ABSTRACT
Phages of Streptococcus thermophilus present a major threat to the production of many fermented dairy products. To date, only a few studies have assessed the biodiversity of S. thermophilus phages in dairy fermentations. In order to develop strategies to limit phage predation in this important industrial environment, it is imperative that such studies are undertaken and that phage-host interactions of this species are better defined. The present study investigated the biodiversity and evolution of phages within an Irish dairy fermentation facility over an 11-year period. This resulted in the isolation of 17 genetically distinct phages, all of which belong to the so-called cos group. The evolution of phages within the factory appears to be influenced by phages from other dairy plants introduced into the factory for whey protein powder production. Modular exchange, primarily within the regions encoding lysogeny and replication functions, was the major observation among the phages isolated between 2006 and 2016. Furthermore, the genotype of the first isolate in 2006 was observed continuously across the following decade, highlighting the ability of these phages to prevail in the factory setting for extended periods of time. The proteins responsible for host recognition were analyzed, and carbohydrate-binding domains (CBDs) were identified in the distal tail (Dit), the baseplate proteins, and the Tail-associated lysin (Tal) variable regions (VR1 and VR2) of many isolates. This supports the notion that S. thermophilus phages recognize a carbohydrate receptor on the cell surface of their host.
IMPORTANCE Dairy fermentations are consistently threatened by the presence of bacterial viruses (bacteriophages or phages), which may lead to a reduction in acidification rates or even complete loss of the fermentate. These phages may persist in factories for long periods of time. The objective of the current study was to monitor the progression of phages infecting the dairy bacterium Streptococcus thermophilus over a period of 11 years in an Irish dairy plant so as to understand how these phages evolve. A focused analysis of the genomic region that encodes host recognition functions highlighted that the associated proteins harbor a variety of carbohydrate-binding domains, which corroborates the notion that phages of S. thermophilus recognize carbohydrate receptors at the initial stages of the phage cycle.
KEYWORDS: Streptococcus, dairy industry, receptor binding protein, genomics, bacteriophage
INTRODUCTION
Streptococcus thermophilus is one of the most extensively employed commercial starter cultures, being widely used in the manufacture of fermented milk products, such as yogurt and various cheeses (1, 2). Phage infection of S. thermophilus starter strains may result in incomplete or failed fermentations, with considerable economic consequences to the dairy industry. Analysis of phage-host interactions is essential in order to derive a detailed understanding of how these problematic phages recognize and infect their host bacteria as a means to prevent or limit phage-mediated problems in the dairy fermentation setting. The initial interaction between S. thermophilus bacteriophages DT1 and MD2 and their hosts has been reported to involve three phage proteins, including the tail tape measure and the host specificity protein (3). The receptor material for these phages is presumed to be a carbohydrate component of the cell wall based on adsorption assays of S. thermophilus phages to differently treated cell wall extracts of their hosts (4).
Several worldwide phage isolation studies have shown that two prevalent groups of S. thermophilus phages exist. These groups are distinguished based on structural protein content and their mode of packaging, which is determined by the recognition of specific sequences, namely, the cos and pac sites, followed by specific or nonspecific cleavage of the DNA (5). The cos phages incorporate cohesive “sticky” ends into their genomes, while pac phages employ a so-called headful DNA packaging system and, therefore, may incorporate additional redundant DNA into their genomes. PCR-based methods targeting the antireceptor gene and the gene encoding the major capsid protein, sometimes in combination with analysis of structural protein content, are two of the primary approaches currently used to identify to which of the subgroups new phage isolates belong (6–8). To date, 64 S. thermophilus phage genomes have been sequenced, approximately half of which utilize the cos packaging mode (i.e., 34 of 64 sequenced phage genomes). Of the remaining 30 S. thermophilus phage genomes, 18 employ the headful packaging or pac method. In addition to the dominantly isolated cos and pac phages, two genetically distinct groups of S. thermophilus phages have recently been described. These include the 5093 group, whose genomes bear greater similarity to prophages of nondairy streptococci than those of dairy streptococcal phages (9), and the 987 group, whose genomes bear similarity to those of lactococcal P335 phages. The mode of DNA packaging of both of these newly described phage groups is not yet described (10). The identification of such novel groups of S. thermophilus phages highlights the importance of continued evaluation of phage biodiversity in dairy fermentation environments to identify resident populations and to develop robust starter strains and strain rotations.
Phage biodiversity surveys in dairy fermentation facilities are widely reported for lactococcal phages (11–18), while studies on S. thermophilus phage biodiversity are comparatively limited (9, 10, 19–21). While these studies have considerable merit in identifying the biodiversity of phages at a given time point, they do not provide temporal insights into the prevalence, maintenance, evolution, and diversification of genetic lineages of phages within the industrial setting. In 2009, a longitudinal study of the evolution of lactococcal lytic 936 group phages in a Canadian cheese factory (22) highlighted that certain genetic lineages are able to survive in the plant for over a year and that genetic diversification was observed between the phages that were isolated over a 9-year period. To our knowledge, no such longitudinal studies of S. thermophilus phages have been published to date.
The success of phages in the dairy environment may be attributed to many factors, including their ability to adapt to host defense mechanisms and their innate resistance to chemical and thermal treatments applied in the dairy industry (23, 24). Significant effort has been invested toward understanding the adaptive responses of phages to host-encoded phage resistance systems in S. thermophilus, particularly with respect to clustered regularly interspaced palindromic repeat (CRISPR) immune systems (25–27). Studies of phage adaptive responses to thermal and chemical treatments have demonstrated the increasing insensitivity of phages of Lactococcus lactis, Lactobacillus delbrueckii, and S. thermophilus to such interventions, thus establishing the requirement for industrial strategies to overcome this issue (24, 28, 29).
In the current study, 17 genetically distinct phages were isolated from an Irish dairy fermentation facility (factory A) over a period of more than a decade (2006 to 2016). The phages were isolated from cheese whey samples produced within the factory. Furthermore, cheese whey (as potential reservoirs of novel phage lineages) that had been introduced into a remote location on the factory site from other producers was also assessed for the presence of phages. The externally derived whey is used in the production of whey protein powders, and such whey protein powders have been demonstrated to be a rich source of dairy phages (30). Furthermore, phages display significant stability in this format, providing an element of risk if whey protein powder is produced at the same site as the primary fermentation. The genomes of these phage isolates were sequenced, and comparative genome analysis revealed two genetic lineages of cos type phages within the cheese production samples. Additionally, a further two genetic lineages exist among phage isolates associated with cheese whey acquired from other cheese-producing facilities that are introduced into the plant for whey protein powder production. Focused analysis of the region encoding the predicted host interacting functions, such as the distal tail (Dit), baseplate (BPP), and the host specificity protein/tail-associated lysin (Tal), highlighted the diversification of these components through the acquisition of carbohydrate-binding domains, which corroborates current thinking that phages of S. thermophilus recognize a carbohydrate receptor on their host cell surface.
RESULTS
Phage isolations.
Between 2006 and 2016, more than 1,000 cheese whey samples from factory A were tested against S. thermophilus P1. This strain is the primary S. thermophilus production strain used to produce a particular Irish hard cheese for a period of approximately 3 to 4 months of the year. From these samples, a single phage type (named STP1) was isolated in 2006. This was considered the starting point of the evolutionary mapping of phages of the production strain P1 in this study. In subsequent years, samples were analyzed for phages, and restriction profile analysis identified the continued presence of the STP1 type phage in the production samples (Table 1) throughout the testing period of a decade, despite the fact that the strain is only in use for at most one-third of the year. Phage isolates with (minor) modifications in their restriction profiles were considered for further testing by host range analysis, multiplex PCR-based typing, and genome sequencing. In addition to studying the phages from the cheese production facility of factory A, this study was aimed at identifying possible sources of different S. thermophilus phage genetic lineages that contribute to the development and evolution of phages in this particular cheese factory. Therefore, cheese whey samples (2,043 samples) acquired from other factories for whey protein powder production were also tested for the presence of phages of S. thermophilus, and their relatedness to those identified in the production plant was assessed. These samples were tested against a panel of 52 S. thermophilus dairy strains that were available within our collection to obtain maximum phage diversity.
TABLE 1.
Details of the samples tested in this study and the phages isolated between 2006 and 2016
| Yr by sample typea | No. of samples | Total no. of tests performedb | No. of phage-positive samples | No. of phages isolated | Isolate genotype (no. of isolates) |
|---|---|---|---|---|---|
| Factory A whey samples | |||||
| 2006 | 105 | NA | 68 | 14 | STP1 (14) |
| 2007 | 98 | NA | 58 | 12 | STP1 (12) |
| 2008 | 97 | NA | 62 | 16 | STP1 (15), STP2 (1) |
| 2009 | 102 | NA | 59 | 14 | STP1 (14) |
| 2010 | 101 | NA | 67 | 12 | STP1 (10), STP2 (2) |
| 2011 | 109 | NA | 65 | 20 | STP1 (16), STP2 (4) |
| 2012 | 115 | NA | 65 | 16 | STP1 (16) |
| 2013 | 106 | NA | 68 | 14 | STP1 (12), STP2 (2) |
| 2014 | 98 | NA | 66 | 12 | STP1 (12) |
| 2015 | 101 | NA | 69 | 16 | STP1 (6), STP2 (2), A0 (1), B0 (1), C0 (1), 9B4 (1), 16B8 (2), 31B4 (2) |
| 2016 | 104 | NA | 64 | 14 | STP1 (12), 7T (2) |
| Total | 1,136 | NA | 711 (63%) | 160 | |
| Externally acquired whey samples | |||||
| 2006 | 94 | 4,888 | 62 | 10 | STP1 (10) |
| 2007 | 98 | 5,096 | 60 | 12 | STP1 (12) |
| 2008 | 96 | 4,992 | 47 | 12 | STP1 (12) |
| 2009 | 92 | 4,784 | 54 | 12 | STP1 (10), STP2 (2) |
| 2010 | 100 | 5,200 | 59 | 14 | STP1 (14) |
| 2011 | 96 | 4,992 | 69 | 12 | STP1 (9), STP2 (3) |
| 2012 | 290 | 15,080 | 207 | 14 | STP1 (4), STP2 (5), B5 (5) |
| 2013 | 306 | 15,912 | 223 | 14 | STP1 (6), B5 (8) |
| 2014 | 295 | 15,340 | 201 | 14 | B5 (10), MM25 (2), M19 (2) |
| 2015 | 305 | 15,860 | 244 | 16 | STP1 (2), 9A (2), L5A1 (2), 7A5 (4), 7T (6) |
| 2016 | 271 | 14,092 | 164 | 16 | A0 (2), B5 (2), STP2 (4), V2 (5), R1 (3) |
| Total | 2,043 | 106,236 | 1,390 (68%) | 146 |
The sampling period for each year was between March and early July. NA, not available.
This refers only to externally acquired samples, as factory A-derived samples were all prescreened on S. thermophilus P1 and only the isolated propagated phages were tested against the panel of 52 S. thermophilus strains. Total number of tests performed as calculated as the number of samples × 52 bacterial strains.
In this study, 17 genetically distinct phages were isolated, eight of which were isolated from factory A-derived cheese whey samples in 2006, 2008, and 2015 (STP1 in 2006, STP2 in 2008, and A0, B0, C0, 9B4, 16B8, and 31B4 in 2015). Between 2008 and 2015, >100 phage isolates with restriction fragment length polymorphism profiles nearly identical to those of STP1 (and, on occasion, STP2) were identified only within the cheese factory itself, highlighting the dominant application of particular starter cultures and the enduring nature of the STP1/STP2 phages in the plant (Table 1). While no novel isolates were observed in the factory during this 7-year period (2008 to 2015), a novel phage genotype (B5) was isolated in the external whey samples in 2012, and additional novel genotypes were identified in the external whey samples in 2014 (MM25 and M19), 2015 (9A, L5A1, 7A5, and 7T), and 2016 (V2 and R1) (see phage list in Table 2). A noteworthy point is the spread of samples across the 11-year period. While an approximately equal number of samples from the factory were tested each year, the number of externally derived samples was increased considerably from 2012 onwards to expand the potential for isolation of novel genotypes. Thus, from 2006 to 2011, approximately 100 externally derived samples per year were tested, while from 2012 to 2016, approximately 300 samples were assessed. This resulted in an increase in the number of genotypes identified, highlighting the benefit of extensive screening. Sixty-three percent of samples from factory A were positive for the presence of phages targeting strain P1, with titers ranging from 102 to 105 PFU · ml−1 in the original whey samples. Sixty-eight percent of the externally derived samples were phage positive against at least one of the 52 strains included in the testing panel, with phage titers ranging from 102 to 108 PFU · ml−1. All phage isolates were identified as cos-type phages by multiplex PCR, with the corresponding PCR product of 170 bp visualized on a 1% agarose gel (data not shown).
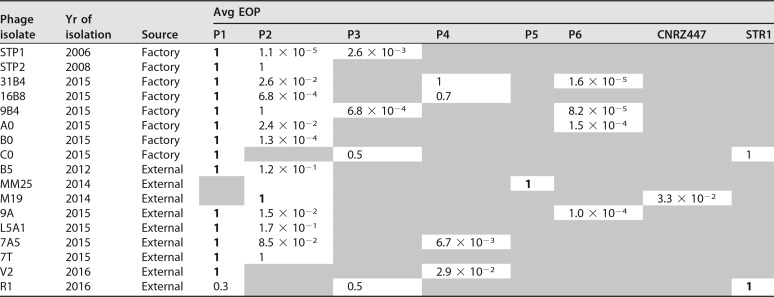
TABLE 2.
Host range highlighting the average EOP of phage isolates relative to the primary host and details of the origin of the isolatesa
The host range of each of the phages was assessed against a panel of 52 S. thermophilus strains. Only those strains that were sensitive to infection by one or more of the phages are presented in the table. Shaded areas indicate strains that were insensitive to infection by the isolated phages. Primary hosts are in bold.
Host range analysis.
All samples derived from the factory were initially tested against S. thermophilus P1 (the strain used in production), and phage isolates were propagated on this strain and subsequently used in a challenge against a collection of 51 additional S. thermophilus strains. The majority of phage isolates from the factory were identified to have a common secondary host (P2), while phage-specific infection profiles were also observed (Table 2). In order to assess the extent of phage biodiversity, whey samples acquired from other factories were screened against the panel of 52 S. thermophilus strains without a prescreening on P1 alone (see Materials and Methods). Many of these samples also contained phages capable of infecting S. thermophilus P1, with six of the nine phages isolated from externally derived whey identified originally on this strain only, and upon production of high-titer lysates (>107 PFU · ml−1), additional hosts were identified (Table 2). Interestingly, phages MM25, M19, and R1, which were identified on primary hosts other than P1 in the phage screen (Table 1), have unique and narrow-host-range profiles. While these phages were not isolated on strain P1, when a high-titer lysate of MM25 was produced on its primary host strain, a very small subpopulation was capable of infecting P1 (at an efficiency of plaquing of 10−6), highlighting their ability to readily adapt to this strain (and others).
Phage lineages.
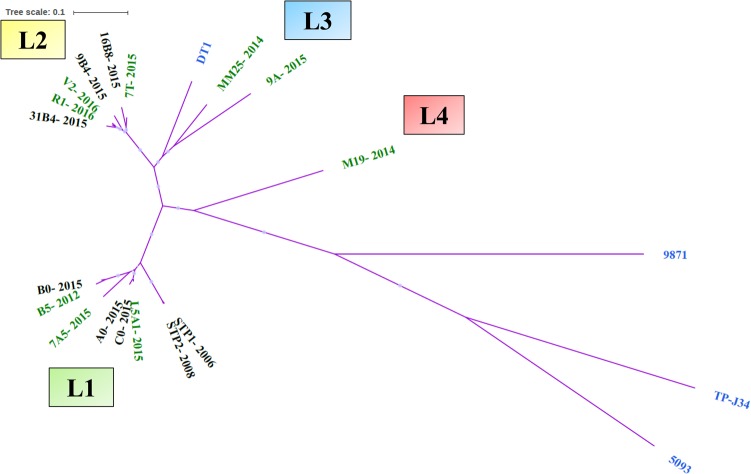
During the 11-year period between 2006 and 2016, more than 300 individual phage isolates were compared (by restriction profiling, data not shown), of which the vast majority (96%) were identified as exhibiting STP1-like profiles (lineage 1) in both the factory- and externally derived whey (Tables 1 and 2). However, a small number of isolates were identified for which clearly distinct restriction profiles and/or host ranges were observed. These were considered for further analysis, resulting in the identification of 17 distinct S. thermophilus phages, which were then subjected to whole-genome analysis (Table S1). Based on phylogenetic analysis of the overall nucleotide sequences, there appears to be four genetic lineages of phages, including two derived from phages within the cheese production facility of factory A (lineages 1 and 2) and two from externally derived whey samples only (lineages 3 and 4) (Fig. 1). The first lineage is that of STP1, the first isolate in 2006, further including phage isolates STP2, A0, B0, C0, L5A1, B5, and 7A5. Phages of this lineage appear to share a similar host range profile and are characterized by their ability to primarily infect strains P1 and P2 (Table 2). Five of these phages originate from the cheese whey from factory A, while three phages were isolated from cheese whey from external sources (B5 in 2012 and L5A1 and 7A5 in 2015) (Fig. 1 and Table 1). In order to perform a more focused analysis of the genetic content of the 17 phage isolates, the most closely related isolates were identified by the alignment of their nucleotide sequences (Fig. 1). This formed the basis of a neighbor mapping of phage genomes following the order identified in the phylogenetic analysis, as displayed in Fig. 2 (lineage 1 and 2 phage isolates) and 3 (lineage 3 and 4 phages that have not yet been observed in factory A). In this analysis, STP1 was the base comparator, as it was the first isolate and served to highlight the major regions of divergence between phages of different lineages and within lineage 1 phages.
FIG 1.
Unrooted phylogenetic tree of the 17 sequenced isolates and a representative cos (DT1), pac (TP-J34), 5093 (5093), and 987 (9871) groups included as references. All phage isolates group most closely to DT1, while distinct subgroups or lineages (L) of the phage isolates are also highlighted in the tree (L1 to L4). Factory-derived phage isolates are indicated in black text, externally derived phage isolates are indicated in green text, and representatives of the four S. thermophilus groups (cos, pac, 5093, and 987) are indicated in blue text.
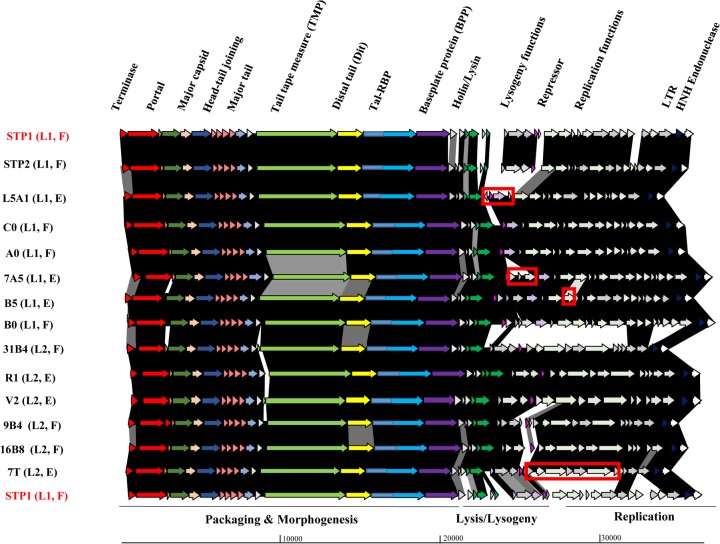
FIG 2.
Schematic representation of the genomes of lineage 1 (STP1, STP2, C0, A0, 7A5, B5, B0, and L5A1) and lineage 2 (7T, 9B4, 31B4, 16B8, R1, and V2) phages. Arrows (indicating protein-encoding regions) joined by shaded boxes indicate genetic regions of similarity, with the black shading indicating 90 to 100%, dark gray indicating 80 to 89%, light gray indicating 50 to 79%, and off-white indicating 30 to 49% aa identity. Arrows of the same color represent genes with a similar function. Gray arrows are indicative of genes encoding proteins of unknown function. The predicted functions of the encoded proteins are presented above the relevant arrows, where known and the functional modules are presented below the schematic. The major region of divergence between 7T (the first lineage 2 isolate) and STP1 (first lineage 1 isolate) is highlighted in the 7T genome representation by a red box. Similarly, regions of genetic novelty associated with isolates B5, 7A5, and L5A1 are highlighted in red boxes. All lineage 2 phage genomes possess a genomic region with greater than 90% identity. The lineage (L1/2) and source (F, factory; E, external) are also presented on the left side of the figure. LTR, long terminal repeat.
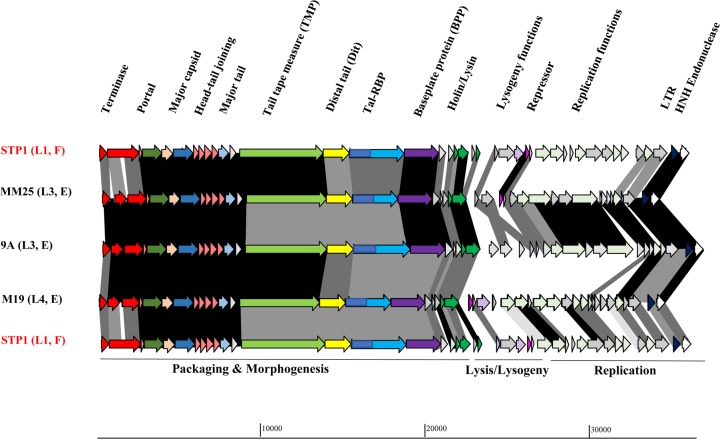
FIG 3.
Schematic representation of the genomes of lineage 3 (MM25 and 9A) and lineage 4 (M19) phages and their comparison to the first isolate of the study (STP1, lineage 1). Arrows (indicating protein-encoding regions) joined by shaded boxes indicate genetic regions of similarity, with the black shading indicating 90 to 100%, dark gray indicating 80 to 89%, light gray indicating 50 to 79%, and off-white indicating 30 to 49% aa identity. Arrows of the same color represent genes with a similar function. Gray arrows are indicative of genes encoding proteins of unknown function. The predicted functions of the encoded proteins are presented above the relevant arrows, where known and the functional modules are presented below the schematic. The lineage (L3/4) and source (F, factory; E, external) are also presented on the left side of the figure.
While lineage 1 phages appear to be highly related, some insertions/deletions and minor rearrangements within the lysogeny and replication modules are observed in comparison to the first isolate, STP1 (Fig. 2). The genome of STP2, isolated in 2008, was essentially identical to that of STP1 but with some point mutations throughout the genome and minor sequence variations at the genomic termini. The remaining lineage 1 phages display a high degree of similarity (Fig. 2). Therefore, minor deletions and genetic rearrangements appear to be the major feature of the evolution of lineage 1 phages, while localized genetic acquisitions were also observed in L5A1, 7A5, and B5 in the lysogeny- and replication-related modules (Fig. 2). It is possible that other companies also use S. thermophilus P1 or closely related strains, thereby explaining the prevalence of this phage lineage in samples derived from external sources (Table 1).
Lineage 2 is represented by phages related to 31B4 and further represented by phage isolates 7T, 9B4, 16B8, V2, and R1 (Fig. 1). These phages appear to be highly similar to the STP1 lineage (lineage 1), most notably in the genomic region encoding the structural components, while having acquired a distinct genomic region within the predicted lysogeny and replication gene modules, with only the very rightward end of the genome displaying similarity between B0 (lineage 1) and 31B4 (lineage 2) (Fig. 2). It seems highly likely that the phages isolated in the factory (31B4, 9B4, and 16B8) have recombined with (one of the) externally derived phages, such as 7T, as 7T was isolated in the externally derived whey prior to the identification of similar restriction profiles and genetically similar isolates in the factory later the same year, possessing an almost identical replication module (Fig. 2). Phage 7T appears to be the ancestor (or its closest relative) of this lineage, since it was the first of this type to be isolated in early 2015 (Table 3), with additional members of this lineage isolated both in the factory- and externally derived whey samples appearing later in 2015 and 2016, respectively (Table 1).
TABLE 3.
Order of appearance and source of first isolates of lineage 1 and 2 phages in factory A
| Phage isolated (lineage) | Date of isolation | Location of first appearance | Reoccurrence in factory | Subsequent factory-isolated related phages |
|---|---|---|---|---|
| STP1 (1) | March 2006 | Factory A | 2007–2016 | STP2 (May 2008), A0, B0, C0 (April 2015) |
| 7T (2) | March 2015 | External factory | 2016 | 9B4, 16B8, 31B4 (June 2015), 7T (2016) |
Lineage 3 consists of MM25 and 9A, both of which were isolated from externally derived whey samples. Lineage 3 contains no members directly derived from factory A. Similarly, lineage 4, which has only one member (M19), was isolated from an externally derived whey sample in 2014. The genetic region encoding the structural elements of phages of lineages 3 and 4 bears significant similarity to those of lineages 1 and 2 (Fig. 3 displays the comparison of lineage 3 and 4 phage genomes to each other and to that of the lineage 1 phage STP1); however, these phages are considerably divergent in their putative replication and lysogeny modules, although there is moderate similarity between lineage 1 and 4 phage isolates in the replication region, suggesting a possible shared ancestry (Fig. 3). Interestingly, the primary host of MM25 and M19 is not P1, indicating that these phages are derived from factories that likely do not employ this strain, although they have demonstrated the ability to adapt to infect P1 (at a frequency of ∼10−6), among other strains. Conversely, 9A was initially isolated on strain P1 and exhibits a host range similar to those of a number of lineage 1 and 2 phages (Table 2). This may indicate that 9A was exposed to strain P1 earlier than MM25, thus providing the phage with the opportunity to adapt. Indeed, phage 9A may have appeared in the factory prior to its first detection in this study (in 2015). The abundance of 9A (and MM25 and M19) may have been below the detection threshold of this study, thus precluding its isolation in previous sampling years. Given the distinct host ranges of these isolates, it is noteworthy that lineage 3 and 4 phages displayed reduced similarity in the genomic region encoding host recognition functions (Fig. 3). Furthermore, these three isolates (MM25, 9A, and M19) were the only members of their kind identified in this study, highlighting the low incidence of these phage genotypes.
Genome organization.
The genomes of phage isolates sequenced in this study are similarly organized, with four identifiable functional modules based on BLASTP analysis, i.e., the structural/packaging, lysis, and lysogenic and replication modules (Fig. 2 and 3). The genomes of the isolated phages are 34.0 to 36.8 kb in length and carry between 39 and 48 predicted open reading frames (ORFs) (Table 2). BLASTN analysis of STP1 highlighted that this phage bears the most significant similarity to the cos-type phages Abc2 and DT1, with almost identical sequences shared across approximately 70% of their respective genomes. Three major functional modules are observed in the genomes of the isolated phages associated with morphogenesis, lysogeny, and replication, and these are highlighted below in more detail. The major region of divergence is contained within the lysogeny and replication modules, as exemplified in Fig. 2 and 3.
The most leftward functional module on the phage genomes (as depicted in Fig. 2 and 3) encodes the predicted structural components and DNA packaging system. Within the structural module of the bioinformatically analyzed phage genomes, it is possible to identify the small and large terminase-encoding genes (terS and terL, respectively), as well as genes specifying the portal, scaffolding, major capsid, head-tail joining, major tail, tail tape measure, and baseplate proteins. Pfam searches with the tail tape measure protein (TMP) sequence revealed two domains at the carboxy terminus, namely, a cysteine histidine-dependent amidohydrolase/peptidase (CHAP) and a soluble lytic transglycosylase (SLT) domain that resemble lytic domains often associated with tail-associated lysins of phages (31, 32). The majority of the structural protein-coding regions are highly conserved between all isolates, with some notable exceptions. Among these, the so-called host specificity proteins encoded by the phage isolates show significant divergence between phages belonging to lineages 1/2 and 3/4 (Fig. 2 to 4), which is likely a reflection of the distinct primary hosts on which these phages were isolated and the different sources of isolation of the phages.
FIG 4.
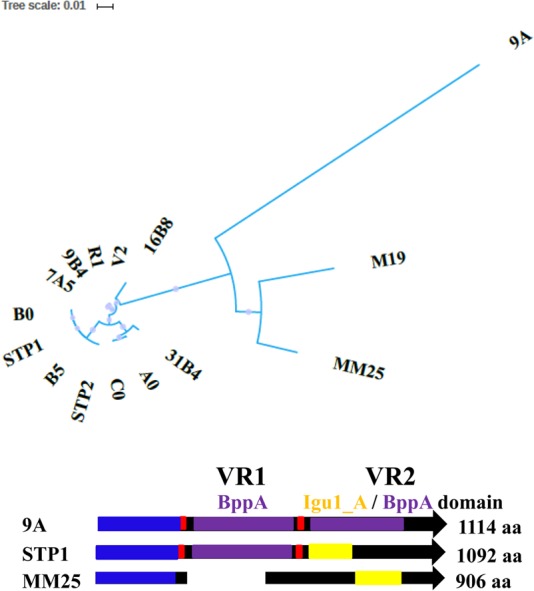
Top, unrooted phylogenetic tree of the Tal-RBPs of the 17 sequenced phages highlighting the disparity of the Tal-RBPs of the lineage 1/2 (left) and 3/4 phages (right). Bottom, schematic depicting the organization of the Tal-RBPs of the sequenced phages using representatives of the group. All Tal-RBPs possess a conserved N-terminal ∼400-aa Tal domain (blue). Phage 9A (lineage 3) is the sole phage encoding a Tal-RBP with two BppA-like (5E7T_B) CBDs (purple), and these constitute the regions described as VR1 and VR2. VR1, where present, is always flanked by collagen repeat motifs (red). The VR2 region may/may not incorporate a 5E7T_B BppA CBD (purple) and/or may represent a distinct CBD, e.g., Igu1_A domain (yellow), or a CBD of unidentified structure. STP1 is representative of the Tal-RBPs of lineage 1 and 2 phages, which all share a similar size and architecture. MM25 is representative of the lineage 4 phages.
The second functional module is represented by the lysis cassette, which typically encompasses holin- and lysin-encoding genes required for progeny phage release at the end of the phage cycle. The vast majority of S. thermophilus phages reported to date encode two lysins, and the phages isolated in this study are no exception to this. The first of the two lysins encoded by these phages possesses an amidase/peptidoglycan hydrolase domain and would be expected to encode a functional lysin. Additionally, the second (where identified) bears similarity to the lysin encoded by the S. thermophilus phages Abc2 and ALQ13.2, for which genome sequence data are available. The stacking of genes involved in lysis may imply that one of the lysins is nonfunctional through mutation or deletion events or that additional genes encoding endopeptidases/lysins were acquired through homologous recombination with other streptococcal phages.
The third gene cassette relates to lysogeny functions with an identifiable repressor for the lytic and lysogenic cycles, while a number of genes encoding proteins of unknown function were identified in several of the phage isolates as well (Fig. 2 and 3). The suggestion that this genomic region is a recombination hot spot may explain the persistence of this genomic region in virulent phages (33). Indeed, in the present study, the lysogeny module is among the most divergent genomic regions within the analyzed genomes. The genomic location of the lysogeny cassette is consistent with that of previously sequenced S. thermophilus phages, i.e., between the lysis and replication modules (34, 35).
The fourth and last module encodes replication-associated proteins, such as DnaC and a single-stranded DNA binding protein. This module is among the most divergent regions of the genomes among the 17 phage isolates (Fig. 3). The observed diversity among the replication regions of these phages is perhaps the most distinctive feature of the four lineages of phages isolated in this study. Lineage 3 and 4 phages display replication modules that are completely distinct from one another (Fig. 3), while that of M19 (lineage 4) bears some observable relationship to those of lineage 1 phages, such as STP1 (Fig. 3). Furthermore, the replication modules of lineage 1 and 2 phages appear to be distinctive, with very limited similarity in this region, as exemplified by B0 and 31B4 shown in Fig. 2. This highlights that among the cos phages isolated in this study, the most significant genomic source of diversity is within the replication module.
Tail tip functions harbor carbohydrate-binding domains.
In Siphoviridae, the first gene following the TMP-encoding gene is the distal tail protein (Dit)-encoding gene (36), which in turn is typically followed by the gene encoding the tail-associated lysin or Tal protein. The Tal protein may encompass several functions, which may or may not include lytic activity, but the ∼400 N-terminal residues have the same topology among many Siphoviridae phages (36). The TMP of the phages that are the subject of the current study is, as mentioned above, predicted to contain lytic activity. The Tal protein encoded by the phages studied in the present study contains, in addition to the topologically conserved N-terminal portion, a previously described host specificity region or receptor binding protein (RBP) domain (3, 37). We therefore refer to this protein as Tal-RBP to represent the apparent dual function of this protein. Finally, the ORF downstream of the Tal-RBP-encoding gene is termed here bpp, based on the notion that it encodes a baseplate protein (BPP). Several ORFs in the DNA region encoding tail components have been found to contribute to sugar recognition in Siphoviridae phages (38–43); thus, the sequences of Dit, Tal-RBP, and BPP were analyzed for potential carbohydrate (or other)-binding domains.
Dit proteins are designated either “classical,” i.e., with a short sequence, or “evolved,” i.e., bearing carbohydrate-binding domains (CBDs) (11, 40). The phages in this study all encode evolved Dit proteins, since they harbor a CBD that shares structural similarity (HHpred score, 99.9%) with the CBD of the evolved Dit from Lactobacillus casei phage J-1 (40).
The Tal-RBPs encoded by the phages isolated in this study range in size from 906 amino acids (aa) (MM25) to 1,114 aa (9A) (Fig. 4). This size range is consistent with previously studied S. thermophilus phage Tal proteins (coined host specificity determinants in reference 37). The Tal-RBPs of S. thermophilus phages are characterized by the presence of a conserved N-terminal region (Tal domain, aa 1 to ∼400), followed by one or two variable regions, named VR1 and VR2, flanked by multiple collagen repeats (Fig. 4) (37). Sequence analysis of Tal-RBPs of the phages isolated in this study revealed that each previously termed variable region is in most cases a predicted CBD, and the variation in size of the Tal-RBP proteins may be accounted for by the presence or absence of such CBDs.
MM25 and M19 encode the smallest Tal-RBPs of 906 aa, and these proteins lack a VR1 region, while at aa positions 488 to 570, a partial 5E7T_B domain, which corresponds to the recently described CBD of a lactococcal phage accessory baseplate protein called BppA (41), is observed. This domain is followed by a CBM4 family Igu1_A domain at positions 570 to 715 based on HHpred analysis in MM25 (Fig. 4). Together, these domains constitute the VR2 domain. In addition, the Tal-RBPs of these phages did not harbor any obvious collagen repeat motifs (G-X-Y). The host-recognizing VR2 regions of MM25 and M19 were distinct from one another, which is consistent with their unique host range profiles. Phages belonging to lineages 1 and 2 described above encode Tal-RBPs that harbor a 5E7T_B (BppA) domain with greater than 97% probability. The presence of the 5E7T_B BppA-like CBD in the Tal-RBPs of lineage 1 and 2 phages (Fig. 4) in this study correlates with the position of the first variable region, VR1, described in phages DT1 and DT2 (37). Figure 4 highlights the phylogeny of the Tal-RBPs of the phages isolated in this study and further illustrates the presence of CBDs and the relative sizes of these proteins in representative lineage 1/2 (STP1), 3 (9A), and 4 (MM25) phage isolates. To further assess if VR1 is typically representative of the BppA CBD in other S. thermophilus phages, the Tal-RBPs of DT1 and DT2 were analyzed using HHpred, revealing the presence of a similar CBD in the same relative position in both Tal-RBPs. This indicates that VR1 is, in fact, a variably present and (apparently) nonessential, yet possibly accessory, BppA-like CBD. In addition, the Tal-RBP of DT2 harbors a second CBD (now known to be a 5E7T_B BppA-like domain with 94% probability) covering part of the position formerly identified as VR2. Similarly, the Tal-RBP of phage 9A harbors two adjacent BppA-like CBDs akin to DT2 and is the largest among the predicted Tal-RBPs encoded by phages in the present study at 1,114 aa, in agreement with the acquisition of multiple CBDs (Fig. 4). The CBDs in the Tal-RBPs of the phages from the present study are also flanked by various numbers of collagen repeat motifs (G-X-Y). Therefore, it is now clear that the variable regions are largely represented by CBDs, and it is possible that the repeat motifs may aid the recombination and insertion of such domains. The VR2 domain includes the host recognition domain (as previously defined) and may additionally include a CBD in some cases, giving rise to the size variation among these proteins (and perhaps the ability to bind to a range of carbohydrate motifs).
In addition to the classical Tal-RBPs (previously termed the host specificity protein) of S. thermophilus phages, their genomes also typically harbor a gene downstream of the Tal-RBP-encoding gene whose product is currently of unknown function, although it is implicated in host interactions. HHpred analysis of this protein in this phage family (e.g., DT1 as a representative of the group of isolates) highlighted the presence of a C-terminal domain with predicted structural similarity to the phage RBP of TP901-1 (97% probability), which is involved in L. lactis cell wall polysaccharide binding. This strongly supports the notion that this highly conserved protein is part of the tail tip and is involved in host interactions.
Morphological analysis.
All phage isolates were identified as cos-type phages related to the well described phage cos-type phage DT1, which exhibits a long noncontractile tail and an isometric head. To confirm that the phages isolated in this study conform to the expected morphological characteristics of these phages, STP1 and MM25 were (randomly) selected for characterization by electron microscopy. STP1 and MM25 were found to possess tails of 253.9 ± 9.7 nm (n = 19) and 247.6 ± 6.2 nm (n = 24), respectively, and heads with diameters of 56.4 ± 1.6 nm (n = 20) and 55.7 ± 2.1 nm (n = 24), respectively (Fig. 5). Given the significant sequence relatedness of the majority of isolates in this study, it is likely that all these phages are representative of the overall group of isolates. Interestingly, a tail-associated “feather-like” appendage protruding from the tail tip was observed for both phages of 46.7 ± 2.3 nm (n = 20) (STP1) and 45.9 ± 2.9 nm (n = 23) (MM25), including a bridging fiber structure of approximately 12 nm (n = 20).
FIG 5.
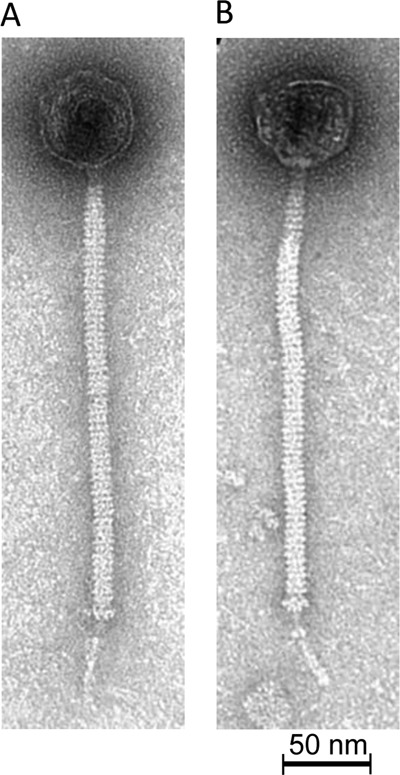
Representative electron micrographs of STP1 (A) and MM25 (B). Both phages display long tails with protruding feather-like appendages from the tail tip.
DISCUSSION
Limited studies pertaining to the diversity, persistence, and evolution of S. thermophilus phages in dairy fermentation plants have been performed (6, 7, 9, 10, 20, 21, 44, 45). Among those that have been performed, limitations, such as sequencing of very localized regions of the phage genomes, such as the VR2 regions (46), and sequencing of limited numbers of phage isolates (33), have caused knowledge of the phage-host interactions in S. thermophilus to lag behind those of their lactococcal counterparts. However, these studies have been very useful in demonstrating that cos phages predominate in dairy samples where S. thermophilus starter strains/cultures are employed. Furthermore, it has been demonstrated that phages are abundant in whey protein powder (30) and that, in particular, S. thermophilus phages are synonymous with modular exchange in terms of their evolutionary development pathways (10, 35, 47). The theory of modular exchange of S. thermophilus phage genomes was clearly demonstrated between the classical cos phages Sfi21 and 7201, among others (33). This highlights that while slow evolutionary drift may occur in this species, homologous recombination of small sets of genes or, indeed, entire modules, such as the replication module in the case of the above-mentioned phages (33) or the morphogenesis module in the case of the 987 phages (10), is a widely observed phenomenon. In the broader context, phage genome evolution has been proposed to proceed via a high- or low-gene-content flux depending on the host species, the lytic or temperate nature of the phages, and the balance of these two types of phages within a given species (48). In S. thermophilus, the incidence of lysogeny is reportedly low (49) and therefore, it would perhaps be expected that these phages would likely follow the low-gene-content flux, as high-gene-content flux is more typically observed among temperate phages. However, this does not appear to be the case for S. thermophilus phages, where modular shuffling and recombination are observed among lytic phages.
In the present study, we report the genomic structure and suggest the evolutionary development of 17 novel S. thermophilus cos phages isolated from whey samples from a single Irish factory, as influenced by whey that was imported into the factory site for processing into whey protein powder. Given that whey protein powder is known to act as a rich reservoir for dairy phages, it is unsurprising perhaps that the phages from the externally derived whey appear to have recombined with phages in the cheese factory itself (30). The identification of novel genotypes in the external whey powders often coincided with the identification of similar genotypes in the factory, indicating the likely entry of the phages from the imported whey into the cheese factory and/or recombination with the existent phage population. This is significant, since there may be a potential for recombination with existing lytic phages in the factory or, to a lesser extent, integrated prophages of P1 (although it is currently not known if P1 harbors prophages in its genome) or other strains employed in the factory. While the incidence of lysogeny is not very high in S. thermophilus, modular exchange of lytic phages is a well-established phenomenon in phages of this species (10, 44, 45, 50). For example, phages 7A5 and L5A1 predate A0, B0, and C0 in the phage isolations during 2015, and there is considerable relatedness between these lineage 1 phages (Fig. 1 and 2). Furthermore, the lineage 2 phage 7T predates 9B4, 16B8, and 31B4, and the genomes of these phages were demonstrated to exhibit novel replication regions relative to lineage 1, 3, or 4 phages (Fig. 2 and 3). Interestingly, the identification of 7T in March 2015 in the externally derived whey samples was followed by the identification of phage isolates with similar DNA restriction profiles in the factory (represented by 9B4, 16B8, and 31B4) later in 2015 (June). Thus, it is likely that lineage 2 phages primarily derived from lineage 1 phages, as their packaging and morphogenesis modules are highly conserved, and homologous recombination and modular exchange with 7T-like phages from the externally derived phages resulted in the presence of lineage 2 phages in the factory. Lineage 3 and 4 phages appear to exhibit a higher degree of genetic novelty and divergence and are, therefore, considered distinct genetic lineages from those represented by lineages 1 and 2. While lineage 3 and 4 isolates MM25 (2014), 9A (2015), and M19 (2014) were characterized as harboring significant diversity in their genetic content, there does not appear to have been a coincidence of phages with similar genotypes in the factory as of yet. These phages were only isolated on two occasions (Table 1); therefore, they are less abundant and prevalent in the processing site, thereby limiting their development and transfer to the cheese factory. However, their presence on the factory site harks a warning to producers to monitor for the continued presence of such phages that may become problematic if they become prevalent or more abundant.
Longitudinal studies such as this one are important to understand the natural evolutionary processes at play in the industrial context and reinforce the notion of evolution of S. thermophilus phages by modular rearrangements and acquisitions while minor evolutionary shifts are also observed (10, 47, 50). Furthermore, the morphological analysis of phages STP1 and MM25 revealed the presence of a feather-like tail appendage (Fig. 5). This feather-like appendage has been observed recently for some cos-type phages (20, 45), although it is not widely described among this group of phages. No obvious genetic element could be identified that is unique to these phages compared to previously sequenced S. thermophilus phage genomes. However, early attempts at imaging CsCl-purified lysates of STP1 were largely unsuccessful due to instability of the phages, and many separated tails and heads were observed, while in addition, the feather-like appendage was not observed (data not shown). Therefore, it is possible that this appendage is a fragile structure that may be destroyed or removed by harsh treatments and/or ultracentrifugation. The preparation of fresh crude lysates that were subsequently diluted to remove media and contaminating background artifacts proved a more effective approach to the analysis of these phages that retain these delicate appendages. Knowledge of the presence of such structures that may play a role in host interactions is vital to developing a detailed understanding of the means by which phages of S. thermophilus recognize and attach to their bacterial hosts.
The Tal-RBP encoded by phages is the primary determinant of host recognition and attachment, and in 2001, the modular arrangement of the host specificity proteins (Tal-RBPs) of seven S. thermophilus phages was analyzed (37). In this study, the Tal-RBPs were determined to be putative RBPs of the phages and were characterized as presenting with up to three domains: (i) a conserved N-terminal region of 491 aa (domain 1); (ii) the VR1 region, which is present in some but not all Tal-RBPs (domain 2); and (iii) the VR2, which is involved in host recognition (domain 3). In the present study, detailed bioinformatic analyses revealed that the VR1 and VR2 regions are often represented by predicted carbohydrate-binding domains. This consolidates the notion that S. thermophilus phages recognize a carbohydrate surface receptor. Furthermore, the presence of a predicted cell wall polysaccharide or teichoic acid-interacting domain in the second baseplate protein (BPP) provides additional insights into the complexity of the interactions of these phages. Structural bioinformatics is a very useful tool to shed light on viral “dark matter,” and this is a noteworthy example of its application. Indeed, it may have implications for the interactions of phages of nondairy streptococci that may employ similar receptor material, and it provides a route of investigation for these phages.
Lactococcal phage-host interactions have become a paradigm for Gram-positive bacteria and their infecting phages, and this is warranted by the extensive application of lactococcal starter strains in the dairy industry and the persistence of their phages in the dairy fermentation setting (41, 51–53). Despite the industrial importance of S. thermophilus starter cultures in the dairy industry, the phage-host interactions of this species have not enjoyed as much attention as their lactococcal counterparts. Therefore, it is essential to generate data on the diversity and evolution of these phages and to unravel the intricacies of their interactions with their respective host bacteria. The finding of presumed CBDs in the tail tip structural proteins of S. thermophilus phages provides clear direction for future studies relating to these phages and the means by which they recognize and attach to their cognate hosts to expand current knowledge on this industrially important subject. Future research will focus on the functional characterization of individual proteins and protein complexes that harbor CBDs to define the role of the individual domains in generalized or specialized binding to the host. The identification of CBDs in phage structural proteins will provide a basis for the targeted analysis of different CBD types to assess the range of binding activities among such phages.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, and media.
Bacterial cultures were grown in M17 broth (Oxoid, Hampshire, UK) supplemented with 0.5% lactose at 42°C. Phages were isolated from dairy whey samples from an Irish cheese production facility (factory A), both those produced in-house as well as those obtained from cheese whey derived from other factories that was destined for whey protein powder production on the premises of factory A. Two phage screening approaches were employed in this study. In the first approach, all samples (>1,000 samples) derived from the cheese factory (factory A) were initially tested against S. thermophilus P1 using the double agar plaque assay method defined by Lillehaug (54), since this is the S. thermophilus strain that is primarily used in the cheese production process of factory A. In all cases, the same master stock of S. thermophilus P1 was used to generate the P1 culture for phage testing (and was sourced from the commercial starter supplier) to ensure that the culture had not changed across the testing period of 11 years. The second approach involved 2,043 whey samples derived from other factories (using unknown starter cultures), which were tested against a panel of 52 S. thermophilus strains obtained from the University College Cork (UCC) strain collection (representing historical dairy isolates; Table S2) to identify potential novel phage isolates that may be introduced into the plant. This second part of the study therefore reflects 106,236 sensitivity assays (52 strains tested against 2,043 samples) performed over the 11-year period. Phages were propagated on relevant hosts (indicated in Table 1) at 42°C with the addition of 10 mM CaCl2, filtered after lysis had occurred, and stored at 4°C until required.
Host range analysis.
All bacteriophages were propagated to a titer of (at least) 107 PFU · ml−1, and their host ranges were subsequently assessed on 51 additional strains from various sources. Host range analysis was performed using the previously described spot test method (55). To verify the results of the spot assays, enumeration of the level of sensitivity of each strain was determined using plaque assays (54). All assays were performed in triplicate, and the efficiency of plaquing (EOP) was calculated and is reported in Table 2. The EOP was defined as the ratio of the average titer of the phage on the test (secondary) host strain to the average titer of the primary propagating host strain.
Multiplex PCR.
To define if isolated phages belong to the cos- or pac-type S. thermophilus phages, multiplex PCR was performed using phage DNA as the template. The multiplex PCR was based on the method of Quiberoni and colleagues using the conserved sequences of the gene encoding the major capsid protein of seven S. thermophilus phages belonging to both the cos and pac subgroups (19). The primer sequences based on the cos phages were 5′-GGTTCACGTGTTTTATGAAAAATGG-3′ (cosFOR), and 5′-AGCAGAATCAGCAAGCAAGCTGTT-3′ (cosREV), with an expected product size of 170 bp, and those based on the pac phages were 5′-GAAGCTATGCGTATGCAAGT-3′ (pacFOR) and 5′-TTAGGGATAAGAGTCAAGTG-3′ (pacREV), with an expected product size of 427 bp. The resulting amplicons were applied to a 1% agarose gel and visualized by UV transillumination.
Phage DNA extraction.
DNA for genome sequencing was extracted from 50 ml of fresh phage lysate (∼108 PFU · ml−1), which was first treated with 1 μg/ml DNase and RNase at 37°C for 30 min. Following centrifugation at 13,200 × g for 15 min, the lysate was transferred to a new tube, after which polyethylene glycol 8000 (PEG 8000) and NaCl were added to final concentrations of 10% and 0.5 M, respectively, and the resulting suspension was then incubated at 4°C overnight. Subsequently, the suspension was centrifuged at 17,700 × g for 15 min and the supernatant removed. The PEG precipitate was resuspended in 5 ml of Tris-EDTA (TE) buffer (pH 9.0) and treated with 120 μl of 20 mg/ml proteinase K for 20 min at 56°C. Potassium acetate was added to a final concentration of 1 M, followed by incubation on ice for 20 min before centrifugation at 13,200 × g for 10 min. The supernatant was then extracted with phenol-chloroform (25:24:1 phenol-chloroform-isoamyl alcohol; Sigma-Aldrich) (at least) twice and the aqueous phase precipitated with 2.5 volumes of ice cold 96% ethanol and 0.1 volume of 3 M sodium acetate (pH 4.8). Subsequent to centrifugation, the pellet was washed in 70% ethanol and resuspended in 100 μl of TE buffer (pH 8.0).
Genome sequencing, assembly, and annotation.
Sequencing of the STP1 genome was conducted using a GS-FLX Titanium sequencer. Ten micrograms of DNA was extracted and verified by NanoDrop quantification, and confirmatory PCR-based identification (ID) tests and restriction profile analysis were conducted on the DNA extract prior to shipment to the contract sequencing facility (Agencourt Bioscience, MA, USA). Chromosomal DNA was mechanically sheared via a HydroShear device (GeneMachines, San Carlos, CA) and fragment size selected (3 kb) on an agarose pulsed-field gel electrophoresis gel, excised, and purified. A similar approach was employed for the sequencing of STP2 but by a different service provider (Macrogen, Seoul, South Korea). The files generated by the 454 FLX instrument were assembled with GSassembler (454 Life Sciences, Branford, CT, USA) to generate a consensus sequence. The assembly was entered into Staden (56), and additional sequencing walks were performed, resulting in a single gapless contig. The remainder of the genomes were sequenced using Illumina MiSeq technology (GenProbio, Parma, Italy). MIRA (Mimicking Intelligent Read Assembly) version 4.0.2 was used for de novo assembly of MiSeq-derived phage genome sequences to generate a consensus sequence. Open reading frames (ORFs) were predicted using a combination of Prodigal version 2.6 and BLASTX (57, 58), followed by manual assessment, curation, and correction of the predicted ORFs. Functional annotations were generated using BLASTP (59) analysis against the nonredundant protein database (nr) provided by the National Center for Biotechnology Information (NCBI; http://blast.ncbi.nlm.nih.gov/Blast.cgi), as well as using the MEGAnnotator pipeline (60). Proposed protein functions were validated by querying protein domain database Pfam (61) and the NCBI Conserved Domain Database (62), and by performing homology prediction searches using HHpred (63). The genomes were searched for the presence of potential tRNA genes using tRNAscan-SE (64). The genomic characteristics of the sequenced phage isolates are presented in Table 2. Quality improvement of the genome sequences involved sequencing of PCR products across the entire genome to ensure correct assembly, double stranding, and the resolution of any remaining base conflicts occurring within homopolymeric tracts. Artemis (65) was employed to inspect the results of the ORF prediction and its associated BLASTP results, which were used for a manual editing effort.
Multiple alignment of nucleotide sequences of the newly sequenced phages isolated in this study and those of previously sequenced members representing the four known S. thermophilus phage groups (DT1 [cos] [66], TP-J34 [pac] [47], 5093 [5093] [9], and 9871 [987] [10]) were performed using the ClustalW software. The alignment was employed to generate an unrooted phylogenetic tree using the iTOL software (http://itol.embl.de/) applying the neighbor-joining method.
Electron microscopy.
Phages STP1 and MM25 were selected as representatives of the phage collection for imaging, and fresh high-titer lysates (at least 109 PFU · ml−1) were produced and diluted 1:100 in SM buffer (100 mM sodium chloride, 8 mM magnesium sulfate, 50 mM Tris-HCl, 10 mM calcium chloride) (67) before imaging. Adsorption of CsCl-purified phages to freshly prepared carbon film floated from a freshly coated mica sheet and negative staining with 2% (wt/vol) uranyl acetate were performed as described previously (68). The film was picked up with a 400-mesh copper grid (Agar Scientific, Essex, UK), and specimens were examined with a Tecnai 10 transmission electron microscope (FEI, Eindhoven, The Netherlands) operated at an acceleration voltage of 80 kV.
Accession number(s).
The newly determined sequence accession numbers were deposited in GenBank under accession numbers MF580759 to MF580775.
Supplementary Material
ACKNOWLEDGMENTS
J. Mahony is supported by a Starting Investigator Research Grant (SIRG) (reference no. 15/SIRG/3430) funded by Science Foundation Ireland (SFI). D. van Sinderen is supported by a Principal Investigator award (reference no. 13/IA/1953) through SFI.
We acknowledge the Irish cheese factory for kindly providing the samples for the phage screening performed in this study.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02855-17.
REFERENCES
- 1.Auclair J, Accolas JP. 1983. Use of thermophilic lactic starters in the dairy industry. Antonie Van Leeuwenhoek 49:313–326. doi: 10.1007/BF00399506. [DOI] [PubMed] [Google Scholar]
- 2.Giraffa G, Paris A, Valcavi L, Gatti M, Neviani E. 2001. Genotypic and phenotypic heterogeneity of Streptococcus thermophilus strains isolated from dairy products. J Appl Microbiol 91:937–943. doi: 10.1046/j.1365-2672.2001.01464.x. [DOI] [PubMed] [Google Scholar]
- 3.Duplessis M, Levesque CM, Moineau S. 2006. Characterization of Streptococcus thermophilus host range phage mutants. Appl Environ Microbiol 72:3036–3041. doi: 10.1128/AEM.72.4.3036-3041.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quiberoni A, Stiefel JI, Reinheimer JA. 2000. Characterization of phage receptors in Streptococcus thermophilus using purified cell walls obtained by a simple protocol. J Appl Microbiol 89:1059–1065. doi: 10.1046/j.1365-2672.2000.01214.x. [DOI] [PubMed] [Google Scholar]
- 5.Fujisawa H, Morita M. 1997. Phage DNA packaging. Genes Cells 2:537–545. doi: 10.1046/j.1365-2443.1997.1450343.x. [DOI] [PubMed] [Google Scholar]
- 6.Binetti AG, Del Rio B, Martin MC, Alvarez MA. 2005. Detection and characterization of Streptococcus thermophilus bacteriophages by use of the antireceptor gene sequence. Appl Environ Microbiol 71:6096–6103. doi: 10.1128/AEM.71.10.6096-6103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Marrec C, van Sinderen D, Walsh L, Stanley E, Vlegels E, Moineau S, Heinze P, Fitzgerald G, Fayard B. 1997. Two groups of bacteriophages infecting Streptococcus thermophilus can be distinguished on the basis of mode of packaging and genetic determinants for major structural proteins. Appl Environ Microbiol 63:3246–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.del Rio B, Binetti AG, Martin MC, Fernandez M, Magadan AH, Alvarez MA. 2007. Multiplex PCR for the detection and identification of dairy bacteriophages in milk. Food Microbiol 24:75–81. doi: 10.1016/j.fm.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Mills S, Griffin C, O'Sullivan O, Coffey A, McAuliffe OE, Meijer WC, Serrano LM, Ross RP. 2011. A new phage on the ‘Mozzarella’ block: bacteriophage 5093 shares a low level of homology with other Streptococcus thermophilus phages. Int Dairy J 21:963–969. doi: 10.1016/j.idairyj.2011.06.003. [DOI] [Google Scholar]
- 10.McDonnell B, Mahony J, Neve H, Hanemaaijer L, Noben JP, Kouwen T, van Sinderen D. 2016. Identification and analysis of a novel group of bacteriophages infecting the lactic acid bacterium Streptococcus thermophilus. Appl Environ Microbiol 82:5153–5165. doi: 10.1128/AEM.00835-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahony J, Oliveira J, Collins B, Hanemaaijer L, Lugli GA, Neve H, Ventura M, Kouwen TR, Cambillau C, van Sinderen D. 2017. Genetic and functional characterisation of the lactococcal P335 phage-host interactions. BMC Genomics 18:146. doi: 10.1186/s12864-017-3537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahony J, Deveau H, Mc Grath S, Ventura M, Canchaya C, Moineau S, Fitzgerald GF, van Sinderen D. 2006. Sequence and comparative genomic analysis of lactococcal bacteriophages jj50, 712 and P008: evolutionary insights into the 936 phage species. FEMS Microbiol Lett 261:253–261. doi: 10.1111/j.1574-6968.2006.00372.x. [DOI] [PubMed] [Google Scholar]
- 13.Szczepańska AK, Hejnowicz MS, Kolakowski P, Bardowski J. 2007. Biodiversity of Lactococcus lactis bacteriophages in Polish dairy environment. Acta Biochim Pol 54:151–158. [PubMed] [Google Scholar]
- 14.Raiski A, Belyasova N. 2009. Biodiversity of Lactococcus lactis bacteriophages in the Republic of Belarus. Int J Food Microbiol 130:1–5. doi: 10.1016/j.ijfoodmicro.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 15.Murphy J, Royer B, Mahony J, Hoyles L, Heller K, Neve H, Bonestroo M, Nauta A, van Sinderen D. 2013. Biodiversity of lactococcal bacteriophages isolated from 3 Gouda-type cheese-producing plants. J Dairy Sci 96:4945–4957. doi: 10.3168/jds.2013-6748. [DOI] [PubMed] [Google Scholar]
- 16.Suárez V, Moineau S, Reinheimer J, Quiberoni A. 2008. Argentinean Lactococcus lactis bacteriophages: genetic characterization and adsorption studies. J Appl Microbiol 104:371–379. [DOI] [PubMed] [Google Scholar]
- 17.Verreault D, Gendron L, Rousseau GM, Veillette M, Masse D, Lindsley WG, Moineau S, Duchaine C. 2011. Detection of airborne lactococcal bacteriophages in cheese manufacturing plants. Appl Environ Microbiol 77:491–497. doi: 10.1128/AEM.01391-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moineau S, Pandian S, Klaenhammer TR. 1993. Restriction/modification systems and restriction endonucleases are more effective on lactococcal bacteriophages that have emerged recently in the dairy industry. Appl Environ Microbiol 59:197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quiberoni A, Tremblay D, Ackermann HW, Moineau S, Reinheimer JA. 2006. Diversity of Streptococcus thermophilus phages in a large-production cheese factory in Argentina. J Dairy Sci 89:3791–3799. doi: 10.3168/jds.S0022-0302(06)72420-1. [DOI] [PubMed] [Google Scholar]
- 20.Szymczak P, Janzen T, Neves AR, Kot W, Hansen LH, Lametsch R, Neve H, Franz CMAP, Vogensen FK. 2017. Novel variants of Streptococcus thermophilus bacteriophages are indicative of genetic recombination among phages from different bacterial species. Appl Environ Microbiol 83:e02748-16. doi: 10.1128/AEM.02748-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Achigar R, Magadan AH, Tremblay DM, Pianzzola MJ, Moineau S. 2017. Phage-host interactions in Streptococcus thermophilus: Genome analysis of phages isolated in Uruguay and ectopic spacer acquisition in CRISPR array. Sci Rep 7:43438. doi: 10.1038/srep43438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rousseau GM, Moineau S. 2009. Evolution of Lactococcus lactis phages within a cheese factory. Appl Environ Microbiol 75:5336–5344. doi: 10.1128/AEM.00761-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madera C, Monjardin C, Suarez JE. 2004. Milk contamination and resistance to processing conditions determine the fate of Lactococcus lactis bacteriophages in dairies. Appl Environ Microbiol 70:7365–7671. doi: 10.1128/AEM.70.12.7365-7371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quiberoni A, Guglielmotti DM, Reinheimer JA. 2003. Inactivation of Lactobacillus delbrueckii bacteriophages by heat and biocides. Int J Food Microbiol 84:51–62. doi: 10.1016/S0168-1605(02)00394-X. [DOI] [PubMed] [Google Scholar]
- 25.Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paez-Espino D, Sharon I, Morovic W, Stahl B, Thomas BC, Barrangou R, Banfield JF. 2015. CRISPR immunity drives rapid phage genome evolution in Streptococcus thermophilus. mBio 6:e00262-15. doi: 10.1128/mBio.00262-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol 190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Binetti AG, Reinheimer JA. 2000. Thermal and chemical inactivation of indigenous Streptococcus thermophilus bacteriophages isolated from Argentinian dairy plants. J Food Prot 63:509–515. doi: 10.4315/0362-028X-63.4.509. [DOI] [PubMed] [Google Scholar]
- 29.Hayes S, Murphy J, Mahony J, Lugli GA, Ventura M, Noben JP, Franz CM, Neve H, Nauta A, Van Sinderen D. 2017. Biocidal inactivation of Lactococcus lactis bacteriophages: efficacy and targets of commonly used sanitizers. Front Microbiol 8:107. doi: 10.3389/fmicb.2017.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner N, Brinks E, Samtlebe M, Hinrichs J, Atamer Z, Kot W, Franz C, Neve H, Heller KJ. 2017. Whey powders are a rich source and excellent storage matrix for dairy bacteriophages. Int J Food Microbiol 241:308–317. doi: 10.1016/j.ijfoodmicro.2016.10.037. [DOI] [PubMed] [Google Scholar]
- 31.Kenny JG, McGrath S, Fitzgerald GF, van Sinderen D. 2004. Bacteriophage Tuc2009 encodes a tail-associated cell wall-degrading activity. J Bacteriol 186:3480–3491. doi: 10.1128/JB.186.11.3480-3491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stockdale SR, Mahony J, Courtin P, Chapot-Chartier MP, van Pijkeren JP, Britton RA, Neve H, Heller KJ, Aideh B, Vogensen FK, van Sinderen D. 2013. The lactococcal phages Tuc2009 and TP901-1 incorporate two alternate forms of their tail fiber into their virions for infection specialization. J Biol Chem 288:5581–5590. doi: 10.1074/jbc.M112.444901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guglielmotti DM, Deveau H, Binetti AG, Reinheimer JA, Moineau S, Quiberoni A. 2009. Genome analysis of two virulent Streptococcus thermophilus phages isolated in Argentina. Int J Food Microbiol 136:101–109. doi: 10.1016/j.ijfoodmicro.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Stanley E, Fitzgerald GF, Le Marrec C, Fayard B, van Sinderen D. 1997. Sequence analysis and characterization of phi O1205, a temperate bacteriophage infecting Streptococcus thermophilus CNRZ1205. Microbiology 143:3417–3429. doi: 10.1099/00221287-143-11-3417. [DOI] [PubMed] [Google Scholar]
- 35.Lucchini S, Desiere F, Brussow H. 1999. The genetic relationship between virulent and temperate Streptococcus thermophilus bacteriophages: whole genome comparison of cos-site phages Sfi19 and Sfi21. Virology 260:232–243. doi: 10.1006/viro.1999.9814. [DOI] [PubMed] [Google Scholar]
- 36.Veesler D, Cambillau C. 2011. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol Mol Biol Rev 75:423–433. doi: 10.1128/MMBR.00014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duplessis M, Moineau S. 2001. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol Microbiol 41:325–336. doi: 10.1046/j.1365-2958.2001.02521.x. [DOI] [PubMed] [Google Scholar]
- 38.Bebeacua C, Tremblay D, Farenc C, Chapot-Chartier MP, Sadovskaya I, van Heel M, Veesler D, Moineau S, Cambillau C. 2013. Structure, adsorption to host, and infection mechanism of virulent lactococcal phage p2. J Virol 87:12302–12312. doi: 10.1128/JVI.02033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spinelli S, Campanacci V, Blangy S, Moineau S, Tegoni M, Cambillau C. 2006. Modular structure of the receptor binding proteins of Lactococcus lactis phages. The RBP structure of the temperate phage TP901-1. J Biol Chem 281:14256–14262. [DOI] [PubMed] [Google Scholar]
- 40.Dieterle ME, Spinelli S, Sadovskaya I, Piuri M, Cambillau C. 2017. Evolved distal tail carbohydrate binding modules of Lactobacillus phage J-1: a novel type of anti-receptor widespread among lactic acid bacteria phages. Mol Microbiol 104:608–620. doi: 10.1111/mmi.13649. [DOI] [PubMed] [Google Scholar]
- 41.Legrand P, Collins B, Blangy S, Murphy J, Spinelli S, Gutierrez C, Richet N, Kellenberger C, Desmyter A, Mahony J, van Sinderen D, Cambillau C. 2016. The atomic structure of the phage Tuc2009 baseplate tripod suggests that host recognition involves two different carbohydrate binding modules. mBio 7:e01781-15. doi: 10.1128/mBio.01781-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, Koc C, Kuhner P, Stierhof YD, Krismer B, Enright MC, Penades JR, Wolz C, Stehle T, Cambillau C, Peschel A, Xia G. 2016. An essential role for the baseplate protein Gp45 in phage adsorption to Staphylococcus aureus. Sci Rep 6:26455. doi: 10.1038/srep26455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koç C, Xia G, Kuhner P, Spinelli S, Roussel A, Cambillau C, Stehle T. 2016. Structure of the host-recognition device of Staphylococcus aureus phage ϕ11. Sci Rep 6:27581. doi: 10.1038/srep27581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Desiere F, Lucchini S, Brussow H. 1998. Evolution of Streptococcus thermophilus bacteriophage genomes by modular exchanges followed by point mutations and small deletions and insertions. Virology 241:345–356. doi: 10.1006/viro.1997.8959. [DOI] [PubMed] [Google Scholar]
- 45.McDonnell B, Mahony J, Hanemaaijer L, Neve H, Noben JP, Lugli GA, Ventura M, Kouwen TR, van Sinderen D. 2017. Global survey and genome exploration of bacteriophages infecting the lactic acid bacterium Streptococcus thermophilus. Front Microbiol 8:1754. doi: 10.3389/fmicb.2017.01754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zinno P, Janzen T, Bennedsen M, Ercolini D, Mauriello G. 2010. Characterization of Streptococcus thermophilus lytic bacteriophages from mozzarella cheese plants. Int J Food Microbiol 138:137–144. doi: 10.1016/j.ijfoodmicro.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 47.Neve H, Zenz KI, Desiere F, Koch A, Heller KJ, Brussow H. 1998. Comparison of the lysogeny modules from the temperate Streptococcus thermophilus bacteriophages TP-J34 and Sfi21: implications for the modular theory of phage evolution. Virology 241:61–72. doi: 10.1006/viro.1997.8960. [DOI] [PubMed] [Google Scholar]
- 48.Mavrich TN, Hatfull GF. 2017. Bacteriophage evolution differs by host, lifestyle and genome. Nat Microbiol 2:17112. doi: 10.1038/nmicrobiol.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koberg S, Mohamed MD, Faulhaber K, Neve H, Heller KJ. 2015. Identification and characterization of cis- and trans-acting elements involved in prophage induction in Streptococcus thermophilus J34. Mol Microbiol 98:535–552. doi: 10.1111/mmi.13140. [DOI] [PubMed] [Google Scholar]
- 50.Lucchini S, Desiere F, Brussow H. 1999. Comparative genomics of Streptococcus thermophilus phage species supports a modular evolution theory. J Virol 73:8647–8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mahony J, Stockdale SR, Collins B, Spinelli S, Douillard FP, Cambillau C, van Sinderen D. 2016. Lactococcus lactis phage TP901-1 as a model for Siphoviridae virion assembly. Bacteriophage 6:e1123795. doi: 10.1080/21597081.2015.1123795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahony J, Randazzo W, Neve H, Settanni L, van Sinderen D. 2015. Lactococcal 949 group phages recognize a carbohydrate receptor on the host cell surface. Appl Environ Microbiol 81:3299–3305. doi: 10.1128/AEM.00143-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chopin A, Bolotin A, Sorokin A, Ehrlich SD, Chopin M. 2001. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res 29:644–651. doi: 10.1093/nar/29.3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lillehaug D. 1997. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J Appl Microbiol 83:85–90. doi: 10.1046/j.1365-2672.1997.00193.x. [DOI] [PubMed] [Google Scholar]
- 55.Dupont K, Vogensen FK, Josephsen J. 2005. Detection of lactococcal 936-species bacteriophages in whey by magnetic capture hybridization PCR targeting a variable region of receptor-binding protein genes. J Appl Microbiol 98:1001–1009. doi: 10.1111/j.1365-2672.2005.02548.x. [DOI] [PubMed] [Google Scholar]
- 56.Staden R, Judge DP, Bonfield JK. 2001. Sequence assembly and finishing methods. Methods Biochem Anal 43:303–322. [DOI] [PubMed] [Google Scholar]
- 57.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gish W, States DJ. 1993. Identification of protein coding regions by database similarity search. Nat Genet 3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 59.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lugli GA, Milani C, Mancabelli L, van Sinderen D, Ventura M. 2016. MEGAnnotator: a user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol Lett 363:fnw049. doi: 10.1093/femsle/fnw049. [DOI] [PubMed] [Google Scholar]
- 61.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. 2004. The Pfam protein families database. Nucleic Acids Res 32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 66.Tremblay DM, Moineau S. 1999. Complete genomic sequence of the lytic bacteriophage DT1 of Streptococcus thermophilus. Virology 255:63–76. doi: 10.1006/viro.1998.9525. [DOI] [PubMed] [Google Scholar]
- 67.Murphy J, Bottacini F, Mahony J, Kelleher P, Neve H, Zomer A, Nauta A, van Sinderen D. 2016. Comparative genomics and functional analysis of the 936 group of lactococcal Siphoviridae phages. Sci Rep 6:21345. doi: 10.1038/srep21345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deasy T, Mahony J, Neve H, Heller KJ, van Sinderen D. 2011. Isolation of a virulent Lactobacillus brevis phage and its application in the control of beer spoilage. J Food Prot 74:2157–2161. doi: 10.4315/0362-028X.JFP-11-262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.