrolides | No | Tilmicosin |
| Triamilides | No | Tulathromycin | |
| Phenicols | NA | No | Florfenicol |
| Tetracyclines | Tetracyclines | No | Chlortetracycline |
| No | Minocycline | ||
| No | Oxytetracycline | ||
| No | Tetracycline | ||
| Drugs for which variability in the log2(MIC) data from the two sample processing approaches was insufficient to perform the statistical equivalence testing | |||
| Aminoglycosides | -Micins | No | Amikacin |
| No | Gentamicin | ||
| -Mycins | No | Neomycin | |
| No | Spectinomycin | ||
| β-Lactams | Penicillins, including amino-, carboxy-, and ureido- | No | Penicillin |
| No | Piperacillin | ||
| Penicillins, including amino-, carboxy-, and ureido-, with β-lactamase inhibitors | Yes | Piperacillin with tazobactam constant | |
| Yes | Ticarcillin with clavulanic acid | ||
| Carbapenems | No | Doripenem | |
| No | Ertapenem | ||
| No | Imipenem | ||
| No | Meropenem | ||
| Monobactams | No | Aztreonam | |
| Cephems | No | Cefepime | |
| No | Ceftazidime | ||
| No | Ceftriaxone | ||
| Macrolides | Macrolides | No | Tylosin |
| Nitrofurans | NA | No | Nitrofurantoin |
| Pleuromutilins | NA | No | Pleuromutilin |
| No | Tiamulin | ||
| Lincosamides | NA | No | Clindamycin |
| Quinolones | Quinolones | No | Nalidixic acid |
| Fluoroquinolones | No | Ciprofloxacin | |
| No | Danofloxacin | ||
| No | Enrofloxacin | ||
| No | Levofloxacin | ||
| Sulfonamides | NA | No | Sulfadimethoxine |
| Yes | Sulfamethoxazole with trimethoprim | ||
| Tetracyclines | Glycylcyclines | No | Tigecycline |
DISCUSSION
The three case studies provided above illustrate the utility of statistical equivalence testing (13, 14) for establishing equivalence of data yielded by two sampling or sample-processing approaches for assessing phenotypic antimicrobial susceptibility in a target bacterial species in a stratum. Advantages of the proposed method include the estimation difference in the average log2(MIC) estimates, Δmin, below which the data from the two approaches are statistically significantly different. This provides data users with flexibility to investigate where practically relevant differences fall relative to the existing differences in the data, as illustrated in Fig. 1. Note that in the proposed method, we determine Δmin between the average log2(MIC) estimates from the two sampling or sample processing approaches using a sequential algorithm that retests the statistical nonequivalence hypothesis over a range of the average log2(MIC) difference values based on the data from the two approaches. For this, we use statistical tests that accommodate censored data with unequal variances and different shapes of the log2(MIC) distributions from the two approaches. This is because the data on an antimicrobial's MIC for a set of bacterial isolates from a stratum are inherently censored (all the isolates with MICs less than or equal to the smallest drug concentration tested are in one category in the beginning and all the isolates with MICs greater than the largest drug concentration tested are in one category in the end of the MIC distribution). The resulting log2(MIC) distributions for commonly tested antimicrobial drugs have various shapes that often do not fit to a normal distribution (7). In the proposed method for the equivalence testing, Welch's t test (30) relaxes the assumption of equal variances in the two compared log2(MIC) distributions and the robust t test (31) further improves handling of overdispersion (e.g., presence of long tails in the distributions). We keep the results obtained using both of these tests in Fig. 2 to 4 for illustrative purposes. In further applications, the robust t test could be recommended for the log2(MIC) distributions that do not follow a normal distribution and demonstrate overdispersion.
In the first case study, we evaluated whether sampling the cecal contents and sampling of ground or trimmed meat in the U.S. cattle processing plants yield equivalent data on antimicrobial susceptibility of nontyphoidal S. enterica subsp. enterica in the processed cattle. The data were collected by the NARMS in 2013-2014 to monitor AMR in the U.S. cattle production chain, and therefore we interpret the results at the same population level. The results showed that S. enterica subpopulations in the cecal content samples may be statistically nonequivalent in their phenotypic antimicrobial susceptibility to S. enterica subpopulations in the ground- or trimmed-meat samples (Fig. 2). Thus, the two sampling approaches yield nonequivalent data for monitoring phenotypic antimicrobial susceptibility in S. enterica in cattle in the processing plants. Possible explanations include decontamination and cross-contamination of cattle carcasses and products within the plant, as well as mixing of different carcass parts for the ground meat (32–34). These processes can reduce the role of the cattle intestinal contents as a source of S. enterica in the meat or ground-meat products.
In the second case study, we evaluated statistical equivalence of the antimicrobial susceptibility estimates for nontyphoidal S. enterica isolates from urine, fecal, and blood samples of humans in the U.S. in 2015. The data were collected by the NARMS to monitor AMR in human enteric pathogens in the United States, and thus again we interpret the results at the same population level. The results demonstrated that susceptibilities of the S. enterica isolates from human blood are nonequivalent to those from urine or feces for several major antimicrobial drug classes, such as aminoglycosides, cephems (β-lactams), macrolides, phenicols, quinolones, and tetracyclines (Fig. 3A and B). Lesser differences were observed between the isolates from urine versus feces (Fig. 3C). The differences of largest magnitude were found for quinolones, with the isolates from human blood being less susceptible than those from urine or feces to the fluoroquinolone ciprofloxacin and the older quinolone nalidixic acid (see Results for more details). This could be due to the common use of fluoroquinolones, e.g., ciprofloxacin, as one of the first-line treatment choices for treating serious infections by nontyphoidal salmonellae in human adults (35–38). Another common treatment choice is cephalosporins (β-lactams), e.g., ceftriaxone (the other choices include combinatory formulations containing β-lactams and β-lactamase inhibitors, aminoglycosides, and, as the last resort, polymyxins and carbapenems [β-lactams]) (35, 38). Considering the data for 2015, susceptibilities to individual cephems of the S. enterica isolates from human blood were less different from (although statistically nonequivalent to) those of the isolates from urine or feces, compared to the differences observed for quinolones (Fig. 3A and B). Notably, across the human nontyphoidal Salmonella isolates, the frequency of those with reduced ciprofloxacin susceptibility has been continuously rising and the frequency of those with reduced ceftriaxone susceptibility has overall increased in the United States since 1996 (38, 39).
In the third case study, we evaluated whether testing four bacterial isolates from a single aliquot of the cattle fecal pad versus testing one bacterial isolate from each of four aliquots taken from locations spread over the longest axis of the pad yield equivalent data on phenotypic antimicrobial susceptibility of fecal E. coli at the population level (n = 32 pads were tested). The results showed that the two sample processing methods yield statistically nonequivalent estimates of E. coli susceptibility to tetracyclines, with smaller but detectable differences for aminopenicillins (β-lactams), aminoglycosides, and macrolides (Fig. 4). These antimicrobial drug classes have been used in food animals in the United States for the longest periods (40). Tetracyclines, penicillins, and aminoglycosides were introduced in the 1940s and macrolides in the 1970s (40). Consequently, multiple genes encoding various degrees of susceptibility to these drug classes have been observed in fecal E. coli and S. enterica isolates from farm animals (41–45). The tetracycline resistance gene pool is especially diverse, with several tens of tet genes described to date for different animal and human hosts (41, 46). Testing E. coli throughout the fecal pad may capture more of the present diversity in the susceptibility to tetracyclines than testing E. coli at a single location in the pad. Also, the statistical power of the equivalence testing depends not only on the sample size but also on variability in the log2(MIC) data. Strongly bimodal (less variable) log2(MIC) distributions for newer antimicrobial drug classes, due to the high frequencies of the highly susceptible bacterial isolates, impede the testing (see Tables 1 to 3 for examples).
Diagnostic microbiologists consider one 2-fold dilution of the antimicrobial drug to be an acceptable variation in the MIC measurement for an individual bacterial isolate in the broth microdilution assay (47). If such variation occurs randomly among the isolates in the two sampling or sample processing approaches, it is accounted for in the variance component of a statistical test of the data (for example, see Materials and Methods). Such random variation does not bias comparisons of the data between the approaches. However, if the variation is nonrandom and has a systematic source, it could bias the comparisons. Consider an extreme case of the MIC being skewed by one 2-fold drug dilution for every isolate obtained from one sampling or sample processing approach but not from the other approach, e.g., if the samples from one approach were examined in one laboratory and the samples from the other approach in another laboratory. The data user believes that the antimicrobial's MIC measurements in one laboratory are consistently one 2-fold dilution higher or lower than the MIC measurements in the second laboratory. In this case, there would be a difference Δ ≥ 1 for the antimicrobial between the average log2(MIC) estimates from the two laboratories. A statistically significant difference beyond that would be manifested as Δmin > 1.
We have included a suggestion for interpreting Δmin > 0.32 as evidence of statistical nonequivalence of the log2(MIC) data for the antimicrobial for the bacterial species in the stratum between the two sampling or sample-processing approaches. This interpretation illustrated in Fig. 2 to 4 is an adaptation of a method for establishing bioequivalence of two drug preparations based on values of a biological drug response variable (16). Other standardized interpretations may be proposed in the future for decision-making on whether the two sampling or sample processing approaches are interchangeable or yield equivalent data on phenotypic antimicrobial susceptibility of the target bacteria (i.e., assess susceptibility in the same bacterial subpopulation) in the stratum.
MATERIALS AND METHODS
Statistical equivalence testing. (i) Rationale for testing the statistical equivalence hypothesis.
Let M1 and M2 be the sampling or sample processing approaches that yield samples Y1 and Y2, respectively, of isolates of the target bacterial species from the stratum. The samples Y1 and Y2 represent subpopulations P1 and P2 of the species in the stratum. The isolate susceptibility to a target antimicrobial is measured (e.g., in our three case studies the susceptibility was measured using the broth microdilution assay and the obtained MICs were log2 transformed for the analysis). The statistics μ1 and μ2 represent the central tendencies of the susceptibility of the unknown source subpopulations P1 and P2. “Conventional” hypothesis testing focuses on rejecting H0 of no statistically significant difference between the central tendencies H0: μ1 − μ2 = 0; Ha: μ1 − μ2 ≠ 0. However, even if H0 is rejected, this provides no proof in favor of Ha. Importantly, testing the conventional H0 delivers no information for what μ1 − μ2 difference signals that the central tendencies of the samples Y1 and Y2 are statistically significantly different.
Equivalence hypothesis.
The equivalence hypothesis testing can provide the sought information on a statistically significant μ1 − μ2. The null and alternative hypotheses are defined as follows (13, 14):
| (1) |
The equivalence hypothesis testing in equation 1 indicates that the samples Y1 and Y2 obtained by the approaches M1 and M2 have equal means up to an acceptable tolerance Δ with a predefined confidence interval 1 − 2α (where α is probability of the type I error). This null hypothesis is rejected if the data provide evidence of the equivalence of the means. Otherwise, the null hypothesis of a statistically significant μ1 − μ2 difference is accepted.
(ii) Student’s t test of the equivalence hypothesis.
The samples Y1 and Y2 are obtained by the sampling or sample processing approaches M1 and M2. The sample means Ȳ1 and Ȳ2 are employed as the point estimators of μ1 and μ2, with standard errors se1 and se2. Therefore, the difference μ1 − μ2 can be estimated by Ȳ1 − Ȳ2 with a standard error , which is equal to
| (2) |
where σ1 and σ2 are the estimates of standard deviations of P1 and P2, respectively, and n1 and n2 are the respective sample sizes of Y1 and Y2. With large sample sizes and the fact that P1 and P2 are known, the sampling distribution of Ȳ1 − Ȳ2 could be estimated through a normal distribution centered at μ1 − μ2 with the standard error given by equation 2. Instead, to avoid introducing extra variability from estimating σ1 and σ2 using sample variances s1 and s2, and to be able to handle the data with different sample sizes, Student’s t test can be used. Student’s t test assumes that samples Y1 and Y2 are both drawn from variables that follow a normal distribution and have equal variances, which can be estimated by a pooled variance combining the sample variances s1 and s2. Applying Student’s t test for samples with unequal variances or sample sizes can lead to unreliable conclusions with large type I and type II error probabilities (48).
(iii) Welch's t test of the equivalence hypothesis.
Welch's t test can handle unequal variances or sample sizes in the data from the two sampling or sample processing approaches (30). The t statistics for Welch's t test of the hypothesis defined in equation 1 are
and H0 is rejected if t1 is less than −tα,df and t2 is greater than tα,df, where df is degrees of freedom.
However, the censored nature of the MIC data results in the presence of aggregated observations in the regions MIC ≤ L and MIC > U, where L and U are the smallest and largest drug concentrations tested, respectively. A long-tail in the left-hand end, MIC ≤ L, and a long-tail in the right-hand end, MIC > U, of the distribution are common (7). Such long-tailed shapes are common in the distributions even after the log2(MIC) transformation (12).
(iv) Robust t test of the equivalence hypothesis.
As noted above, the log2(MIC) distributions most often do not follow a normal distribution. To improve robustness of the equivalence hypothesis testing and handle the long tails in the log2(MIC) distributions, we built upon Welch's t test to use the trimmed data and Winsorized variance. This is known as the robust t test (31). Let Y1 and Y2 be the ordered sample data, e.g., ; then under H0:
where Ȳ1,tg and Ȳ2,tg are the trimmed (indicated by t) means and g is the number of the trimmed data points from each of the sides of the ordered Y1 and Y2 defined as
and the Winsorized (indicated by w) sum of squares and are defined as
The trimmed t statistics and each follow a t distribution with the degrees of freedom df (49)
where c is given by
Thus, H0 is rejected if .
(v) Power analysis of the statistical equivalence testing.
The power of testing statistical equivalence of the average log2(MIC) estimates for each antimicrobial drug from the two sampling or sample processing approaches for the target bacterial species in the stratum can be computed (using De Morgan's law [50]) as follows:
| (3) |
To compute the right-hand side of equation 3, we use the noncentral t distribution at and its cumulative distribution and a similar distribution of :
The test can be considered to have an acceptable power if the power is ≥0.80, following published guidelines (51, 52).
Determining Δmin from 95% confidence interval of the difference of the means.
The threshold difference Δmin is the minimum Δ in equation 1 at which the two sampling or sample processing approaches still yield statistically nonequivalent estimates of susceptibility to the antimicrobial drug of the bacterial species in the sampled stratum. This threshold value for each antimicrobial and statistical test (Welch's t test or robust t test) was found from the data via a sequential algorithm repeating the test over a range of the difference values, starting from a large value [e.g., a 4-log difference between the average log2(MIC) estimates] and then shrinking Δ by a small step (0.01 log2) and retesting the H0 of a statistically significant difference between the average log2(MIC) estimates from the two approaches, until reaching the Δmin value below which the H0 could no longer be rejected.
Microbiological procedures. (i) Case studies 1 and 2.
Microbiological procedures used by the NARMS are described in the program's Manual of Laboratory Methods (53). Phenotypic susceptibility of the S. enterica isolates to antimicrobials is determined in the broth microdilution assay using the Sensititre system (TREK Diagnostic Systems Inc., Cleveland, OH), in accordance with the manufacturer recommendations and the Clinical and Laboratory Standards Institute (CLSI) guidelines (47, 53). The assays for the S. enterica isolates from cattle processing plants in 2013-2014 were performed using the Sensititre plate format CMV3AGNF as of those years, which included antimicrobials listed in Table 1. The assays for the nontyphoidal S. enterica isolates from humans in 2015 were performed using the Sensititre CMV3AGNF plate format as of that year and an additional plate format containing broad-spectrum β-lactams; the tested antimicrobials are listed in Table 2.
(ii) Case study 3: sampling.
Fresh fecal pads (n = 32) were collected (the entire pad was lifted from the ground without mixing, placed into a sterile plastic bag, and transported while being kept horizontal) from different beef and dairy cattle at research facilities at Kansas State University during May to June 2016. The animals that were sampled had not received antimicrobial drugs in the preceding week but 1 to 3 weeks prior had received either antimicrobials (macrolides or tetracyclines to treat limited bovine respiratory disease or as a part of a research study) or feed supplemented with copper or zinc (these feed additives can coselect AMR in the animal fecal bacteria [11, 26–29]). This animal selection was done to ensure that the cattle fecal E. coli would have detectable but not uniformly high levels of AMR, to facilitate statistical analyses of the data. A collected fecal pad weighed 1.2 kg on average (5th and 95th percentiles: 0.4, 2.0). The average (5th, 95th percentile) pad dimensions were a length of 24 (18, 31) cm, a width of 18 (12, 25) cm, and a height of 3 (5, 9) cm.
(iii) Sample processing.
On each fecal pad, four locations along the longest axis of the pad—its length—were marked using a sterile plastic loop. The locations were spread along the pad length (depending on the length) equidistantly ∼3 to 5 cm (∼1.5 to 2 in.) apart. Feces at the four locations were opened to the depth of ∼1 cm using sterile tools (to avoid the possibility of culturing E. coli that may have accidentally contaminated the pad exterior). One fecal aliquot of ∼1 g was aseptically collected from the bottom of the opening at each of the four locations. The locations were counted left to right. A random number was generated from a Uniform (1,4). When the aliquot from the location with the number corresponding to the generated random number was plated on a MacConkey agar plate, four E. coli colonies were obtained from the plate for isolation. When each of the aliquots from the other three locations on the pad was plated on a MacConkey agar plate, one E. coli isolate was obtained from the plate for isolation.
(iv) Microbiological procedures.
Each fecal aliquot of ∼1 g was diluted in 10 ml of buffered peptone water (PBS) and vortexed gently until fully mixed. Of the supernatant, 100 μl was diluted 1:10 in sterile PBS and 100 μl was diluted 1:100 in sterile PBS. Of each of the dilutions, 100 μl was plated on a MacConkey agar plate and incubated at 37.5°C for 24 h. For the randomly selected aliquot on the pad, from the MacConkey plate with well-separated colonies, 4 typical coliform colonies chosen from different parts of the plate (convenience randomization) were each streaked on a tryptic soy broth supplemented with 5% sheep blood agar plate (BAP) and incubated at 37.5°C for 24 h. For each of the other three aliquots from the pad, from the MacConkey plate with well-separated colonies, one typical coliform colony (chosen via convenience randomization) was streaked on a BAP plate and incubated at 37.5°C for 24 h. Presumptive E. coli colonies from each BAP plate were subjected to the indole test, and the indole-producing ones were identified as E. coli. When needed, additional coliform colonies from the MacConkey plate were replated for isolation and subjected to the indole test to obtain the sought number of E. coli isolates from the fecal aliquot (i.e., one or four isolates). Phenotypic susceptibility to antimicrobials of each E. coli isolate was determined in the broth microdilution assay following the Sensititre plate manufacturer instructions and in accordance with the CLSI recommendations (47, 53). The Sensititre plate formats GN4F and BOPO6F as of 2016 were used. The strain E. coli ATCC 25922 was used for the quality control of the assays, along with the positive- and negative-control wells. The assay results on the plates were read on the Sensititre ARIS automated reading instrument (TREK Diagnostic Systems Inc., Cleveland, OH).
Software.
The NARMS 2013 to 2015 data (publicly available) and the data for the fecal pads were gathered in Microsoft Office Excel (Microsoft, Inc., Redmond, WA). The data were imported into R 3.4, in which the statistical analyses were performed. All the figures were made in Python 3 (Python Software Foundation).
ACKNOWLEDGMENTS
This work was supported by the Kansas Bioscience Authority via its support for the Institute of Computational Comparative Medicine at Kansas State University. As well, contributions of V.V. were supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R15GM126503.
The manuscript content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Johnson AP. 2015. Surveillance of antibiotic resistance. Philos Trans R Soc Lond B Biol Sci 370:20140080. doi: 10.1098/rstb.2014.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. 2015. Global antimicrobial resistance surveillance system: manual for early implementation, p 1–2 World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/188783/1/9789241549400_eng.pdf?ua=1 Accessed 6 January 2018. [Google Scholar]
- 3.CLSI. 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard, 4th ed CLSI document VET01-A4. CLSI, Wayne, PA. [Google Scholar]
- 4.Wagner BA, Dargatz DA, Morley PS, Keefe TJ, Salman MD. 2003. Analysis methods for evaluating bacterial antimicrobial resistance outcomes. Am J Vet Res 64:1570–1579. doi: 10.2460/ajvr.2003.64.1570. [DOI] [PubMed] [Google Scholar]
- 5.van de Kassteele J, van Santen-Verheuvel MG, Koedijk FD, van Dam AP, van der Sande MA, de Neeling AJ. 2012. New statistical technique for analyzing MIC-based susceptibility data. Antimicrob Agents Chemother 56:1557–1563. doi: 10.1128/AAC.05777-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stegeman JA, Vernooij JC, Khalifa OA, Van den Broek J, Mevius DJ. 2006. Establishing the change in antibiotic resistance of Enterococcus faecium strains isolated from Dutch broilers by logistic regression and survival analysis. Prev Vet Med 74:56–66. doi: 10.1016/j.prevetmed.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Mazloom R, Jaberi-Douraki M, Comer J, Volkova V. 2018. Potential information loss due to categorization of MIC frequency distributions. Foodborne Pathog Dis 15:44–54. doi: 10.1089/fpd.2017.2301. [DOI] [PubMed] [Google Scholar]
- 8.Thornsberry C, Jones ME, Hickey ML, Mauriz Y, Kahn J, Sahm DF. 1999. Resistance surveillance of Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis isolated in the United States, 1997–1998. J Antimicrob Chemother 44:749–759. doi: 10.1093/jac/44.6.749. [DOI] [PubMed] [Google Scholar]
- 9.Sahm DF, Jones ME, Hickey ML, Diakun DR, Mani SV, Thornsberry C. 2000. Resistance surveillance of Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis isolated in Asia and Europe, 1997–1998. J Antimicrob Chemother 45:457–466. doi: 10.1093/jac/45.4.457. [DOI] [PubMed] [Google Scholar]
- 10.Wagner BA, Dargatz DA, Salman Morley PS, Wittum TE, Keefe TJ. 2002. Comparison of sampling techniques for measuring the antimicrobial susceptibility of enteric Escherichia coli recovered from feedlot cattle. Am J Vet Res 63:1662–1670. doi: 10.2460/ajvr.2002.63.1662. [DOI] [PubMed] [Google Scholar]
- 11.Agga GE, Scott HM, Amachawadi RG, Nagaraja TG, Vinasco J, Bai J, Norby B, Renter DG, Dritz SS, Nelssen JL, Tokach MD. 2014. Effects of chlortetracycline and copper supplementation on antimicrobial resistance of fecal Escherichia coli from weaned pigs. Prev Vet Med 114:231–246. doi: 10.1016/j.prevetmed.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 12.Zawack K, Li M, Booth JG, Love W, Lanzas C, Grohn YT. 2016. Monitoring antimicrobial resistance in the food supply chain and its implications for FDA policy initiatives. Antimicrob Agents Chemother 60:5302–5311. doi: 10.1128/AAC.00688-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wellek S. 2010. Testing statistical hypotheses of equivalence and noninferiority, 2nd ed Chapman and Hall/CRC Press, Boca Raton, FL. [Google Scholar]
- 14.Anderson SF, Maxwell SE. 2016. There's more than one way to conduct a replication study: beyond statistical significance. Psychol Methods 21:1–12. doi: 10.1037/met0000051. [DOI] [PubMed] [Google Scholar]
- 15.Love WJ, Zawack KA, Booth JG, Grhn YT, Lanzas C. 2016. Markov networks of collateral resistance: National Antimicrobial Resistance Monitoring System surveillance results from Escherichia coli isolates, 2004–2012. PLoS Comput Biol 12:e1005160. doi: 10.1371/journal.pcbi.1005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.FDA CDER. 2001. Guidance for industry. Statistical procedures for bioequivalence studies. US Food and Drug Administration, Silver Spring, MD: https://www.fda.gov/downloads/drugs/guidances/ucm070244.pdf Accessed 1 July 2017. [Google Scholar]
- 17.Karp B, Tate H, Plumblee JR, Dessai U, Whichard JM, Thacker EL, Hale KR, Wilson W, Friedman CR, Griffin PM, McDermott P. 2017. National Antimicrobial Resistance Monitoring System: two decades of advancing public health through integrated surveillance of antimicrobial resistance. Foodborne Pathog Dis 14:545–557. doi: 10.1089/fpd.2017.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.FDA Center for Veterinary Medicine. 2016. National Antimicrobial Resistance Monitoring System (NARMS) 2014: integrated report. The National Antimicrobial Resistance Monitoring System: enteric bacteria. US Food and Drug Administration, Silver Spring, MD: https://www.fda.gov/downloads/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/UCM528861.pdf Accessed 1 October 2017. [Google Scholar]
- 19.Garcia LS. (ed). 2010. Clinical microbiology procedures handbook. 3rd ed ASM Press, Washington, DC. [Google Scholar]
- 20.WHO. 2003. Manual for identification and antimicrobial susceptibility testing of bacterial pathogens of public health importance in the developing world. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/68554/1/WHO_CDS_CSR_RMD_2003.6.pdf Accessed 1 October 2017. [Google Scholar]
- 21.Ahmed S, Olsen JE, Herrero-Fresno A. 2017. The genetic diversity of commensal Escherichia coli strains isolated from non-antimicrobial treated pigs varies according to age group. PLoS One 12:e0178623. doi: 10.1371/journal.pone.0178623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson MA, Whitlock JE, Harwood VJ. 2006. Diversity and distribution of Escherichia coli genotypes and antibiotic resistance phenotypes in feces of humans, cattle, and horses. Appl Environ Microbiol 72:6914–6922. doi: 10.1128/AEM.01029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caugant DA, Levin BR, Selander RK. 1981. Genetic diversity and temporal variation in the E. coli population of a human host. Genetics 98:467–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aslam M, Nattress F, Greer G, Yost C, Gill C, McMullen L. 2003. Origin of contamination and genetic diversity of Escherichia coli in beef cattle. Appl Environ Microbiol 69:2794–2799. doi: 10.1128/AEM.69.5.2794-2799.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aslam M, Greer GG, Nattress FM, Gill CO, McMullen LM. 2004. Genetic diversity of Escherichia coli recovered from the oral cavity of beef cattle and their relatedness to faecal E. coli. Lett Appl Microbiol 39:523–527. doi: 10.1111/j.1472-765X.2004.01620.x. [DOI] [PubMed] [Google Scholar]
- 26.Agga GE, Scott HM, Vinasco J, Nagaraja TG, Amachawadi RG, Bai J, Norby B, Renter DG, Dritz SS, Nelssen JL, Tokach MD. 2015. Effects of chlortetracycline and copper supplementation on the prevalence, distribution, and quantity of antimicrobial resistance genes in the fecal metagenome of weaned pigs. Prev Vet Med 119:179–189. doi: 10.1016/j.prevetmed.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 27.Amachawadi RG, Scott HM, Vinasco J, Tokach MD, Dritz SS, Nelssen JL, Nagaraja TG. 2015. Effects of in-feed copper, chlortetracycline, and tylosin on the prevalence of transferable copper resistance gene, tcrB, among fecal enterococci of weaned piglets. Foodborne Pathog Dis 12:670–678. doi: 10.1089/fpd.2015.1961. [DOI] [PubMed] [Google Scholar]
- 28.Amachawadi RG, Scott HM, Aperce C, Vinasco J, Drouillard JS, Nagaraja TG. 2015. Effects of in-feed copper and tylosin supplementations on copper and antimicrobial resistance in faecal enterococci of feedlot cattle. J Appl Microbiol 118:1287–1297. doi: 10.1111/jam.12790. [DOI] [PubMed] [Google Scholar]
- 29.Feldpausch JA, Amachawadi R, Tokach MD, Scott HM, Dritz SS, Nagaraja TG, Goodband RD, Woodworth JC, DeRouchey JM. 2016. Effects of dietary copper, zinc, and ractopamine hydochloride on finishing pig growth performance, carcass characteristics, and antimicrobial susceptibility of enteric bacteria. J Anim Sci 94:3278–3293. doi: 10.2527/jas.2016-0340. [DOI] [PubMed] [Google Scholar]
- 30.Welch BL. 1947. The generalisation of student's problems when several different population variances are involved. Biometrika 34:28–35. [DOI] [PubMed] [Google Scholar]
- 31.Yuen KK. 1974. The 2-sample trimmed t for unequal population variances. Biometrika 61:165–170. [Google Scholar]
- 32.Koohmaraie M, Scanga JA, De La Zerda MJ, Koohmaraie B, Tapay L, Beskhlebnaya V, Mai T, Greeson K, Samadpour M. 2012. Tracking the sources of Salmonella in ground beef produced from nonfed cattle. J Food Prot 75:1464–1468. doi: 10.4315/0362-028X.JFP-11-540. [DOI] [PubMed] [Google Scholar]
- 33.Koohmaraie M, Arthur TM, Bosilevac JM, Guerini M, Shackelford SD, Wheeler TL. 2005. Post-harvest interventions to reduce/eliminate pathogens in beef. Meat Sci 71:79–91. doi: 10.1016/j.meatsci.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 34.Sofos JN, Kochevar SL, Bellinger GR, Buege DR, Hancock DD, Ingham SC, Morgan JB, Reagan JO, Smith GC. 1999. Sources and extent of microbiological contamination of beef carcasses in seven United States slaughtering plants. J Food Prot 62:140–145. doi: 10.4315/0362-028X-62.2.140. [DOI] [PubMed] [Google Scholar]
- 35.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf Accessed 1 February 2017. [Google Scholar]
- 36.Hohmann EL. 2001. Nontyphoidal salmonellosis. Clin Infect Dis 32:263–269. doi: 10.1086/318457. [DOI] [PubMed] [Google Scholar]
- 37.Onwuezobe IA, Oshun PO, Odigwe CC. 2012. Antimicrobials for treating symptomatic non-typhoidal Salmonella infection. Cochrane Database Syst Rev 2012:CD001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Centers for Disease Control and Prevention National Antimicrobial Resistance Monitoring System (NARMS). 2016. 2014 human isolates surveillance report. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/narms/pdf/2014-annual-report-narms-508c.pdf Accessed 18 February 2018. [Google Scholar]
- 39.FDA Center for Veterinary Medicine. 2017. National Antimicrobial Resistance Monitoring System (NARMS) 2015 integrated report. US Food and Drug Administration, Silver Spring, MD: https://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/ucm059103.htm Accessed 8 February 2018. [Google Scholar]
- 40.Volkova VV, DeMars Z. 2017. Short history of regulations and approved indications of antimicrobial drugs for food animals in the USA. J Vet Pharmacol Ther 40:211–217. doi: 10.1111/jvp.12376. [DOI] [PubMed] [Google Scholar]
- 41.Bryan A, Shapir N, Sadowsky MJ. 2004. Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl Environ Microbiol 70:2503–2507. doi: 10.1128/AEM.70.4.2503-2507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, Zhao S. 2016. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother 60:5515–5520. doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyson GH, McDermott PF, Li C, Chen Y, Tadesse DA, Mukherjee S, Bodeis-Jones S, Kabera C, Gaines SA, Loneragan GH, Edrington TS, Torrence M, Harhay DM, Zhao S. 2015. WGS accurately predicts antimicrobial resistance in Escherichia coli. Antimicrob Agents Chemother 70:2763–2769. doi: 10.1093/jac/dkv186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santamaría J, Lopez L, Soto CY. 2011. Detection and diversity evaluation of tetracycline resistance genes in grassland-based production systems in Colombia, South America. Front Microbiol 2:252. doi: 10.3389/fmicb.2011.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cormier AC, Chalmers G, McAllister TA, Cook S, Zaheer R, Scott HM, Booker C, Read R, Boerlin P. 2016. Extended-spectrum-cephalosporin resistance genes in Escherichia coli from beef cattle. Antimicrob Agents Chemother 60:1162–1163. doi: 10.1128/AAC.02516-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts MC. 2005. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett 245:195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 47.CLSI. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 10th ed CLSI document M07-A10. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 48.Ruxton GD. 2006. The unequal variance t-test is an underused alternative to Student's t-test and the Mann-Whitney U test. Behav Ecol 17:688–690. doi: 10.1093/beheco/ark016. [DOI] [Google Scholar]
- 49.Yuen KK, Dixon WJ. 1973. Approximate behavior and performance of 2-sample trimmed t. Biometrika 60:369–374. [Google Scholar]
- 50.Hurley PJ. 2014. A concise introduction to logic, 12th ed Cengage Learning, Stamford, CT. [Google Scholar]
- 51.Cohen J. 1992. A power primer. Psychol Bull 112:155–159. doi: 10.1037/0033-2909.112.1.155. [DOI] [PubMed] [Google Scholar]
- 52.Cohen J. 1988. Statistical power analysis for the behavioral sciences, 2nd ed Lawrence Erlbaum Associates, New York, NY. [Google Scholar]
- 53.National Antimicrobial Resistance Monitoring System (NARMS). 2016. Manual of Laboratory Methods, 3rd ed US Food and Drug Administration, Silver Spring, MD: https://www.fda.gov/downloads/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/UCM528831.pdf Accessed 1 October 2017. [Google Scholar]