ABSTRACT
Leptothrix ochracea is known for producing large volumes of iron oxyhydroxide sheaths that alter wetland biogeochemistry. For over a century, these delicate structures have fascinated microbiologists and geoscientists. Because L. ochracea still resists long-term in vitro culture, the debate regarding its metabolic classification dates back to 1885. We developed a novel culturing technique for L. ochracea using in situ natural waters and coupled this with single-cell genomics and nanoscale secondary-ion mass spectrophotometry (nanoSIMS) to probe L. ochracea's physiology. In microslide cultures L. ochracea doubled every 5.7 h and had an absolute growth requirement for ferrous iron, the genomic capacity for iron oxidation, and a branched electron transport chain with cytochromes putatively involved in lithotrophic iron oxidation. Additionally, its genome encoded several electron transport chain proteins, including a molybdopterin alternative complex III (ACIII), a cytochrome bd oxidase reductase, and several terminal oxidase genes. L. ochracea contained two key autotrophic proteins in the Calvin-Benson-Bassham cycle, a form II ribulose bisphosphate carboxylase, and a phosphoribulose kinase. L. ochracea also assimilated bicarbonate, although calculations suggest that bicarbonate assimilation is a small fraction of its total carbon assimilation. Finally, L. ochracea's fundamental physiology is a hybrid of those of the chemolithotrophic Gallionella-type iron-oxidizing bacteria and the sheathed, heterotrophic filamentous metal-oxidizing bacteria of the Leptothrix-Sphaerotilus genera. This allows L. ochracea to inhabit a unique niche within the neutrophilic iron seeps.
IMPORTANCE Leptothrix ochracea was one of three groups of organisms that Sergei Winogradsky used in the 1880s to develop his hypothesis on chemolithotrophy. L. ochracea continues to resist cultivation and appears to have an absolute requirement for organic-rich waters, suggesting that its true physiology remains unknown. Further, L. ochracea is an ecological engineer; a few L. ochracea cells can generate prodigious volumes of iron oxyhydroxides, changing the ecosystem's geochemistry and ecology. Therefore, to determine L. ochracea's basic physiology, we employed new single-cell techniques to demonstrate that L. ochracea oxidizes iron to generate energy and, despite having predicted genes for autotrophic growth, assimilates a fraction of the total CO2 that autotrophs do. Although not a true chemolithoautotroph, L. ochracea's physiological strategy allows it to be flexible and to extensively colonize iron-rich wetlands.
KEYWORDS: Leptothrix, biofouling, chemolithotrophy, iron-oxidizing bacteria, mixotrophs
INTRODUCTION
In iron-rich systems, just a relatively few Leptothrix ochracea cells can generate enough iron oxyhydroxide extracellular sheaths that they can alter elemental biogeochemical cycling in wetlands and water systems. L. ochracea's role in the ecosystem is counter to the common assumption that a microbe's environmental influence is by dint of numbers; that is, the sheer cell number that any particular population is capable of producing should directly correlate with its ecological influence (1–4). Thus, L. ochracea is a prime example of an organism that has a major habitat impact without being numerically dominant (5–9). A previous study showed that in a centimeter-thick freshwater iron mat composed almost entirely of L. ochracea-like sheaths, the zone of active L. ochracea cell growth occurred only along the leading edge in a band 50 to 100 μm thick and with a cell density up to ∼4 × 107 cells cm−3 (8). As a result, >90% of the sheaths are empty in an actively accreting iron mat. Sheath production creates a porous Fe oxyhydroxide matrix (here referred to as iron mats) that influences water flow and chemistry, providing large surface areas of reactive, poorly crystalline ferrihydrite for sorption of organic carbon, trace metals, and anions such as arsenate and phosphate. It is also a substrate for other bacteria to colonize, with mats containing up to 109 cells cm−3 (dry weight basis) (10, 11). Based on these observations, L. ochracea's capacity to influence the environment is likely derived from its ability to oxidize ferrous iron and to rapidly translocate within its environment. However, fundamental questions about L. ochracea's metabolism and physiology, and how these contribute to success in oxic iron-rich waters, remain unanswered.
Despite L. ochracea having been described over 100 years ago, three fundamental aspects of its physiology remain a mystery: (i) its source of carbon assimilation (heterotroph, autotroph, or mixotroph), (ii) its electron donor for proton motive force generation (litho- or organotrophy), and (iii) its energy fueling source for chemotrophy (12–14). Circumstantial evidence suggests that L. ochracea is most likely a chemolithoautotroph due to a strong association with waters with high Fe(II) concentrations (typically ∼90 μM) where other chemolithoautotrophic iron-oxidizing bacteria (FeOB) live. Its production of copious quantities of Fe oxides with relatively small biomass accumulation is consistent with growth on a substrate that yields little free energy (∼−90 kJ mol−1) (15, 16). On the other hand, recent phylogenetic analysis confirms that L. ochracea is a close relative of the Leptothrix-Sphaerotilus group of sheathed Fe- and Mn-oxidizing bacteria (17). Leptothrix-Sphaerotilus members are represented by several species of pure cultures. While these species can oxidize Fe(II), they are unable to use it as an electron donor and instead grow heterotrophically on minimal media with a single carbon source (e.g., pyruvate or galactose). The non-L. ochracea members of the Leptothrix-Sphaerotilus group are rarely found in the iron mats where L. ochracea flourishes, and L. ochracea cannot be enriched using currently established Leptothrix cultivation techniques (17, 18). Interestingly, despite numerous efforts, it has not been possible to enrich L. ochracea on a medium used to isolate other chemolithotrophic FeOB, such as Gallionella spp. and Sideroxydans spp. Thus, acquiring reliable biochemical and genomic data from L. ochracea in order to understand what allows this organism to dominate in freshwater iron seeps has been challenging.
The availability of single-cell techniques to examine the biology of uncultured microorganisms has allowed microbiologists to develop a deeper understanding of the lifestyle and physiology of previously inaccessible organisms (19, 20). We set out to define the fundamental physiology of L. ochracea by determining the metabolic pathways responsible for carbon assimilation and energy fueling using several complementary approaches, including single-cell genomics, measurement of H13CO3 uptake, and analysis of in vitro enrichments. By more precisely defining L. ochracea's fundamental physiology and comparing it to that of other neutrophilic iron-oxidizing bacteria, we can determine what makes these cells succeed in these iron-rich environments.
RESULTS
Energy fueling.
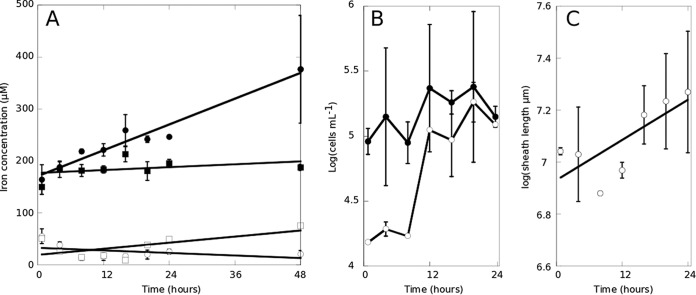
Our goal was to establish the role that Fe(II) plays in the growth of L. ochracea by quantifying rates of growth and iron oxidation and to identify putative electron transport chain (ETC) genes. To do this, we modified the classic enrichment and isolation techniques for chemolithoautotrophic FeOB (21) by using an opposing gradient of Fe(II) and O2. If FeS was not included, L. ochracea cells did not multiply and sheaths did not extend, indicating that Fe(II) was required for growth. Conversely, growth of L. ochracea was not observed in a mineral salt growth medium containing Fe(II) that is used for routine culture of obligate lithotrophic FeOB (21); instead, 0.22-μm-filtered natural site water was required for growth. Using 0.22-μm-filtered natural water, Fe(III) production was associated with sheath production, and there was little accumulation of Fe oxides not associated with the sheaths (Fig. 1). A two-way analysis of variance (ANOVA), Levene's test, and a Kruskal-Wallis rank sum test confirmed that the median and average sheath lengths changed with time and that the increase was significant. Variances among the data were not significant, further supporting that the sheath production was associated with the incubations. The addition of sodium azide inhibited Fe oxide accumulation, sheath production, and L. ochracea cell growth, indicating that Fe(II) oxidation was biological (Fig. 1A; Table 1). If incubations were started within 30 min after field collection, L. ochracea cells did not exhibit a lag phase and showed consistent exponential growth for 24 to 48 h. The density of L. ochracea cells was never greater than 2.4 × 105 cells ml−1. Under these conditions, the Fe(III) concentrations of L. ochracea cells incubated with filter-sterilized water from the environment increased between 4.5- and 8.9-fold compared to those in azide-inhibited samples (Table 1). While there was variance in the amount of biotic Fe oxidation from six measured growth curves (Table 1), overall biotic iron oxidation accounted for 71 to 89% of total iron oxidation in these experiments. Tracking of cell growth during incubation revealed that sheathed, filamentous cells had a generation time of 5.7 h, the Fe(III) concentration doubled every 44 h, and L. ochracea cells doubled their sheath length every ∼12 h at a rate of 125 μm r−1 (Fig. 1) Sustained growth or attempts at isolation via dilution were unsuccessful. However, in some cases enrichments could be grown and transferred three or four times (data not shown).
FIG 1.
Growth of L. ochracea in xenic culture. Cultures were monitored for Fe(II) (open symbols) and Fe(III) (closed symbols) with 5 μM sodium azide (squares) or without azide (circles) (A), for growth of all cells (closed symbols) and L. ochracea cells (open symbols) (B), and for total sheath length (C).
TABLE 1.
Increases in iron oxidation, numbers of total and sheathed filamentous cells, and sheath length for growth curves
| Growth curve | Fold increase over azide-killed controls |
Biotic oxidation (%) | Doubling time (h) |
|||
|---|---|---|---|---|---|---|
| Fe(II) | Fe(III) | Total cellse | Sheathed filamentous cellse | Sheath lengthf | ||
| 3a | 0.5 | 1.8 | 46 | 17.2 | 36.8 | |
| 4b | −0.4 | 8.9 | 89 | 24.9 | 5.7 | 12.9 |
| 6Ac | 0.9 | 5.9 | 83 | |||
| 6B | 0.6 | 3.5 | 71 | |||
| 7d | 1.4 | 7.9 | 87 | |||
| 7 + HCO3c | 0.8 | 4.6 | 78 | |||
This growth curve was performed during the Gallionella-Leptothrix transition documented previously (7); all other growth curves were performed under conditions where Leptothrix was dominant.
Growth curve for data collected for Fig. 1.
Growth curve for data collected for Fig. 4.
Growth curve for data collected for oxygen measurements.
No data were collected for growth curves 6A, 6B, 7, and 7 plus bicarbonate.
No data were collected for growth curves 3, 6A, 6B, 7, and 7 plus bicarbonate.
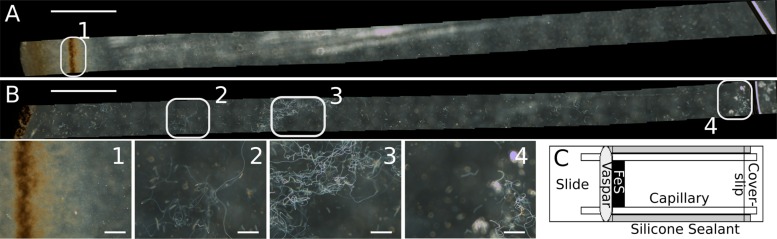
The use of microslides and test tube microcosms allowed for the growth of L. ochracea enrichments under trackable and relatively controlled conditions in the laboratory and allowed us to visualize and compare L. ochracea's growth to that of Sideroxydans lithotrophicus ES-1. L. ochracea growth appears not to be constrained to a tight oxic/anoxic boundary (Fig. 2). Direct microscopic observation of L. ochracea in opposing gradients of O2 and Fe(II) in microslides revealed a patchy growth distribution as assessed by visualization of areas of active sheath production (Fig. 2, numbers). Even at 200 μM O2 (80% oxygen saturation), L. ochracea enrichments generated sheaths and cells and accumulated biologically associated Fe(III) oxides when measured in tube microcosms (Table 1, growth curve 7). This was in contrast to an axenic strain of S. lithotrophicus ES-1 that formed, as expected, a narrow band of dense growth close to the FeS source and presumably at the oxic/anoxic interface when observed in the microslides. Based on these observations, we concluded that with regard to oxic/anoxic gradients L. ochracea's growth was more flexible than that of S. lithotrophicus.
FIG 2.
(A and B) A mosaic of micrographs showing growth patterns of iron-oxidizing bacteria. Modified growth microslides show the growth of Sideroxydans lithotrophicus ES-1 grown axenically in MWMM medium (A) and of Leptothrix ochracea grown xenically in filter sterilized natural waters (B). Close-ups show that S. lithotrophicus growth forms a tight band of iron-oxides (1) and that L. ochracea growth is more diffuse (2, 3, and 4). Size bars for microslides are 0.5 mm (A and B) and 20 μm (1 to 4). (C) The microslide is made of a glass slide with 2 capillary slides, silicone sealant, and a coverslip slide. FeS with agarose is then added and sealed with Vaspar, and finally the slide is filled with sterile filtered water and inoculated.
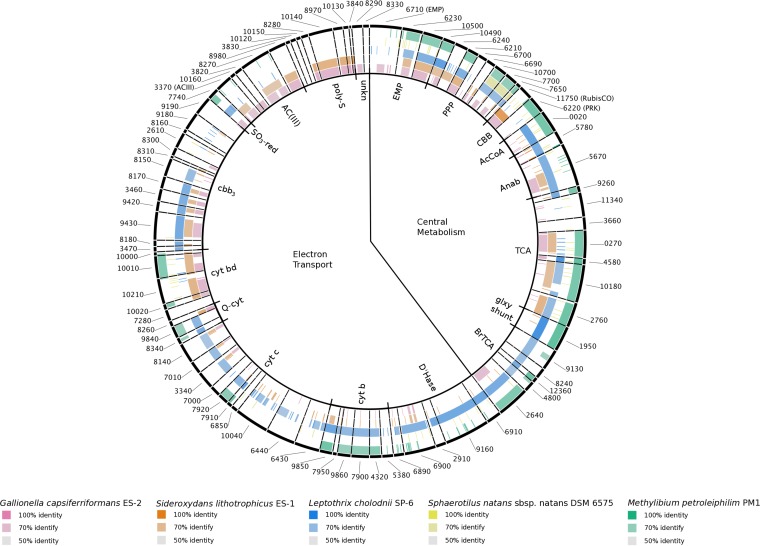
The L. ochracea single amplified genome (SAG) (17) yielded a partial draft assembly of 0.87 Mb carrying 1,269 genes (1,222 putative protein-coding genes) with a GC content of 57%. Based on analysis of tRNAs, the total genome size is approximately 2.2 Mb. This is much smaller than the 4.9-Mb and 4.6-Mb genomes for Leptothrix cholodnii and Sphaerotilus natans, respectively. A smaller genome is consistent with L. ochracea having a much more limited metabolic repertoire than either L. cholodnii or S. natans and is in line with the genome sizes of other chemolithotrophic FeOB (2.3 to 3.0 Mb). Previous analysis of the phylogenetically conserved genes gyrB and rrnA from the L. ochracea genome confirmed that its closest phylogenetic relative is L. cholodnii (17), and the subsequent availability of a genome for S. natans further confirmed that L. ochracea is most closely related to this taxonomic group (Fig. 3).
FIG 3.
Genes coding for proteins putatively involved in central metabolism and the electron transport chain. Protein sequences from L. ochracea's partial genome are compared to those in other taxa generated using the BLAST Ring Generator. L. ochracea is compared to Gallionella capsiferriformans ES-2 (rose), Sideroxydans lithotrophicus ES-1 (orange), L. cholodnii SP-6 (blue), Sphaerotilus natans (yellow), and Methylibium petroleiphilum PM1 (green). The intensity of color describes the percent sequence identity, and these are grouped clockwise around the ring starting at the top with central metabolism, with the Emden-Meyerhof-Parnas (EMP) pathway, pentose phosphate pathway (PPP), Calvin-Benson-Bassham (CBB) cycle, acetyl coenzyme A (AcCoA) connector, anapleurotic anabolic (Anab) reactions, tricarboxylic acid (TCA) cycle, horseshoe or branched TCA (BrTCA) cycle, and glyoxylate shunt (glxy_shnt) for central metabolism. The putative electron transport chain enzymes include dehydrogenases (D'Hase), cytochrome b subunits (b/b6), cytochrome c (cyt c), ubiquinol-cytochrome c reductase (Q-cytc), cytochrome bd (cyt bd), cytochrome cbb3 (cbb3) sulfite reductase (SO3-red), alternative complex III (ACIII), polysulfide reductase (poly-S), and proteins of unknown function.
Genes thought to be involved in the electron transport chain (ETC) from L. ochracea's partial single amplified genome were identified using the Integrated Microbial Genomes (IMG) interface (22). Individual ETC genes were identified by comparing sequences to those important in the ETC (23–26) and by looking for any putative proteins that contained transmembrane regions and redox-reactive prosthetic groups. A total of 57 different genes were identified as encoding potential ETC-associated proteins (Fig. 3). Several putative ETC-associated genes (Fig. 3) are unique to lithotrophic FeOB, are not found in Leptothrix-Sphaerotilus species, and have been implicated in iron oxidation (23–27). These genes included those for a molybdopterin oxidoreductase and a cytochrome bd oxidase (Fig. 3; see Fig. S1 in the supplemental material), which are proposed to be part of the forward electron transport chain and involved in electron transfer and as a terminal oxidase, respectively (23, 24). The proposed reverse ETC in the iron-oxidizing bacterium Mariprofundus ferrooxydans PV-1 includes the molybdopterin oxidoreductase, a quinone, and an NADH dehydrogenase. While M. ferrooxydans had a cytochrome b/c complex, the expression levels were low (23). There is little support in the L. ochracea genome for the reverse ETC. The molybdopterin oxidoreductase is proposed to be used in both forward and reverse electron transport, making it difficult to assign it to only one pathway. Quinones are made through biosynthesis rather than transcription and translation, and without membrane fractions, the structures are difficult to compare. Finally, the genome lacks an NADH dehydrogenase gene, possibly because it was not captured in the sequencing effort or because the cell simply lacks NADH dehydrogenase. L. ochracea's cytochrome b/c was similar to those in L. cholodnii and Methylibium petroleiphilum. In addition to the cytochrome b/c gene, L. ochracea's genome contained genes for several ETC components that were similar to those in L. cholodnii SP-6, including several formate dehydrogenases, a cytochrome c, and a cytochrome cbb3 (Fig. 3). With the aforementioned gene combinations, L. ochracea has genes implicated in iron oxidation, which supports an iron-oxidizing lithotrophic metabolism, as well as several possible electron transport pathways, which suggests a branched ETC, and a formate dehydrogenase.
Potential carbon assimilation routes.
A continued unknown in L. ochracea's physiology is its source of carbon assimilation. To identify possible routes of carbon assimilation, we performed labeled-uptake experiments and subsequently visualized carbon uptake in individual cells using nanoscale secondary-ion mass spectroscopy (nanoSIMS) and compared key genes, pathways, and gene neighborhoods from a single-cell genome sequence.
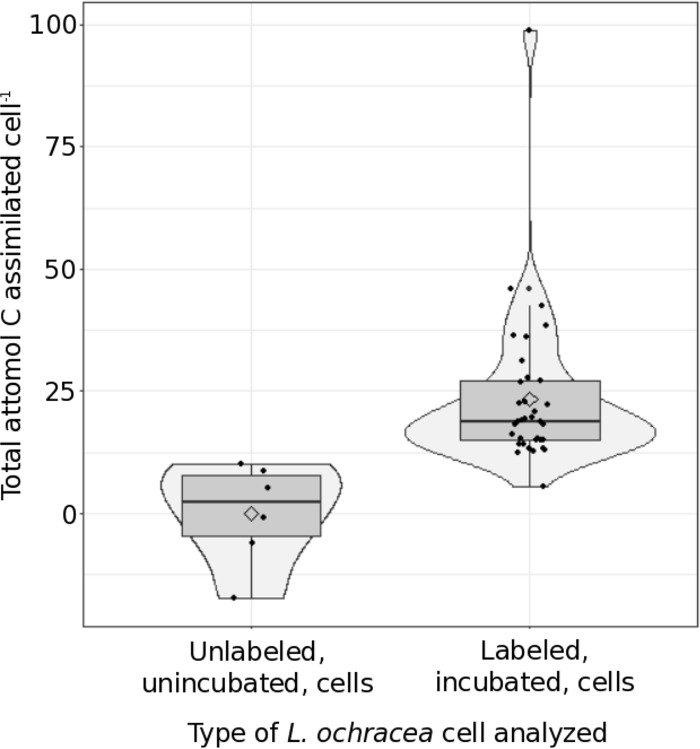
We employed a culture-independent method using nanoSIMS to determine individual 13C bicarbonate uptake in L. ochracea cells. First, during incubation, the dissolved inorganic carbon (DIC) dropped from 270 μM to 249 μM (from 0 to 14 h, respectively) and then from 88 μM to 73 μM (from 24 to 42.75 h). The mean L. ochracea cell 13C/12C ratios were significantly higher than the mean sheath 13C/12C ratios (P = 0.015) after using the conservative Bonferroni correction for multiple comparisons. This relationship suggests that labeled inorganic carbon was incorporated into L. ochracea cells and rules out abiotic adsorption or natural variability. Relative to the mean value in unincubated L. ochracea cells, 33 of 34 cells incubated with 13C-bicarbonate showed some assimilation of the 13C label. These L. ochracea cells assimilated between 12.41 and 98.76 amol C cell−1, with a mean of 24.00 ± 16.03 amol C cell−1 and a median of 18.81 amol C cell−1 (Fig. 4). Total carbon assimilation rates ranged from 0.29 to 2.31 amol cell−1 h−1, with a mean of 0.56 ± 0.37 amol cell−1 h−1 and a median of 0.44 amol cell−1 h−1. If a single cell has 2,160 amol C cell−1 (28) and that cell had divided at least once, then a cell that had assimilated 98.76 amol of CO2 cell−1 would have incorporated up to 9% of its total cell carbon from CO2, a fraction of the cell's total carbon needs.
FIG 4.
Carbon assimilation by individual L. ochracea cells. A box plot with a violin plot overlay is shown, with the total attomoles C cell−1 assimilated for individual L. ochracea cells that either had been incubated for 42.75 h in 1 mM labeled (10%) bicarbonate (labeled, incubated) or had not been incubated and fixed for analysis (unlabeled, unincubated). Uptake was gathered for individual cells using nanoSIMS analysis and is described in Text S1 in the supplemental material. Samples had been incubated for 42.75 h with 1 mM labeled (10%) bicarbonate. The layered box plot shows the bulk of the data and the median (horizontal line), and the violin plot overlay shows the density/histogram distribution of the data per group and the mean (diamond) per group.
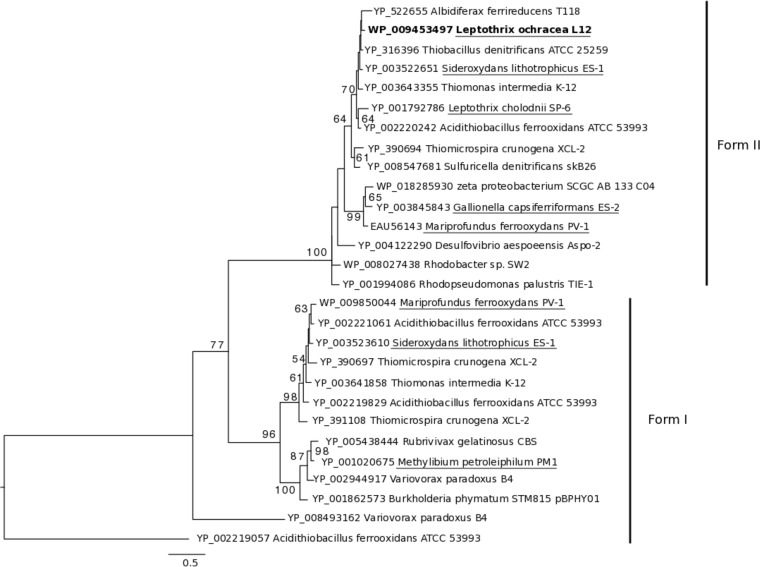
Cultivated neutrophilic Fe(II)-oxidizing chemolithoautotrophs assimilate carbon via the Calvin-Benson-Bassham (CBB) pathway. The L. ochracea SAG contained the two key CBB-specific genes in the nine-enzyme pathway, those for ribulose 1,5-bisphosphate carboxylase (RubisCO) (E.C. 4.1.1.39) and phosphoribulokinase (PRK) (E.C. 2.7.1.19). Based on phylogenetic analysis, the translated RubisCO gene clustered with form II RubisCO sequences from other FeOB, some sulfur-oxidizing chemolithoautotrophs, and L. cholodnii SP-6 (Fig. 5). Seven of the nine enzymes in the CBB pathway are reversible and are the same as in the oxidative pentose phosphate pathway (PPP) (RubisCO and PRK are not). L. ochracea has six of these seven genes, two of which overlap with the Embden-Meyerhof-Parnas (EMP) pathway (Fig. 3; see Table S1 in the supplemental material). L. ochracea also has genes from the branched tricarboxylic acid pathway and key anaplerotic reactions (Fig. 3). The arrangement of the shared CBB/PPP-associated genes in the genome is notable, as they are more similar to those in L. cholodnii SP-6 and Sphaerotilus natans.
FIG 5.
Phylogenetic relationship of RubisCO protein sequences from lithotrophic, photomixotrophic, and heterotrophic organisms. The tree was generated using maximum likelihood with protein sequences obtained from pure cultures or a single amplified genome with 1,000 bootstraps. Only those bootstraps above 50 are displayed on the tree. L. ochracea L12's RubisCO sequence is in bold, comparison genera from Fig. 3 are underlined, and all gene accession numbers are included.
DISCUSSION
An essential question for an ecological engineer is what physiological traits allow these organisms to adapt to their environment and significantly alter the environmental landscape. As demonstrated here, just a few L. ochracea cells oxidize Fe(II) and generate prodigious amounts of Fe(III)-oxyhydroxide coated sheaths to obtain their energy. This is an energy acquisition strategy that L. ochracea shares with the lithoautotrophic Gallionellaceae FeOB. In addition to iron(II) oxidation, L. ochracea and the Gallionella-like FeOB share the capacity for autotrophy. However, unlike in the Gallionella-like FeOB, the measured carbon assimilation rates do not meet all of L. ochracea's carbon demand, indicating that L. ochracea may assimilate organic carbon as well. This is consistent with the findings of a successional study in an Fe(II)-rich wetland where L. ochracea supplanted Gallionella (7). An increase in dissolved organic carbon was identified as an important factor associated with dominance by L. ochracea. In addition, previous studies have shown that L. ochracea-like sheaths can dominate over stalk-producing Gallionella spp. in relatively O2-rich environments (5, 7, 29). These physiological studies are consistent with genomic traits of L. ochracea's central metabolism that include an amalgamation of the traits from the Gallionella-like FeOB and the Leptothrix-Sphaerotilus genera. An FeOB with the characteristics of the Leptothrix-Sphaerotilus genera, such as the ability to assimilate organic carbon, the lack of a requirement for the reverse electron transport pathway to generate reducing power, and the use of a branched electron transport pathway, would have the ability to live in a broad habitat range and dominate in these ecosystems.
Lithotrophy and electron transport.
Based on our results, we speculate that aerobic iron oxidation is L. ochracea's primary route for energy generation. The energetics of lithotrophic Fe oxidation inherently require FeOB to oxidize generous quantities of Fe(II). The resultant Fe(III) oxyhydroxides are bound to organic polymers that form sheaths, stalks, or sheath/stalk hybrid structures that are proposed to be used for positioning the cells in the gradient or to aid motility (8, 16, 30–33). Lithoautotrophic organisms must shunt electrons through reverse electron transport to convert inorganic carbon to organic carbon, thus reducing the energy yields from the inorganic electron donor (34). L. ochracea oxidizes Fe(II) at higher rates than other autotrophic FeOB, doubles at higher rates than other autotrophic FeOB (5.7 h versus ∼8 to 16 h for lithotrophic FeOB) (35, 36), and does not assimilate enough inorganic carbon to support obligate autotroph growth.
L. ochracea, based on physiological and genomic analysis, likely uses iron to generate a proton motive force to support ATP production but does not appear to require iron oxidation for NADPH production. First, microcosm incubations link sheathed filamentous cell production to biological iron oxidation. Second, L. ochracea contains several genes found in other neutrophilic FeOB that are thought to be important for neutrophilic Fe oxidation (23, 25–27, 37). The L. ochracea genome codes for a cytochrome bd, a cytochrome cbb3, and an alternative complex III, proposed to be involved in the forward direction of the iron-oxidizing electron transport pathway present in most of the known FeOB (23, 24, 38). In particular, the alternative complex III, including a homolog of actB, a molybdopterin oxidoreductase gene, is highly conserved by both homology and gene order between both freshwater and marine FeOB. L. ochracea alternative complex III genes share homology and gene order with those of known FeOB (Fig. 3), although the gene cassette in L. ochracea is truncated due to the fragmented nature of the single-cell assembly. L. ochracea also contains a cytochrome bd oxidase protein complex that is absent in Leptothrix-Sphaerotilus species but shares substantial similarity with those of other FeOB (23, 25, 27, 37) (see Fig. S1 in the supplemental material). We also note that we did not find obvious evidence for horizontal gene transfer (HGT), such as transposable elements or genomic islands; however, this could be due to the incompleteness of the L. ochracea single amplified genome. A final line of evidence for a mixed growth strategy is that L. ochracea has an absolute requirement for organic-rich waters (7). This suggests that the cells could obtain NADPH or NADH from glycolysis and the tricarboxylic acid cycle and would not be required to use an energy-expensive reverse electron transport pathway. These results strongly support L. ochracea's role as an iron-oxidizing bacterium but one with an overall metabolic strategy different from that of other lithoautotrophic FeOB.
L. ochracea can also catalyze iron oxidation over a wide range of oxygen tensions. Cells are able to grow throughout the water column and at greater oxygen tensions, as observed by the shallow oxygen gradients present in in iron mats dominated by L. ochracea (5, 7, 8) and the presence of L. ochracea cells concentrated in the outer tips of the iron mats (8) and as seen in the modified microslides (Fig. 2) (39, 40). Second, L. ochracea's genome encodes multiple putative terminal oxidases, and these are likely to be expressed under varied oxygen tensions. Finally, the water pH is slightly lower and the recalcitrant organic carbon concentration is higher when L. ochracea is vigorously growing than when the twisted stalks of Gallionella spp. dominate (7). A decrease in pH and addition of chelation elements from high-molecular-weight carbon can alter the iron oxidation kinetics such that higher concentrations of oxygen cause less competition for biotic iron oxidation, allowing cells to grow throughout the water column. These lines of evidence support L. ochracea's ability to use oxygen at varied concentrations, the presence of a branched electron transport chain, and the capacity for L. ochracea to adapt to changing environmental conditions.
Carbon assimilation.
Single L. ochracea cells assimilated 13C-bicarbonate, and L. ochracea's genome contained key CBB pathway genes for CO2 fixation. These results are consistent with the capacity to assimilate inorganic carbon but also reveal fundamental questions about L. ochracea's trophic strategy. First, the nanoSIMS data indicate that L. ochracea assimilates inorganic carbon at a fraction of the rates and total assimilation percentage of other autotrophs. For comparison, rates of DIC incorporation for three different species of individual autotrophic green and purple sulfur bacterial cells determined using single-cell techniques were 1 to 30, 100 to 300, and 3,000 to 20,000 amol cell−1 h−1 and accounted for ∼ 85% of total carbon assimilation (3), almost 10 times that of L. ochracea. Doubling times for bulk L. ochracea enrichments were between 0.24 and 1.5 days (Table 1), in comparison to pure culture doubling times of 0.33 and 0.5 days for the chemolithoautotrophic Sideroxydans lithotrophicus ES-1 and Gallionella capsiferriformans ES-2, respectively (35). This comparison illustrates that individual L. ochracea cells have much lower inorganic carbon assimilation rates than other autotrophs. Second, L. ochracea contains two key irreversible genes in the CBB pathway that are used in carbon assimilation, those for RubisCO and PRK. However, it is not clear if L. ochracea's RubisCO enzyme functions like those in other lithoautotrophic bacteria (23, 25). The phylogenetic tree of RubisCO protein sequences places L. ochracea's sequence with sequences from organisms that obtain their energy and carbon through a diverse set of pathways rather than with sequences from obligate chemolithoautotrophic iron-oxidizing bacteria (Fig. 5). The gene neighborhoods and operons housing the PPP in L. ochracea are more similar in content and order to those in other heterotrophic sheathed filamentous bacteria than to those in obligate chemolithoautotrophic iron-oxidizing bacteria. Because bacterial genes are arranged in operons and the translation efficiency is likely influenced by the overall structure of a polycistronic message (41), neighborhood structure is preserved evolutionarily, suggesting that this pathway may have a different metabolic role in the Leptothrix-Sphaerotilus than in the lithoautotrophic FeOB. Together, both uptake experiments and the genome analysis suggest an alternative role for carbon assimilation and for RubisCO in L. ochracea.
One possible interpretation of these results is the use of RubisCO for mixotrophy. Mixotrophy, or the simultaneous uptake of organic C and fixation of CO2, is well known among chemolithotrophic bacteria. For example, under energy limitation, obligate sulfur-oxidizing chemolithoautotrophs can assimilate up to 20 to 30% of their organic carbon from external sources (42, 43). Certain facultative chemolithotrophic bacteria employ a flexible carbon assimilation strategy alternating between heterotrophic and autotrophic growth (44–46). Genetically, autotrophs and heterotrophs should share many core metabolic pathways (for example, the PPP, CBB cycle, EMP pathway, and tricarboxylic acid [TCA] cycle), and thus it is challenging to determine if a pathway is for heterotrophic or autotrophic carbon assimilation. Evidence hints at L. ochracea's ability to assimilate organic carbon in combination with inorganic carbon. L. ochracea cells have doubling times faster than that of the obligate autotrophic G. capsiferriformans or S. lithotrophicus. This is remarkable since L. ochracea can putatively use the CBB energy-intensive pathway to assimilate CO2, but at assimilation rates lower than those of other autotrophs. Moreover, L. ochracea may need to support its growth with both inorganic and organic carbon assimilation, because L. ochracea cells cooccur with the increased concentrations of DOC (7). This example of mixotrophy would give L. ochracea the ecological edge in such environments. Without well-documented environmental, physiological, regulatory, or genetic markers for blended trophic strategies, it is difficult to predict the conditions and the balance between organic and inorganic carbon assimilation.
An alternative conclusion to the low carbon assimilation and presence of RubisCO is that L. ochracea is using the CBB cycle to balance the reducing power of the cell when growing in the presence of high-molecular-weight, recalcitrant organic carbon or when growing in excess electron-reducing power. Rhodopseudomonas palustris has the ability to degrade aromatic, recalcitrant organic carbon, and many of these compounds are reduced compared to the biomass of the cell (47). When growing phototrophically with reduced carbon substrates, R. palustris uses the CBB pathway to balance internal cell redox (48). L. ochracea is common in the iron seeps, fens, and bogs where highly recalcitrant, structurally undefined complex organic compounds are typically present (7, 49, 50). Interestingly, the genome of L. cholodnii, a close relative of L. ochracea, has putative genes and pathways for degrading highly aromatic, complex organic matter (E. J. Fleming, unpublished observations), although homologous genes were not found in the partial genome of L. ochracea. Ecologically, L. ochracea populations dominate iron seeps that have waters with a high spectral slope, a proxy for high-molecular-weight carbon, and with high ferrous iron concentrations (7). When L. ochracea is growing in high Fe concentrations, it (like phototrophs) is in an environment with excess electrons and reducing power. Use of the CBB pathway could offer an additional route to shunt excess reducing power and gain some carbon (48). High-molecular-weight, aromatic organic carbon is difficult to degrade. If an organism can supplement its biomass by fixing inorganic carbon, the organism may gain an ecological advantage. The use of RubisCO as a redox-balancing enzyme or as a pathway to supplement organic carbon assimilation provides L. ochracea with a mechanism for cells to adapt to changing environmental conditions.
Conclusion.
The results and data gathered from the genomic content, growth habits, and habitat preferences suggest that Leptothrix ochracea is a hybrid of the sheath-forming metal oxidizing bacteria and the Gallionella-like FeOB that is possibly a mixotroph. This combination of traits allows for it to be metabolically flexible and to compete in a changing environment versus organisms that are more restricted in their metabolic potential.
MATERIALS AND METHODS
Single-cell sequencing and annotation.
To establish L. ochracea's genomic potential, a single amplified genome of L. ochracea cells was obtained as previously described (17). Briefly, cells were collected from actively growing mats from the Lakeside Drive field site, labeled with a Syto9 nucleic acid stain, and then sorted into individual wells of a 384-well plate using fluorescence-activated cell sorting. The cells were lysed, and genomes were amplified with multiple-displacement amplification (MDA). After screening the 16S rRNA sequences of all amplified genomes, the single amplified genome matching the dominant L. ochracea genotype was increased in quantity using secondary MDA. The SAG draft assembly was generated at the DOE Joint Genome Institute (JGI) using a hybrid approach of 454-titanium and Solexa technologies. Further sorting of samples to yield additional L. ochracea single amplified genomes failed, due to the potentially fragile nature of the L. ochracea cells and their tendency to break upon manipulation, especially if cells are frozen after sampling and then thawed for cell sorting. Raw sequence data were filtered for known sequencing and library preparation artifacts and then screened and trimmed according to the k-mers present in the data set. Reads representing highly abundant k-mers were removed such that no k-mers with a coverage of more than 30× were present after filtering. Reads with an average k-mer depth of less than 2× were removed. To perform assembly, filtered Illumina reads were assembled using Velvet version 1.1.04 (51). The VelvetOptimiser script (version 2.1.7) was used with default optimization functions (n50 for k-mer choice and total number of base pairs in large contigs for cov_cutoff optimization). One- to 3-kbp simulated paired-end reads were then created from Velvet contigs using the wgsim software. Finally, the normalized Illumina reads were assembled together with simulated read pairs using Allpaths-LG (version 41043) (52).
Genes were identified using Prodigal (53). The predicted coded DNA sequences (CDS) were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant (nr), UniProt, TIGRFam, Pfam, KEGG, COG, and InterPro databases. The tRNAScan-SE tool (54) was used to identify tRNA genes, whereas rRNA genes were identified by searches against models of the rRNA genes built from SILVA (55). Other noncoding RNAs, such as the RNA components of the protein secretion complex and RNase P, were identified by searching genomes for the corresponding Rfam profiles using INFERNAL (56). Additional gene prediction analysis and manual functional annotation were performed within the Integrated Microbial Genomes (IMG) platform (22) developed by the Joint Genome Institute, Walnut Creek, CA, USA (http://img.jgi.doe.gov).
For comparative purposes the genome of Leptothrix cholodnii SP-6 was also sequenced at the JGI using standard sequencing protocols for pure cultures of bacteria as described previously (25). The total genome size of L. ochracea was estimated using the number of predicted total tRNAs present in the sequenced L. ochracea L12 genome compared to the number found in L. cholodnii SP-6 (57, 58).
The sequences are publicly available at the Department of Energy's Joint Genome Institute's Integrated Microbial Genomes (IMG) platform under IMG Genome ID number 2506520002 at http://img.jgi.doe.gov.
Comparative genomics.
To identify L. ochracea's most basic assimilatory and dissimilatory pathways, proteins potentially involved in central metabolism, electron transfer, and oxygen metabolism were identified using the IMG system with the basic local alignment search tool (BLAST), the annotation generated at JGI, and genome-genome similarity analysis tools. Protein sequences were compared to those of proteins found in the IMG database, and all sequences of interest were also subjected to BLAST analysis against the NCBI nr database (see Table S1 in the supplemental material). Homology between sequences was assessed by gene neighborhood similarity, alignment, evolutionary distance, and maximum-likelihood phylogeny. Gene neighborhood similarity, identification of signal peptides, and transmembrane regions were assessed using IMG. Detailed analysis focused on the publically available genomes of three phylogenetically closely related species/genera (L. cholodnii SP-6, Sphaerotilus natans, and Methylibium petroleiphilum PM1), as well as two genera of distantly related lithotrophic FeOB isolated from freshwater neutrophilic iron seeps (Sideroxydans lithotrophicus ES-1 and Gallionella capsiferriformans ES-2).
With the assumption that an organism's metabolically associated genomic profile is the sum of its genetic material, we used the BLAST Ring Image Generator (BRIG) (59) to organize, distill, and compare L. ochracea's metabolic profile in a clear and simplified manner. Because the focus of this study was to determine the fundamental physiology in L. ochracea, we focused on the putative gene products related to central metabolic pathways and electron transport chain components from L. ochracea's partial genome. L. ochracea genes were clustered according to function and assembled as a Multi-FASTA file, and then this FASTA file was used as a reference “genome.” Default settings were used for comparisons (59).
Three separate phylogenetic trees were generated to visualize the maximum-likelihood phylogenetic relationships for (i) ribulose bisphosphate carboxylase (RuBisCO), (ii) cytochrome bd, and (iii) molybdopterin oxidoreductase. To build each phylogenetic tree, the sequences were aligned using Clustal Omega (60), and then the maximum likelihood for each protein relationship was calculated in RAxML 8.0.0 (61) using either 100 or 1,000 rapid bootstrap analyses and site-specific evolutionary rates with a BLOSUM62 substitution model. The final tree was evaluated and optimized under gamma substitution rates up to an accuracy of 0.1 log likelihood.
Collection of L. ochracea.
L. ochracea cannot be maintained in pure culture or artificial media but can be grown in enrichments with 0.22-μm-filtered natural waters (described below). All experiments were performed with freshly collected L. ochracea sheaths and cells from a neutrophilic freshwater iron mat on Lakeside Drive, Boothbay Harbor, ME. The site, chemical characterization, and makeup and dynamics of the Fe-oxidizing communities were described previously (17). For the present study, samples of actively growing iron mats were collected from the uppermost 0.5 cm of the mat during periods of active L. ochracea growth (7, 17, 62). Natural water samples that were used to establish laboratory growth experiments were collected at the same time by submerging a polypropylene bottle below the surface of the water, excluding all gas bubbles, and capping the bottle.
In vitro enrichment assembly.
All experiments to test L. ochracea's physiology were set up using static enrichments incubated at an ∼8° angle. Enrichments were set up with either Bellco 150- by 20-mm glass Balch tubes (holds 24 ml) with size 0 butyl rubber stoppers or 85- by 13-mm glass Pyrex Vista tubes (holds 4.5 ml) with size 000 butyl rubber stoppers. The butyl rubber stoppers were triple boiled. Initially, ∼0.5 ml of an autoclave-sterilized FeS-tap water (1:1) slurry containing 1% high-melting-point agarose (described in reference 21) was distributed at the tip of each tube and allowed to harden. The freshly collected natural water samples were sterile filtered through a 0.2-μm filter. The inocula for the growth curves/enrichments were prepared by 1:100 dilution of the L. ochracea iron mat sample into the filtered water to a yield concentration of approximately 1 × 104 L. ochracea cells ml−1. The cell number was confirmed by microscopy using fluorescence in situ hybridization (FISH) (detailed below). The inoculum was gently mixed and then divided again so that various treatments could be added to each bottle. For experiments where 13C-labeled bicarbonate was added, the final concentration was 1 mM bicarbonate, and when a killed control was included in the experiment, the final concentration was 5 μM azide. Aliquots from each bottle were added to glass tubes (24 or 4.5 ml [large or small, respectively]), and rubber stoppers were applied so as to leave a small air bubble. After inverting the tubes twice, they were incubated in the dark at 26°C at an ∼8° angle.
Analysis of 13C-bicarbonate assimilation in L. ochracea using nanoSIMS.
To determine whether L. ochracea cells had the capacity to fix CO2, 24-ml tube microcosms were used to enrich L. ochracea in natural waters augmented with 10% 13C-bicarbonate label in a 1 mM final bicarbonate concentration. From each 24-ml sample, 2 ml was taken for dissolved inorganic carbon (DIC) analysis, 1 ml was preserved in 2% (final concentration) glutaraldehyde to preserve the total cells and sheaths produced (Electron Microscopy sciences, Hatfield, PA, USA), 1 ml was used for iron [Fe(II), Fe(III), and total iron] determination, and the remainder was preserved with paraformaldehyde (PFA). After PFA preservation, the material was concentrated by centrifugation at 3,000 × g for 5 min, washed in a modified phosphate-buffered saline (PBS) solution (20× phosphate, 1× saline) twice, and split into two fractions. In one fraction, samples were preserved with 1:1 PBS–100% electron microscopy (EM) grade ethanol (Sigma-Aldrich, St. Louis, MO, USA) and stored at −20°C. In the second fraction, iron oxides were dissolved using a 2.5% (final strength) solution of freshly mixed 50 g liter−1 dithionite, 0.2 M sodium citrate, and 0.35 M acetic acid (63). The cells were then rinsed in modified PBS and then preserved and stored in 1:1 PBS–100% EM grade ethanol and stored at −20°C. Both types of samples were tested for the presence of L. ochracea cells using a modified catalyzed reporter deposition (CARD) technique adapted for fluorescence in situ hybridization (FISH) as described previously (17) and modified from reference 64. Briefly, samples were prehybridized for an hour at 46°C with sheared salmon sperm, yeast RNA, and Denhardt's reagent, and then the standard method of hybridization was performed using formamide to modulate stringency followed, by salt washes. This was performed with both the Lepto175 probe (ATC CAC AGA TCA CAT GCG) (17) and the CARD-FISH probe EUB338 (GCT GCC TCC CGT AGG AGT) (65).
For the nanoscale secondary-ion mass spectroscopy (nanoSIMS) analysis, preserved samples (inoculum and samples that had been incubated with 13C-bicarbonate after 42.75 h) were vacuum filtered to evenly distribute the sample onto ∼5-mm-diameter, 0.2-μm-pore-size gold-palladium-sputtered polycarbonate filters (Millipore, Darmstadt, Germany) and dipped in a thin coat of 0.1% agarose. L. ochracea cells on the filter were then located by using either the Lepto175 CARD-FISH probe or the EUB 338 CARD-FISH probe to examine key species characteristics, such as filamentous sheathed cells. Sections containing L. ochracea cells were marked with a Leica DM6000 B microscope with microdissection capabilities (Wetzlar, Germany). A nanoSIMS 50L (Cameca, Gennevilliers, France) at the MPI Bremen was used for isotopic ratio measurements. The marked areas of interest were rastered with a Cs+ primary ion beam with a current between 2 and 3 pA over a raster size between 3 by 3 μm and 40 by 40 μm and an image size of 256 by 256 pixels. The dwell time for all measurements was 1 ms pixel−1. Secondary ion images for 12C−, 13C−, 19F−, 12C14N−, 32S−, and 56Fe16O− were recorded simultaneously from the samples as previously described (3). Multiple scans of the targeted area were made until either the cells and sheaths were vaporized or enough scans had been obtained to measure isotope concentrations of the L. ochracea cells. In total, 12 successful measurements were obtained from the inoculum (n = 2) and the 42.75-h incubations (n = 10).
Analysis of nanoSIMS data.
The data were processed using the MATLab-based (MathWorks, Natick, MA) software Look@NanoSIMS described previously (66) (http://nanosims.geo.uu.nl/nanosims-wiki/doku.php/nanosims:lans). Sample analyses were performed as described in the Look@NanoSIMS manual for a typical data processing session. Raw data were loaded, the scans containing errors were discarded, any drift was corrected, and all remaining scans were accumulated to one image. Several regions of interest (ROIs) were identified using the Lepto176-, EUB338-, or DAPI (4′,6′-diamidino-2-phenylindole)-labeled images or using the 12C14N, 32S, or 19F signal. These ROIs were L. ochracea cells, sheath material devoid of cells, non-L. ochracea cells, and background filter free of material, which were then processed within the analysis program. The background filter from each set of scans served as the 13C/12C standard ratio for the 13C/12C ROIs that had been acquired at the same time.
To eliminate ROIs with statistically significant uncertainty levels and to identify the ROIs that assimilated 13C, the data were subjected to several tests (see Text S1 in the supplemental material). Those ROIs with too great an uncertainty were eliminated from further analysis. The distribution of and relationship between the remaining ROIs had a natural and significant distribution, and these data were then used to calculate the total carbon assimilation for each cell (see Text S1 and Fig. S3 and S4 in the supplemental material). Statistical total carbon assimilation by incubated cells was then compared to that by unincubated cells.
Growth curves.
For growth experiments, cell enrichments were sacrificed for analysis at each time point. Six growth curves were performed using enrichments from the Lakeside Drive field site described previously (7). At Lakeside Drive the FeOB community changes from being dominated by Gallionella spp. to being dominated by Leptothrix spp., and the transition is accompanied by a shift in chemical characteristics. The first growth curve (growth curve 3) was performed during the Leptothrix-Gallionella transition (7), and the other five were performed when Leptothrix dominated. Prior to invasive sampling, oxygen measurements were performed by inserting a Clark-type O2 minielectrode (World Precision Instruments, Sarasota, FL, USA) into the tube and recording the O2 concentration. The electrode was calibrated with air-sparged or N2-sparged water. Soluble Fe(II) in the medium was measured directly, and Fe(III) was reduced with hydroxylamine and then measured by the ferrozine method (67) as previously described (7). Dissolved inorganic carbon was measured (68), and sheath length was determined as previously described (7). Briefly, 10-μl subsamples were evenly spread within 10-mm-diameter circles on counting slides (Electron Microscopy Sciences, Hatfield, PA, USA) and imaged with an Olympus BX60 microscope (Center Valley, PA, USA) coupled with a Qicam camera and QCapture Pro software (Surrey, BC, Canada). Ten fields per circle were imaged. The area of sheath in each image was traced using a Wacom Intous4 Pen Tablet (Vancouver, WA, US). To determine the number of total cells and filamentous cells within sheaths, a similar imaging protocol was used, except samples were stained with Syto13 (Invitrogen-Life Technologies, Grand Island, NY, USA) and the cells counted directly within a microscope field viewed by epifluorescence microscopy. FISH using the Lepto175 FISH probe (17) was used to confirm that sheathed filamentous cells in enrichments were L. ochracea.
To determine whether sheath production was statistically significant, several statistical tests were performed. A two-way ANOVA was used to confirm that the average sheath production changes over time with an F(6, 132) test statistic of 2.024 and a P value of 0.0668. Furthermore, a Levene test was conducted to test for homogeneity of variance, without assuming a normal distribution of the data. This test indicated that the variances of px across time groups were not statistically different, with an F(6, 133) test statistic of 1.16 and a P value of 0.33. Finally, a follow-up Kruskal Wallis rank sum test was used to test whether the median sheath formation is equal across groups (without having to assume a normal distribution, making it less sensitive to outliers). This test also indicated that at least one group had a different median sheath length and that the difference was significant (chi-square [6] = 33.14, P < 0.0001).
In general, the Fe(II) concentrations were 34 μM to 430 μM and remained relatively constant over the duration of the incubations, indicating that the cultures did not become Fe(II) limited. Incubation of L. ochracea enrichments in which natural waters were replaced with modified Wolfe's mineral medium (MWMM) (21) used to grow the FeOB S. lithotrophicus and G. capsiferriformans did not result in the growth of L. ochracea cells or production of sheaths, suggesting that the natural waters retain some key component required for L. ochracea's growth. All subsequent incubations were performed using natural waters from Lakeside Drive with a pH of 5.2 ± 0.2, an Fe2+ concentration of 89 ± 70, and a spectral slope of 1.128 ± 0.107, as described by Fleming et al. (7).
Visualizing L. ochracea's relationship to oxygen.
To directly visualize growth of L. ochracea in opposing gradients of iron and oxygen without disruption of the sheaths, a modified growth microslide was constructed (Fig. 2). These slides are similar to those described previously (40), with the following modifications. Two pairs of glass capillaries (Vitrotubes; Vitrocom, Mountain Lakes, NJ, USA) were glued together (side to side) with a thin layer of silicone sealant (RTV silicone adhesive sealant; Permatex, Hartford, CT, USA). Each pair of glass capillaries was laid lengthwise on a standard borosilicate glass slide with a 70-mm-long size 1 coverslip placed on top, and silicone sealant was used to seal both sides to make a watertight seal. After the sealant had cured for 24 h, the slides were immersed in water, boiled for 5 min, and allowed to cool and dry. This procedure was repeated three times to rid the slides of any remaining acetate from the silicone sealant. The slides were sterilized by autoclaving, and then a drop of sterile 1% agarose with FeS was added to seal one end. After the agarose-FeS hardened, it was coated with a small amount of sterile Vaspar (1:1 Vaseline-paraffin wax) as an O2 barrier. For an inoculum, an iron mat was prepared in natural water medium as described above. For comparative purposes, a pure culture of Sideroxydans lithotrophicus ES-1 was prepared to the same cell density as L. ochracea in the same medium. The microslides were inoculated with a 1.5-in 22-gauge needle so that an even dispersion of medium-cells filled the slide to the top of the coverslip with the open end being left exposed to the air. The microslides were incubated at an 8° angle and then visualized and imaged nondisruptively using the 20× and 40× phase-contrast objectives of an Olympus microscope. Longitudinal image mosaics were assembled in ImageJ (http://imagej.nih.gov/ij/) with the MosaicJ plugin. The presence of L. ochracea cells in sheaths was further confirmed by visualizing the Syto13-stained cells under fluorescence microscopy.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by a grant from the National Science Foundation (IOS-0951077). The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231. Work was also funded by the California State University Program for Education and Research in Biotechnology through the New Investigator Grant Program.
The funders had no role in the study design, data collection and interpretation, or decision to submit this work for publication.
We thank Lynne Goodwin and Stephanie Malafatti for help with genome sequencing and assembly. We appreciate the contribution to the gene characterization by the students in the 2015 and 2016 spring CSU, Chico, undergraduate bacterial physiology class (BIOL 412). Amelia Emerson isolated the DNA from Leptothrix cholodnii SP-6. We acknowledge help from Niculina Musat, Rachel Foster, Daniela Tienken, Tomas Vagner, Andreas Krupke, Danny Ionescu, and Robin Sutka in preparing and running the samples using nanoSIMS. We thank the MPG for the support of the nanoSIMS facility at the MPI Bremen. We appreciate help from Joyce McBeth, Jochen Nuester, and Carlton Rasuchenburg in logistics and experimental analysis. Finally, we thank Erin Field, Jeremy Jacquot, Adam Mumford, Jeffery Bell, David Stachura, Christian Fosen, Troy Cline, the CSU, Chico, faculty writing group, and three anonymous reviewers for helpful comments on the manuscript.
E.J.F. and D.E. designed the experiments, E.J.F. performed the research, M.M.M.K., T.W., S.L., and A.S. contributed new reagents and tools, E.J.F., D.E., R.A.D., and T.W. analyzed the data, and E.J.F., T.W., and D.E. wrote the paper.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02239-17.
REFERENCES
- 1.Denef VJ, Mueller RS, Banfield JF. 2010. AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J 4:599–610. doi: 10.1038/ismej.2009.158. [DOI] [PubMed] [Google Scholar]
- 2.Dejonghe W, Boon N, Seghers D, Top EM, Verstraete W. 2001. Bioaugmentation of soils by increasing microbial richness: missing links. Environ Microbiol 3:649–657. doi: 10.1046/j.1462-2920.2001.00236.x. [DOI] [PubMed] [Google Scholar]
- 3.Musat N, Halm H, Winterholler B, Hoppe P, Peduzzi S, Hillion F, Horreard F, Amann R, Jørgensen BB, Kuypers MMM. 2008. A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc Natl Acad Sci U S A 105:17861–17866. doi: 10.1073/pnas.0809329105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jousset A, Bienhold C, Chatzinotas A, Gallien L, Gobet AEL, Kurm V, Küsel K, Rillig MC, Rivett DW, Salles JF, van der Heijden MGA, Youssef NH, Zhang X, Wei Z, Gera Hol WH. 2017. Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J 11:853–862. doi: 10.1038/ismej.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emerson D, Revsbech N. 1994. Investigation of an iron-oxidizing microbial mat community located near Aarhus, Denmark: field studies. Appl Environ Microbiol 60:4022–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haaijer SCM, Harhangi HR, Meijerink BB, Strous M, Pol A, Smolders AJP, Verwegen K, Jetten MSM, Op Den Camp HJM. 2008. Bacteria associated with iron seeps in a sulfur-rich, neutral pH, freshwater ecosystem. ISME J 2:1231–1242. doi: 10.1038/ismej.2008.75. [DOI] [PubMed] [Google Scholar]
- 7.Fleming EJ, Cetinić I, Chan CS, King DW, Emerson D. 2014. Ecological succession among iron-oxidizing bacteria. ISME J 8:804–815. doi: 10.1038/ismej.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan CS, Mcallister SM, Leavitt AH, Glazer BT, Krepski ST, Emerson D. 2016. The architecture of iron microbial mats reflects the adaptation of chemolithotrophic iron oxidation in freshwater and marine environments. Front Microbiol 7:796. doi: 10.3389/fmicb.2016.00796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruun A-M, Finster K, Gunnlaugsson H, Nørnberg P, Friedrich MW. 2010. A comprehensive investigation of iron cycling in a freshwater seep including microscopy, cultivation and molecular community analysis. Geomicrobiol J 27:15–34. doi: 10.1080/01490450903232165. [DOI] [Google Scholar]
- 10.Fru EC, Piccinelli P, Fortin D. 2012. Insights into the global microbial community structure associated with iron oxyhydroxide minerals deposited in the aerobic biogeosphere. Geomicrobiol J 29:587–610. doi: 10.1080/01490451.2011.599474. [DOI] [Google Scholar]
- 11.Rentz JA, Turner I, Ullman J. 2009. Removal of phosphorus from solution using biogenic iron oxides. Water Res 43:2029–2035. doi: 10.1016/j.watres.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 12.Winogradsky S. 1922. Eisenbakterien als Anogoxydanten. Zentralb Bakteriol Parasitenkd Infektionskrankh Hyg 57:1–21. [Google Scholar]
- 13.van Veen W, Mulder E, Deinema M. 1978. The Sphaerotilus-Leptothrix group of bacteria. Microbiol Rev 42:329–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghiorse WC. 1984. Biology of iron- and manganese-depositing bacteria. Annu Rev Microbiol 38:515–550. doi: 10.1146/annurev.mi.38.100184.002503. [DOI] [PubMed] [Google Scholar]
- 15.Sobolev D, Roden EE. 2004. Characterization of a neutrophilic, chemolithoautotrophic Fe(II)-oxidizing β-proteobacterium from freshwater wetland sediments. Geomicrobiol J 21:1–10. doi: 10.1080/01490450490253310. [DOI] [Google Scholar]
- 16.Emerson D, Fleming EJ, McBeth JM. 2010. Iron-oxidizing bacteria: an environmental and genomic perspective. Annu Rev Microbiol 64:561–583. doi: 10.1146/annurev.micro.112408.134208. [DOI] [PubMed] [Google Scholar]
- 17.Fleming EJ, Langdon AE, Martinez-Garcia M, Stepanauskas R, Poulton NJ, Masland EDP, Emerson D. 2011. What's new is old: resolving the identity of Leptothrix ochracea using single-cell genomics, pyrosequencing and FISH. PLoS One 6:e17769. doi: 10.1371/journal.pone.0017769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spring S. 2006. The Genera Leptothrix and Sphaerotilus, p 758–777. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (ed), The prokaryotes, 3rd ed Springer, New York, NY. [Google Scholar]
- 19.Stepanauskas R. 2012. Single-cell genomics: an individual look at microbes. Curr Opin Microbiol 15:613–620. doi: 10.1016/j.mib.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Musat N, Musat F, Weber PK, Pett-Ridge J. 2016. Tracking microbial interactions with NanoSIMS. Curr Opin Biotechnol 41:114–121. doi: 10.1016/j.copbio.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Emerson D, Merrill-Floyd M. 2005. Enrichment and isolation of iron-oxidizing bacteria at neutral pH. Methods Enzymol 397:112–123. doi: 10.1016/S0076-6879(05)97006-7. [DOI] [PubMed] [Google Scholar]
- 22.Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, Pillay M, Ratner A, Huang J, Woyke T, Huntemann M, Anderson I, Billis K, Varghese N, Mavromatis K, Pati A, Ivanova NN, Kyrpides NC. 2014. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res 42:D560–D567. doi: 10.1093/nar/gkt963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barco RA, Emerson D, Sylvan JB, Orcutt BN, Jacobson Meyers ME, Ramírez GA, Zhong JD, Edwards KJ. 2015. New insight into microbial iron oxidation as revealed by the proteomic profile of an obligate iron-oxidizing chemolithoautotroph. Appl Environ Microbiol 81:5927–5937. doi: 10.1128/AEM.01374-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato S, Ohkuma M, Powell DH, Krepski ST, Oshima K, Hattori M, Shapiro N, Woyke T, Chan CS. 2015. Comparative genomic insights into ecophysiology of neutrophilic, microaerophilic iron oxidizing bacteria. Front Microbiol 6:1265. doi: 10.3389/fmicb.2015.01265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emerson D, Field EK, Chertkov O, Davenport KW, Goodwin L, Munk C, Nolan M, Woyke T. 2013. Comparative genomics of freshwater Fe-oxidizing bacteria: implications for physiology, ecology, and systematics. Front Microbiol 4:254. doi: 10.3389/fmicb.2013.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Field EK, Sczyrba A, Lyman AE, Harris CC, Woyke T, Stepanauskas R, Emerson D. 2015. Genomic insights into the uncultivated marine Zetaproteobacteria at Loihi Seamount. ISME J 9:857–870. doi: 10.1038/ismej.2014.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He S, Tominski C, Kappler A, Behrens SF, Roden EE. 2016. Metagenomic analyses of the autotrophic Fe(II)-oxidizing, nitrate-reducing enrichment culture KS. Appl Environ Microbiol 82:2656–2668. doi: 10.1128/AEM.03493-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Troussellier M, Bouvy M, Courties C. 1997. Variation of carbon content among bacterial species under starvation condition. Aquat Microb Ecol 13:113–119. doi: 10.3354/ame013113. [DOI] [Google Scholar]
- 29.Rentz JA, Kraiya C, Luther G III, Emerson D. 2007. Control of ferrous iron oxidation within circumneutral microbial iron mats by cellular activity and autocatalysis. Environ Sci Technol 41:6084–6089. doi: 10.1021/es062203e. [DOI] [PubMed] [Google Scholar]
- 30.Sobolev D, Roden EE. 2002. Evidence for rapid microscale bacterial redox cycling of iron in circumneutral environments. Antonie Van Leeuwenhoek 81:587–597. doi: 10.1023/A:1020569908536. [DOI] [PubMed] [Google Scholar]
- 31.Chan CS, Fakra SC, Emerson D, Fleming EJ, Edwards KJ. 2011. Lithotrophic iron-oxidizing bacteria produce organic stalks to control mineral growth: implications for biosignature formation. ISME J 5:717–727. doi: 10.1038/ismej.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan CS, De Stasio G, Welch SA, Girasole M, Frazer BH, Nesterova MV, Fakra S, Banfield JF. 2004. Microbial polysaccharides template assembly of nanocrystal fibers. Science 303:1656–1658. doi: 10.1126/science.1092098. [DOI] [PubMed] [Google Scholar]
- 33.Vesenka J, Havu J, Hruby K, Emerson D. 2018. A model for sheath formation coupled to motility in Leptothrix ochracea. Geomicrobiol J 35:366–374. doi: 10.1080/01490451.2017.1370516. [DOI] [Google Scholar]
- 34.Nelson DC, Hagen K. 1995. Physiology and biochemistry of symbiotic and free-living chemoautotrophic sulfur bacteria. Integr Comp Biol 35:91. [Google Scholar]
- 35.Emerson D, Moyer C. 1997. Isolation and characterization of novel iron-oxidizing bacteria that grow at circumneutral pH. Appl Environ Microbiol 63:4784–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hallbeck L, Pedersen K. 1990. Culture parameters regulating stalk formation and growth rate of Gallionella ferruginea. Microbiology 136:1675. [Google Scholar]
- 37.Singer E, Emerson D, Webb EA, Barco RA, Kuenen JG, Nelson WC, Chan CS, Comolli LR, Ferriera S, Johnson J, Heidelberg JF, Edwards KJ. 2011. Mariprofundus ferrooxydans PV-1 the first genome of a marine Fe(II) oxidizing Zetaproteobacterium. PLoS One 6:e25386. doi: 10.1371/journal.pone.0025386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiu BK, Shingo K, McAllister SM, Field EK, Clara CS. 2017. Novel pelagic iron-oxidizing Zetaproteobacteria from the Chesapeake Bay oxic-anoxic transition zone. Front Microbiol 8:1280. doi: 10.3389/fmicb.2017.01280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Druschel GK, Emerson D, Sutka R, Suchecki P, Luther G III. 2008. Low-oxygen and chemical kinetic constraints on the geochemical niche of neutrophilic iron(II) oxidizing microorganisms. Geochim Cosmochim Acta 72:3358–3370. doi: 10.1016/j.gca.2008.04.035. [DOI] [Google Scholar]
- 40.Krepski ST, Emerson D, Hredzak-Showalter PL, Luther GW III, Chan CS. 2013. Morphology of biogenic iron oxides records microbial physiology and environmental conditions: toward interpreting iron microfossils. Geobiology 11:457–471. doi: 10.1111/gbi.12043. [DOI] [PubMed] [Google Scholar]
- 41.Burkhardt DH, Rouskin S, Zhang Y, Li G-W, Weissman JS, Gross CA. 2017. Operon mRNAs are organized into ORF-centric structures that predict translation efficiency. Elife Sci 6:e22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuenen JG, Veldkamp H. 1973. Effects of organic compounds on growth of chemostat cultures of Thiomicrospira pelophila, Thiobacillus thioparus and Thiobacillus neapolitanus. Arch Mikrobiol 94:173–190. doi: 10.1007/BF00416691. [DOI] [PubMed] [Google Scholar]
- 43.Kelly DP, Wood AP, Gottschal JC. 1979. Autotrophic metabolism of formate by Thiobacillus strain A2. J Gen Microbiol 114:1–13. doi: 10.1099/00221287-114-1-1. [DOI] [Google Scholar]
- 44.Smith AL, Kelly DP. 1979. Competition in the chemostat between an obligately and a facultatively chemolithotrophic Thiobacillus. Microbiology 115:377–384. [Google Scholar]
- 45.Beudeker RF, Gottschal JC, Kuenen JG. 1982. Reactivity versus flexibility in thiobacilli. Antonie Van Leeuwenhoek 48:39–51. doi: 10.1007/BF00399485. [DOI] [PubMed] [Google Scholar]
- 46.Gottschal JC, Nanninga HJ, Kuenen JG. 1981. Growth of Thiobacillus A2 under alternating growth conditions in the chemostat. J Gen Microbiol 126:85–96. [Google Scholar]
- 47.Harwood CS, Gibson J. 1988. Anaerobic and aerobic metabolism of diverse aromatic compounds by the photosynthetic bacterium Rhodopseudomonas palustris. Appl Environ Microbiol 54:712–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKinlay JB, Harwood CS. 2010. Carbon dioxide fixation as a central redox cofactor recycling mechanism in bacteria. Proc Natl Acad Sci U S A 107:11669–11675. doi: 10.1073/pnas.1006175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams MR, Wessel BM, Filoso S. 2016. Sources of iron (Fe) and factors regulating the development of flocculate from Fe-oxidizing bacteria in regenerative streamwater conveyance structures. Ecol Eng 95:723–737. doi: 10.1016/j.ecoleng.2016.06.120. [DOI] [Google Scholar]
- 50.Shotyk W. 1988. Review of the inorganic geochemistry of peats and peatland waters. Earth Sci Rev 25:95–176. doi: 10.1016/0012-8252(88)90067-0. [DOI] [Google Scholar]
- 51.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gnerre S, Maccallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, Sharpe T, Hall G, Shea TP, Sykes S, Berlin AM, Aird D, Costello M, Daza R, Williams L, Nicol R, Gnirke A, Nusbaum C, Lander ES, Jaffe DB. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A 108:1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Makarova KS, Aravind L, Galperin MY, Grishin NV, Tatusov RL, Wolf YI, Koonin EV. 1999. Comparative genomics of the Archaea (Euryarchaeota): evolution of conserved protein families, the stable core, and the variable shell. Genome Res 9:608–628. [PubMed] [Google Scholar]
- 57.Woyke T, Xie G, Copeland A, González JM, Han C, Kiss H, Saw JH, Senin P, Yang C, Chatterji S, Cheng J-F, Eisen JA, Sieracki ME, Stepanauskas R. 2009. Assembling the marine metagenome, one cell at a time. PLoS One 4:e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wasmund K, Schreiber L, Lloyd KG, Petersen DG, Schramm A, Stepanauskas R, Jørgensen BB, Adrian L. 2014. Genome sequencing of a single-cell of the widely distributed marine subsurface Dehalococcoidia, phylum Chloroflexi. ISME J 8:383–397. doi: 10.1038/ismej.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol System Biol 7:1–6. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emerson D, Weiss J. 2004. Bacterial iron oxidation in circumneutral freshwater habitats: findings from the field and the laboratory. Geomicrobiol J 21:405–414. doi: 10.1080/01490450490485881. [DOI] [Google Scholar]
- 63.Thamdrup B, Finster K, Hansen JW, Bak F. 1993. Bacterial disproportionation of elemental sulfur coupled to chemical reduction of iron or manganese. Appl Environ Microbiol 59:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pernthaler A, Pernthaler J, Amann R. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microbiol 68:3094–3101. doi: 10.1128/AEM.68.6.3094-3101.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amann R, Krumholz L, Stahl D. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol 172:762. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Polerecky L, Adam B, Milucka J, Musat N, Vagner T, Kuypers MMM. 2012. Look@NanoSIMS—a tool for the analysis of nanoSIMS data in environmental microbiology. Environ Microbiol 14:1009–1023. doi: 10.1111/j.1462-2920.2011.02681.x. [DOI] [PubMed] [Google Scholar]
- 67.Stookey LL. 1970. Ferrozine—a new spectrophotometric reagent for iron. Anal Chem 42:779–781. doi: 10.1021/ac60289a016. [DOI] [Google Scholar]
- 68.Hall POJ, Aller RC. 1992. Rapid, small-volume, flow injection analysis for SCO2, and NH4+ in marine and freshwaters. Limnol Oceanogr 37:1113–1119. doi: 10.4319/lo.1992.37.5.1113. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.