ABSTRACT
The structural variation of the bacterial community associated with particulate matter (PM) was assessed in an urban area of Beijing during hazy and nonhazy days. Sampling for different PM fractions (PM2.5 [<2.5 μm], PM10 [<10 μm], and total suspended particulate) was conducted using three portable air samplers from September 2014 to February 2015. The airborne bacterial community in these samples was analyzed using the Illumina MiSeq platform with bacterium-specific primers targeting the 16S rRNA gene. A total of 1,707,072 reads belonging to 6,009 operational taxonomic units were observed. The airborne bacterial community composition was significantly affected by PM fractions (R = 0.157, P < 0.01). In addition, the relative abundances of several genera significantly differed between samples with various haze levels; for example, Methylobacillus, Tumebacillus, and Desulfurispora spp. increased in heavy-haze days. Canonical correspondence analysis and permutation tests showed that temperature, SO2 concentration, relative humidity, PM10 concentration, and CO concentration were significant factors that associated with airborne bacterial community composition. Only six genera increased across PM10 samples (Dokdonella, Caenimonas, Geminicoccus, and Sphingopyxis) and PM2.5 samples (Cellulomonas and Rhizobacter), while a large number of taxa significantly increased in total suspended particulate samples, such as Paracoccus, Kocuria, and Sphingomonas. Network analysis indicated that Paracoccus, Rubellimicrobium, Kocuria, and Arthrobacter were the key genera in the airborne PM samples. Overall, the findings presented here suggest that diverse airborne bacterial communities are associated with PM and provide further understanding of bacterial community structure in the atmosphere during hazy and nonhazy days.
IMPORTANCE The results presented here represent an analysis of the airborne bacterial community associated with particulate matter (PM) and advance our understanding of the structural variation of these communities. We observed a shift in bacterial community composition with PM fractions but no significant difference with haze levels. This may be because the bacterial differences are obscured by high bacterial diversity in the atmosphere. However, we also observed that a few genera (such as Methylobacillus, Tumebacillus, and Desulfurispora) increased significantly on heavy-haze days. In addition, Paracoccus, Rubellimicrobium, Kocuria, and Arthrobacter were the key genera in the airborne PM samples. Accurate and real-time techniques, such as metagenomics and metatranscriptomics, should be developed for a future survey of the relationship of airborne bacteria and haze.
KEYWORDS: airborne bacterial community, PM2.5, PM10, TSP, haze, high-throughput sequencing
INTRODUCTION
Particulate matter (PM) is a complicated mixture composed of inorganic substances, organic compounds, and biological components (1). Numerous studies have demonstrated that PM plays an important role in air pollution, visibility reduction, and climate change (2–6). PM varies considerably in size from 0.02 to 100 μm (7). It is necessary to study the characteristics of total suspended particulates (TSP), PM10 (particulate size, <10 μm), and PM2.5 (particulate size, <2.5 μm), as the various PM fractions show significantly different compositions (8). Previous evidence suggests that exposure to high levels of PM pollutants, particularly PM2.5, is linked to increased human morbidity and mortality, as well as a shortened life span (9, 10).
Bacteria associated with PM are present in the atmosphere in the form of spores, vegetative cells, or dividing cells (11, 12). Before removal from the atmosphere by precipitation or direct deposition onto surfaces, small bacteria can reside in the air for several days and be transported over long distances (13). Because of their important role in the atmosphere, there are many studies on the bacterial communities associated with PM. However, most previous studies have focused on the bacterial diversity associated with PM using traditional methods of identification of culturable isolates (14–24). However, the culture method cannot reflect the actual diversity of airborne bacteria because of its selectivity.
With recent advances in high-throughput sequencing, more accurate diversity and community composition of PM-associated airborne bacteria have been revealed in different regions, including Beijing (China) (25, 26); Hong Kong (China) (27); Chicago (IL), Cleveland (OH), and Detroit (MI) (28); Denver and Greely (CO) (29); Boulder (CO) (30); Washington, DC (31); Colorado Front Range (USA) (32); Milan (Italy) (33, 34); the McMurdo Dry Valleys (Antarctica) (12); and the Pyrenees (Spain) (35). However, to our knowledge, there has been no study on the bacterial communities in various PM fractions (TSP, PM10, and PM2.5) during hazy and nonhazy days.
As a metropolis with a population of over 20 million, Beijing has long been suffering from serious haze due to the accumulation of PM (25, 36–40). Under stable weather conditions, PM can accumulate over a long period to form heavy haze (41). High relative humidity (rH) and low temperatures accelerate the chemical transformation to aerosols (42). Regional transport has also been found to play an important role during haze episodes. Ge et al. (43) indicated that the contribution of regional transport to haze was higher than that of local emissions in the Beijing-Tianjin-Hebei region during haze episodes. Secondary organic aerosols and secondary inorganic aerosols, which are the major contributors to PM, have multiple environmental effects, especially visibility degradation, in Beijing (44–46). Huang et al. (44) found that the major sources of haze were coal burning, dust, traffic, biomass burning, and cooking. Airborne bacterial communities associated with PM in Beijing have been characterized (25, 26). However, these studies were based on a limited number of samples or lacked long-term monitoring.
This study used high-throughput sequencing to investigate the PM-associated airborne bacterial community in Beijing during the autumn and winter. The aims of the research were to (i) examine how airborne bacterial diversity and community composition are associated with PM, (ii) determine whether the bacterial community composition differs between various types of PM or between samples from different haze levels, (iii) identify the genera associated with PM, and (iv) determine which environmental factors influence the bacterial community composition.
RESULTS
Richness and diversity of airborne bacteria.
We obtained 3,616,411 raw reads and 2,911,649 valid reads after demultiplexing and quality filtering (see Table S1 in the supplemental material). After subsampling, a total of 1,707,072 reads (17,782 reads per sample) were clustered into 6,009 unique operational taxonomic units (OTUs) (97% similarity). Alpha diversity was estimated by the Chao1 index, Good's coverage estimator, and Shannon index (Table S1). The rarefaction curves indicated that high-throughput sequencing captured the dominant phylotypes, and Shannon index curves showed high diversity in the airborne bacterial community (Fig. S1). On average, there were 370 (range, 111 to 904), 903 (range, 216 to 1,786), and 1,146 (range, 255 to 1,970) OTU in PM2.5 samples (PM2.5S), PM10 samples (PM10S), and TSP samples (TSPS), respectively, which suggests that the number of OTUs increases as the size fraction increases. The highest values of Chao1 and Shannon index were observed in TSPS, while the lowest ones were found in PM2.5S. Moreover, one-way analysis of variance (ANOVA) showed significant differences between the various PM fractions (P < 0.05), which indicated significantly higher richness and diversity in bigger-PM-fraction samples. However, no significant differences in Chao1 and Shannon index values were found between the hazy and nonhazy samples (Fig. S2).
Airborne bacterial community composition.
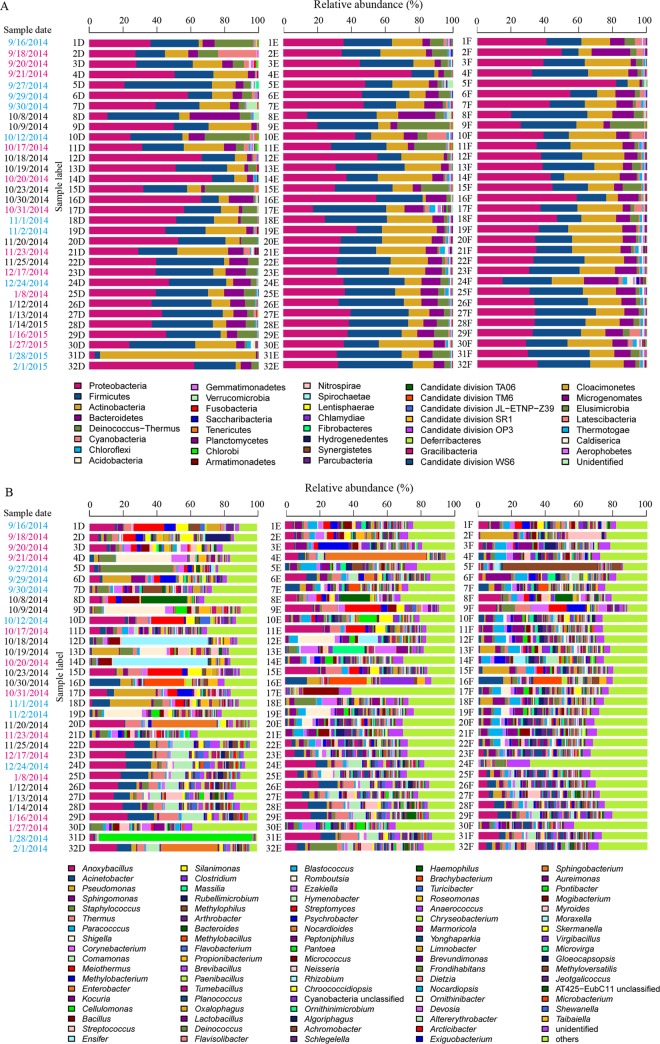
We found that the airborne bacterial community was dominated by the Proteobacteria (38.5%), including Gammaproteobacteria (15.0%), Alphaproteobacteria (14.3%), and Betaproteobacteria (8.5%), followed by the Firmicutes (26.8%), Actinobacteria (17.2%), Bacteroidetes (7.7%), and Deinococcus-Thermus (5.2%) (Table S3 and Fig. 1A). The dominant orders were the Bacillales (16.1%) and Clostridiales (6.7%), which belong to Firmicutes; Pseudomonadales (7.4%), Burkholderiales (5.6%), and Rhizobiales (5%), which belong to Proteobacteria; and Micrococcales (7.9%), which belongs to Actinobacteria (Table S3). Anoxybacillus (6.6%) was the most abundant genus, followed by Acinetobacter (3.8%), Pseudomonas (2.7%), Sphingomonas (2.5%), Staphylococcus (2.5%), Thermus (2.5%), and Paracoccus (2.5%) (Table S3 and Fig. 1B).
FIG 1.
Dominant bacterial groups identified at the phylum level (A) and at the genus level (B). Blue text, pink text, and black text for the sample dates refer to non-, light-, and heavy-haze sample days, respectively. D, E, and F represent PM2.5, PM10, and TSP, respectively. The category “others” includes all genera for which relative abundances are lower than 2%.
Airborne bacterial communities with different PM fractions.
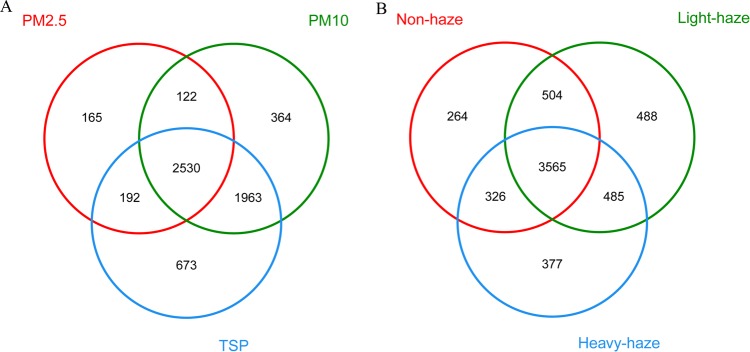
The Venn diagram presented in Fig. 2A shows that there were 2,530 OTU shared among PM2.5S, PM10S, and TSPS. Most genera were distributed differently in three PM fractions, except for Methylobacterium and Streptococcus, which had a similar relative abundances in PM samples. Anoxybacillus and Acinetobacter were the most abundant genera in both PM2.5S and PM10S but not in TSPS. Moreover, Pseudomonas and Thermus were the third most abundant genera in PM2.5S and PM10S, respectively. In TSPS, Paracoccus, Sphingomonas, and Anoxybacillus were the most abundant genera (Table S4). In addition, some genera were more abundant in certain PM fractions, for example, Paracoccus, Sphingomonas, and Kocuria were richer in PM10S and TSPS (Fig. S3).
FIG 2.
Venn diagrams illustrating the number of unique and shared OTUs among the three PM fractions (PM2.5S, PM10S, and TSPS) (A) and the three haze levels (non-, light, and heavy haze) (B).
The analysis of similarity (ANOSIM; R = 0.157, P < 0.01) and principal-component analysis (PCoA) indicated that there was a significant effect of PM fractions on airborne bacterial community composition, especially between PM2.5S and TSPS (Fig. S4A). The linear discriminant analysis effect size (LEfSe) analysis demonstrated significant differences in the various bacterial taxa associated with PM fractions. However, only six genera increased in total across PM10S (Dokdonella, Caenimonas, Geminicoccus, and Sphingopyxis) and PM2.5S (Cellulomonas and Rhizobacter). Using LEfSe, we identified a large number of taxa that were significantly increased in TSPS, such as Paracoccus, Kocuria, and Sphingomonas, indicating that more richness and differences existed in TSPS (Fig. S5).
Airborne bacterial communities with different haze levels.
The Venn diagram presented in Fig. 2B shows that there were 3,565 OTU shared among non-, light-, and heavy-haze days. More unique OTUs were observed in light-haze samples than in non- and heavy-haze samples. Several genera were abundant in samples with all three levels of haze. For example, Anoxybacillus was the most abundant genus in three fractions of samples. Acinetobacter was the second most abundant in light- and heavy-haze samples, while it was the fourth most abundant in nonhazy samples. However, some genera showed higher abundance in specific samples. For instance, Methylophilus was a relatively abundant genus (>3%) in nonhazy samples but showed low abundance (<1%) in both light- and heavy-haze samples (Table S5).
A heatmap diagram (Fig. S3) was used to illustrate the distributions of dominant bacterial genera in PMs collected from various-haze-level days. For example, Streptococcus increased as the haze became heavier; whereas Sphingomonas spp. decreased during heavy-haze days. Methylophilus spp. increased during nonhazy days.
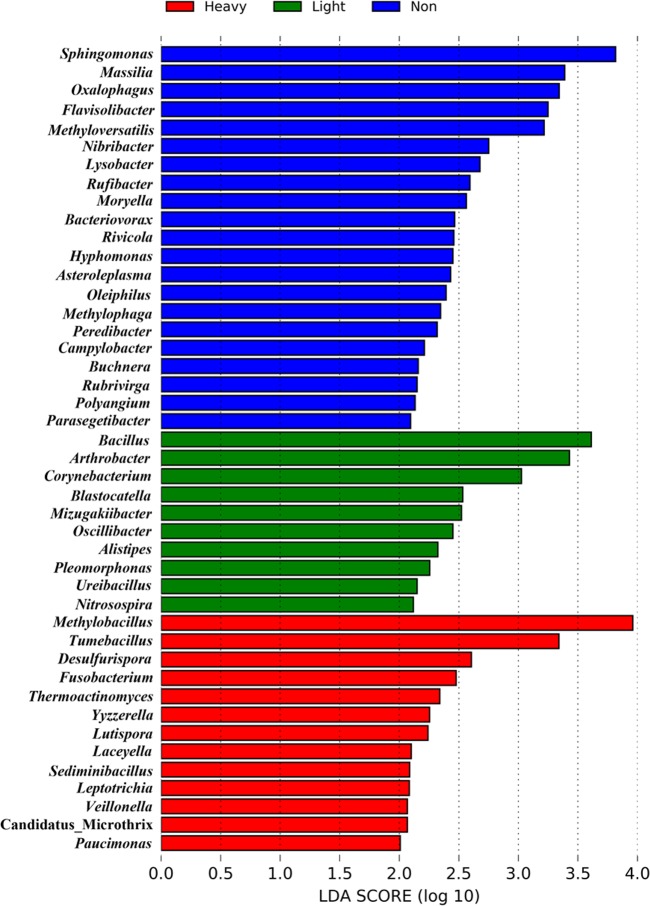
Though there was no significant differences in bacterial communities among non-, light-, and heavy-haze samples (R = 0.017, P > 0.05; Fig. S4B), several genera with significant different relative abundances were observed using LEfSe analysis. We found that Methylobacillus, Tumebacillus, and Desulfurispora increased in heavy-haze samples. Conversely, Sphingomonas, Massilia, and Oxalophagus increased in nonhaze samples (Fig. 3).
FIG 3.
LEfSe analysis illustrating differentially abundant bacterial genera among samples with different haze levels. LDA scores (only those genera that obtain a log LDA score of >2 are ultimately considered) can be interpreted as the degree of consistent difference in relative abundance between genera in non-, light-, and heavy-haze samples.
The effects of environmental factors on the airborne bacterial community.
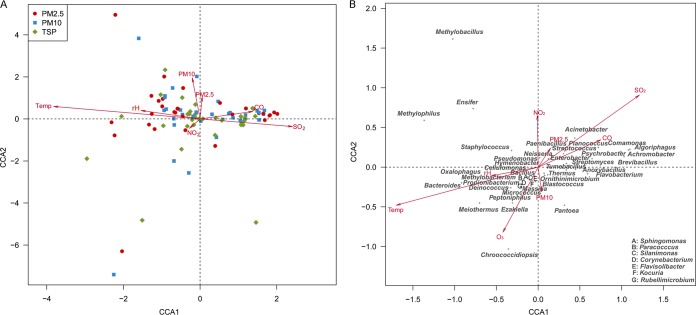
The canonical correspondence analysis (CCA) (Fig. 4A) and permutation test (Table S6) were performed to examine the relationships between the environmental factors and bacterial community composition. Five environmental factors, including temperature (r2 = 0.72, P < 0.01), SO2 concentration (r2 = 0.29, P < 0.01), rH (r2 = 0.12, P < 0.01), PM10 (r2 = 0.1, P < 0.01), and CO concentration (r2 = 0.09, P < 0.05) showed significant correlations with airborne bacterial community composition.
FIG 4.
Canonical correspondence analysis showing the relationships between environmental factors and airborne bacterial community composition (A), and the relationships between environmental factors and dominant genera (B).
Figure 4B shows the distributions of bacterial genera in relation to environmental factors. Genera, such as Methylophilus, Meiothermus, Oxalophagus, Ensifer, and Propionibacterium, were positively correlated with temperature and rH at the lower concentrations of SO2 and CO. In contrast, Algoriphagus, Comamonas, Achromobacter, Brevibacillus, Planococcus, and the two most abundant genera (Acinetobacter and Anoxybacillus) were negatively correlated with temperature and rH at the higher concentrations of SO2 and CO.
Airborne bacterial cooccurrences within the networks.
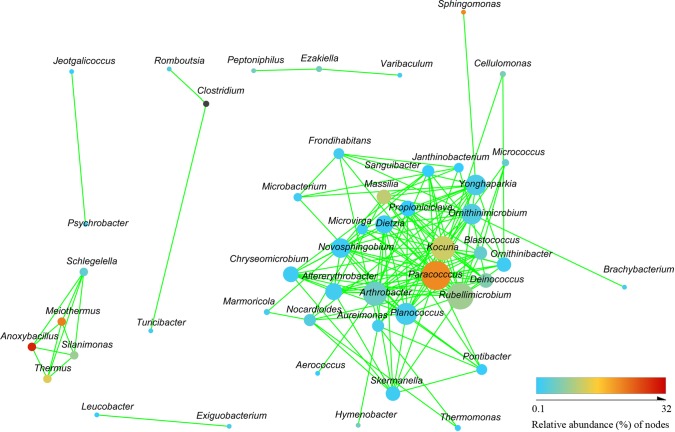
To study the possible interactions among airborne bacteria in the PM samples, the 122 dominant genera (relative abundance, >0.1%) were used to construct a network. In the resulting network, each node is a bacterial genus (Fig. 5), and edges indicate significant cooccurrent (green) or mutual exclusion (red) interactions. The network included 48 nodes and 155 edges. However, all the networks generated had exclusively cooccurrent interactions and never exhibited mutual exclusions. Topological properties are always used in network analysis to describe the complex pattern of interrelationships. The average network distance between all pairs of nodes (average path length) was 1.97. The clustering coefficient (the degree to which they tend to cluster together) was 0.46. The heterogeneity value (the possibility of hubs) of this network was 0.85. Paracoccus (21 edges), Rubellimicrobium (19 edges), and Kocuria (17 edges), which increased in bigger-PM samples, were the key genera in the airborne PM samples. Arthrobacter (17 edges), which increased in light-haze samples, was the key genus.
FIG 5.
Network analysis indicating interactions among dominant bacteria.
DISCUSSION
As a result of our chosen sampling and sequencing method, we observed higher diversity and richness than most previous culture-independent studies in airborne bacterial communities (26, 28–30, 32, 47); however, similar diversity patterns were observed in the surface atmosphere in the United States (29). The increased size of the PM fraction increased the diversity and richness of airborne bacterial communities. In addition, no significant differences in airborne bacterial diversity and richness were observed between hazy and nonhazy days. To our knowledge, no similar studies of airborne bacterial diversity in various haze levels have been reported, as previous studies have largely focused on viable bacterial concentration. Wei et al. (48) reported that heavily polluted areas had similar concentrations of viable bioaerosol particles with near-pristine environments. The PM can act as carriers and supply the nutrition, which leads to increases to the airborne microbial concentration. However, bacteria could also be inactivated by atmospheric pollutants with the increase of PM (49). Thus, the increase in PM may have a variety of effects on the airborne bacterial diversity and richness.
The PM-associated bacterial community composition in Beijing found in this study is qualitatively similar to that identified from other air surveys (28, 34, 35, 50). The high abundance of human-associated Gammaproteobacteria (Acinetobacter, Pseudomonas, etc.) and plant-associated Alphaproteobacteria (Sphingomonas, Paracoccus, etc.) may be because of the features of region in a large city (28, 34). Public places, such as parks, hospitals, and the theater, are not too far from our sampling site, which would strongly contribute to defining the dominant phylum. Spore-forming bacteria, most of which are classified as Firmicutes and Actinobacteria, were also found to be increased in the atmospheric environments of Texas (51), Colorado (50), Hong Kong (27), Milan (34), Antarctica (12), and the Arctic (52). This suggests that these groups could be the result of the selection for spore-forming bacteria during long-range transport (12). Anoxybacillus was the most abundant genus in this study and also dominated the airborne bacterial community in Antarctica (12). Previous studies indicated that Anoxybacillus spp., isolated from hot springs, are thermophilic bacteria and might be highly mobile as spores (12, 53). In addition, Acinetobacter spp. are ubiquitous in soil and water, resistant to dry conditions, and easily transported by air (54). Hervàs et al. (55) proposed Acinetobacter spp. to be indicators of airborne transport from the Sahara. The ability to be transported and retain DNA integrity in the atmosphere may contribute to the dominance of these genera.
The PM fraction influenced the composition of the airborne bacterial community in this study. Various genera were distributed in different PM fractions, probably because of their different sources. In previous studies, road and soil dust, coal combustion, biomass burning, and vehicle exhausts contributed most sources of PM in haze episodes in Beijing (56, 57). Coal combustion and biomass burning were the most important sources of fine particles, and soil dust contributed most to coarse particles (56, 57). For example, one strain of Cellulomonas was isolated from coal mine soil (58), and two strains of Cellulomonas were isolated from medium-added coal tar as the sole source of carbon and energy (59). Therefore, Cellulomonas spp. may be able to live with coal or biomass ashes, which explained the significant increase in Cellulomonas spp. in PM2.5S compared with other fractions. In addition, most genera that significantly increased in coarse particles, such as Dokdonella, Caenimonas, Paracoccus, and Kocuria, were consistently sourced from soil (53). In addition, the features of each bacterial genus may influence the bacterial community in different sizes of PM. We suggest that spore-forming bacteria, such as Anoxybacillus and Bacillus spp., decrease in larger-PM fractions because of the small size of spores. Microorganisms that can easily aggregate will form larger PM. For example, certain species of Sphingomonas can aggregate by polar fimbriae (53), which may increase Sphingomonas spp. in TSPS. Furthermore, pathogens, allergens, and toxins in fine-PM fractions are more likely to penetrate deeper into the human lung, triggering serious diseases (60). In particular, potential opportunistic pathogens were also observed in these samples. Pseudomonas and Acinetobacter spp. were in great abundance in PM2.5S; thus, these genera are known to have members that are potential pathogens of respiratory tract disease (61, 62).
Compared to large bacteria, small bacteria, such as spores, can easily float in the air and increase in concentration during haze events, perhaps because of low wind speed on hazy days. Moreover, spore-forming bacteria with high resistance to pollution can also survive in harsh environments, such as surfaces of soil, rocks, and plants, and are the origin of airborne bacteria. This may explain why Methylobacillus and Tumebacillus spp. are rich during hazy days. In addition, previous studies have reported that most of the haze pollution in Beijing originates from traffic, coal burning, biomass burning, and cooking (44, 63). Therefore, the thermophilic sulfate-reducing bacterium Desulfurispora (64), the numbers of which increased on hazy days, may retain intact DNA in extreme hot experiments, such as coal burning areas. Sphingomonas spp., which were common in previous studies on airborne bacterial communities (55, 65–67) and decreased in hazy days in this study, are sensitive to pollution. It is therefore difficult to retain DNA integrity in samples, possibly because these bacteria are non-spore forming. Though several taxa were found to be related to haze levels, no significant difference was observed in the bacterial communities between hazy and nonhazy samples. In contrast, we observed a significant difference in the fungal community between hazy and nonhazy samples in our previous study (68). We speculate that fungi, which are than bacteria, cannot be easily suspended in the air during hazy days because of the weak convection.
Based on our results, temperature had the highest correlation with bacterial community composition, which is similar to the results from previous studies (16, 18, 21, 22). According to Alghamdi et al. (18), temperature may help enhance the chemical reactions of PM to form more toxic compounds. In addition, the effect of temperature could also be interpreted as having an indirect effect on microbial growth on the surfaces of road, soil, and plants, as it has a drying effect and is a factor in favor of aerosolization of settled particles influencing the structure of bacterial community. PM10 is significantly correlated with bacterial community composition, which indicates that PM10 might influence the colonization of the bacteria. The increase of the PM concentration may heighten the opportunity for the colonization of the bacteria on the surface of rocks, urban roads, and so on. In addition, there are many other factors influencing the bacterial community in the atmosphere, such as wind speed and sunlight duration. Therefore, airborne bacterial communities should be surveyed in the long term, and their relationships with environmental factors require further study.
Cooccurrence patterns identified using network analysis can reflect the interactions between microbes (69). Similar methods have been applied to study bacterium-bacterium (70, 71), bacteriophage-bacterium (65), and bacterium-fungus (66) interactions. Mutual exclusions in the network in the current study were not found, suggesting a positive nature among microbes in the airborne PM samples. The network properties offer the possibility for quick and easy comparisons among complex data sets from different ecosystem types to explore how the general traits of a certain habitat type may influence the composition of microbial communities (69). We calculated the average path length for the network, and the value was 1.97, indicating that approximately two edges separate average nodes. This value was lower than those in environmental interactions in marine networks (bacteria and archaea, 3.05; bacteria, protist, and virus, 3.00; and bacteria and phage, 3.10) (65, 67, 72) and soil networks (bacteria, 5.53) (69). Hubs are thought to describe key nodes that, if removed, would lead to large changes in the community (67). Thus, key genera might be Paracoccus, Rubellimicrobium, Kocuria, and Arthrobacter. The four genera that were rich in the samples of larger-PM fractions may have a characteristic in which they gather other bacteria to form bigger PM.
In summary, the results presented here represent an analysis of the airborne bacterial community associated with PM and advance our understanding of the diversity and composition of these communities. We observed the shift in bacterial community composition with PM fraction sizes, but no significant difference was found among haze levels. Our results also indicated that multiple factors influenced the airborne bacterial community composition. More attention should be given to airborne bacteria and their role in the atmosphere. Metagenomics and metatranscriptomics will provide further details on the potential and actual functions of airborne bacteria with higher accuracy; however, analysis of such data remains technically challenging. Consequently, an accurate real-time technique for survey of airborne bacteria needs to be developed in future.
MATERIALS AND METHODS
Sample collection.
PM2.5 samples (PM2.5S; samples containing only PM smaller than 2.5 μm), PM10 samples (PM10S; samples containing only PM smaller than 10 μm), and TSP samples (TSPS; samples containing total suspended particulates) were collected from the roof of the Conference Building at Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences (39°52′43″N, 116°23′21″E, ∼8 m above the ground, ∼400 m from Temple of Heaven Park), an area without major industrial pollution sources nearby. We collected a total of 96 PM samples over 32 days (collecting only one sample per day for each PM fraction, which represents 32 samples for each PM fraction [i.e., 96 in total for all 3]) within the period of September 2014 to February 2015 during different haze levels (Table S1). Sampling was conducted by portable ambient air samplers (AirMetrics, USA), with one sampler each for PM2.5S, PM10S, and TSPS (three samplers in total). The samplers were positioned at a distance of 5 m from each other and operated at the same time. Ambient air was drawn at an average flow rate of 5 liters/min for 24 h per sampling day.
The PM samples were collected on 47-mm quartz aerosol collection filters (Pall, USA). Prior to sampling, all the filters were sterilized by autoclaving at 121°C for 20 min. The filter holders of the samplers were cleaned with 75% ethanol, and all the tools used for changing new filters were autoclaved every day to avoid contamination. After sampling, the filters were kept in a 2-ml centrifuge tube after cutting into small pieces with scissors and stored at −20°C (68).
The air quality index (AQI) (73), was used to indicate the haze level. We defined a day with AQI lower than 100 as a nonhazy day, that with an AQI in the range of 100 to 200 as a light-haze day, and that with an AQI higher than 200 as a heavy-haze day.
Environmental factors, such as concentrations of PM2.5, PM10, CO, SO2, and NO2, were recorded every 6 h in each sampling day, and the average value was used as the data set for analysis from the monitoring data of Temple of Heaven Park site (∼800 m from the sampling site) of the Beijing Municipal Environmental Monitoring Center (http://zx.bjmemc.com.cn/). Temperature (temp) and relative humidity (rH) were recorded according to the reports of the Chinese National Meteorological Center (http://www.nmc.cn/). The environmental factors are listed in Table S2.
DNA extraction and PCR amplification.
Sample filters in the 2-ml centrifuge tubes were removed from the fridge, and the sample filters were directly transferred into PowerBead tubes (Mo Bio, USA). The samples were then heated to 65°C for 10 min, followed by vortexing for 10 min. The remaining steps of the DNA extraction were performed according to the PowerSoil DNA isolation protocol. The V3–V4 region of the bacterial rRNA gene was amplified by PCR (95°C for 3 min, followed by 27 cycles at 95°C for 30 s, 55°C for 40 s, and 72°C for 45 s, and a final extension at 72°C for 10 min) using primers 338F (5′-barcode-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (74). PCR amplification was conducted using high-fidelity TransStart FastPfu DNA polymerase (Transgen, China). The PCRs were performed in triplicate in 20-μl mixtures containing 4 μl of 5× FastPfu buffer, 2 μl of 2.5 mM dinucleoside triphosphates (dNTPs), 0.8 μl of each primer (5 μM), 0.4 μl of FastPfu polymerase, and 2 to 4 μl (according to the concentration of DNA) of template DNA.
Illumina MiSeq sequencing.
Amplicons were purified using the AxyPrep DNA gel extraction kit (Axygen, USA), according to the manufacturer's instructions, and quantified using QuantiFluor-ST (Promega, USA). Purified amplicons were pooled in equimolar amounts and paired-end sequenced (2 × 250 bp) on an Illumina MiSeq platform, according to the standard protocols.
Processing of sequencing data.
Raw read sequences were demultiplexed and quality filtered using QIIME (version 1.8.0) (75), with the following criteria: (i) the 300-bp reads were truncated at any site receiving an average quality score of <20 over a 50-bp sliding window, discarding the truncated reads that were shorter than 50 bp; (ii) exact barcode matching, a two-nucleotide mismatch in primer matching, with reads containing ambiguous characters being removed; and (iii) only paired-end reads with overlap longer than 10 bp were assembled according to their overlap sequence. Reads that could not be assembled were discarded.
Statistical analyses.
All samples were subsampled at same sequence depth (17,782 reads). Operational taxonomic units (OTUs) were clustered with 97% similarity cutoff using UPARSE (version 7.1). Chimeric sequences were identified and removed using UCHIME. The OTUs were used as a basis for calculating α-diversity and β-diversity metrics. The taxonomy of each 16S rRNA gene sequence was analyzed using the RDP Classifier (76) against the SILVA (SSU115) 16S rRNA database using a confidence threshold of 70% (77). Statistical analyses of the OTU diversity of each air sample (Chao1 index, Good's coverage estimator, and Shannon's index) were performed using the QIIME 1.8.0 software (75). Statistical comparisons of Chao1 and Shannon indices among different samples were made by one-way analysis of variance (ANOVA) using the Statistical Package for the Social Sciences (SPSS, version 17.0). An analysis of similarity (ANOSIM) was performed using the QIIME 1.8.0 software to determine whether various fractions of PM samples and haze levels had significantly different bacterial community compositions. Bar plots, Venn diagrams, canonical correspondence analysis (CCA), principal-coordinate analysis (PCoA), and heatmaps were created in the R software (version 3.7.0). Linear discriminant analysis effect size (LEfSe; https://huttenhower.sph.harvard.edu/galaxy/) was applied to identify differentially abundant bacterial taxa among samples with different PM fractions and haze levels (78). Only those taxa that obtained a log linear discriminant analysis (LDA) score of >2 were ultimately considered. The network was generated using the CoNet plugin (version 1.0b7) for Cytoscape (version 3.6.0) on the basis of the nonparametric Spearman correlation coefficients, with a minimal cutoff threshold ρ of 0.6 (P < 0.01, Bonferroni corrected) (79, 80). We present correlation data for dominant genera (relative abundance, >0.1%) that were detected in airborne bacteria.
Accession number(s).
The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database under accession number SRP068265.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the National Major New Drug Creation Program (grants 2014ZX09201001-011 and 2018ZX09711001-007-001), CAMS Innovation Fund for Medical Science (grants 2016-I2M-2-002 and 2016-I2M-2-003), the National Infrastructure of Microbial Resources (grant NIMR-2018-3), the National Natural Science Foundation of China (NSFC) (grants 81373452 and 81621064), and the 863 Program (grant 2014AA021504). Li-Yan Yu is supported by Xiehe Scholar.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00004-18.
REFERENCES
- 1.Van Dingenen R, Raes F, Putaud J-P, Baltensperger U, Charron A, Facchini MC, Decesari S, Fuzzi S, Gehrig R, Hansson H-C, Harrison RM, Hüglin C, Jones AM, Laj P, Lorbeer G, Maenhaut W, Palmgren F, Querol X, Rodriguez S, Schneider J, ten Brink H, Tunved P, Tørseth K, Wehner B, Weingartner E, Wiedensohler A, Wåhlin P. 2004. A European aerosol phenomenology—1: physical characteristics of particulate matter at kerbside, urban, rural and background sites in Europe. Atmos Environ 38:2561–2577. doi: 10.1016/j.atmosenv.2004.01.040. [DOI] [Google Scholar]
- 2.Querol X, Alastuey A, Ruiz CR, Artiñano B, Hansson HC, Harrison RM, Buringh E, ten Brink HM, Lutz M, Bruckmann P, Straehl P, Schneider J. 2004. Speciation and origin of PM10 and PM2.5 in selected European cities. Atmos Environ 38:6547–6555. doi: 10.1016/j.atmosenv.2004.08.037. [DOI] [Google Scholar]
- 3.Wang L, Xu J, Yang J, Zhao X, Wei W, Cheng D, Pan X, Su J. 2012. Understanding haze pollution over the southern Hebei area of China using the CMAQ model. Atmos Environ 56:69–79. doi: 10.1016/j.atmosenv.2012.04.013. [DOI] [Google Scholar]
- 4.Shimadera H, Hayami H, Morino Y, Ohara T, Chatani S, Hasegawa S, Kaneyasu N. 2013. Analysis of summertime atmospheric transport of fine particulate matter in Northeast Asia. Asia-Pac. J Atmos Sci 49:347–360. doi: 10.1007/s13143-013-0033-y. [DOI] [Google Scholar]
- 5.Booth BB, Dunstone NJ, Halloran PR, Andrews T, Bellouin N. 2012. Aerosols implicated as a prime driver of twentieth-century North Atlantic climate variability. Nature 484:228–232. doi: 10.1038/nature10946. [DOI] [PubMed] [Google Scholar]
- 6.Randles CA, Colarco PR, da Silva A. 2013. Direct and semi-direct aerosol effects in the NASA GEOS-5 AGCM: aerosol-climate interactions due to prognostic versus prescribed aerosols. J Geophys Res 118:149–169. doi: 10.1029/2012JD018388. [DOI] [Google Scholar]
- 7.Dowd S. 2000. Aeromicrobiology, p 91–122. In Maier RM, Pepper IL, Gerba CP (ed), Environmental microbiology. Academic Press, San Diego, CA. [Google Scholar]
- 8.Zhang WJ, Sun YL, Zhuang GS, Xu DQ. 2006. Characteristics and seasonal variations of PM2.5, PM10, and TSP aerosol in Beijing. Biomed Environ Sci 19:461. [PubMed] [Google Scholar]
- 9.Andrew H, Abraham JL, Bret J, Berry CL. 2003. Toxicologic and epidemiologic clues from the characterization of the 1952 London smog fine particulate matter in archival autopsy lung tissues. Environ Health Perspect 111:1209–1214. doi: 10.1289/ehp.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pope CA III, Ezzati M, Dockery DW. 2009. Fine-particulate air pollution and life expectancy in the United States. N Engl J Med 2009:376–386. doi: 10.1056/NEJMsa0805646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalogerakis N, Paschali D, Lekaditis V, Pantidou A, Eleftheriadis K, Lazaridis M. 2005. Indoor air quality—bioaerosol measurements in domestic and office premises. J Aerosol Sci 36:751–761. doi: 10.1016/j.jaerosci.2005.02.004. [DOI] [Google Scholar]
- 12.Bottos EM, Woo AC, Zawar-Reza P, Pointing SB, Cary SC. 2014. Airborne bacterial populations above desert soils of the McMurdo Dry Valleys, Antarctica. Microb Ecol 67:120–128. doi: 10.1007/s00248-013-0296-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burrows SM, Butler T, Jöckel P, Tost H, Kerkweg A, Pöschl U, Lawrence MG. 2009. Bacteria in the global atmosphere–part 2: modeling of emissions and transport between different ecosystems. Atmos Chem Phys 9:9281–9297. doi: 10.5194/acp-9-9281-2009. [DOI] [Google Scholar]
- 14.Kim KY, Kim YS, Kim D, Kim HT. 2011. Exposure level and distribution characteristics of airborne bacteria and fungi in Seoul metropolitan subway stations. Ind Health 49:242–248. doi: 10.2486/indhealth.MS1199. [DOI] [PubMed] [Google Scholar]
- 15.Raisi L, Aleksandropoulou V, Lazaridis M, Katsivela E. 2012. Size distribution of viable, cultivable, airborne microbes and their relationship to particulate matter concentrations and meteorological conditions in a Mediterranean site. Aerobiologia 29:233–248. doi: 10.1007/s10453-012-9276-9. [DOI] [Google Scholar]
- 16.Haas D, Galler H, Luxner J, Zarfel G, Buzina W, Friedl H, Marth E, Habib J, Reinthaler FF. 2013. The concentrations of culturable microorganisms in relation to particulate matter in urban air. Atmos Environ 65:215–222. doi: 10.1016/j.atmosenv.2012.10.031. [DOI] [Google Scholar]
- 17.Fang Z. 2014. Characteristic and concentration distribution of culturable airborne bacteria in residential environments in Beijing, China. Aerosol Air Qual Res 14:943–953. doi: 10.4209/aaqr.2013.04.0109. [DOI] [Google Scholar]
- 18.Alghamdi MA, Shamy M, Redal MA, Khoder M, Awad AH, Elserougy S. 2014. Microorganisms associated particulate matter: a preliminary study. Sci Total Environ 479–480:109–116. doi: 10.1016/j.scitotenv.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Fu H, Wang W, Liu J, Meng Q, Wang W. 2015. Characteristics of bacterial and fungal aerosols during the autumn haze days in Xi'an, China. Atmos Environ 122:439–447. doi: 10.1016/j.atmosenv.2015.09.070. [DOI] [Google Scholar]
- 20.Oh HJ, Jeong NN, Chi WB, Seo JH, Jun SM, Sohn JR. 2015. Characterization of particulate matter concentrations and bioaerosol on each floor at a building in Seoul, Korea. Environ Sci Pollut Res Int 22:16040–16050. doi: 10.1007/s11356-015-4810-2. [DOI] [PubMed] [Google Scholar]
- 21.Niazi S, Hassanvand MS, Mahvi AH, Nabizadeh R, Alimohammadi M, Nabavi S, Faridi S, Dehghani A, Hoseini M, Moradi-Joo M, Mokamel A, Kashani H, Yarali N, Yunesian M. 2015. Assessment of bioaerosol contamination (bacteria and fungi) in the largest urban wastewater treatment plant in the Middle East. Environ Sci Pollut Res Int 22:16014–16021. doi: 10.1007/s11356-015-4793-z. [DOI] [PubMed] [Google Scholar]
- 22.Dong L, Qi J, Shao C, Zhong X, Gao D, Cao W, Gao J, Bai R, Long G, Chu C. 2016. Concentration and size distribution of total airborne microbes in hazy and foggy weather. Sci Total Environ 541:1011–1018. doi: 10.1016/j.scitotenv.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Yuan H, Zhang D, Shi Y, Li B, Yang J, Yu X, Chen N, Kakikawa M. 2017. Cell concentration, viability and culture composition of airborne bacteria during a dust event in Beijing. J Environ Sci (China) 55:33–40. doi: 10.1016/j.jes.2016.03.033. [DOI] [PubMed] [Google Scholar]
- 24.Okubo T, Osaki T, Nozaki E, Uemura A, Sakai K, Matushita M, Matsuo J, Nakamura S, Kamiya S, Yamaguchi H. 2017. Walker occupancy has an impact on changing airborne bacterial communities in an underground pedestrian space, as small-dust particles increased with raising both temperature and humidity. PLoS One 12:e0184980. doi: 10.1371/journal.pone.0184980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao C, Jiang W, Wang B, Fang J, Lang J, Tian G, Jiang J, Zhu TF. 2014. Inhalable microorganisms in Beijing's PM2.5 and PM10 pollutants during a severe smog event. Environ Sci Technol 48:1499–1507. doi: 10.1021/es4048472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei K, Zou Z, Zheng Y, Li J, Shen F, Wu CY, Wu Y, Hu M, Yao M. 2016. Ambient bioaerosol particle dynamics observed during haze and sunny days in Beijing. Sci Total Environ 550:751–759. doi: 10.1016/j.scitotenv.2016.01.137. [DOI] [PubMed] [Google Scholar]
- 27.Woo AC, Brar MS, Chan Y, Lau MCY, Leung FCC, Scott JA, Vrijmoed LLP, Zawar-Reza P, Pointing SB. 2013. Temporal variation in airborne microbial populations and microbially-derived allergens in a tropical urban landscape. Atmos Environ 74:291–300. doi: 10.1016/j.atmosenv.2013.03.047. [DOI] [Google Scholar]
- 28.Bowers RM, Sullivan AP, Costello EK, Collett JL Jr, Knight R, Fierer N. 2011. Sources of bacteria in outdoor air across cities in the midwestern United States. Appl Environ Microbiol 77:6350–6356. doi: 10.1128/AEM.05498-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowers RM, Clements N, Emerson JB, Wiedinmyer C, Hannigan MP, Fierer N. 2013. Seasonal variability in bacterial and fungal diversity of the near-surface atmosphere. Environ Sci Technol 47:12097–12106. doi: 10.1021/es402970s. [DOI] [PubMed] [Google Scholar]
- 30.Be NA, Thissen JB, Fofanov VY, Allen JE, Rojas M, Golovko G, Fofanov Y, Koshinsky H, Jaing CJ. 2015. Metagenomic analysis of the airborne environment in urban spaces. Microb Ecol 69:346–355. doi: 10.1007/s00248-014-0517-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emerson JB, Keady PB, Brewer TE, Clements N, Morgan EE, Awerbuch J, Miller SL, Fierer N. 2015. Impacts of flood damage on airborne bacteria and fungi in homes after the 2013 Colorado Front Range flood. Environ Sci Technol 49:2675–2684. doi: 10.1021/es503845j. [DOI] [PubMed] [Google Scholar]
- 32.Bowers RM, McLetchie S, Knight R, Fierer N. 2011. Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. ISME J 5:601–612. doi: 10.1038/ismej.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertolini V, Gandolfi I, Ambrosini R, Bestetti G, Innocente E, Rampazzo G, Franzetti A. 2013. Temporal variability and effect of environmental variables on airborne bacterial communities in an urban area of Northern Italy. Appl Microbiol Biotechnol 97:6561–6570. doi: 10.1007/s00253-012-4450-0. [DOI] [PubMed] [Google Scholar]
- 34.Franzetti A, Gandolfi I, Gaspari E, Ambrosini R, Bestetti G. 2011. Seasonal variability of bacteria in fine and coarse urban air particulate matter. Appl Microbiol Biotechnol 90:745–753. doi: 10.1007/s00253-010-3048-7. [DOI] [PubMed] [Google Scholar]
- 35.Barberán A, Henley J, Fierer N, Casamayor EO. 2014. Structure, inter-annual recurrence, and global-scale connectivity of airborne microbial communities. Sci Total Environ 487:187–195. doi: 10.1016/j.scitotenv.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 36.Sun Y, Zhuang G, Tang AA, Wang Y, An Z. 2006. Chemical characteristics of PM2.5 and PM10 in haze-fog episodes in Beijing. Environ Sci Technol 40:3148–3155. doi: 10.1021/es051533g. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Mauzerall DL, Zhu T, Liang S, Ezzati M, Remais JV. 2010. Environmental health in China: progress towards clean air and safe water. Lancet 375:1110–1119. doi: 10.1016/S0140-6736(10)60062-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Q, He K, Huo H. 2012. Policy: cleaning China's air. Nature 484:161–162. [DOI] [PubMed] [Google Scholar]
- 39.Cheng Z, Jiang J, Fajardo O, Wang S, Hao J. 2013. Characteristics and health impacts of particulate matter pollution in China (2001–2011). Atmos Environ 65:186–194. doi: 10.1016/j.atmosenv.2012.10.022. [DOI] [Google Scholar]
- 40.Gao M, Jia R, Qiu T, Han M, Song Y, Wang X. 2015. Seasonal size distribution of airborne culturable bacteria and fungi and preliminary estimation of their deposition in human lungs during non-haze and haze days. Atmos Environ 118:203–210. doi: 10.1016/j.atmosenv.2015.08.004. [DOI] [Google Scholar]
- 41.Xiangde XU, Shi X, Zhang S, Ding G, Miao Q, Li Z. 2006. Aerosol influence domain of Beijing and peripheral city agglomeration and its climatic effect. Sci Bull 51:2016–2026. doi: 10.1007/s11434-006-2066-4. [DOI] [Google Scholar]
- 42.Zheng B, Zhang Q, Zhang Y, He KB, Wang K, Zheng GJ, Duan FK, Ma YL, Kimoto T. 2015. Heterogeneous chemistry: a mechanism missing in current models to explain secondary inorganic aerosol formation during the January 2013 haze episode in North China. Atmos Chem Phys 14:2031–2049. doi: 10.5194/acp-15-2031-2015. [DOI] [Google Scholar]
- 43.Ge BZ, Xu XB, Lin WL, Li J, Wang ZF. 2012. Impact of the regional transport of urban Beijing pollutants on downwind areas in summer: ozone production efficiency analysis, Tellus B Chem Phys Meteorol 64:1–13. [Google Scholar]
- 44.Huang RJ, Zhang Y, Bozzetti C, Ho KF, Cao JJ, Han Y, Daellenbach KR, Slowik JG, Platt SM, Canonaco F, Zotter P, Wolf R, Pieber SM, Bruns EA, Crippa M, Ciarelli G, Piazzalunga A, Schwikowski M, Abbaszade G, Schnelle-Kreis J, Zimmermann R, An Z, Szidat S, Baltensperger U, El Haddad I, Prevot AS. 2014. High secondary aerosol contribution to particulate pollution during haze events in China. Nature 514:218–222. doi: 10.1038/nature13774. [DOI] [PubMed] [Google Scholar]
- 45.Quan J, Tie X, Zhang Q, Liu Q, Li X, Gao Y, Zhao D. 2014. Characteristics of heavy aerosol pollution during the 2012-2013 winter in Beijing, China. Atmos Environ 88:83–89. doi: 10.1016/j.atmosenv.2014.01.058. [DOI] [Google Scholar]
- 46.Yang H, Chen J, Wen J, Tian H, Liu X. 2016. Composition and sources of PM2.5 around the heating periods of 2013 and 2014 in Beijing: implications for efficient mitigation measures. Atmos Environ 124:378–386. doi: 10.1016/j.atmosenv.2015.05.015. [DOI] [Google Scholar]
- 47.DeLeon-Rodriguez N, Lathem TL, Rodriguez RL, Barazesh JM, Anderson BE, Beyersdorf AJ, Ziemba LD, Bergin M, Nenes A, Konstantinidis KT. 2013. Microbiome of the upper troposphere: species composition and prevalence, effects of tropical storms, and atmospheric implications. Proc Natl Acad Sci U S A 110:2575–2580. doi: 10.1073/pnas.1212089110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei K, Zheng Y, Li J, Shen F, Zou Z, Fan H, Li X, Wu C-Y, Yao M. 2015. Microbial aerosol characteristics in highly polluted and near-pristine environments featuring different climatic conditions. Sci Bull 60:1439–1447. doi: 10.1007/s11434-015-0868-y. [DOI] [Google Scholar]
- 49.Vaitilingom M, Deguillaume L, Vinatier V, Sancelme M, Amato P, Chaumerliac N, Delort AM. 2013. Potential impact of microbial activity on the oxidant capacity and organic carbon budget in clouds. Proc Natl Acad Sci U S A 110:559–564. doi: 10.1073/pnas.1205743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fierer N, Liu Z, Rodriguez-Hernandez M, Knight R, Henn M, Hernandez MT. 2008. Short-term temporal variability in airborne bacterial and fungal populations. Appl Environ Microbiol 74:200–207. doi: 10.1128/AEM.01467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brodie EL, DeSantis TZ, Parker JPM, Zubietta IX, Piceno YM, Andersen GL. 2007. Urban aerosols harbor diverse and dynamic bacterial populations. Proc Natl Acad Sci U S A 104:299–304. doi: 10.1073/pnas.0608255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harding T, Jungblut AD, Lovejoy C, Vincent WF. 2011. Microbes in high arctic snow and implications for the cold biosphere. Appl Environ Microbiol 77:3234–3243. doi: 10.1128/AEM.02611-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garrity G, Vos PD, Jones D, Kreig N, Ludwig W, Rainey FA, Schleifer KH, Whitman WB. 2009. Bergey's manual of systematic bacteriology, 2nd ed Volume 3: the Firmicutes. Springer, New York, NY. [Google Scholar]
- 54.Bergogne-Bérézin E, Towner KJ. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev 9:148–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hervàs A, Camarero L, Reche I, Casamayor EO. 2009. Viability and potential for immigration of airborne bacteria from Africa that reach high mountain lakes in Europe. Environ Microbiol 11:1612–1623. doi: 10.1111/j.1462-2920.2009.01926.x. [DOI] [PubMed] [Google Scholar]
- 56.Tan J, Duan J, Zhen N, He K, Hao J. 2016. Chemical characteristics and source of size-fractionated atmospheric particle in haze episode in Beijing. Atmos Res 167:24–33. doi: 10.1016/j.atmosres.2015.06.015. [DOI] [Google Scholar]
- 57.Gao J, Tian H, Cheng K, Lu L, Zheng M, Wang S, Hao J, Wang K, Hua S, Zhu C, Wang Y. 2015. The variation of chemical characteristics of PM2.5 and PM10 and formation causes during two haze pollution events in urban Beijing, China. Atmos Environ 107:1–8. doi: 10.1016/j.atmosenv.2015.02.022. [DOI] [Google Scholar]
- 58.Shi Z, Luo G, Wang G. 2012. Cellulomonas carbonis sp. nov., isolated from coal mine soil. Int J Syst Evol Microbiol 62:2004–2010. doi: 10.1099/ijs.0.034934-0. [DOI] [PubMed] [Google Scholar]
- 59.Andreolli M, Lampis S, Brignoli P, Vallini G. 2015. Bioaugmentation and biostimulation as strategies for the bioremediation of a burned woodland soil contaminated by toxic hydrocarbons: a comparative study. J Environ Manage 153:121–131. doi: 10.1016/j.jenvman.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 60.Brook RD, Franklin B, Cascio W, Hong Y, Howard G, Lipsett M, Luepker R, Mittleman M, Samet J, Smith SC Jr, Tager I, Expert Panel on Population and Prevention Science of the American Heart Association. 2004. Air pollution and cardiovascular disease: a statement for healthcare professionals from the Expert Panel on Population and Prevention Science of the American Heart Association. Circulation 109:2655–2671. doi: 10.1161/01.CIR.0000128587.30041.C8. [DOI] [PubMed] [Google Scholar]
- 61.Bjarnsholt T, Jensen PO, Fiandaca MJ, Pedersen J, Hansen CR, Andersen CB, Pressler T, Givskov M, Hoiby N. 2009. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr Pulmonol 44:547–558. doi: 10.1002/ppul.21011. [DOI] [PubMed] [Google Scholar]
- 62.Forster DH, Daschner FD. 1998. Acinetobacter species as nosocomial pathogens. Eur J Clin Microbiol 17:73–77. doi: 10.1007/BF01682159. [DOI] [PubMed] [Google Scholar]
- 63.Sun Y, Jiang Q, Wang Z, Fu P, Li J, Yang T, Yin Y. 2014. Investigation of the sources and evolution processes of severe haze pollution in Beijing in January 2013. J Geophys Res 119:4380–4398. doi: 10.1002/2014JD021641. [DOI] [Google Scholar]
- 64.Kaksonen AH, Spring S, Schumann P, Kroppenstedt RM, Puhakka JA. 2007. Desulfurispora thermophila gen. nov., sp. nov., a thermophilic, spore-forming sulfate-reducer isolated from a sulfidogenic fluidized-bed reactor. Int J Syst Evol Microbiol 57:1089–1094. doi: 10.1099/ijs.0.64593-0. [DOI] [PubMed] [Google Scholar]
- 65.Soffer N, Zaneveld J, Vega Thurber R. 2015. Phage-bacteria network analysis and its implication for the understanding of coral disease. Environ Microbiol 17:1203–1218. doi: 10.1111/1462-2920.12553. [DOI] [PubMed] [Google Scholar]
- 66.de Menezes AB, Prendergast-Miller MT, Richardson AE, Toscas P, Farrell M, Macdonald LM, Baker G, Wark T, Thrall PH. 2015. Network analysis reveals that bacteria and fungi form modules that correlate independently with soil parameters. Environ Microbiol 17:2677–2689. doi: 10.1111/1462-2920.12559. [DOI] [PubMed] [Google Scholar]
- 67.Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY, Chow CE, Sachdeva R, Jones AC, Schwalbach MS, Rose JM, Hewson I, Patel A, Sun F, Caron DA, Fuhrman JA. 2011. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J 5:1414–1425. doi: 10.1038/ismej.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yan D, Zhang T, Su J, Zhao LL, Wang H, Fang XM, Zhang YQ, Liu HY, Yu LY. 2016. Diversity and composition of airborne fungal community associated with particulate matters in Beijing during haze and non-haze days. Front Microbiol 7:487. doi: 10.3389/fmicb.2016.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barberán A, Bates ST, Casamayor EO, Fierer N. 2012. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J 6:343–351. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Faust K, Raes J. 2012. Microbial interactions: from networks to models. Nat Rev Microbiol 10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 71.Rodriguez-Lanetty M, Granados-Cifuentes C, Barberan A, Bellantuono AJ, Bastidas C. 2013. Ecological inferences from a deep screening of the complex bacterial consortia associated with the coral, Porites astreoides. Mol Ecol 22:4349–4362. doi: 10.1111/mec.12392. [DOI] [PubMed] [Google Scholar]
- 72.Chow CE, Kim DY, Sachdeva R, Caron DA, Fuhrman JA. 2014. Top-down controls on bacterial community structure: microbial network analysis of bacteria, T4-like viruses and protists. ISME J 8:816–829. doi: 10.1038/ismej.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y, Zhuang G, Sun Y, An Z. 2006. The variation of characteristics and formation mechanisms of aerosols in dust, haze, and clear days in Beijing. Atmos Environ 40:6579–6591. doi: 10.1016/j.atmosenv.2006.05.066. [DOI] [Google Scholar]
- 74.Huws SA, Edwards JE, Kim EJ, Scollan ND. 2007. Specificity and sensitivity of eubacterial primers utilized for molecular profiling of bacteria within complex microbial ecosystems. J Microbiol Methods 70:565–569. doi: 10.1016/j.mimet.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 75.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Amato KR, Yeoman CJ, Kent A, Righini N, Carbonero F, Estrada A, Gaskins HR, Stumpf RM, Yildirim S, Torralba M, Gillis M, Wilson BA, Nelson KE, White BA, Leigh SR. 2013. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J 7:1344–1353. doi: 10.1038/ismej.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C. 2012. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol 8:e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T. 2012. A travel guide to Cytoscape plugins. Nat Methods 9:1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.